Abstract
Bacteriophage PM2 is the only described member of the Corticoviridae family. It is an icosahedral dsDNA virus with a membrane residing underneath the protein coat. PM2 infects some gram-negative Pseudoalteromonas spp. In the present study, we mapped the viral promoters and showed that the PM2 genome consists of three operons. Four new virus genes were assigned based on their function in transcription. Proteins P15 and P16 are shown to repress early transcription, and proteins P13 and P14 are shown to activate late transcription events. The early regulatory region, containing genes for proteins P15 and P16, as well as the newly identified early promoter region in PM2, has significant sequence similarity with the Pseudoalteromonas pAS28 plasmid. P14, the transcription activator for the structural genes, has a zinc finger motif homologous to archaeal and eukaryotic TFIIS-type regulatory factors.
Bacteriophage PM2 is the only described member of the Corticoviridae family (1, 15). It was isolated, together with its host Pseudoalteromonas espejiana BAL-31 (originally Pseudomonas sp. strain BAL-31 [16, 18]) from seawater off the coast of Chile. Pseudoalteromonas sp. strain ER72M2, is an alternative host for PM2 (27).
PM2, the first bacteriophage shown to contain lipids as part of the virion (10), has an icosahedrally organized capsid of ca. 60 nm in diameter surrounding a membranous inner core (28). In this way, it resembles bacteriophage PRD1, a tectivirus, which has the same overall structural organization (4, 5, 9). Inside the membrane core resides the phage genome. The genome is circular and the most tightly supercoiled dsDNA molecule known (17, 20). For this reason, it has been widely used as a substrate in different enzymatic assays. The virion has nine structural proteins (28). Protein P1 (37.5 kDa) forms the receptor-binding spike structure at the vertices of the virion, and P2 (30.2 kDa) is the coat protein. Proteins P3, P4, P5, P6, P7, P8, and P9 are associated with the membrane (reference 28 and references therein).
The nucleotide sequence of the 10,079-bp PM2 genome has revealed 21 putative genes (33). The genes shown to encode a protein were given a Roman numeral, and the rest were assigned as open reading frames (ORFs) with a lowercase letter. The genes I to IX, coding for the structural proteins, were confirmed by comparing the predicted amino acid sequences to the amino-terminal sequences of the virion proteins (27, 28, 33). Gene XII, coding for the nonstructural protein P12, was identified based on the conserved sequence motifs that are similar to replication initiation proteins of other bacteriophages, such as φX174 and P2, and initiation proteins of cyanobacterial and archaeal plasmids (24).
Bacterial viruses express their genes in a specific time sequence, regulated primarily by transcription. This regulation is achieved by either covalent or noncovalent modification of the host RNA polymerase (RNAP; σα2ββ′) or by involvement of DNA-binding proteins recognizing regulatory elements. The need for additional protein activators appears to be restricted to promoters exhibiting weak sequence conservation in the −10 and −35 recognition hexanucleotides specific for the sigma subunits. The covalent modification of the RNAP occurs, for example, by ADP ribosylation of the α-subunit by the phage-encoded ADPR transferase as exemplified with T4 (36), which employs the core subunits of Escherichia coli RNAP throughout its transcription. In another example, bacteriophage T7, E. coli RNAP subunit β′ is the principal target for phosphorylation during the early viral gene transcription. The modification finally results in the inactivation of the host RNAP and leads to degradation of the E. coli genome (14). This is possible since bacteriophage T7 encodes its own RNA polymerase specific for the subsequent transcription of the phage proteins (35). Noncovalent modifications of the RNAP are carried out by proteins that bind to the polymerase core. The best-characterized members of this group are the sigma-like initiator factors, which positively regulate gene expression by specifically altering promoter recognition (40). Alternatively, transcription can be controlled by DNA-binding proteins, which interact with and enhance the activity of RNAP (34).
To describe the PM2 system in more detail, we investigated the regulation of gene expression. The primer extension studies were carried out by using mRNA molecules obtained from PM2-infected Pseudoalteromonas cells. Due to the lack of genetic tools in Pseudoalteromonas, a reporter gene expression system was set up in E. coli in which it was extensively used to probe the promoter regions and their regulation. The genome contains three operons, which are regulated by phage-encoded factors. One of the activators has a putative zinc finger motif similar to the eukaryotic TFIIS, RNAP A subunit 12, RNAP B subunit 9, and their archaeal homolog RpoM. Two identified repressors are highly homologous to Pseudoalteromonas plasmid pAS28 gene products.
MATERIALS AND METHODS
Bacteria, plasmids, and phages.
E. coli HMS174 (11) and HB101 (39) were used as cloning and expression hosts. Cells were grown in Luria-Bertani (LB) medium (39). When appropriate, antibiotics were added at the following concentrations: ampicillin (AMP) at 150 μg/ml, kanamycin (KAN) at 25 μg/ml, and chloramphenicol (CHL) at 10 μg/ml. Bacteriophage PM2 was grown on Pseudoalteromonas sp. strain ER72M2 in SB broth (27). The plasmids used in the present study are listed in Table 1. The plasmid pALHI used as an expression vector is a derivative of pDMI,1 (12). The 145-bp fragment from pDS12/RBSII/ΔCAT (8) containing the promoter and a multiple cloning site was digested with HindIII and XhoI restriction enzymes and ligated into HindIII- and SspI-digested pDMI,1. The resulting linear plasmid was converted into blunt-ended DNA by Klenow treatment and then ligated into the circular form by using T4 DNA ligase. Promoter reporter vectors pMG2 and pMG3 contain a promoterless CHL resistance gene (the CHL acetyltransferase [CAT] gene) (19). The CAT gene is preceded by the multiple cloning site and is in the opposite orientation in pMG2 and pMG3.
TABLE 1.
Plasmids used in this study
| Plasmid | Descriptiona | Replicon | Selective markerb | Source or reference |
|---|---|---|---|---|
| Plasmids used in CHL resistance assay | ||||
| pMG2/pMG3 | Promoter reporter vector | pMB1 | AMP (CHL) | 7, 19 |
| pSU18 | Cloning vector | p15A | CHL | 6 |
| pRM50 | pSU18 BamHI Ω(PM2 1583-77) | p15A | CHL | This study |
| pRM52 | pSU18 BamHI Ω(PM2 850-3719) | p15A | CHL | This study |
| pRM950 | pMG2 SmaI Ω(PM2 77-1583) | pMB1 | AMP (CHL) | This study |
| pRM950.2 | pMG2 SmaI Ω(PM2 1583-77) | pMB1 | AMP (CHL) | This study |
| pRM952 | pMG2 SmaI Ω(PM2 850-3719) | pMB1 | AMP (CHL) | This study |
| pRM952.2 | pMG2 SmaI Ω(PM2 3719-850) | pMB1 | AMP (CHL) | This study |
| pRM958 | pMG2 Δ(XbaI-SacI) Ω(PM2 1129-1358) | pMB1 | AMP (CHL) | This study |
| pRM975 | pMG2 Δ(XbaI-SacI) Ω(PM2 5272-5405) | pMB1 | AMP (CHL) | This study |
| pRM984 | pMG2 Δ(XbaI-BamHI) Ω(PM2 4782-5818) | pMB1 | AMP (CHL) | This study |
| pRM1054 | pMG3 Ω(PM2 1583-551, 542-77) | pMB1 | AMP (CHL) | This study |
| pRM1059 | pMG3 Δ(SacI-XbaI) Ω(PM2 1358-1129) | pMB1 | AMP (CHL) | This study |
| pRM992 | pMG2 Δ(XbaI-SacI) Ω(PM2 77-1583), P1193 mutated | pMB1 | AMP (CHL) | This study |
| pRM993 | pMG2 Δ(XbaI-SacI) Ω(PM2 77-1583), P1253 mutated | pMB1 | AMP (CHL) | This study |
| pRM994 | pMG2 Δ(XbaI-SacI) Ω(PM2 3719-850), P1207 mutated | pMB1 | AMP (CHL) | This study |
| pRM1092 | pMG3 Δ(SacI-XbaI) Ω(PM2 1583-77), P1193 mutated | pMB1 | AMP (CHL) | This study |
| pRM1094 | pMG3 Δ(SacI-XbaI) Ω(PM2 850-3719), P1207 mutated | pMB1 | AMP (CHL) | This study |
| Plasmids used to study viral transcriptional activators | ||||
| pALHI | Expression vector | p15A | KAN | This study |
| pRM513 | pALHI Δ(EcoRI-BamHI) Ω(PM2 3702-3910) | p15A | KAN | This study |
| pRM521 | pALHI Δ(EcoRI-BamHI) Ω(PM2 561-77) | p15A | KAN | This study |
| pRM531 | pALHI Δ(EcoRI-BamHI) Ω(PM2 4196-4643) | p15A | KAN | This study |
| pRM533 | pALHI Δ(EcoRI-BamHI) Ω(PM2 3892-4212) | p15A | KAN | This study |
| pRM590 | pALHI Δ(EcoRI-BamHI) Ω(PM2 3702-4212) | p15A | KAN | This study |
| pRM591 | pALHI Δ(EcoRI-BamHI) Ω(PM2 3702-4643) | p15A | KAN | This study |
| pRM595 | pALHI Δ(EcoRI-BamHI) Ω(PM2 1145-77) | p15A | KAN | This study |
| pRM596 | pALHI Δ(EcoRI-BamHI) Ω(PM2 1346-1822) | p15A | KAN | This study |
| pSN100 | pALHI Ω(PM2 1145-551, 100-77) | p15A | KAN | This study |
| pSN102 | pALHI Ω(PM2 1346-1397, 1564-1822) | p15A | KAN | This study |
| pSN104 | pALHI Ω(PM2 1346-1600, 1796-1822) | p15A | KAN | This study |
| pSN106 | pALHI Ω(PM2 1145-1087, 852-551, 100-77) | p15A | KAN | This study |
| pSN108 | pALHI Ω(PM2 1145-847, 553-551, 100-77) | p15A | KAN | This study |
Nucleotide coordinates of bacteriophage PM2 genome as previously described (33). Ω, insertion; Δ, deletion.
The selective marker indicated in parentheses is only expressed when a promoter sequence is inserted in front of it.
DNA cloning.
DNA fragments of the PM2 genome containing either ORFs a, b, c, and d or ORFs c, d, and e, along with gene XII, were cut out from plasmids pRM50 and pRM52, respectively, and converted into blunt-ended DNA by Klenow treatment. The fragments were ligated into the SmaI-digested pMG2 vector, and the orientation of the fragments in the resulting plasmids (pRM950, pRM950.2, and pRM952, pRM952.2 [Table 1]) was confirmed by sequencing. The DNA corresponding to the rest of the PM2 genome was cloned as eight overlapping DNA fragments in the pMG2 vector (data not shown). The fragments were amplified by PCR with specific primers and PM2 genomic DNA as a template.
ORF a in pRM50 was mutated by introducing an 8-bp deletion (bp 550 to 543) resulting in a frame shift, thus preventing the production of ORF a encoded protein. In vitro mutagenesis was performed by PCR with specific primers containing overlapping, complementary ends. Prior to transformation into HMS174 cells, methylated template DNA was digested with 10 U of DpnI for 2 h at 37°C. In the PCR, a new ApaI restriction enzyme site was introduced and subsequently used for screening the resulting clones. The fragment with the desired deletion was recloned into pMG3 (pRM1054 [Table 1]) and used in the CHL resistance assay (i.e., the CAT assay).
For the analysis of transcriptional regulators, ORFs were cloned into the pALHI vector. ORFs f, g, and h were cloned both separately and in different combinations (pRM513, pRM531, pRM533, pRM590, and pRM591 [Table 1]). Similarly, DNA fragments containing ORFs a, b, and c, as well as ORFs d and e, were cloned into pALHI, resulting in plasmids pRM595 and pRM596, respectively (Table 1). ORF a was deleted by PCR from plasmid pRM595, simultaneously introducing an XbaI site for screening (pSN100). Plasmid pSN100 (ORFs b and c) was used as a template for PCR, where ORF b or c was deleted one at a time (and a SacI site was created), resulting in plasmids pSN108 and pSN106, respectively (Table 1). Accordingly, ORFs d and e were deleted (and an XbaI site was introduced for screening) from plasmid pRM596 one at the time, resulting in plasmids pSN102 and pSN104, respectively (Table 1).
Site-directed mutagenesis.
PCR was used to carry out site-directed mutagenesis of the promoter regions. Two primers with complementary sequences were used to produce a circular product with desired changes. The following changes were made: the −35 consensus sequences of P1193 (TTGACC) and P1253 (TTGGCT) were changed to GGGCCC, resulting in an ApaI restriction site. Plasmid pRM50 was used as a template. The sequence of the −10 region of the promoter P1207 (TATAAT) was changed accordingly to GGGCCC by using plasmid pRM52 as a template. Amplification was performed in 50 μl of standard buffer with 2.5 U of native Pfu DNA polymerase (Stratagene), 25 pmol of primers, and a 250 μM concentration of each deoxynucleoside triphosphate. The altered fragments were recloned into the pMG2 and pMG3 vectors (Table 1). These plasmids were used in the CAT assay.
Promoter activity assay.
Overnight cultures of cells, carrying the PM2 genome fragments in plasmid pMG2 or pMG3, were diluted into fresh LB medium supplemented with AMP and grown to a density of 107 to 108 CFU/ml. Cells were plated on LB medium, LB medium plus AMP, or LB medium plus CHL, and colonies were counted after 20 to 36 h of incubation at 37°C. The same assay was performed with cells containing two different plasmids: the genome fragment in pMG2 or pMG3 containing an AMP resistance marker and putative activator genes in pALHI containing a KAN resistance marker. The expression of the gene products from pALHI at a density of 105 to 106 CFU/ml was induced by the addition of 50 μM IPTG (isopropyl-β-d-thiogalactopyranoside). Growth on LB medium plus AMP plus KAN was allowed to continue until cultures reached a density of 107 to 108 CFU/ml, after which the cells were plated on LB medium, LB medium plus AMP plus KAN, or LB medium plus CHL plus IPTG (100 μM). Colonies were counted as described above.
Isolation of total RNA from infected cells.
The chemically defined enterobacterial medium (37) was modified to support the growth for Pseudoalteromonas sp. strain ER72M2. For this, the concentration of NaCl was changed from 50 to 400 mM, and the concentration of CaCl2 was changed from 0.0005 to 10 mM, concentrations that correspond to the concentrations in the rich SB medium used for this bacterium (15, 27). Similarly, based on the SB medium, MgSO4 (50 mM final) and KCl (10 mM final) were added to the enterobacterial medium. Pseudoalteromonas sp. strain ER72M2 was grown in the defined medium at 28°C to a density of 6 × 108 CFU/ml, and the culture was infected with PM2 by using a multiplicity of infection of 30. Then, 10-ml samples were obtained (5, 10, 15, 30, and 50 min after infection), and the RNA was isolated as described earlier (19) and further purified by using the Qiagen RNeasy Midi Kit. Purified RNA was stored at −80°C until analysis.
Primer extension.
The 25- to 30-nucleotide (nt) specific oligonucleotides hybridizing to the PM2 genome were used in the assay. Oligonucleotides were labeled at their 5′ ends (39) with [γ-32P]ATP and T4 polynucleotide kinase (both from Amersham Pharmacia Biotech), after which the unbound label was removed by using Sephadex G25 columns (Amersham Pharmacia Biotech). Primer extension reactions were performed essentially as described previously (19). Total RNA from PM2 infected cells (15 to 30 μg) was hybridized to the labeled oligonucleotide (1 pmol) in 40 mM PIPES (pH 6.8)-1 mM EDTA (pH 8.0)-0.4 M NaCl-80% formamide. The mixture was heated to 65°C for 5 min and allowed to slowly cool to the appropriate hybridization temperature calculated from the oligonucleotide sequence. After additional incubation at the same temperature for 60 min, nucleic acids were precipitated and resuspended in RT buffer (Amersham Pharmacia Biotech) supplemented with 5 mM MgCl2 and a 1 mM concentration of each deoxynucleoside triphosphate. The reverse transcriptase reaction was started by the addition of 30 U of avian myeloblastosis virus reverse transcriptase (Amersham Pharmacia Biotech), and the reaction was allowed to proceed at 37°C for 30 min. Template RNA was digested with RNase A, and the extension products were precipitated with ethanol. The DNA pellet was dissolved in gel electrophoresis loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol FF) and then analyzed in a sequencing (7% acrylamide, 8.3 M urea) gel. DNA ladders from sequencing reactions were used as molecular weight markers. The sequencing reactions were performed by using PM2 genome as a template, and the corresponding oligonucleotides for the primer extension reactions as primers.
RESULTS
Primer extension revealed four mRNA start sites in the PM2 genome.
Instead of using the standard rich medium prepared from extracts (SB broth [27]), we optimized the PM2 infection in defined medium (see Materials and Methods) to obtain a maximal amount of mRNA for primer extension experiments.
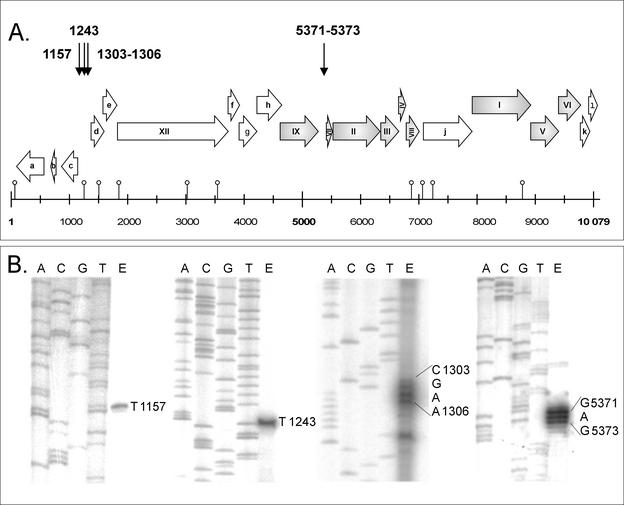
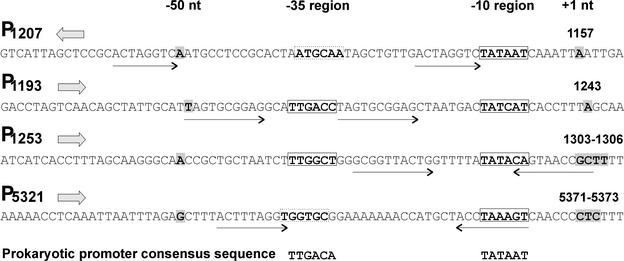
Oligonucleotides were designed to hybridize ca. 50 bp downstream of the initiation codon of ORFs c, d, f, g, j, and k; genes I, II, V, VI, VII, and IX; and within gene XII (Fig. 1). Total RNA samples isolated at different time points of the PM2 life cycle were combined and used as templates. When the resulting products were analyzed in sequencing gels, four putative mRNA start sites were resolved at positions 1157, 1243, 1303 to 1306, and 5371 to 5373 (nucleotide coordinates refer to the PM2 genome sequence; AF155037 [33]; Fig. 1). The corresponding putative promoter sequences (P1207, P1193, P1253, and P5321, respectively) are shown in Fig. 2. All observed start sites are appropriately positioned downstream from a sequence region similar to that of the E. coli −10 hexameric consensus (21), whereas the −35 region is not as recognizable in P1207 and P5321. In the case of promoters P1193, and P1253, 18 bp are spaced between the two hexameric consensus sequence motifs.
FIG. 1.
Primer extension analysis of mRNA start sites in the PM2 genome. The numbers refer to the nucleotide coordinates of the PM2 genome (AF155037). (A) PM2 genome map linearized at a unique EcoRII site. The genes (assigned by Roman numerals) and ORFs (marked with lowercase letters) (28, 33) are presented with arrows indicating the direction of transcription. Positions of the predicted hairpin structures are indicated by an open circle upon the scale bar (33). The genes confirmed to code for structural proteins are shaded gray. The mRNA start sites determined by primer extension are marked with downward arrows at the top of the figure. (B) Results of the primer extension reactions (lane E) analyzed parallel to the sequencing reactions in a sequencing gel.
FIG. 2.
Alignment of the four PM2 promoter sequences revealed by primer extension. Promoters are named with P after the nucleotide coordinate at the −50 position. Orientation of the promoter in the PM2 genome (presented in Fig. 1) is indicated by a solid arrow. The +1 (mRNA start site) and −50 nt are shaded in gray. Predicted hexameric promoter consensus sequences −35 and −10 are boxed either with solid lines (well conserved) or dashed lines (less conserved). Repeat sequences in the promoter regions are indicated with arrows.
A CHL resistance assay was used to measure promoter activity.
Since no cloning and expression system is available for Pseudoalteromonas, the CAT assay was performed in E. coli. For this, the PM2 genome was cloned as overlapping fragments into promoter reporter vectors pMG2 and pMG3, where expression of the CHL resistance marker was obtained only if the cloned insert had promoter activity. E. coli cells carrying the plasmids were plated both on LB medium plus AMP and on LB medium plus CHL, and the colonies were counted. The cell count without any selection was also determined. The promoter activity of the cloned fragments was determined as a proportion of the number of CHL-resistant colonies of those obtained in the presence of AMP (scoring the presence of the reporter plasmid).
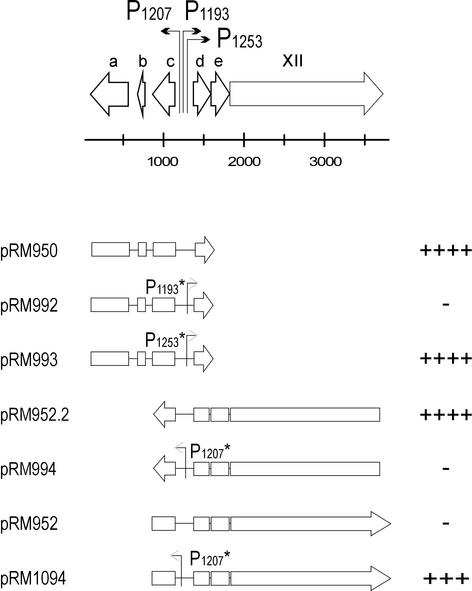
Most of the cloned PM2 genomic fragments could not promote CHL resistance in the initial assays. Activity was recorded only with plasmids pRM950 (containing fragment 77-1583 from PM2 genome; Table 1) and pRM952.2 (3719-850; Table 1). Both of these constructs carry three of the putative promoters—P1207, P1193, and P1253—determined by primer extension. No CAT activity was observed with fragments containing the fourth putative promoter, P5321 (data not shown).
Activity of P1207 represses P1193.
The functionality of the three closely positioned promoters P1207, P1193, and P1253 was analyzed by site-directed mutagenesis. On the basis of the primer extension results, P1207 promotes leftward transcription, whereas P1193 and P1253 promote rightward transcription (Fig. 3). Promoters P1207 and P1193 are organized back to back so that their −35 consensus sequences and −50 nt partially overlap. Promoters were mutated individually as described in Materials and Methods, and the CAT assay was performed by using the corresponding wild-type (wt) genomic fragments as controls.
FIG. 3.
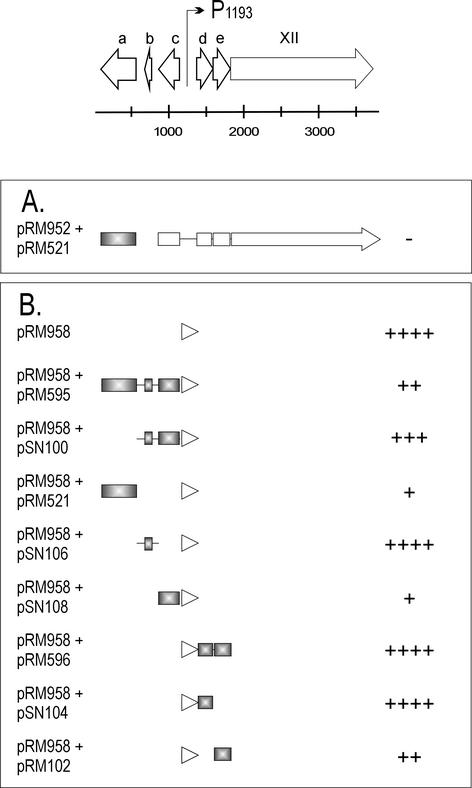
Functional analysis of the PM2 genome region from nt 77 to 3719 by using the CAT assay. The putative protein coding ORFs are presented with open arrows, and promoters are indicated by bent arrows at the top of the figure. Below are shown the genomic fragments from this region cloned into the CAT reporter plasmid, and the corresponding plasmids are denoted at the left. Open boxes and arrows indicate the ORFs. The arrowhead points to the direction of transcription in the CAT fusion construct. Mutated promoters are indicated with an asterisk after the promoter name. On the right, the pluses and minuses represent the number of CHL-resistant colonies in the CAT assay: the cell count on LB medium-plus-AMP plates was compared to the cell count on LB medium-plus-CHL plates. ++++, All colonies were CHL resistant; +++, the number of CHL-resistant colonies was 1 order of magnitude lower; ++, the number of CHL-resistant colonies was 2 orders of magnitude lower; +, the number of CHL-resistant colonies was 3 orders of magnitude lower than on the LB medium; −, no CHL-resistant colonies were obtained.
Rightward transcription was promoted by P1193 only, since changing the sequence of P1193 (pRM992) by site-directed mutagenesis resulted in CHL sensitivity, whereas mutating the sequence of P1253 (pRM993) did not affect CAT activity compared to the control plasmid pRM950 with the wt promoter (Fig. 3). This finding suggested that P1253 might not be a functional promoter, although it was revealed by primer extension. The palindromic sequence centered at the position 1290 has a potential to form a strong hairpin loop (−ΔG = 17.2 kcal/mol) (Fig. 1) (33), which can cause immature termination of the extension reaction, leading to a false-positive result in mRNA start site determination.
The function of the leftward promoter (P1207) was examined with plasmid pRM952.2, which showed high activity in the CAT assay (Fig. 3). When the −10 consensus sequence of P1207 was mutated, the resulting plasmid, pRM994, lost the ability to promote CAT activity. Interestingly, if the orientation of the wt fragment was inverted (pRM952), signaling the activity of the rightward promoters, no CAT activity was detected. However, the ability of this construct to promote CHL resistance was achieved by mutating the −10 sequence of P1207 (pRM1094, Fig. 3). This indicated that the rightward promoter P1193 is only active when the leftward promoter P1207 is shut down. This is not in concordance with the result obtained with plasmid pRM950 (Fig. 3), for which the activity of P1193 could be detected in the presence of active leftward P1207.
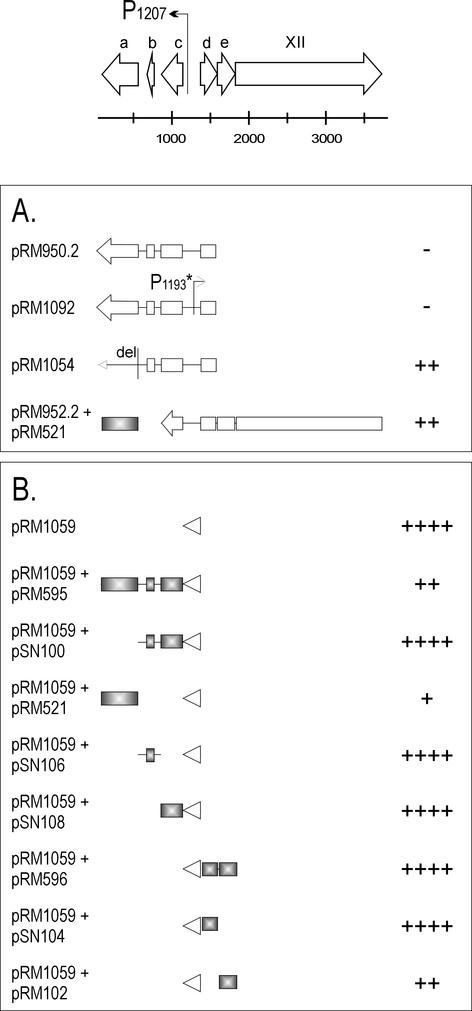
The product of ORF a is a repressor of its own promoter P1207.
The activity of the leftward promoter P1207 could not be detected with plasmid pRM950.2 (Fig. 4A). Further, activity could not be restored by mutating the rightward promoter P1193 (pRM1092). The fact that the activity of P1207 was not dependent on the loss of function of the rightward promoter and the inconsistent results obtained with pRM950 and pRM952 (see above and Fig. 3) led us to hypothesize that one of the gene products under P1207 regulation could self-regulate its promoter activity by repression. The candidates for this repression were ORFs a, b, and c. The first 8 nt of ORF a were deleted to make a frameshift mutation. Indeed, knocking out the product of ORF a (pRM1054) resulted in the activation of promoter P1207 (Fig. 4A), which indicated that the ORF a-encoded factor is a repressor of the leftward promoter. This observation was confirmed in a coexpression experiment, in which ORF a was expressed from the pALHI vector and the promoter activity was recorded with the promoter reporter vector. The CAT activity obtained with the plasmid pRM952.2 (Fig. 3) was reduced when the ORF a-expressing plasmid pRM521 was in the same cell (Fig. 4A). The products of the other ORFs of the operon (b and c) did not have any repressor function on P1207, as shown below.
FIG. 4.
Analysis of the promoter P1207 with different potential regulators by the CAT assay. For an explanation of the genomic region at the top of the figure, see the legend to Fig. 3. (A) Promoter activity was assayed with long genomic fragments containing several ORFs possibly coding for regulators. (B) The 230-bp promoter region (presented by an arrowhead) was cloned into the CAT reporter vector and the potential regulator genes (colored gray) in another, compatible vector. The plasmid constructs are indicated at the left. For the level of the CAT expression (plus and minus signs), see Fig. 3 legend.
Analysis of other possible regulators for P1207.
The short, noncoding, 230-bp region between ORFs c and d containing the promoter P1207 (and P1193) was cloned into the promoter reporter vector pMG3. Resulting plasmid pRM1059 showed activity in the CAT assay (Fig. 4B). The effect of overexpressed ORF a, b, c, d, or e products on the promoter activity was examined by coexpressing them individually and in different combinations with plasmid pRM1059. The repressor effect of the ORF a product on promoter P1207 also was clearly seen in this experiment (pRM1059 plus pRM595 and pRM1059 plus pRM521; Fig. 4B). The effect was obtained both with the ORF a product alone and with the products of ORFs a, b, and c together. ORFs b and c together and separately (pSN100, pSN106, and pSN108, respectively) did not have any effect on P1207 activity (Fig. 4B). A second repressor effect on P1207 was recorded with the ORF e-encoded product (pSN102). The activity of P1207 was reduced when coexpressed with pRM1059. However, this result could not be confirmed with a longer DNA fragment expressing ORFs d and e together (pRM596; Fig. 4B).
The product of ORF a has also an effect on rightward promoter P1193.
First, plasmid pRM952 (signaling the functionality of the rightward promoter P1193), which showed no activity in the CAT assay (Fig. 3), was tested together with ORF a, but this did not result in activation of P1193 (Fig. 5A). The effect of other possible regulators on P1193 was studied with the coexpression experiments (Fig. 5B). It was shown, surprisingly, that the product of ORF a also had repressive effect on P1193, strongly diminishing its activity (pRM958 plus pRM595 and pRM958 plus pRM521; Fig. 5B). The effect of the ORF a product on P1193 was unexpected and might indicate that it is a DNA-binding protein. The close packing of the two early promoters might result in repression of both promoters upon binding of the ORF a product.
FIG. 5.
Analysis of the promoter P1193 with potential regulator proteins. For the explanation of the genomic region at the top of the figure, see the legend to Fig. 3. (A) Promoter activity is assayed with long genomic fragments containing several ORFs possibly coding for regulators.(B) The 230-bp promoter region (indicated by an arrowhead) was cloned into the CAT reporter vector, and the potential regulator genes (shaded gray) were cloned into another, compatible vector. The plasmid constructions are indicated at the left. For the level of the CAT expression (plus and minus signs), see the legend to Fig. 3.
In contrast to the result obtained with P1207, overexpression of ORF c with P1193 resulted in a clear decrease in promoter activity (pRM958 plus pSN108; Fig. 5B). ORFs a and c were classified as genes XV and XVI, respectively, and the corresponding proteins were named P15 and P16. As in the case of P1207 (see above), the product of ORF e somewhat repressed the activity of P1193 when tested as a single gene in the coexpression experiment (pRM958 plus pSN102). Similar to the results obtained with P1207, this could not be confirmed with a construct carrying both ORFs e and d (pRM958 plus pRM596; Fig. 5B). ORF e might have repressor activity on both P1207 and P1193, but since the results were obtained with the one plasmid system only, we do not yet give a gene status for ORF e.
The early regulation region encoding repressor proteins P15 and P16 is similar to a Pseudoalteromonas plasmid.
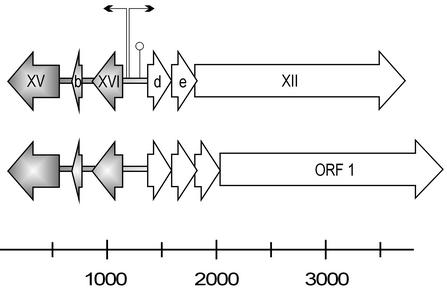
Alignment of the maintenance region sequence of plasmid pAS28, isolated from Pseudoalteromonas sp. strain A28 (AB009311 [26]) with the PM2 early region revealed similar genome organization (Fig. 6). The 1.2-kb region containing genes XV and XVI and ORF b is highly homologous to pAS28 (33). The identities of proteins P15 and P16 and the product of ORF b (at the amino acid level) to the corresponding gene products in pAS28 were 76, 72, and 57%, respectively. The intergenic regions were less similar, but the putative −10 and −35 consensus sequences of promoters also can be found in pAS28 matching the position of those in PM2 (data not shown). Even a palindromic sequence, which can cause a hairpin structure similar to the sequence found in PM2 P1253, is found in pAS28. The homology between PM2 and pAS28 might extend even further. The PM2 ORF d product has some similarity (28% identity) to the centrally located small ORFs transcribed rightward in pAS28. Deletion of ORF 1 (coding for an 81-kDa protein; the counterpart of PM2 gene XII) from pAS28 prevents plasmid replication (26). On the basis of this observation, the ORF 1 product has been classified as a replication protein, although it does not possess any sequence similarity to proteins in the data bank (26), even to PM2 P12.
FIG. 6.
Alignment of the PM2 genome region from nt 77 to 3719 with the maintenance region from Pseudoalteromonas plasmid pAS28 (accession no. AB009311) showing similar organization. Genes and ORFs are denoted by open arrows indicating the direction of transcription. The two promoters (bent arrows) and the possible secondary structure (○) in the noncoding region of PM2 are marked. The gray shaded areas in PM2 and pAS28 are homologous to each other.
P5321, silent in the CAT assay, can be activated by phage-encoded factors.
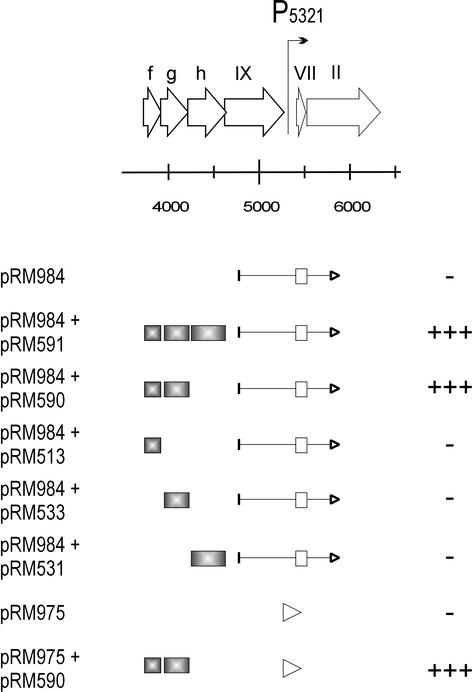
Promoter P5321, which is located in the middle of the linearized genome map preceding the genes for the phage structural proteins, was not active in the CAT assay (pRM984; Fig. 7). The possible regulatory effect of the nearby ORFs was tested by coexpression experiments. For this, the candidate genes (ORFs f, g, and h) were cloned separately and in different combinations into the pALHI expression vector, and the resulting constructs were expressed with pRM984 (Fig. 7). None of these putative gene products alone was able to activate the promoter. However, coexpression of ORFs f and g (pRM590) or ORFs f, g, and h (pRM591) with plasmid pRM984 resulted in increased colony count on plates containing LB medium plus CHL (Fig. 7). The result that ORFs f and g together but not separately are able to activate P5321 was confirmed by using the 134-bp promoter fragment (pRM975; Fig. 7). Consequently, ORFs f and g were classified as proteins P13 and P14, respectively, and the corresponding genes were named XIII and XIV.
FIG. 7.
Analysis of the promoter P5321 with the potential activators by the CAT assay. The PM2 genome region from nt 3716 to 6332 is illustrated at top of the figure. Genes and ORFs are depicted with open arrows indicating the direction of transcription. The location of the promoter P5321 in the genome is indicated with a bent arrow. Below are indicated the genomic fragments, which were used in the promoter activity assay. ORFs and genes are marked with boxes, and an arrowhead points to the direction of transcription in the CAT fusion construct. The plasmids are denoted on the left. The assay was performed with either a long genomic fragment or the 134-bp promoter region cloned into the CAT reporter vector (pRM984 and pRM975, respectively). The possible regulators (shaded gray) were cloned into another compatible vector. For the level of the CAT expression (plus and minus signs), see the legend to Fig. 3.
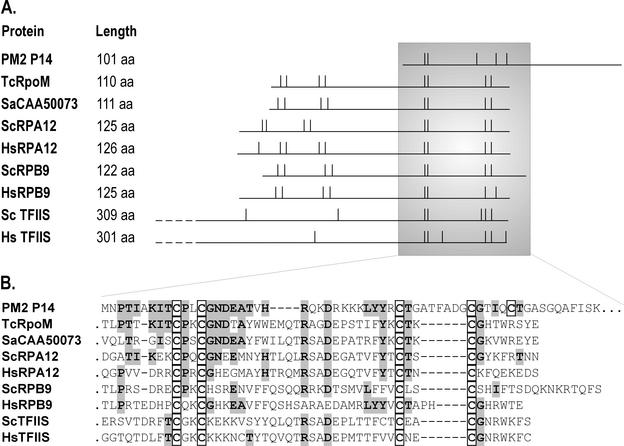
A database search (2, 3) revealed that P14 contains a putative zinc finger motif similar to archaeal and eukaryotic transcription factors (Fig. 8) (41). The closest amino acid sequence similarity was detected between the N-terminal region of P14 and the C-terminal domain of archaeal Thermococcus celer transcription-associated protein (PSI-BLAST e-score of 5e-10) (25). Other proteins containing this conserved zinc finger motif are small subunits of eukaryotic RNA polymerases I (A12) and II (B9), as well as the eukaryotic transcription factor TFIIS. Transcription initiation factor TFIIB also contains an analogous zinc finger structure (42). A database search revealed no sequence similarities for protein P13, which together with P14, was shown to activate promoter P5321.
FIG. 8.
Comparison of PM2 protein P14 with some archaeal and eukaryotic transcription regulators with zinc finger motifs. (A) The polypeptides are aligned (horizontal lines) so that the cysteine residues match (vertical lines). Aligned proteins PM2 P14, as well as HsTFIIS and ScTFIIS, have only one zinc finger domain (indicated by two cysteine pairs), whereas the others contain two. (B) The amino acid (aa) sequence from the shadowed area in panel A is shown. Identical amino acids between P14 and related sequences are shaded gray, and the cysteines of the zinc finger domain are boxed. Note that there is an alternative way to align the second cysteine pair in P14. Abbreviations: Tc, Thermococcus celer; Sa, Sulfolobus acidocaldarius; Sc, Saccharomyces cerevisiae; Hs, Homo sapiens; RpoM, archaeal RNA polymerase subunit M; RPA12 and RPB9, eukaryotic RNA polymerase I and II subunits, respectively, TFIIS, eukaryotic general transcription factor. Accession numbers: AAD43545 (PM2 P14), Q56254 (TcRpoM), CAA50073 (transcription-associated protein from Sa), AAA34992 (ScRPA12), AAG50159 (HsRPA12), P27999 (ScRPB9), P36954 (HsRPB9), P07273 (ScTFIIS), NP_006747 (HsTFIIS).
DISCUSSION
The genetic approach to reveal the transcription regulation system of PM2 used (i) primer extension results obtained from PM2 infection in the native host and (ii) extensive promoter activity experiments carried out in E. coli since there are no genetic tools available for the Pseudoalteromonas-PM2 system. Although the CAT reporter assay can be considered a qualitative method, the results obtained were in good agreement with those obtained in primer extension experiments. In addition, the reporter assays were always carried out with a number of control experiments verifying the qualitative results obtained. For quantitative promoter activity measurements, a different experimental setup is required, but this is outside the scope of this investigation.
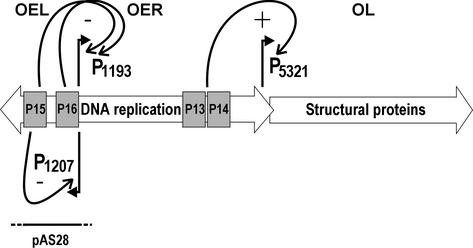
The leftward transcribing early operon (OEL, under P1207) was shown to code for two transcriptional repressors (Fig. 9). Protein P15 controls its own promoter and P16 controls the rightward early operon (OER, under P1193). These two functional promoters could be narrowed to a 230-bp DNA fragment. Primer extension results revealing the third early promoter P1253 have not been verified but are explained by the presence of a strong palindromic DNA structure centered at this position. Expression of the late operon (OL) for the structural proteins is controlled by proteins P13 and P14 (Fig. 9). The late promoter region was mapped to a 134-bp DNA fragment.
FIG. 9.
Proposed temporal sequence of PM2 transcription. The three functional promoters are depicted. Promoter P1207 controlling the early genes (operon OEL) is repressed by protein P15. The genes for the DNA replication proteins (operon OER) are under the control of P1193. Proteins P15 and P16 have repressor effects on this promoter. Transcription factors P13 and P14 are needed to switch on the late promoter P5321 for the production of the structural components of the virus particle (operon OL). Homology to the Pseudoalteromonas plasmid pAS28 is indicated with a bar.
We previously reported that an ∼1.2-kb region in the PM2 genome is homologous to the Pseudoalteromonas plasmid pAS28 regulatory region (26, 33) (Fig. 9). A more detailed comparison revealed high homology in the coding regions of genes XV and XVI and somewhat less homology in ORF b. The noncoding regions do not have clear sequence similarity. However, the consensus sequences of promoters could be observed in pAS28 in the area corresponding to the positions of the PM2 early promoters. Gene XII for the replication initiation protein does not seem to be related to that of pAS28. Although the accurate borders of recombination events are difficult to define, it appears that the early autoregulated operon in PM2 might be a moron (for the definition, see reference 23) sharing the same origin with pAS28.
The operon OL is positively regulated (activated) by proteins P13 and P14 encoded from the early operon (OER). These proteins work in concert since they had no effect when tested alone. Intriguingly, data bank searches revealed that P14 has a zinc finger motif similar to the transcription factors of eucaryal and archaeal origin, more closely resembling the archaeal counterparts (Fig. 8). Several eukaryotic transcription initiation factors have been identified from archaeal species (32), TFIIB from Pyrococcus woesei being the first one (38). In addition, bacterial transcription regulators also have been described from Archaea (13, 29, 30). By comparing the sequences of known transcription-associated proteins from Eucarya and Bacteria with the sequenced archaeal genomes, it was shown that, among the 280 predicted transcription-associated proteins, 51 had homologs only in Eucarya, 168 had homologs only in Bacteria, and the remaining 61 had homologs in both domains representing the group of universal transcription factors. The archaeal factors present only in Eucarya included, among others, the RNAP A subunit 12 and RNAP B subunit 9 and the general transcription factor TFIIS. These are thought to be the homologs of the archaeal polymerase subunit M (RpoM) (31). We report here, for the first time, a TFIIS-type factor from a virus replicating in Bacteria. Whether gene XIV, coding for this factor, has been acquired by PM2 via horizontal gene transfer or whether the transcription machineries operating in the three domains of life are more related than previously expected remains to be evaluated.
The analysis of the genome structure and function of PM2 revealed a mosaic arrangement and clearly indicated horizontal gene transfer events between Pseudoalteromonas phages and plasmids. This finding is in line with the observations made when tailed double-stranded DNA phage genomes were compared (22, 23). The homology revealed between bacterial, archaeal, and eucaryal transcription factors not previously observed could point to more uniform transcription mechanisms than are presently considered.
Acknowledgments
This work was supported by a research grant 172904 (J.K.H.B.) and research grants 162993 and 164298 (D.H.B.; Finnish Centre of Excellence Programme [2000-2005]) from the Academy of Finland. R.H.M. is a fellow of the Viikki Graduate School in Biosciences.
The technical assistance of Sara Nummi is gratefully acknowledged.
REFERENCES
- 1.Ackermann, H.-W. 2000. Family Corticoviridae, p. 117-120. In M. H. V. van Regenmortel, C. M. Fauquet, and D. H. L. Bishop (ed.), Virus taxomony: classification and nomenclature of virus. Academic Press, Inc., San Diego, Calif.
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bamford, D. H., and H.-W. Ackermann. 2000. Family Tectiviridae, p. 111-116. In M. H. V. van Regenmortel, C. M. Fauquet, and D. H. L. Bishop (ed.), Virus taxomony: classification and nomenclature of virus. Academic Press, Inc., San Diego, Calif.
- 5.Bamford, J. K. H., A.-L. Hänninen, T. M. Pakula, P. M. Ojala, N. Kalkkinen, M. Frilander, and D. H. Bamford. 1991. Genome organization of membrane-containing bacteriophage PRD1. Virology 183:658-676. [DOI] [PubMed] [Google Scholar]
- 6.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 7.Brosius, J. 1984. Plasmid vectors for the selection of promoters. Gene 27:151-160. [DOI] [PubMed] [Google Scholar]
- 8.Bujard, H., R. Gentz, M. Lanzer, D. Stueber, M. Mueller, I. Ibrahimi, M.-T. Haeuptle, and B. Dobberstein. 1987. A T5 promoter-based transcription-translation system for the analysis of proteins in vitro and in vivo. Methods Enzymol. 155:416-433. [DOI] [PubMed] [Google Scholar]
- 9.Butcher, S. J., D. H. Bamford, and S. D. Fuller. 1995. DNA packaging orders the membrane of bacteriophage PRD1. EMBO J. 14:6078-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camerini-Otero, R. D., and R. M. Franklin. 1972. Structure and synthesis of a lipid-containing bacteriophage. XII. The fatty acids and lipid content of bacteriophage PM2. Virology 49:385-393. [DOI] [PubMed] [Google Scholar]
- 11.Campbell, J. L., C. C. Richardson, and F. W. Studier. 1978. Genetic recombination and complementation between bacteriophage T7 and cloned fragments of T7 DNA. Proc. Natl. Acad. Sci. USA 75:2276-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Certa, U., W. Bannwarth, D. Stüber, R. Gentz, M. Lanzer, S. le Grice, F. Guillot, I. Wendler, G. Hunsmann, H. Bujard, and J. Mous. 1986. Subregions of a conserved part of the HIV gp41 transmembrane protein are differentially recognized by antibodies of infected individuals. EMBO J. 5:3051-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen-Kupiec, R., C. Blank, and J. A. Leigh. 1997. Transcriptional regulation in Archaea: in vivo demonstration of a repressor binding site in a methanogen. Proc. Natl. Acad. Sci. USA 94:1316-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunn, J. J., and F. W. Studier. 1983. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 166:477-535. [DOI] [PubMed] [Google Scholar]
- 15.Espejo, R. T., and E. S. Canelo. 1968. Properties of bacteriophage PM2: a lipid-containing bacterial virus. Virology 34:738-747. [DOI] [PubMed] [Google Scholar]
- 16.Espejo, R. T., and E. S. Canelo. 1968. Properties and characterization of the host bacterium of bacteriophage PM2. J. Bacteriol. 95:1887-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espejo, R. T., E. S. Canelo, and R. L. Sinsheimer. 1969. DNA of bacteriophage PM2: a closed circular double-stranded molecule. Proc. Natl. Acad. Sci. USA 63:1164-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gauthier, G., M. Gauthier, and R. Christen. 1995. Phylogenetic analysis of the genera Alteromonas, Shewanella and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int. J. Syst. Bacteriol. 45:755-761. [DOI] [PubMed] [Google Scholar]
- 19.Grahn, A. M., J. K. H. Bamford, M. C. O'Neill, and D. H. Bamford. 1994. Functional organization of the bacteriophage PRD1 genome. J. Bacteriol. 176:3062-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray, H. B. J., W. B. Upholt, and J. Vinograd. 1971. A buoyant method for the determination of superhelix density of closed circular DNA. J. Mol. Biol. 62:1-19. [DOI] [PubMed] [Google Scholar]
- 21.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendrix, R. W., J. G. Lawrence, G. F. Hatfull, and S. Casjens. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504-508. [DOI] [PubMed] [Google Scholar]
- 24.Ilyina, T. V., and E. V. Koonin. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 20:3279-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaine, B. P., I. J. Mehr, and C. R. Woese. 1994. The sequence, and its evolutionary implications, of a Thermococcus celer protein associated with transcription. Proc. Natl. Acad. Sci. USA 91:3854-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato, J., J. Amie, Y. Murata, A. Kuroda, A. Mitsutani, and H. Ohtake. 1998. Development of a genetic transformation system for an alga-lysing bacterium. Appl. Environ. Microbiol. 64:2061-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kivelä, H. M., R. H. Männistö, N. Kalkkinen, and D. H. Bamford. 1999. Purification and protein composition of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262:364-374. [DOI] [PubMed] [Google Scholar]
- 28.Kivelä, H. M., N. Kalkkinen, and D. H. Bamford. 2002. Bacteriophage PM2 has a protein capsid surrounding a spherical lipid-protein core. J. Virol. 76:8169-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyrpides, N. C., and C. A. Ouzounis. 1995. The eubacterial transcriptional activator Lrp is present in the archaeon Pyrococcus furiosus. Trends Biochem. Sci. 20:140-141. [DOI] [PubMed] [Google Scholar]
- 30.Kyrpides, N. C., and C. A. Ouzounis. 1997. Bacterial sigma 70 transcription factor DNA-binding domains in the archaeon Methanococcus jannaschii. J. Mol. Evol. 45:706-707. [PubMed] [Google Scholar]
- 31.Kyrpides, N. C., and C. A. Ouzounis. 1999. Transcription in Archaea. Proc. Natl. Acad. Sci. USA 96:8545-8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langer, D., J. Hain, P. Thuriaux, and W. Zillig. 1995. Transcription in Archaea: similarity to that in Eucarya. Proc. Natl. Acad. Sci. USA 92:5768-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Männistö, R. H., H. M. Kivelä, L. Paulin, D. H. Bamford, and J. K. H. Bamford. 1999. The complete genome sequence of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262:355-363. [DOI] [PubMed] [Google Scholar]
- 34.Mencía, M., M. Salas, and F. Rojo. 1993. Residues of the Bacillus subtilis phage φ29 transcriptional activator required both to interact with RNA polymerase and to activate transcription. J. Mol. Biol. 233:695-704. [DOI] [PubMed] [Google Scholar]
- 35.Molineux, I. J. 2001. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 40:1-8. [DOI] [PubMed] [Google Scholar]
- 36.Mosig, G., and F. Eiserling. 1988. Phage T4 structure and metabolism. III. Regulation of transcription. p. 542-548. In R. Calendar (ed.), The bacteriophages, vol. 2. Plenum Press, Inc., New York, N.Y.
- 37.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ouzounis, C., and C. Sander. 1992. TFIIB, an evolutionary link between the transcription machineries of archaebacteria and eukaryotes. Cell 71:189-190. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell (ed.). 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Stewart, C. 1988. Bacteriophage SPO1. V. Regulation of SPO1 gene action, p. 484-501. In R. Calendar (ed.), The bacteriophages, vol. 1. Plenum Press, Inc., New York, N.Y.
- 41.Wang, B., D. N. Jones, B. P. Kaine, and M. A. Weiss. 1998. High-resolution structure of an archaeal zinc ribbon defines a general architectural motif in eukaryotic RNA polymerases. Structure 6:555-569. [DOI] [PubMed] [Google Scholar]
- 42.Zhu, W., Q. Zeng, C. M. Colangelo, L. M. Lewis, M. F. Summers, and R. A. Scott. 1996. The N-terminal domain of TFIIB from Pyrococcus furiosus forms a zinc ribbon. Nat. Struct. Biol. 3:122-124. [DOI] [PubMed] [Google Scholar]