Abstract
Mycothiol is the major thiol present in most actinomycetes and is produced from the pseudodisaccharide 1d-myo-inosityl 2-acetamido-2-deoxy-α-d-glucopyranoside (GlcNAc-Ins). A transposon mutant of Mycobacterium smegmatis shown to be GlcNAc-Ins and mycothiol deficient was sequenced to identify a putative glycosyltransferase gene designated mshA. The ortholog in Mycobacterium tuberculosis, Rv0486, was used to complement the mutant phenotype.
Mycothiol (MSH) (Fig. 1) is the principal thiol found in mycobacteria and most other actinomycetes. It appears to function in many ways like glutathione, which is not produced by actinomycetes (15). This suggested that MSH metabolism might provide suitable targets for new drugs directed against Mycobacterium tuberculosis (13, 20, 21). Support for this possibility came from studies of Mycobacterium smegmatis chemical mutants blocked in MSH biosynthesis. MSH mutants were shown to have enhanced sensitivity to hydrogen peroxide and various toxins, including rifampin and other antibiotics, but were found to be resistant to isoniazid (16, 17). The enzymes involved in MSH biosynthesis therefore appear to have potential as new drug targets. In this study we identified a glycosyltransferase gene involved in an initial step of MSH biosynthesis.
FIG. 1.
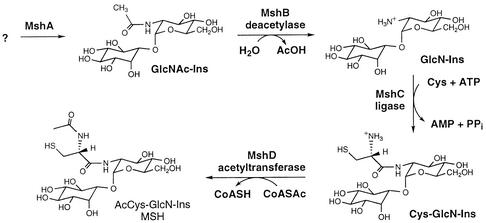
Structure and biosynthetic pathway for MSH. M. tuberculosis genes encoding MshB (Rv1170; GlcNAc-Ins deacetylase) (14), MshC (Rv2130c; ATP-dependent l-Cys:GlcN-Ins ligase) (18), and MshD (Rv0819; acetyltransferase, mycothiol synthase) (9) have been identified.
The pathway of MSH biosynthesis involves at least four enzymes, the last three of which are known (Fig. 1). To facilitate identification of MSH biosynthesis genes, a Tn5 transposon library enriched for MSH-deficient mutants was produced by selecting for resistance to both kanamycin and isoniazid (9); the latter resistance has been established as a phenotype which is characteristic of MSH-deficient strains (16, 17). One of three MSH-deficient mutants isolated proved to be defective in the final step of MSH biosynthesis and was used to identify mshD (9). Here we characterized a second mutant, tentatively designated mshA::Tn5 because it was found to produce no measurable amount of the pseudodisaccharide precursors of MSH, 1d-myo-inosityl 2-acetamido-2-deoxy-α-d-glucopyranoside (GlcNAc-Ins) and 1d-myo-inosityl 2-amino-2-deoxy-α-d-glucopyranoside (GlcN-Ins) (Table 1), which suggested that it was defective in MshA. This is the same phenotype as that of M. smegmatis chemical mutant strain 49 (14, 16). In order to obtain a high-quality sequence at the site of insertion, it was necessary to subclone SalI- or PstI-digested genomic DNA from mshA::Tn5 into pUC18 as previously described (9).
TABLE 1.
Levels of MSH and its precursors in mshA mutants and their complements
| M. smegmatis strains | Cellular level (μmol/g [residual dry wt]) ofa:
|
||
|---|---|---|---|
| GlcNAc-Insb | GlcN-Insb | MSHc | |
| mc2155 | ≤0.2 | 1.0 ± 0.2 | 10 ± 3 |
| mshA::Tn5 | ≤0.01 | ≤0.01 | ≤0.01 |
| mshA::Tn5 pAL0486 | |||
| Clone 1d | 0.10 ± 0.02 | 0.49 ± 0.05 | 9.4 ± 0.4 |
| Clone 2d | ≤0.05 | 0.64 ± 0.03 | 9.6 ± 0.2 |
| 49 | ≤0.001 | ≤0.001 | ≤0.01 |
| 49::pAL0486 | |||
| Clone 1d | 0.25 ± 0.09 | 1.3 ± 0.2 | 11.5 ± 0.3 |
| Clone 2d | 0.15 ± 0.04 | 0.84 ± 0.08 | 11.1 ± 0.3 |
Mean and range for duplicate samples of exponential cells cultured in Middlebrook 7H9 medium containing 1.0% glucose and 0.05% Tween 80 with 75 μg of hygromycin per ml and 20 μg of kanamycin per ml as appropriate.
Determined by the method described by Buchmeier et al. (3).
Determined by the method described by Koledin et al. (9).
Cells were cultured as described above to an optical density at 600 nm 0.5, transferred to Middlebrook 7H9 medium without glucose, and induced for 20 h at 23°C with 0.2% acetamide.
The SalI clone produced 205 bp of sequence, and the PstI clone produced 822 bp of sequence, both at the same insertion site; the PstI clone corresponded exactly to the complement of bases 507551 to 508372 of contig 3311 of the unfinished M. smegmatis genome in The Institute for Genomic Research database (http://www.tigr.org). Since the M. smegmatis sequence has not been annotated yet, the experimental sequence was used to search the M. tuberculosis H37Rv GenBank genome database by using tblastx. This resulted in identification of a 236-amino-acid sequence from open reading frame Rv0486 having 84% identity with the translated experimental sequence. Thus, the reading frame for the M. smegmatis sequence was established, and a downstream stop codon defined the termination site for the M. smegmatis gene. The start position for the gene was thought to be defined by a GTG codon found upstream from the end of the region with high identity to the M. tuberculosis sequence and downstream from an in-frame stop codon. This resulted in identification of the sequence for MshA, as shown in Fig. 2. The M. smegmatis and M. tuberculosis MshA sequences are 75% identical in a 446-residue overlap.
FIG. 2.
Alignment of the sequences of MshA from M. smegmatis (MshA-Msmeg) and M. tuberculosis (MshA-Mtub) with the sequences of SpsA from Anaebaena sp. strain PCC7120 (SpsA-Anab) and PimB (Rv0557) from M. tuberculosis (PimB-Mtub). The site of the G32D mutation in M. smegmatis mutant 49 is indicated by an asterisk. See the text for a discussion of specifically designated residues and domains; the numbers are numbers in the M. tuberculosis MshA sequence.
To verify that Rv0486 encodes the enzyme activity missing in the transposon and chemical mutants, the gene was cloned into the pALACE vector (an Escherichia coli-Mycobacterium shuttle plasmid; Hygr; induced by acetamide and derived from pACE [6] to introduce an N-terminal His6 tag) to produce pAL0486, which was used to transform mshA::Tn5 and mutant 49. M. tuberculosis H37Rv genomic DNA was prepared as described previously (2). Open reading frame Rv0486 was amplified by PCR by using this DNA and primers 486 PAL 3 (5′CATATGCACGGTCGGCAAGGAGG3′) and 486 PAL 5 (5′AGGATCCATGGCAGGTGTGCGGCAC3′). These primers were designed to contain NdeI and BamHI restriction sites, respectively. PCR was performed as described previously (17), and the appropriate PCR product was ligated into pALACE to obtain plasmid pAL0486. The pAL0486 plasmid was electroporated into the MSH-deficient mutants, and selection was performed on plates containing hygromycin for mutant 49 and hygromycin plus kanomycin for mshA::Tn5 (16).
In each case, two clones were selected from the hygromycin plates for growth in liquid culture to produce cells for analysis of MSH and its precursors. The results (Table 1) demonstrated that full restoration of MSH production to wild-type levels occurred with acetamide-induced cultures, and the levels of GlcNAc-Ins and GlcN-Ins increased to normal or nearly normal levels in both mutants. This confirmed that the loss of MSH biosynthesis capacity in mutant mshA::Tn5 resulted solely from inactivation of the mshA gene and demonstrated that mutant 49 was defective in the mshA gene.
To ascertain the nature of the defect in the mshA gene of mutant 49, the mshA genes from the parent strain, M. smegmatis mc2155, and mutant 49 were cloned and sequenced. The M. smegmatis sequence obtained from The Institute for Genomic Research was used to design primers upstream of the start codon (primer 49seq 5′; 5′GCAACGAGAAGGCCGTCGAACT3′) and downstream of the 3′ region (primer 49seq 3′; 5′GTCCTCGATGATCTTCCTGACA3′) of the mshA gene. The primers were used to amplify the DNA from M. smegmatis mc2155 and from two different colonies of mutant 49, and each amplified band was cloned into pCR2.1 (Invitrogen). After we ensured by restriction digestion that the M. smegmatis mshA gene had been cloned, the DNA was sequenced by using primers 49seq 5′ and 49seq 3′, as well as the universal primers T7 and M13R. To sequence the internal region of the homolog and confirm the missense mutation, the same procedure was followed with primers 49MED 5′ (5′GCGTGGCGGTGTTGTCGGTA3′) and 49MED 3′ (5′GACCAGTTGTTCGCGGCTCT3′). Comparison of the sequences revealed a single base pair change in the mutant which converted a GGC codon to GAC. This resulted in a change in the amino acid at position 32 of the M. smegmatis sequence from glycine in mc2155 to aspartic acid in mutant 49 (Fig. 2).
The sensitivity of M. smegmatis parent strain mc2155, transposon mutant mshA::Tn5, and chemical mutant 49 to isoniazid was tested by using Estrips (Oxoid) and assessing inhibition of growth after incubation for 2 to 3 days at 37°C. The MICs of isoniazid determined were 1, >250, and >250 μg/ml, respectively. The MICs of isoniazid for mc2155 and mutant 49 are similar to those obtained previously (2 and >50 μg/ml) by plating on antibiotic-containing media (16). When the isoniazid sensitivities of the complemented mshA::Tn5 and strain 49 mutants were tested on plates containing 1% acetamide but lacking glucose, the MICs were 2.7 and 28 μg/ml, respectively, demonstrating that there was substantial reversion to the parental phenotype. The mechanistic basis for the isoniazid resistance has not been established, but there is evidence indicating that the isoniazid sensitivity is specifically linked to MSH rather than a generally high level of cellular thiol (9).
The results described above establish that the mshA gene in M. smegmatis is essential for the production of GlcNAc-Ins and therefore for the synthesis of MSH. The M. tuberculosis mshA gene, Rv0486, was previously identified by Campbell et al. (4) (who listed it as unknown protein MTCY20G9.12) as a member of glycosyltransferase family 4 in the CAZy database (http://afmb.cnrs-mrs.fr/CAZY/). This family includes a number of sucrose synthases, sucrose phosphate synthases (Sps), mannosyl transferases, and GlcNAc transferases. The most likely homologs for MshA seemed to be the GlcNAc transferases PigA, which is involved in glycosyl phosphatidylinositol anchor biosynthesis (8, 10), and RfaK, which transfers GlcNAc from UDP to lipopolysaccharide (12), but full sequence alignments revealed only 18 and 19% identities between these sequences and the sequence of M. tuberculosis MshA, respectively. Higher levels of identity were obtained with sucrose and sucrose phosphate synthases and with the mannosyl transferase PimB. Figure 2 includes sequences for SpsA from Anabaena sp. strain PCC7120 (5) and PimB (Rv0557) from M. tuberculosis (19) (GenBank accession numbers AJ302071 and NP_215071, respectively). SpsA has 29% overall sequence identity with M. tuberculosis MshA, whereas PimB has 24% identity. Cumino et al. (5) identified two highly conserved motifs present in the glycosyltransferase domains of sucrose phosphate synthases and sucrose synthases, designated box I and box II, as shown in Fig. 2. M. tuberculosis MshA is identical to SpsA at 6 of 12 box I residues and at 17 of 25 box II residues. The first Gly of box I is changed to Asp in M. smegmatis mutant 49. This results in complete loss of the ability to synthesize GlcNAc-Ins, GlcN-Ins, and MSH in vivo, showing that this region is critical to the activity of MshA. The box II domain contains the E-X7-E signature (7) universally found in the retaining glycosyl transferases of CAZy family 4, and the first residue of this signature, E353 (M. tuberculosis numbering), has been shown to be essential for activity of the mannosyl transferase AceA from Acetobacter xylinum (1). PimB is less similar to SpsA than MshA is, but there are some additional residues that were identified by Kremer et al. (11) as highly conserved in mannosyl transferases that are conserved in MshA and SpsA. These include Lys278, which has also been shown to be essential for activity in AceA (1), as well as Gly241, Asp243, and Arg273, for which no function has been established yet (11). The homology of MshA with known CAZy family 4 glycosyltransferases clearly indicates that MshA belongs to this group and is the glycosyltransferase required for biosynthesis of the pseudodisaccharide GlcNAc-Ins (Fig. 1). However, there has been no clear indication of the nature of the substrate sugar donor or sugar acceptor.
The most obvious candidate for the biochemical reaction catalyzed by MshA involves transfer of GlcNAc from UDP-GlcNAc to myo-inositol, but we have not been able to detect production of GlcNAc-Ins using these substrates and either crude extracts of M. smegmatis or recombinant His6 MshA purified from mutant 49::pAL0486 on an Ni2+ affinity column. Thus, the reaction either requires some special conditions not yet identified or utilizes some other combination of substrates. This is the subject of continuing studies.
Acknowledgments
This work was supported by grants to R.C.F. from the National Institute of Allergy and Infectious Diseases (grant AI49174) and the Fogarty International Center (grant TW00976), and to Y.A. from the British Columbia Lung Association and the TB Veterans Association.
Y.A. is a Canadian Institute of Health Research Foundation-BC Lung Association Scholar. We thank Koen De Smet for kindly providing the pALACE shuttle plasmid.
REFERENCES
- 1.Abdian, P. L., A. C. Lellouch, C. Gautier, L. Ielpi, and R. A. Geremia. 2000. Identification of essential amino acids in the bacterial α-mannosyltransferase AceA. J. Biol. Chem. 275:40568-40575. [DOI] [PubMed] [Google Scholar]
- 2.Av-Gay, Y., S. Jamil, and S. J. Drews. 1999. Expression and characterization of the Mycobacterium tuberculosis serine/threonine protein kinase pknB. Infect. Immun. 67:5676-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchmeier, N., G. L. Newton, T. Koledin, and R. C. Fahey. 2003. Association of mycothiol with protection of Mycobacterium tuberculosis from toxic oxidants and antibiotics. Mol. Microbiol. 47:1723-1732. [DOI] [PubMed]
- 4.Campbell, J. A., G. J. Davies, V. Bulone, and B. Henrissat. 1997. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326:929-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cumino, A., L. Curatti, L. Giarrocco, and G. L. Salerno. 2002. Sucrose metabolism: Anabaena sucrose-phosphate synthase and sucrose-phosphate phosphatase define minimal functional domains shuffled during evolution. FEBS Lett. 517:19-23. [DOI] [PubMed] [Google Scholar]
- 6.De Smet, K. A. L., K. E. Kempsell, A. Gallagher, K. Duncan, and D. B. Young. 1999. Alteration of a single amino acid residue reverses fosfomycin resistance of recombinant MurA from Mycobacterium tuberculosis. Microbiology 145:3177-3184. [DOI] [PubMed] [Google Scholar]
- 7.Geremia, R. A., E. A. Petroni, L. Ielpi, and B. Henrissat. 1996. Towards a classification of glycosyltransferases based on amino acid sequence similarities: prokaryotic α-mannosyltransferases. Biochem. J. 318:133-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinoshita, T., and N. Inoue. 2000. Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biosynthesis. Curr. Opin. Chem. Biol. 4:632-638. [DOI] [PubMed] [Google Scholar]
- 9.Koledin, T., G. L. Newton, and R. C. Fahey. 2002. Identification of the mycothiol synthase gene (mshD) encoding the acetyltransferase producing mycothiol in actinomycetes. Arch. Microbiol. 178:331-337. [DOI] [PubMed] [Google Scholar]
- 10.Kostova, Z., D. M. Rancour, A. K. Menon, and P. Orlean. 2000. Photoaffinity labelling with P3-(4-azidoanilido)uridine 5′-triphosphate identifies Gpi3p as the UDP-GlcNAc-binding subunit of the enzyme that catalyses formation of GlcNAc-phosphatidylinositol, the first glycolipid intermediate in glycosylphosphatidylinositol synthesis. Biochem. J. 350:815-822. [PMC free article] [PubMed] [Google Scholar]
- 11.Kremer, L., S. S. Gurcha, P. Bifani, P. G. Hitchen, A. Baulard, H. R. Morris, A. Dell, P. J. Brennan, and G. S. Besra. 2002. Characterization of a putative α-mannosyltransferase involved in phosphatidylinositol trimannoside biosynthesis in Mycobacterium tuberculosis. Biochem. J. 363:437-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacLachlan, P. R., S. K. Kadam, and K. E. Sanderson. 1991. Cloning, characterization, and DNA sequence of the rfaLK region for lipopolysaccharide synthesis in Salmonella typhimurium LT2. J. Bacteriol. 173:7151-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newton, G. L., K. Arnold, M. S. Price, C. Sherrill, S. B. delCardayré, Y. Aharonowitz, G. Cohen, J. Davies, R. C. Fahey, and C. Davis. 1996. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newton, G. L., Y. Av-Gay, and R. C. Fahey. 2000. N-Acetyl-1-d-myo-inosityl-2-amino-2-deoxy-α-d-glucopyranoside deacetylase (MshB) is a key enzyme in mycothiol biosynthesis. J. Bacteriol. 182:6958-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newton, G. L., and R. C. Fahey. 2002. Mycothiol biochemistry. Arch. Microbiol. 178:388-394. [DOI] [PubMed] [Google Scholar]
- 16.Newton, G. L., M. D. Unson, S. J. Anderberg, J. A. Aguilera, N. N. Oh, S. B. delCardayré, J. Davies, Y. Av-Gay, and R. C. Fahey. 1999. Characterization of a Mycobacterium smegmatis mutant defective in 1-d-myo-inosityl-2-amino-2-deoxy-α-d-glucopyranoside and mycothiol biosynthesis. Biochem. Biophys. Res. Commun. 255:239-244. [DOI] [PubMed] [Google Scholar]
- 17.Rawat, M., G. L. Newton, M. Ko, G. J. Martinez, R. C. Fahey, and Y. Av-Gay. 2002. Mycothiol-deficient mutants in Mycobacterium smegmatis are hypersensitive to alkylating agents, free radicals, and antibiotics. Antimicrob. Agents Chemother. 46:3348-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sareen, D., M. Steffek, G. L. Newton, and R. C. Fahey. 2002. ATP-dependent l-cysteine:1d-myo-inosityl 2-amino-2-deoxy-α-d-glucopyranoside ligase:mycothiol biosynthesis enzyme MshC is related to class I cysteinyl-tRNA synthetases. Biochemistry 41:6885-6890. [DOI] [PubMed] [Google Scholar]
- 19.Schaeffer, M. L., K. H. Khoo, G. S. Besra, D. Chatterjee, P. J. Brennan, J. T. Belisle, and J. M. Inamine. 1999. The pimB gene of Mycobacterium tuberculosis encodes a mannosyltransferase involved in lipoarabinomannan biosynthesis. J. Biol. Chem. 274:31625-31631. [DOI] [PubMed] [Google Scholar]
- 20.Spies, H. S., and D. J. Steenkamp. 1994. Thiols of intracellular pathogens. Identification of ovothiol A in Leishmania donovani and structural analysis of a novel thiol from Mycobacterium bovis. Eur. J. Biochem. 224:203-213. [DOI] [PubMed] [Google Scholar]
- 21.Young, D. B., and K. Duncan. 1995. Prospects for new interventions in the treatment and prevention of mycobacterial disease. Annu. Rev. Microbiol. 49:641-673. [DOI] [PubMed] [Google Scholar]