Abstract
The virB-encoded type IV transport complex of Agrobacterium tumefaciens mediates the transfer of DNA and proteins into plant cells, as well as the conjugal transfer of IncQ plasmids, such as RSF1010, between Agrobacterium strains. While several studies have indicated that there are physical interactions among the 11 VirB proteins, the functional significance of the interactions has been difficult to establish since all of the proteins are required for substrate transfer. Our previous studies, however, indicated that although all of the VirB proteins are required for the capacity of a strain to serve as an RSF1010 donor, only a subset of these proteins in the recipient is necessary to increase the conjugal frequency by 3 to 4 logs. The roles of particular groups of VirB proteins in this increased recipient activity were examined in the study reported here. Examination of the expression of subgroups of virB genes revealed that translation of virB6 is necessary for expression of downstream open reading frames. Expression of limited subsets of the VirB proteins in a recipient strain lacking the Ti plasmid revealed that the VirB7 to VirB10 proteins yield a subcomplex that is functional in the recipient assay but that the VirB1 to VirB4 proteins, as a group, dramatically increase this activity in strains expressing VirB7 to VirB10. Finally, the membrane distribution and cross-linking patterns of VirB10, but not of VirB8 or VirB9, in a strain expressing only VirB7 to VirB10 are significantly altered compared to the patterns of the wild type. These characteristics are, however, restored to the wild-type status by coexpression of VirB1 to VirB3. Taken together, these results define subsets of type IV transport complex proteins that are critical in allowing a strain to participate as a recipient in virB-mediated conjugal RSF1010 transfer.
Type IV transport complexes are utilized to transport macromolecules out of gram-negative bacteria in a wide variety of circumstances, including conjugal transfer of plasmids between bacteria, transfer of virulence factors into host cells, and secretion of virulence factors into the extracellular environment (for reviews see references 19, 23, and 83). Included among the pathogens that utilize type IV secretion systems are Helicobacter pylori, Campylobacter jejuni, Legionella pneumophila, Bordetella pertussis, Brucella suis, and Bartonella henselae. Agrobacterium tumefaciens, a pathogen that causes tumor formation on a wide variety of plants, utilizes the archetypal type IV transporter, the VirB complex (45, 65, 72, 77, 78), to transfer the T-DNA segment of the resident Ti (tumor-inducing) plasmid, as well as virulence proteins, into host cells. Specifically, a protein-single-stranded DNA intermediate (VirD2-T strand) and the proteins VirE2 and VirF are substrates for VirB transporter activity (14, 24, 53, 58, 66, 74, 85). Once in the plant cells, the T-DNA is ultimately integrated into the nuclear genome and expressed, resulting in the tumorous phenotype of transformed cells (16, 36). Besides T-DNA and protein transfer to host plant cells, the VirB complex can direct the conjugal transfer of an IncQ plasmid between A. tumefaciens strains or from A. tumefaciens into plant cells (11, 18). The processing of RSF1010 to form a transferable intermediate depends on its own gene products, the Mob proteins, and the oriT site (18). However, transfer of this plasmid from A. tumefaciens to either plant cells or other bacteria relies on the same VirB transfer machinery used for T-DNA transfer (11, 17, 33, 81).
The virB operon, along with virD4, encodes 12 membrane-associated proteins that are postulated to form the macromolecular transfer apparatus (for reviews see references 21, 86, and 87). This operon is regulated by a two-component regulatory system, VirA and VirG, that induces vir gene expression in response to plant compounds such as acetosyringone (AS) (15, 82). Most of the VirB proteins are required for virulence; the only exception is the protein encoded by virB1, which, nevertheless, is necessary for maximal transfer efficiency (13). The VirB proteins and VirD4 are thought to form a complex that spans the inner and outer membranes. Most models suggest that VirB6, VirB7, VirB8, VirB9, and VirB10 form the core of the VirB complex. VirB6 has multiple membrane-spanning domains (27) that suggest that it may anchor the export apparatus to a specific location in the inner membrane, although recent data suggest that its activities are more critical for T-pilus formation than for transport complex integrity (37). VirB7, an outer membrane lipoprotein, interacts with itself (37, 61) and with VirB9 via disulfide bonds between unique reactive cysteine residues present in each protein (3, 9, 70). Several lines of evidence indicate that the VirB7-VirB9 complex, in turn, is necessary for VirB10 stabilization and complex formation (9, 10, 31). Recent evidence indicates the presence of a high-molecular-weight subassembly containing these proteins that can be solubilized from isolated membranes (44). In the yeast two-hybrid assay, interactions among VirB8, VirB9, and VirB10 were observed (28), as were interactions between VirB proteins that should result in a contiguous complex from the cytoplasm to the outer membrane (76). Finally, immunofluorescence and immunoelectron microscopic methods (46) have shown that VirB8, VirB9, and VirB10 are concentrated at several foci on the cell surface, which represent potential sites of complex formation and/or potential activity.
While the VirB7 to VirB10 proteins have been proposed to be the core of the transport complex, all of the VirB proteins are required for the formation of the T pilus (34, 48). This extracellular structure is primarily composed of processed VirB2 proteins (47), but it also includes VirB5 (62) and VirB7 (61) as minor components. Interestingly, a subassembly of these proteins, distinct from the VirB7-VirB8-VirB9-VirB10 subassembly, can be isolated from membrane preparations of wild-type strain C58 (44). The T pilus is probably involved in contact with the host cell, although it may also provide a conduit for DNA transport (49). VirB4 and VirB11 are membrane localized and have ATP binding sites that are necessary for virulence (12, 25, 60, 71), and VirD4, also required for virulence, is membrane localized and has a nucleoside triphosphate binding domain (6, 56). These ATPases could be involved either directly in the transport process or in construction of the transport apparatus.
The proposed VirB complex is being intensively investigated, particularly with regard to the localization of individual VirB proteins, interactions between VirB proteins, and possible interactions between transported substrates and particular VirB proteins. However, the complex nature of the VirB transporter has made it difficult to relate physical characteristics to function. Our previous studies demonstrated that the presence of the Ti plasmid in a recipient Agrobacterium cell increases the frequency of RSF1010 conjugal transfer by as much as 3 to 4 logs (17). Moreover, while all of the VirB proteins are required for a strain to serve as an RSF1010 donor, only a subset of these proteins is necessary for increased recipient activity. Specifically, several of the VirB proteins are not required for recipient activity, including VirD4 and VirB11 (17), as well as the ATPase activity of VirB4, although the VirB4 protein is required (25). Moreover, point mutations in VirB9 that quantitatively affect the export activity of the VirB complex have a similar quantitative effect on recipient activity (17). The latter finding indicates that at least some of the structural requirements of the type IV complex are the same for donor and recipient activities.
These results are unexpected since previous studies indicated that if anything, the presence of conjugal plasmids can inhibit a strain from serving efficiently as a recipient by preventing either mating pair formation or signaling to initiate donor conjugal DNA synthesis (i.e., surface and entry exclusion) (1). However, the properties of bacterial cells that make them efficient recipients in conjugal transfer are poorly defined, particularly in systems other than F plasmid transfer (29). Studies of the F plasmid have demonstrated that ompA mutants of Escherichia coli strains are poor recipients for this plasmid but are not affected as recipients for other types of plasmids (32, 38, 67). Other experiments have indicated that mutations in the lipopolysaccharide genes can have either positive or negative effects on recipient activity, depending on the specific type of lipopolysaccharide that ends up being produced (4, 63). Even in these cases, however, most of these mutations do not affect mating carried out on solid surfaces rather than in liquid (2).
The VirB-mediated increase in the capacity of an Agrobacterium strain to serve as a recipient thus provides a unique opportunity to examine mechanisms involved in conjugal DNA transfer across various membrane and wall systems. Characterization of the role played by the VirB proteins in this process requires an understanding of the particular proteins involved and whether interactions among them are required. Here we describe a study in which various subsets of VirB proteins were expressed in a strain lacking a Ti plasmid. Examination of the resultant strains revealed that (i) expression of genes downstream of virB6 is dependent on translation of this gene and (ii) expression of the VirB7 to VirB10 proteins in a strain leads to a 5- to 10-fold increase in the capacity of the strain to serve as a recipient in the conjugal transfer of RSF1010 between Agrobacterium strains, but wild-type activity is observed only when the VirB1 to VirB4 proteins are also present. In addition, membrane localization and/or the cross-linking patterns of the VirB7, VirB8, VirB9, and VirB10 proteins were examined in this study. The data show that the membrane localization and chemical cross-linking of VirB10, but not those of VirB7, VirB8, and VirB9, are dramatically altered in cells expressing only VirB7 to VirB10 but are restored to the wild-type parameters by coexpression of VirB1 to VirB3.
MATERIALS AND METHODS
Strains and growth media.
The strains and plasmids used in this study are shown in Table 1. E. coli strains used for cloning procedures were grown in Luria-Bertani (LB) media at 37°C. A. tumefaciens strains were routinely grown at 25°C in LB broth or in AB induction medium (ABIM) (84) under appropriate antibiotic selection conditions. The antibiotic concentrations used in liquid and solid media were as follows: spectinomycin, 50 and 100 μg/ml, respectively; carbenicillin, 30 and 100 μg/ml, respectively; kanamycin, 10 and 50 μg/ml, respectively; and tetracycline, 3 and 5 μg/ml, respectively.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| A. tumefaciens strains | ||
| A136 | C58 heat cured of pTiC58 | 35 |
| A348 | A136 carrying pTiA6 | 35 |
| PC1001 to PC1011 | Complete deletion of each virB open reading frame in pTiA6 of A348 | 13 |
| E. coli DH5α | F− φ80dlacZΔMI5 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | GIBCO BRL |
| Plasmids | ||
| pAB123 | KpnI fragment containing VirA, from pVRA5, cloned into KpnI site of pMutG; Ampr | This study |
| pAD1287 | 10.5-kb NdeI-XhoI-encoded virB operon cloned into pBSII | 3 |
| pCB35 | PstI-BamHI virB7 to virB10 fragment in pSW213; IncP Tetr | 10 |
| pED31 | 0.6-kb EcoRI fragment virB promoter cassette in pBS KS+: Ampr | 79 |
| pED33 | Broad-host-range cloning vector with virB promoter; IncP Kanr | 79 |
| pHP45Ω Tc | Modified pBR322 with a Tetr cartridge flanked by polylinkers; Ampr Tetr | 59 |
| pJB20 | IncW Ampr Tetr | 10 |
| pJB31 | RSF1010 derivative broad-host-range vector; IncQ Specr | 10 |
| pMutG | Broad-host-range plasmid carrying Ri origin of replication and virG from pTiA6; Ampr | 52 |
| pTrcB1 | virB1 PCR fragment cloned downstream of trc promoter in pTrc262 | |
| pVIK219 | Cosmid plasmid containing virB genes from pTiA6 | 43 |
| pVRA5 | 4.6-kb KpnI fragment from pTiA6 containing virA cloned into pUC9; Ampr | 50 |
| pYW15C | Broad-host-range expression vector: IncW Ampr | 75 |
| pYW12 | pJB20 derivative with lac promoter followed by multiple cloning site from pUC19; IncW Ampr | 75 |
| pZL5 | XhoI virB promoter fragment from pED31, containing the polylinker, cloned into ScaI site of pJB20; IncW Tetr | This study |
| virB gene constructs | ||
| pZL3 | virB1 to virB4, NdeI (106) to AatII (4721), containing virBa | This study |
| promoter cloned into pJB20 ScaI; IncW Tetr Ampr | ||
| pZL12 | virB3 and virB4, HpaI (262) to EcoRI (3103), cloned into pZL5 SmaI site, under control of virB promoter; IncW Kanra | This study |
| pZL24 | virB2 to virB4, PvuII (1255) to BstEII (5518), cloned into pZL5 SmaI site, under control of virB promoter; IncW Tetra | This study |
| pZL36 | virB7 to virB10, Pst I (5641) to BamHI (9154) from pCB35, cloned into pED33 EcoRV/SmaI site; IncP Tetra | This study |
| pZL42 | virB1 to virB5, NdeI (106) to BstEII (5518) from pVIK219, containing virB promoter, cloned into pYW15c KpnI site; pYW15C Ampr gene inactivated by insertion of Tetr ge ne from pHP45Ω Tc; IncW Tetra | This study |
| pZL48 | virB6 to virB10, NotI (5155) to EcoRV (9331) from pAD1287, cloned into pED33 KpnI site; IncP Kanra | This study |
| pZL49 | virB1 to virB3, including the promoter region; pZL51 cut with BglII and XbaI, removing virB4 to virB11 and leaving the NdeI (106) to BglII (2204) fragment carrying vir B1 to virB3; IncW Tetra | This study |
| pZL51 | virB1 to virB11, including its promoter region, from pAD1287 cloned into pYW12 XbaI/KpnI site; pYW12 Ampr gene inactivated by insertion of Tetr gene from pHP45 Ω Tc; IncW Tetr | This study |
| virB6 mutants | ||
| pZL48-D1 | pZL48 derivative with deletion from amino acid 205 to amino acid 224 of VirB6 | This study |
| pZL48-D2 | pZL48 derivative with deletion from amino acid 151 to amino acid 237 of VirB6 | This study |
| pZL48-D3 | pZL48 derivative with deletion from amino acid 37 to amino acid 134 of VirB6 | This study |
| pZL48-D4 | pZL48 derivative with deletion from amino acid 1 to amino acid 134 of VirB6 | This study |
| pZL48-D5 | pZL48 derivative with deletion from amino acid 37 to amino acid 134 and from amino acid 151 to amino acid 237 of VirB6 | This study |
| pZL48-m1 | pZL48 derivative with a change of Ser-133 of VirB6 to Pro | This study |
| pZL48-m2 | pZL48 derivative with a change of Glu-134 of VirB6 to Gly | This study |
| pZL48-m12 | pZL48 derivative with changes of Ser-133 and Glu-134 of VirB6 to Pro and Gly, respectively | This study |
| pZL48-XbaI | Frameshift mutation of virB6 of pZL48 by insertion of an XbaI linker (dGCTCTAGAGC) in RsrII site of virB6 (position 5718) | This study |
The numbers in parentheses are the base positions in relation to the SalI site of the virB sequence described by Ward et al. (78).
Plasmid construction.
The IncP plasmid pED33 (79) contains a virB promoter followed by a polylinker and was used in pZL48 and pZL36 expressing virB6 to virB10 and virB7 to virB10, respectively (Table 1). The IncW plasmid pJB20 (10) and its derivatives pYW15c and pYW12 (75) served as vectors for virB1 to virB4, virB1 to virB5, and virB1 to virB11 (pZL3, pZL42, and pZL51, respectively). pZL5 was constructed by cloning the XhoI virB promoter- and polylinker-containing fragment from pED31 (79) into pJB20 This plasmid was used to express virB2 to virB4 in plasmid pZL24 and virB3 and virB4 in plasmid pZL12. All plasmids carrying the virB genes were tested for the capacity to complement the appropriate nonpolar deletions in the PC10XX series (13), and all of the plasmids except pZL36 were positive (see below). pAB123 was constructed by cloning the 4.6-kb KpnI virA-containing fragment of pTiA6 from pVRA5 (50) into pMutG (52), an IncRi derivative.
Mutagenesis of virB6.
Internal in-frame deletions of virB6 were generated by overlap extension PCR mutagenesis as described previously (51). PCR was carried out with Pfu DNA polymerase (Stratagene) and the following primers: T3 (5′-CCA AGC GCG CAA TTA ACC CTC ACT AAA GGG-3′) (from pBS II), B6 (5′-GTT GGG CAG GCT AAC TAC CA-3′) (positions 5544 to 5563), AB6 (5′-CCT AGC CCC GTT CAA CCT GAG-3′) (positions 6198 to 6177), AB8 (5′-CGT CAT GGT GCG CCC TGG CCT A-3′) (positions 7078 to 7100), M12 reverse (5′-GTC GTT CAT CGG ACC GAT TCC GGG TGC GAT-3′; underlined bases carry mutations), D1 forward (5′-CTT ATC ACC ATC-GCG CTC ACC CTC ATG CTT GGT-3′) (positions 5910 to 5922 and 5982 to 6003), D1 reverse (5′-GAG GGT GAG CGC-GAT GGT GAT AAG TTG CCC GAT-3′) (positions 5994 to 5982 and 5922 to 5901), D2 forward (5′-CAA GGG GCA CAG-ACG ACC GCG GCC AAG ATC AT-3′), D2 reverse (5′-GGC CGC GGT CGT-CTG TGC CCC TTG GAA AGC AAG-3′), D3 forward (5′-GCG GTG AGT GCG-ATC GGT CCG ATG AAC GAC CAG-3′), D3 reverse (5′-CAT CGG ACC GAT-CGC ACT CAC CGC CTC CTG GAT-3′), D4 forward (5′-CAG GTC CAA TCG-ATC GGT CCG ATG AAC GAC CAG-3′), and D4 reverse (5′-CAT CGG ACC GAT-CGA TTG GAC CTG AAC TTG GCT-3′). The deletions within virB6 were constructed by two rounds of PCR in which two internal mutagenic primers and two primers (B6, AB8) outside the coding region were used. For example, the internal mutagenic primers D1 forward and D1 reverse were designed to create an in-frame deletion from amino acids 205 to 224 of VirB6. D1 forward and D1 reverse each contain 12 bases that are 5′ to the deletion and 21 bases that are 3′ to the deletion. In the first round of amplification PCRs were carried out with primers D1 forward and AB8 and primers D1 reverse and B6. The product of each reaction was gel purified with a QIAquick gel extraction kit (Qiagen), the products were mixed, and the 12-base overhangs from the products of the first reactions caused the two fragments to base pair. Primers B6 and AB8 were then added to the mixture, and a second round of PCR amplification resulted in production of full-length virB6 carrying the deletion. This amplified PCR product was then digested with RsrII and NcoI and exchanged with the wild-type gene fragment of virB6 in pZL48. By using the same strategy VirB6 with an in-frame deletion of amino acids 151 to 237 was produced with the mutagenic primers D2 forward and D2 reverse and outside primers B6 and AB8. The final PCR fragment containing the deletion was digested with RsrII and NcoI and exchanged with the wild-type gene fragment of virB6 in pZL48, resulting in pZL48-D2. Similarly, VirB6 with an internal deletion from amino acid 37 to amino acid 134 was created by using two internal primers, D3 forward and D3 reverse, and outside primers T3 and AB6. The PCR fragment carrying the virB6 deletion was digested by XhoI and RsrII and was exchanged either with the wild-type fragment of virB6 on pZL48, which resulted in pZL48-D3, or with the same fragment in the incomplete virB6 (deletion from amino acid 151 to amino acid 237) of pZL48-D2, which resulted in pZL48-D5. Two point mutations (Ser to Pro at position 133, Glu to Gly at position 133) were introduced into VirB6 by utilizing the mutant primer AB6M12 (5′-GTC GTT CAT CGG ACC GAT TCC GGG TGC GAT-3′; mutated bases underlined). A 0.6-kb PCR product was amplified from pZL48 with primers AB6M12 and T3, digested by XhoI and RsrII, and exchanged with the wild-type virB6 gene on pZL48, yielding pZL48-M12. A frameshift mutation in virB6 was engineered by digesting pZL48 with RsrII, filling in with the Klenow fragment, adding a 10-bp XbaI linker (New England Biolabs), and ligating to obtain pZL48-XbaI. All mutations were confirmed by sequencing.
Immunoblot analysis and protein cross-linking.
Equal numbers of cells grown at 25°C in ABIM with or without the vir gene inducer AS (Aldrich) were collected, resuspended in sodium dodecyl sulfate (SDS) sample buffer (12% sucrose, 4% SDS, 0.1 M Tris-HCl[pH 6.8], 5 mM EDTA, 0.04% bromophenol blue, 0.1 M dithiothreitol[DTT]), resolved by electrophoresis in SDS-10 to 12% polyacrylamide gel electrophoresis (PAGE) using polyacrylamide gels (acrylamide/bisacrylamide ratio, 29:1). The gels were transferred to a polyvinylidene difluoride membrane (Amersham) and probed with antibodies against VirB8, VirB9, and VirB10 as described previously (10, 51). For analysis of VirB7, samples were electrophoresed on SDS-12% PAGE polyacrylamide gels with an acrylamide/bisacrylamide ratio of 19:1 in Tris-Tricine-SDS buffer (Bio-Rad). Protein cross-linking was performed with whole cells by using BS3 (Pierce Chemical Co.) as described previously (10).
Expression analysis by RT-PCR.
Total RNA isolation was carried out by an acid-phenol method (55), as follows. Portions (30 ml) of cultures of Agrobacterium strains were induced by growing them in ABIM with 200 μM AS for 6 h to 8 h (optical density at 600 nm [OD600], 0.4 to 0.6) and then harvested. Each cell pellet was resuspended in 1.6 ml of buffer (20 mM sodium acetate [pH 5.5], 1 mM EDTA) and then added to an SDS-acid phenol solution (110 μl of 10% SDS, 1 ml of phenol; pH 5.5) at 65°C and incubated for 7 min; this was followed by centrifugation at 16,060 × g for 5 min at 25°C. The aqueous phase was extracted with 1 ml of phenol-CH2Cl2 (1:1) and then with 1 ml of CH2Cl2 (preequilibrated in 10 mM Tris [pH 7.0]), and the RNA was precipitated with 2.5 volumes of 100% ethanol. The RNA pellet was washed with 70% ethanol, resuspended in diethyl pyrocarbonate-treated H2O, quantified by determining the OD260, and used to synthesize the first strand of cDNA. To ensure linearity of the reverse transcription (RT)-PCR, first-strand cDNA synthesis was performed with 0.3, 0.1, and 0.03 μg of each pool of RNA. The cDNAs were synthesized by using a SuperScript first-strand system for RT-PCR kit (Invitrogen) as recommended by the supplier and a total volume of 20 μl. The RT reaction mixture was precipitated with 2.5 volumes of 100% ethanol supplemented with 1 μl of a 20-mg/ml glycogen solution (Roche). After centrifugation the pellets were dissolved in 40 μl of H2O and subjected to PCR. The PCR mixtures (total volume, 50 μl) contained 1 μl of the RT reaction mixture, virB8 primers (forward primer [starting from the end of virB7 gene], 5′-CAGCTCCGCAATTCGGTGGAC-3′; reverse primer [starting from the beginning of virB9], 5′-TCAGCCTCCGCACCAGTCGC), and virB1 primers for internal control (forward primer, 5′-GCTTCACTGGCAAGATCACA-3′; reverse primer, 5′GTGCAATTTGCTTCTCGTCAAT-3′), and the PCR program consisted of 94°C for 20 s, 60°C for 30 s, and 72°C for 40 s for 20 cycles. Twenty-microliter portions of the PCR mixtures were analyzed by electrophoresis in 1.5% agarose gels.
Membrane localization of VirB8, VirB9, and VirB10.
VirB protein localization in the inner or outer membranes was determined by sucrose density gradient fractionation as described previously (9, 26, 31). Briefly, cells (ca. 400 ml) were induced in ABIM with 100 μM AS overnight at 25°C, harvested, washed three times with 50 mM sodium phosphate buffer (pH 7.6), and resuspended in phosphate buffer containing 20% (wt/vol) sucrose (cell concentration, 1 g/10 ml). The cells were lysed by three passes through a French press at 16,000 lb/in2, and 1 mM EDTA and 0.01% phenylmethylsulfonyl fluoride protease inhibitor were added just before lysis. Four milligrams of lysozyme was added to each sample, and the samples were incubated on ice for 30 min. After an initial low-speed centrifugation (Sorvall-SS34, 15 min, 13,000 rpm) 0.2 M (final concentration) KCl was added to the cleared cell lysates, which were then subjected to ultracentrifugation in a Beckman L7 ultracentrifuge for 90 min at 45,000 rpm. The insoluble pellet was dispersed in phosphate buffer containing 0.1 M DTT, 5 mM EDTA, and 20% (wt/vol) sucrose, the concentration was adjusted to 2 to 3 mg of protein/ml, and the pellet was layered on top of a two-step sucrose gradient (1 ml of 70% [wt/vol] sucrose, 2.9 ml of 53% [wt/vol] sucrose). After centrifugation at 4°C for 17 h at 33,000 rpm in a Beckman SW 50.1 rotor, fractions (0.3 ml) were collected from the top of the gradient. NADH oxidase was used as a marker for the inner membrane (57). The protein concentration was determined by using the Bio-Rad (Bradford) protein assay reagent and procedures. Five microliters of each fraction was resolved on an SDS-10 to 12% PAGE gel, which was blotted and subsequently probed with VirB8, VirB9, and VirB10 antibodies (see below).
Conjugation assays. Mating between donor and recipient strains of A. tumefaciens was performed as described previously (17). Donor strain A348 containing RSF1010 derivative IncQ plasmid pJB31 and recipient strain A136(pAB123) expressing various virB genes or A348(pAB123) were grown in LB broth overnight and then transferred into ABIM with 250 μM AS to an OD600 of 0.2 and grown for 6 h at 25°C. Donor and recipient bacteria were then mixed at a ratio of 5:1, and 5 μl of each conjugation mixture was spotted on solid (1.5% Bacto Agar) ABIM with 500 μM AS in a 24-well plate. After 3 days of incubation at 25°C, the cells were resuspended in 1 ml of 0.9% NaCl. The cell suspensions were diluted as appropriate and plated onto LB agar plates supplemented with spectinomycin, carbenicillin, and spectinomycin plus carbenicillin to recover donors, recipients, and transconjugants, respectively. Colonies were counted after 3 days of growth at 25°C. A statistical analysis was carried out by using a two-way mixed model analysis of variance with replication, with date as a random effect and treatments as fixed effects. The Tukey-Kramer test for unplanned comparisons (69) was used to test for differences in the conjugation frequency by using the software program JMPIn 4.0.4 (SAS Institute; http://www.duxbury.com/statistics_d/). Data were collected from eight different experiments, and each recipient strain was tested at least three times. Three separate conjugations were performed with each recipient strain tested in an experiment.
Virulence assays.
The virulence of various A. tumefaciens strains was assayed with Kalanchoe daigremontiana by wounding the youngest expanded leaves with 2-cm needle scratches and inoculating them with ∼10−6 CFU. Tumor formation was monitored 14, 21, and 28 days after inoculation. Virulence assays in which tobacco leaf square transformation was used were carried out as described previously (7). Briefly, overnight cultures of agrobacteria were diluted to an OD600 of 0.5 and cocultivated with Nicotiana tabacum cv. Havana 425 leaf squares on hormone-free MS medium with 300 μM AS. After 2 days, the leaf squares were transferred to hormone-free MS medium containing vancomycin (200 μg/ml) and timentin (200 μg/ml). Tumors were scored and photographed 12 days after the start of the cocultivation. A total of 14 to 16 leaf squares were tested for each strain in an experiment, and all strains were tested at least three times.
RESULTS
Construction of plasmids to express subsets of VirB proteins.
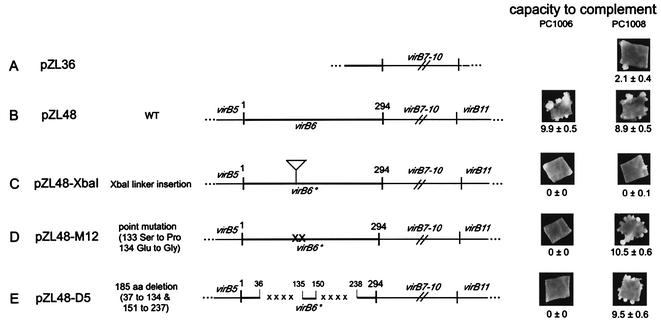
To characterize the requirements for the different VirB proteins in recipient activity, we constructed plasmids that expressed subsets of the various virB genes from the native virB promoter (Table 1). These plasmids were electroporated into strain A136(pAB123) lacking the Ti plasmid, which also carried the virA-virG regulatory system necessary for inducible expression from the virB promoter. They were also tested for the capacity to complement nonpolar deletions in each of the appropriate reading frames of the PC10XX strains (e.g., PC1001 carries a nonpolar deletion of virB1). All plasmids carrying any combination of the virB1, virB2, virB3, virB4, and virB5 genes were capable of restoring virulence to the corresponding PC10XX strain. For example, pZL3, carrying virB1 to virB4, restored the capacities of PC1001, PC1002, PC1003, and PC1004 to induce tumors on Kalanchoe leaves (data not shown). Immunoblot analysis indicated that VirB4 is produced by all plasmids carrying the virB4 gene (data not shown). In contrast, however, virulence assays performed with Kalanchoe leaves and tobacco leaf explants demonstrated that pZL36, designed to express virB7 to virB10, only weakly complemented PC1007, PC1008, PC1009, and PC1010 (Fig. 1A and data not shown). pZL48, carrying virB6 to virB10, was capable of complementing each of these deletion strains, as well as PC1006 (Fig. 1B and data not shown), suggesting that the presence of VirB6 might be important in the activities or accumulation of VirB7 to VirB10.
FIG. 1.
Maps and complementation activities of various virB constructs. Plasmids were constructed as described in Table 1 and the text. The capacity to complement either the PC1006 (virB6 deletion) or PC1008 (virB8 deletion) strain of A. tumefaciens was tested by utilizing the tobacco leaf explant assay system described in Materials and Methods. A representative leaf explant was photographed in each case. The number of tumors per leaf explant (mean ± standard error; n = 14) is indicated below each photograph. In this experiment wild-type strain A348 yielded 11.7 ± 0.8 tumors/leaf explant, and PC1006 and PC1008 yielded no tumors. Wt. wild type; aa, amino acid.
Translation of virB6 sequences is required for VirB7 to VirB10 accumulation.
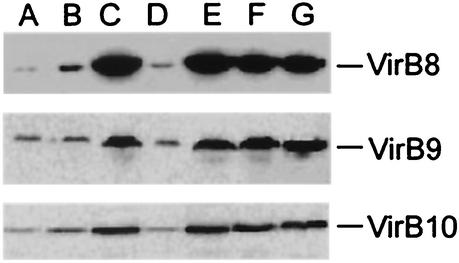
Strain A136(pZL36) did not accumulate large quantities of VirB8, VirB9, and VirB10, whereas A136(pZL48) did (Fig. 2A and C), suggesting that the VirB6 protein may stabilize these proteins. However, strain PC1008(pZL36) did not accumulate VirB8, whereas PC1008(pZL48) did (data not shown). This indicated that production of the VirB6 protein by PC1008 could not support accumulation of VirB8 from the virB7-virB8-virB9-virB10 construct that was in trans. One possible explanation for this is that virB6 needed to be cis to virB7 to virB10 for the latter genes to be properly expressed. We tested the hypothesis that virB6 and virB7 to virB10 must be cotranslated in order to achieve high-level expression of VirB7 to VirB10. A frameshift mutation was introduced into the virB6 coding sequence via a 10-bp XbaI linker at the RsrII site (amino acid 138) of virB6 in pZL48, resulting in a theoretical protein consisting of 188 amino acids, compared to the 295-amino-acid wild-type VirB6 (Fig. 1C). Complementation tests of the virB deletion strains with the resulting plasmid, pZL48-XbaI, showed that virulence was not restored in PC1006 and, importantly, was only partially restored in PC1007, PC1008, PC1009, and PC1010 (Fig. 1C and data not shown). Importantly, expression of VirB8, VirB9, and VirB10 was reduced when this plasmid was present in strain A136(pAB123) (Fig. 2D). Similarly, pZL48-D4, from which the sequences encoding the first 134 amino acids, including the start codon, were deleted, exhibited poor expression of VirB8, VirB9, and VirB10, could not complement PC1006, and barely complemented PC1008 for virulence (data not shown). In contrast, two point mutations (Ser to Pro at position 133, Glu to Gly at position 134) incorporated into virB6 of pZL48 resulted in plasmid pZL48-M12, which could not complement PC1006 for virulence but could complement PC1008 (Fig. 1D) and was able to support VirB8, VirB9, and VirB10 accumulation (Fig. 2F). To further characterize the possible effects of the VirB6 protein on the VirB7 to VirB10 proteins, two separate internal portions of VirB6 were deleted in frame and tested individually and together. None of the internal deletions resulted in a form of VirB6 that could complement the PC1006 mutant strain in virulence tests on tobacco leaves (Fig. 1E). However, every in-frame deletion tested could fully restore virulence in the PC1008 mutant strain (Fig. 1E and data not shown), as well as in PC1007, PC1009, and PC1010 (data not shown), and the strains were able to accumulate VirB8, VirB9, and VirB10 (Fig. 2E and data not shown). Thus, the forms of VirB6 having internal deletions had completely lost the virulence function but had no negative effects on downstream expression of virB7 to virB10.
FIG. 2.
Immunoblot analysis of various virB constructs expressed in A136(pAB123). Lane A, pZL36 (virB7 to virB10); lane B, pZL36 and pZL3 (virB7 to virB10, virB1 to virB4); lane C, pZL48 (virB6 to virB10); lane D, pZL48-XbaI (virB6 to virB10 with XbaI linker insertion in virB6); lane E, pZL48-D2 (virB6 to virB10 with deletion from amino acid 151 to amino acid 237 of VirB6); lane F, pZL48-m12 (virB6 to virB10 with point mutations at amino acids 133 and 134 of VirB6); lane G, A348 (wild-type Ti plasmid).
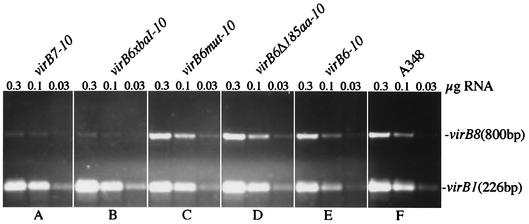
The fact that functional VirB6 protein was not required (e.g., pZL48-D2) for accumulation of VirB8 to VirB10 means that intact VirB6 protein is not required for this cis effect of translated virB6. One possible explanation for these results is that accumulation of virB7 to virB10 mRNA is affected by translation of cis virB6 sequences. To test this possibility, RNA was prepared from AS-induced strains carrying various versions of virB6 to virB10 (see above) and, on a separate plasmid, virB1 to virB4, also expressed from a virB promoter (pZL3), as an internal control. Additionally, RNA preparations were made from the Ti plasmid-free strain A136 and from A348, which carries the wild-type Ti plasmid pTiA6. RT-PCR was carried out by using primers that amplified an 800-bp fragment of virB8 or a 226-bp fragment of virB1. The results of these experiments indicated that while virB1 was amplified equivalently in all RNA samples used except, as expected, from A136 samples, the amount of the RT-PCR product from virB8 was drastically reduced in the samples that did not contain a translatable form of virB6 (Fig. 3). Strains carrying pZL36 and pZL48-XbaI (Fig. 3A and B) both yielded very reduced levels of the virB8 RT-PCR product. In contrast, strains carrying the translatable virB6 gene with either point mutations or deletions (Fig. 3C and D) or wild-type virB6 (Fig. 3E) yielded levels of this product equivalent to the level obtained with the Ti plasmid of A348 (Fig. 3F).
FIG. 3.
RT-PCR analysis of virB8 transcript levels of Agrobacterium virB6 mutant strains. RT reactions were performed with 0.3, 0.1, and 0.03 μg of RNA isolated from AS-induced cells as described in Materials and Methods. virB1 was used as an internal control during RT-PCR. (A) A136(pAB123, pZL36, pZL3) (virB7 to virB10, virB1 to virB4); (B) A136(pAB123, pZL48-XbaI, pZL3) (virB6xbaI-10, virB1 to virB4); (C) A136(pAB123, pZL48-m12, pZL3) (virB6mut-10, virB1 to virB4); (D) A136(pAB123, pZL48-D5, pZL3) (virB6Δ185aa-10, virB1 to virB4); (E) A136(pAB123, pZL48, pZL3) (virB6 to virB10, virB1 to virB4); (F) A348.
VirB proteins required for minimal and maximal increases in recipient activity.
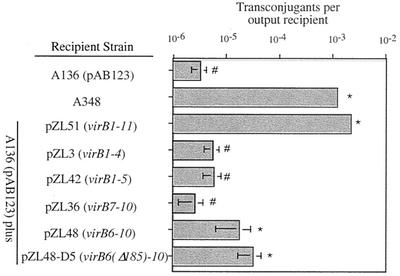
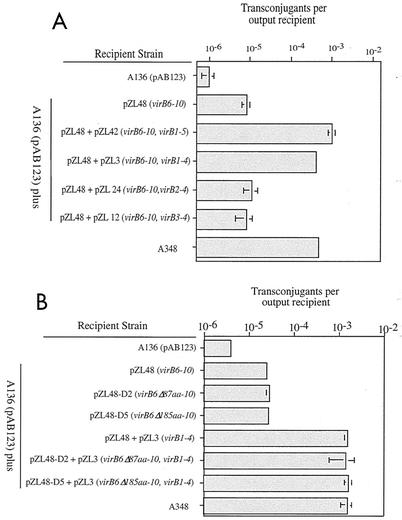
After the plasmids capable of expressing various VirB proteins in strain A136 were characterized, the roles of these proteins in the recipient assay were examined. Various strains were mated with wild-type donor strain A348 carrying the IncQ derivative plasmid pJB31 as described above. In the first set of experiments, individual subsets of the VirB proteins were tested. Data from eight different experiments, not all of which included each strain, were analyzed by a two-way, mixed model analysis of variance with replication. With a type I error rate of 5%, the treatment effect (different VirB proteins in the recipient) was very significant (F = 209; P < 0.0001; df, 7 and 99). The Tukey-Kramer test for unplanned comparisons (69) was used to test for differences between strains carrying different VirB proteins. These assays demonstrated that VirB1 to VirB11, when expressed in A136, resulted in recipient activity equivalent to that of the wild-type Ti plasmid-containing strain A348 (Fig. 4), thus demonstrating that no other Ti plasmid genes are required for this activity. Plasmids designed to express VirB1 to VirB4, VirB1 to VirB5, VirB6 to VirB10, and VirB7 to VirB10 were also tested. Strains carrying pZL48 (encoding VirB6 to VirB10) or pZL48-D5 (encoding VirB7 to VirB10) yielded recipient activity that was 5- to 10-fold greater than, and significantly different from, the activity of the A136 control (Fig. 4). In contrast, A136 strains carrying plasmids pZL3 (virB1 to virB4), pZL42 (virB1 to virB5), and pZL36 (virB7 to virB10, which were poorly expressed [see above]) did not exhibit significant differences in recipient activity compared to A136. Thus, VirB7 to VirB10 represent the subset of VirB proteins tested that provides minimal recipient activity.
FIG. 4.
Recipient activities of strain A136(pAB123) carrying plasmids expressing various subsets of the VirB proteins. The adjusted mean numbers of transconjugants per output recipient (from the Tukey-Kramer analysis [see Materials and Methods]) are indicated along with 2 standard errors. An asterisk indicates that a value is significantly different than the value obtained for A136(pAB123) at a 95% confidence level; a number sign indicates that a value is not significantly different from the value obtained for A136 (see Materials and Methods for details concerning statistics).
Increasingly complex sets of the VirB proteins were then tested in order to determine the minimal group necessary for wild-type recipient activity. The results demonstrated that wild-type activity was present when either VirB1 to VirB5 (pZL42) or VirB1 to VirB4 (pZL3) was expressed along with VirB6 to VirB10 (from pZL48) (Fig. 5A). In contrast, when strains carrying pZL48 as well as VirB2 to VirB4 or VirB3 and VirB4 were tested, no such stimulation was observed (Fig. 5A). Similarly, no stimulation of recipient activity was obtained when either VirB1 (pTrcB1) (62) or VirB1 to VirB3 (pZL49) were tested in strains expressing VirB6 to VirB10 (data not shown), demonstrating the importance of the VirB1, VirB2, VirB3, and VirB4 proteins. These results are also consistent with the results of a previous study (17) that demonstrated that strains having nonpolar deletions of VirB2 or VirB3 in an otherwise wild-type Ti plasmid exhibit low-level recipient activity similar to that observed in this study for strains expressing only VirB7 to VirB10. To test the role of VirB6 in the observed synergistic interaction between VirB6 to VirB10 and VirB1 to VirB4, we examined strains carrying pZL48-D2 and strains carrying pZL48-D5 (with 87- and 185-amino-acid deletions in VirB6, respectively). Results identical to those obtained with pZL48 were obtained; the maximal recipient activity was observed when pZL3 (virB1 to virB4) was present along with pZL48-D2 or pZL48-D5 (Fig. 5B). These results demonstrate that the VirB6 protein plays no role in the increased capacity of a strain to serve as a recipient in conjugal transfer of IncQ plasmids between Agrobacterium strains and that VirB1 to VirB4 along with VirB7 to VirB10 are required for maximal recipient activity.
FIG. 5.
Recipient activities of strain A136(pAB123) containing pZL48 (virB6 to virB10) (A) or pZL48-D2 (virB6Δ87aa-10) or pZL48-D5 (virB6Δ185aa-10) (B) with or with out other virB genes, as indicated. The mean numbers of transconjugants per output recipient are indicated. The error bars (some of which are not visible due to scale) indicate the standard errors for three separate conjugations performed on the same day. Similar results were observed in two other experiments.
Localization and cross-linking of VirB10 are dramatically affected by VirB1 to VirB3.
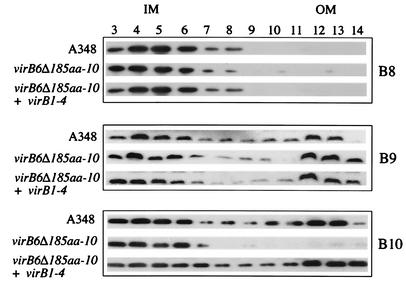
The results described above indicate that VirB7, VirB8, VirB9, VirB10 comprise the minimal subset of VirB proteins that can increase the recipient activity of strain A136 but that expression of VirB1 to VirB4 is necessary to achieve maximal stimulation of this activity. This suggests that the VirB1-VirB2-VirB3-VirB4 subset of proteins affects the VirB7-VirB8-VirB9-VirB10 subset in some fashion (or vice versa), resulting in increased biological activity. To determine whether the VirB1 to VirB4 proteins affect VirB7 to VirB10, we characterized the membrane distribution of these VirB proteins, as well as their capacity to interact with themselves or each other as revealed by cross-linking studies. In the first set of experiments, membranes were isolated from strains grown in ABIM containing AS (to induce expression of the vir genes), and sucrose density gradients were used to separate the inner and outer membranes, as described in Materials and Methods. The sucrose gradient fractions were then subjected to SDS-PAGE and immunoblot analysis by using anti-VirB8, anti-VirB9, and anti-VirB10 as probes. In the case of wild-type strain A348, VirB8 localized exclusively to the inner membrane, whereas VirB9 and VirB10 were found to be distributed in both of these fractions (Fig. 6), a pattern of distribution that has been observed previously (30, 31, 64, 73). In the case of pZL48-D5 (which expressed only VirB7 to VirB10) the distribution of VirB8 and VirB9 was unchanged compared to the wild-type distribution. However, the distribution of VirB10 was dramatically altered in this case, and this protein was found predominantly in the inner membrane (Fig. 6). We next sought to determine which other VirB proteins may be necessary for VirB10 to be distributed in a wild-type fashion in the inner and outer membrane systems. In these experiments plasmids carrying various combinations of VirB1, VirB2, VirB3, and VirB4 were tested. Analysis of membrane fractions from a strain producing VirB1 to VirB4 as well as VirB7 to VirB10 [A136(pAB123, pZL3, pZL48-D5)] demonstrated that VirB10 was distributed in both the inner and outer membranes (Fig. 6), similar to the wild-type distribution. A similar result was obtained when VirB1 to VirB3 were expressed along with VirB7 to VirB10 (data not shown). However, if only VirB2 to VirB4 or VirB3 and VirB4 were expressed along with VirB7 to VirB10, VirB10 was distributed in the membrane fractions in the aberrant fashion observed when VirB7 to VirB10 were expressed by themselves (data not shown).
FIG. 6.
Membrane localization of VirB8, VirB9, and VirB10 from A348, A136(pAB123, pZL48-D5, pZL3) (virB6Δ185aa-10, virB1 to virB4) and A136(pAB123, pZL48-D5) (virB6Δ185aa-10). Membrane fractions were separated by sucrose density gradient as described in Materials and Methods. Membrane fractions 3 to 14 (from the top to the bottom of the gradient) were subjected to SDS-PAGE, blotted, and probed with anti-VirB8, anti-VirB9, or anti-VirB10. Similar results were observed in three other experiments. IM, inner membrane; OM, outer membrane.
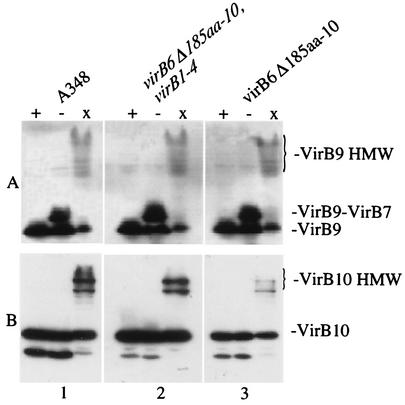
Distribution of the VirB9 and VirB10 proteins in both membrane fractions despite their predicted localization to the outer and inner membranes, respectively, has been proposed to be the result of protein interactions within the complex (21, 31, 73). The capacities of the various VirB proteins to interact with each other or with themselves were therefore examined by using two different methods. First, the interactions of VirB7 and VirB9 through disulfide bond formation were examined by immunoblot analysis of reduced or nonreduced samples. These experiments revealed that the strain expressing only VirB7 to VirB10 accumulated VirB7-VirB9 heterodimers in a wild-type fashion (Fig. 7A). In a second series of experiments, the previously established capacity of VirB9 and VirB10 to form high-molecular-weight aggregates after cross-linking with BS3 (10, 80) was tested. To examine the cross-linking patterns of these proteins, whole cells were treated with BS3 as described above, and samples were then subjected to SDS-PAGE and immunoblot analysis. As expected, high-molecular-weight forms of VirB9 and VirB10 were found in samples prepared from cross-linked cells of wild-type strain A348. In the strain expressing only VirB7 to VirB10 [A136(pAB123, pZL48-D5)] the abundance of cross-linked VirB9 was similar to that in the wild-type strain (Fig. 7A). In contrast, the levels of high-molecular-weight forms of VirB10 in the cross-linked samples were dramatically diminished in this strain (Fig. 7B). While the overall abundance of VirB10 was decreased, scanning of the blots revealed a disproportionately lower abundance of the cross-linked forms of this protein; in four separate experiments, there was an average fivefold change in the ratio of monomers to multimers. Interestingly, as was the case for its membrane distribution, the abundance of cross-linked VirB10 was restored in strains expressing VirB7 to VirB10 if VirB1 to VirB4 (Fig. 7B) or VirB1 to VirB3 (data not shown) were also expressed. However, strains coexpressing the VirB2 to VirB4 proteins or the VirB3 and VirB4 proteins along with VirB7 to VirB10 did not restore the cross-linking capacity of VirB10 to the wild-type situation (data not shown).
FIG. 7.
Capacity of VirB9 and VirB10 to form high-molecular-weight complexes in the presence or absence of VirB1 to VirB4. Samples from AS-induced strains were added to loading buffer with DTT (lanes +), added to loading buffer without DTT (lanes −), or cross-linked with BS3 and added to loading buffer with DTT (lanes x). Extracts from equivalent numbers of cells were then resolved by SDS-10% PAGE, blotted, and probed with anti-VirB10 (A) or anti-VirB9 (B) antibodies. Gel 1, A348; gel 2, A136(pAB123, pZL48-D5, pZL3) (virB6Δ185aa-10, virB1 to virB4); gel 3, A136(pAB123, pZL48-D5) (virB6Δ185aa-10).
DISCUSSION
The objective of this study was to determine whether particular subsets of the VirB proteins are required for the virB-mediated increase in the capacity of a strain to serve as a recipient in virB-mediated conjugal transfer of plasmid RSF1010 between agrobacteria. In particular, we sought to determine whether subsets that had previously been speculated to be functionally important in the VirB donor complex (e.g., VirB7 to VirB10 [22]) were functional in the recipient phenotype. Development of constructs to test the role of the VirB6 to VirB10 proteins in the recipient assay revealed that translation through the virB6 open reading frame to sequences upstream of virB7 is critical to the expression of downstream virB genes. This is clearly the case, for example, when expression of virB7 to virB10 from constructs that lack (pZL36) or contain (pZL48) such translatable virB6 sequences is studied (Fig. 1 and 2). When translation of virB6 does not start (e.g., pZL48-D4 or pZL36) or is disrupted by a nonsense mutation (pZL48-XbaI), then both the levels of the VirB7 to VirB10 proteins and the levels of virB7 to virB10 mRNA are significantly reduced (Fig. 2 and 3, respectively). In-frame deletions of virB6 sequences do not have such an effect on downstream expression, demonstrating that the intact VirB6 protein is not required for accumulation of virB7 to virB10 mRNA or the VirB7 to VirB10 proteins. The regulatory mechanisms responsible for the effect on RNA accumulation are not clear. However, the results of an analysis of strain PC1006, having a precise deletion of the entire coding sequence of virB6 and the nontranslated sequence upstream of virB7 (13), are consistent with important effects of the sequences immediately 5′ to the virB7 gene. The deletion in this strain results in virB7 translation that uses the ribosome binding site and upstream sequence 5′ to virB6. This has a clear negative effect on the accumulation of VirB7 and VirB8 compared to the wild-type accumulation (data not shown), as well as on VirB9 and VirB10 (13). These results suggest that the sequences upstream of virB7 cannot be completely replaced by those upstream of virB6 in terms of virB7 to virB10 expression. Thus, translation through virB6 and utilization of the intergenic region between virB6 and virB7 are crucial in the accumulation of virB7 to virB10 mRNA. These findings are consistent with models proposing that translation of polycistronic messages is necessary for stability of the mRNA molecule (see reference 20 for an analysis of decay models).
Utilizing strains expressing different subsets of the VirB proteins, we tested their activities in the recipient assay. The data obtained demonstrate that (i) VirB6 is not involved in the functions necessary for recipient activity, (ii) the VirB7-VirB8-VirB9-VirB10 subset is the minimal subset of VirB proteins tested in the recipient that can increase conjugal DNA transfer, and (iii) a synergistic interaction (either direct or indirect) among the VirB1 to VirB4 and VirB7 to VirB10 proteins yields significant biological activity. The first conclusion indicates that VirB6 is involved in a donor-specific function of this type IV transport complex. For example, it could be involved in substrate recognition and/or targeting to the transport complex or in the energy transduction necessary for substrate movement out of the cell. Examination of the membrane localization of VirB8, VirB9, and VirB10 indicated that these proteins are distributed normally in the cell envelope in cells lacking VirB6 (Fig. 6). As expected from previous studies (70), VirB7-VirB9 heterodimer formation does not require VirB6. We show here that VirB6 is not required for the capacity of VirB9 or VirB10 to form wild-type high-molecular-weight forms upon chemical cross-linking (Fig. 7). These results, therefore, support a model in which VirB6 has donor-specific activity but is not involved in the structural integrity of the VirB7-VirB8-VirB9-VirB10 subassembly that is proposed to be the transport complex core.
The observation that expression of VirB7 to VirB10 by themselves increases recipient activity (Fig. 4) provides the first experimental evidence that this group of proteins, often proposed as the transenvelope channel of the VirB complex (22), is by itself capable of transport or transport-related activity. Previous experiments (17) demonstrated that nonpolar deletion of virB7, virB8, virB9, or virB10 in an otherwise wild-type Ti plasmid completely eliminates recipient activity. This, along with the data presented in this paper, suggests that VirB7 to VirB10 constitute the minimal subset of VirB proteins that can support recipient activity. The mechanism of this activity is not yet known (see below). The biological activity of VirB7 to VirB10 is consistent with the recent observation that a high-molecular-weight subassembly consisting of VirB7 to VirB10 exists in the membrane of wild-type cells (44). Several studies have previously indicated that there are specific interactions between members of this group of proteins (3, 9, 10, 28, 70). Mutations that disrupt such interactions also disrupt recipient activity (17), indicating that there is a relationship between core structure and recipient function. However, the fact that the VirB1 to VirB4 proteins, as a group, are required along with VirB7 to VirB10 for maximal recipient activity (Fig. 5) indicates that a synergistic interaction between these two subsets of proteins is crucial for the biological activity of the complex.
Several studies have revealed interactions between specific members of the VirB1-VirB2-VirB3-VirB4 subset with members of the VirB7-VirB8VirB9-VirB10 subset (8, 61, 76). The experiments presented in this report show that in strains expressing VirB7 to VirB10 and VirB1 to VirB3, there is wild-type membrane localization of VirB10 and there are wild-type levels of high-molecular-weight forms of VirB10, as revealed by chemical cross-linking. While the possible role of VirB2 and VirB3 in this activity is being investigated, the results indicate that at least VirB1 is required for wild-type VirB10 localization and interaction with itself or other proteins in cross-linking studies. A recent study showed that VirB1 interacts with VirB10 in a yeast two-hybrid interaction assay (76). One possibility is that the peptidoglycanase activity (54) of VirB1 interacting with VirB10 is necessary to provide an appropriate periplasmic environment for VirB10 to assemble. These results are also consistent with a model (21, 31, 73) in which tight associations of VirB10 with other components of the VirB complex are necessary for both appropriate localization and optimal biological activity. Previous studies (10) demonstrated that VirB7 and VirB9 are necessary for wild-type levels of high-molecular-weight forms of VirB10 in response to chemical cross-linking. Thus, the VirB1-VirB2-VirB3 subset may function through direct effects on VirB10 or indirectly by facilitating interaction of the VirB7-VirB9 heterodimer with VirB10.
Interestingly, our results demonstrate that VirB4 is not necessary for wild-type membrane localization and cross-linking of the VirB7 to VirB10 proteins but is required, along with VirB1 to VirB3, for maximal recipient activity. This could be the result of interactions of VirB4 with either of these groups. Previous studies (25) showed that VirB4 can dimerize and can support recipient activity in the absence of its ATPase activity. Additionally, VirB4 interaction with VirB10 has been demonstrated by using the yeast two-hybrid system (76). One possibility that emerged from the present study is that VirB10 needs to be in an appropriate structure and location in order to interact with VirB4 in a fashion that supports recipient activity. In contrast to interactions of VirB7 to VirB10 with themselves or other VirB proteins, there is relatively little evidence concerning interactions among VirB1, VirB2, VirB3, and VirB4. One candidate protein with which VirB4 may interact is VirB3; nonpolar mutations in VirB4 result in lower levels of VirB3 and the loss of VirB3 associated with the outer membrane (42). The observation that nonpolar deletions of virB1, virB2, or virB3 result in lower levels of VirB4 (13) is consistent with a model in which these proteins interact, although other possibilities for this result have not been ruled out. VirB1interactions with VirB4 in the yeast two-hybrid system have also been observed (76) and may be important in recipient function.
Our data provide evidence for the functionality of the VirB1 to VirB4 and VirB7 to VirB10 proteins in a transport process, in this case the movement of DNA into the bacterial cell during conjugation. In general, the role of the recipient cell in conjugal plasmid transfer is poorly understood, except for F plasmid transfer in liquid mating (29). Thus, there is little precedence for the remarkable increase in recipient activity (3 to 4 logs) that results from the presence of specific proteins (in this case, parts of the VirB complex). Increases in either mating pair formation or DNA transfer efficiency could be responsible for the phenotype, and experiments to distinguish between these possibilities are in progress. We have, however, recently demonstrated that the presence of the IncP plasmid RK2 also increases the capacity of Agrobacterium cells to serve as recipients in this assay (unpublished observations). This suggests that the VirB complex may reveal a transport capacity of type IV systems that is normally obscured in other conjugal plasmid systems that have evolved surface and/or entry exclusion as a means of blocking recipient activity (1). Consistent with this model, Hofreuter et al. (40, 41) recently demonstrated that natural transformation competence in Helicobacter pylori is mediated by homologues to VirB4 and to VirB7 to VirB10, further implicating them as components of a DNA transport channel. Similarly, Bacon et al. (5) found that mutations in comB3 (related to virB10 of the Ti plasmid) of Campylobacter jejuni resulted in an 80% decrease in competence for natural transformation, while the VirB11 homologue was not involved in this activity. These results have led to the proposal that natural competence in these bacteria is an inverse of the conjugation process (39, 68). The recipient activity mediated by the VirB subsets described here provides a unique opportunity to examine how recipient cells are involved in conjugal DNA transfer and how type IV complexes may play a role in this process.
Acknowledgments
We thank Christian Baron for providing pTrcB1, Anath Das for providing pAD1287, Pat Zambryski for providing pMutG, Peter Christie for providing anti-VirB7 antibodies, and Arlene Wise, Colleen McCullen, Mark Jacobs, Mecky Pohlschröder, and Fevzi Daldal for reading early versions of the manuscript. Special thanks go to Heidi Kuehne and Warren Ewens for help with the statistical analysis.
This work was supported by NSF grant MCB 9817149.
REFERENCES
- 1.Achtman, M., N. Kennedy, and R. Skurray. 1977. Cell-cell interactions in conjugating Escherichia coli: role of traT protein in surface exclusion. Proc. Natl. Acad. Sci. USA 74:5104-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achtman, M., S. Schwuchow, R. Helmuth, G. Morelli, and P. A. Manning. 1978. Cell-cell interactions in conjugating Escherichia coli: Con− mutants and stabilization of mating aggregates. Mol. Gen. Genet. 164:171-183.
- 3.Anderson, L. B., A. V. Hertzel, and A. Das. 1996. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc. Natl. Acad. Sci. USA 93:8889-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anthony, K. G., C. Sherburne, R. Sherburne, and L. S. Frost. 1994. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol. Microbiol. 13:939-953. [DOI] [PubMed] [Google Scholar]
- 5.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balzer, D., W. Pansegrau, and E. Lanka. 1994. Essential motifs of relaxase (TraI) and TraG proteins involved in conjugative transfer of plasmid RP4. J. Bacteriol. 176:4285-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banta, L. M., R. D. Joerger, V. R. Howitz, A. M. Campbell, and A. N. Binns. 1994. Glu-255 outside the predicted ChvE binding site in VirA is crucial for sugar enhancement of acetosyringone perception by Agrobacterium tumefaciens. J. Bacteriol. 176:3242-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baron, C., M. Llosa, S. Zhou, and P. C. Zambryski. 1997. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens, is processed to a C-terminal secreted product, VirB1∗. J. Bacteriol. 179:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baron, C., Y. R. Thorstenson, and P. C. Zambryski. 1997. The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J. Bacteriol. 179:1211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beaupré, C. F., J. Bohne, E. M. Dale, and A. N. Binns. 1997. Interactions between VirB9 and VirB10 proteins involved in movement of DNA from Agrobacterium tumefaciens to plant cells. J. Bacteriol. 179:78-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beijersbergen, A., A. D. Dulk-Ras, R. A. Schilperoort, and P. J. J. Hooykaas. 1992. Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science 256:1324-1327. [DOI] [PubMed] [Google Scholar]
- 12.Berger, B. R., and P. J. Christie. 1993. The Agrobacterium tumefaciens virB4 gene product is an essential virulence protein requiring an intact nucleoside triphosphate-binding domain. J. Bacteriol. 175:1723-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger, B. R., and P. J. Christie. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176:3646-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binns, A. N., C. F. Beaupré, and E. M. Dale. 1995. Inhibition of VirB-mediated transfer of diverse substrates from Agrobacterium tumefaciens by the IncQ plasmid RSF1010. J. Bacteriol. 177:4890-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binns, A. N., and V. R. Howitz. 1994. The genetic and chemical basis of recognition in the Agrobacterium:plant interaction. Curr. Top. Microbiol. Immunol. 192:119-138. [DOI] [PubMed] [Google Scholar]
- 16.Binns, A. N., and M. F. Thomashow. 1988. Cell biology of Agrobacterium infection and transformation of plants. Annu. Rev. Microbiol. 42:575-606. [Google Scholar]
- 17.Bohne, J., A. Yim, and A. N. Binns. 1998. The Ti plasmid increases the efficiency of Agrobacterium tumefaciens as a recipient in virB-mediated conjugal transfer of an IncQ plasmid. Proc. Natl. Acad. Sci. USA 95:7057-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchanan-Wolloston, V., J. E. Passiatore, and F. Cannon. 1987. The mob and oriT mobilization functions of a bacterial plasmid promote its transfer to plants. Nature (London) 328:172-175. [Google Scholar]
- 19.Cao, T. B., and M. H. Saier. 2001. Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: organismal distribution, structural constraints and evolutionary conclusions. Microbiology 147:3201-3214. [DOI] [PubMed] [Google Scholar]
- 20.Carrier, T. A., and J. D. Keasling. 1997. Mechanistic modeling of prokaryotic mRNA decay. J. Theor. Biol. 189:195-209. [DOI] [PubMed] [Google Scholar]
- 21.Christie, P. J. 1997. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 179:3085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:350-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christie, P. J., J. E. Ward, S. C. Winans, and E. W. Nester. 1988. The Agrobacterium tumefaciens virE2 gene product is a single-stranded DNA-binding protein that associates with T-DNA. J. Bacteriol. 170:2659-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dang, T. A., X. R. Zhou, B. Graf, and P. J. Christie. 1999. Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on the assembly and function of the T-DNA transporter. Mol. Microbiol 32:1239-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang, T. A. T., and P. J. Christie. 1997. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J. Bacteriol. 179:453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das, A., and Y.-H. Xie. 1998. Construction of transposon Tn3phoA: its application in defining the membrane topology of the Agrobacterium tumefaciens DNA transfer proteins. Mol. Microbiol. 27:405-414. [DOI] [PubMed] [Google Scholar]
- 28.Das, A., and Y.-H. Xie. 2000. The Agrobacterium tumefaciens T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 182:758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dreiseikelmann, B. 1994. Translocation of DNA across bacterial membranes. Microbiol. Rev. 58:293-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez, D., G. M. Spudich, X.-R. Zhou, and P. J. Christie. 1996. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J. Bacteriol. 178:3168-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finberg, K. E., T. R. Muth, S. P. Young, J. B. Maken, S. M. Heitritter, A. N. Binns, and L. M. Banta. 1995. Interactions of VirB9, -10 and -11 with the membrane fraction of Agrobacterium tumefaciens: solubility studies provide evidence for tight associations. J. Bacteriol. 177:4881-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frost, L. S., and J. Simon. 1993. Studies on the pili of the promiscuous plasmid RP4, p. 47-65. In C. I. Kado and J. H. Crosa (ed.), Molecular mechanisms of bacterial virulence. Kluwer Academic Publishers, Amsterdam, The Netherlands.
- 33.Fullner, K. J. 1998. Role of Agrobacterium virB genes in transfer of T complexes and RSF1010. J. Bacteriol. 180:430-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fullner, K. J., J. C. Lara, and E. W. Nester. 1996. Pilus assembly by Agrobacterium T-DNA transfer genes. Science 273:1107-1109. [DOI] [PubMed] [Google Scholar]
- 35.Garfinkel, D. J., R. B. Simpson, L. W. Ream, F. F. White, M. P. Gordon, and E. W. Nester. 1981. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell 27:143-153. [DOI] [PubMed] [Google Scholar]
- 36.Gelvin, S. B. 2000. Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51:223-256. [DOI] [PubMed] [Google Scholar]
- 37.Hapfelmeier, S., N. Domke, P. C. Zambryski, and C. Baron. 2000. VirB6 is required for stabilization of VirB5 and VirB3 and formation of VirB7 homodimers in Agrobacterium tumefaciens. J. Bacteriol. 182:4505-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Havekes, L., W. Hoeksta, and H. Kempen. 1977. Relation between F, R100 and R144 Escherichia coli KL-12 donor strains in mating. Mol. Gen. Genet. 155:185-189. [DOI] [PubMed] [Google Scholar]
- 39.Hofreuter, D., and R. Haas. 2002. Natural transformation in Helicobacter pylori: DNA transport in an unexpected way (response). Trends Microbiol. 10:162.. [DOI] [PubMed] [Google Scholar]
- 40.Hofreuter, D., S. Odenbreit, and R. Haas. 2001. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 41:379-391. [DOI] [PubMed] [Google Scholar]
- 41.Hofreuter, D., S. Odenbreit, G. Henke, and R. Haas. 1998. Natural competence for DNA transformation in Helicobacter pylori: identification and genetic characterization of the comB locus. Mol. Microbiol. 28:1027-1038. [DOI] [PubMed] [Google Scholar]
- 42.Jones, A. L., K. Shirasu, and C. I. Kado. 1994. The product of the virB4 gene of Agrobacterium tumefaciens promotes accumulation of VirB3 protein. J. Bacteriol. 176:5255-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knauf, V. C., and E. W. Nester. 1982. Wide host range cloning vectors—a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid 8:45-54. [DOI] [PubMed] [Google Scholar]
- 44.Krall, L., U. Wiedemann, G. Unsin, S. Weiss, N. Domke, and C. Baron. 2002. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 99:11405-11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuldau, G. A., G. DeVos, J. Owen, G. McCaffrey, and P. Zambryski. 1990. The virB operon of Agrobacterium tumefaciens pTiC58 encodes 11 open reading frames. Mol. Gen. Genet. 221:256-266. [DOI] [PubMed] [Google Scholar]
- 46.Kumar, R. B., Y. H. Xie, and A. Das. 2000. Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins: VirB8 is essential for the assembly of the transport pore. Mol. Microbiol. 36:608-617. [DOI] [PubMed] [Google Scholar]
- 47.Lai, E.-M. 1998. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 180:2711-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai, E. M., O. Chesnokova, L. M. Banta, and C. I. Kado. 2000. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J. Bacteriol. 182:3705-3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai, E. M., and C. I. Kado. 2000. The T-pilus of Agrobacterium tumefaciens. Trends Microbiol. 8:361-369. [DOI] [PubMed] [Google Scholar]
- 50.Lee, K., M. W. Dudley, K. M. Hess, D. G. Lynn, R. D. Joerger, and A. N. Binns. 1992. Mechanisms of activation of Agrobacterium virulence genes: identification of phenol-binding proteins. Proc. Natl. Acad. Sci. USA 89:8666-8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu, Z., M. Jacobs, D. A. Schaff, C. A. McCullen, and A. N. Binns. 2001. ChvD, a chromosomally encoded ATP-binding cassette transporter-homologous protein involved in regulation of virulence gene expression in Agrobacterium tumefaciens. J. Bacteriol. 183:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLean, B. G., E. A. Greene, and P. C. Zambryski. 1994. Mutants of Agrobacterium VirA that activate vir gene expression in the absence of the inducer acetosyringone. J. Biol. Chem. 269:2645-2651. [PubMed] [Google Scholar]
- 53.Melchers, L. S., M. J. Maroney, A. den Dulk-Ras, D. V. Thompson, H. A. J. van Vuuren, R. A. Schilperoort, and P. J. J. Hooykaas. 1990. Octopine and nopaline strains of Agrobacterium tumefaciens differ in virulence; molecular characterization of the virF locus. Plant Mol. Biol. 14:249-259. [DOI] [PubMed] [Google Scholar]
- 54.Mushegian, A. R., K. J. Fullner, E. V. Koonin, and E. W. Nester. 1996. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc. Natl. Acad. Sci. USA 93:7321-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nuyts, S., L. Van Mellaert, P. Lambin, and J. Anné. 2001. Efficient isolation of total RNA from Clostridium without DNA contamination. J. Microbiol. Methods 44:235-238. [DOI] [PubMed] [Google Scholar]
- 56.Okamoto, S., A. Toyoda-Yamamoto, K. Ito, I. Takebe, and Y. Machida. 1991. Localization and orientation of the VirD4 protein of Agrobacterium tumefaciens in the cell membrane. Mol. Gen. Genet. 228:24-32. [DOI] [PubMed] [Google Scholar]
- 57.Osborn, M. J., J. E. Gander, E. Parisi, and J. Carson. 1972. Mechanism of assembly of the outer membrane of Salmonella typhimurium. J. Biol. Chem. 247:3962-3972. [PubMed] [Google Scholar]
- 58.Otten, L., H. De Greve, J. Leemans, R. Hain, P. Hooykaas, and J. Schell. 1984. Restoration of virulence of vir region mutants of Agrobacterium tumefaciens strain B6S3 by coinfection with normal and mutant Agrobacterium strains. Mol. Gen. Genet. 195:159-163. [Google Scholar]
- 59.Prentki, P., and H. P. Krisch. 1984. In vitro mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 60.Rashkova, S., G. M. Spudich, and P. J. Christie. 1997. Characterization of membrane and protein interaction determinants of the Agrobacterium tumefaciens VirB11 ATPase. J. Bacteriol. 179:583-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sagulenko, V., E. Sagulenko, S. Jakubowski, E. Spudich, and P. J. Christie. 2001. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J. Bacteriol. 183:3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt-Eisenlohr, H., N. Domke, C. Angerer, G. Wanner, P. C. Zambryski, and C. Baron. 1999. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 181:7485-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sherburne, C., and D. E. Taylor. 1997. Effect of lipopolysaccharide mutations on recipient ability of Salmonella typhimurium for incompatibility group H plasmids. J. Bacteriol. 179:952-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shirasu, K., and C. I. Kado. 1993. Membrane location of the Ti plasmid VirB proteins involved in the biosynthesis of a pilin-like conjugative structure on Agrobacterium tumefaciens. FEMS Microbiol. Lett. 111:287-294. [DOI] [PubMed] [Google Scholar]
- 65.Shirasu, K., P. Morel, and C. I. Kado. 1990. Characterization of the virB operon of Agrobacterium tumefaciens: nucleotide sequence and protein analysis. Mol. Microbiol. 4:1153-1163. [DOI] [PubMed] [Google Scholar]
- 66.Simone, M., C. A. McCullen, L. E. Stahl, and A. N. Binns. 2001. The carboxy-terminus of VirE2 from Agrobacterium tumefaciens is required for its transport to host cells by the virB-encoded type IV transport system. Mol. Microbiol 41:1283-1293. [DOI] [PubMed] [Google Scholar]
- 67.Skurray, R. A., R. E. W. Hancock, and P. Reeves. 1974. Con− mutants of Escherichia coli K12 lacking a major cell wall protein and defective in conjugation and adsorption of bacteriophage. J. Bacteriol. 119:726-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smeets, L. C., and J. G. Kusters. 2002. Natural transformation in Helicobacter pylori: DNA transport in an unexpected way. Trends Microbiol. 10:159-162. [DOI] [PubMed] [Google Scholar]
- 69.Sokal, R. R., and F. J. Rohlf. 1995. Biometry. W. H. Freeman and Co., New York, N.Y.
- 70.Spudich, G. M., D. Fernandez, X.-R. Zhou, and P. J. Christie. 1996. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc. Natl. Acad. Sci. USA 93:7512-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stephens, K. M., C. Rousch, and E. Nester. 1995. Agrobacterium tumefaciens VirB11 protein requires a consensus nucleotide-binding site for function in virulence. J. Bacteriol. 177:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thompson, D. V., L. S. Melchers, R. A. Idler, R. A. Schilperoort, and P. J. J. Hooykaas. 1988. Analysis of the complete nucleotide sequence of the Agrobacterium tumefaciens virB operon. Nucleic Acids Res. 16:4621-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thorstenson, Y. R., G. A. Kuldau, and P. C. Zambryski. 1993. Subcellular localization of seven VirB proteins of Agrobacterium tumefaciens: implications for the formation of a T-DNA transport structure. J. Bacteriol. 175:5233-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vergunst, A. C., B. Schrammeijer, A. den Dulk-Ras, C. M. T. de Vlaam, T. J. G. Regensburg-Tuïnk, and P. J. J. Hooykaas. 2000. VirB/D4 dependent protein translocation from Agrobacterium into plant cells. Science 290:979-982. [DOI] [PubMed] [Google Scholar]
- 75.Wang, Y., A. Mukhopadhyay, V. R. Howitz, A. N. Binns, and D. G. Lynn. 2000. Construction of an efficient expression system for Agrobacterium tumefaciens based on the coliphage T5 promoter. Gene 242:105-112. [DOI] [PubMed] [Google Scholar]
- 76.Ward, D. V., O. Draper, J. R. Zupan, and P. C. Zambryski. 2002. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. USA 99:11493-11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ward, J. E., D. E. Akiyoshi, D. Regier, A. Datta, M. P. Gordon, and E. W. Nester. 1988. Characterization of the virB operon from an Agrobacterium tumefaciens Ti plasmid. J. Biol. Chem. 263:5804-5814. [PubMed] [Google Scholar]
- 78.Ward, J. E., D. E. Akiyoshi, D. Regier, A. Datta, M. P. Gordon, and E. W. Nester. 1990. Characterization of the virB operon from an Agrobacterium tumefaciens Ti plasmid. J. Biol. Chem. 265:4768.. [PubMed] [Google Scholar]
- 79.Ward, J. E., Jr., E. M. Dale, P. J. Christie, E. W. Nester, and A. N. Binns. 1990. Complementation analysis of Agrobacterium tumefaciens Ti plasmid virB genes by use of a vir promoter expression vector: virB9, virB10, and vir B11 are essential virulence genes. J. Bacteriol. 172:5187-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ward, J. E., Jr., E. M. Dale, E. W. Nester, and A. N. Binns. 1990. Identification of a VirB10 protein aggregate in the inner membrane of Agrobacterium tumefaciens. J. Bacteriol. 172:5200-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ward, J. E., Jr., E. M. Dale, and A. N. Binns. 1991. Activity of the Agrobacterium T-DNA transfer machinery is affected by virB gene products. Proc. Natl. Acad. Sci. USA 88:9350-9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Winans, S. C. 1992. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol. Rev. 56:12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Winans, S. C., D. L. Burns, and P. J. Christie. 1996. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 4:64-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Winans, S. C., R. A. Kerstetter, and E. W. Nester. 1988. Transcriptional regulation of the virA and virG genes of Agrobacterium tumefaciens. J. Bacteriol. 170:4047-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yusibov, V. M., T. R. Steck, V. Gupta, and S. B. Gelvin. 1994. Association of single-stranded transferred DNA from Agrobacterium tumefaciens with tobacco cells. Proc. Natl. Acad. Sci. USA 91:2994-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu, J., P. M. Oger, B. Schrammeijer, P. J. Hooykaas, S. K. Farrand, and S. C. Winans. 2000. The bases of crown gall tumorigenesis. J. Bacteriol. 182:3885-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zupan, J. R., D. Ward, and P. Zambryski. 1998. Assembly of the VirB transport complex for DNA transfer from Agrobacterium tumefaciens to plant cells. Curr. Opin. Microbiol. 1:649-655. [DOI] [PubMed] [Google Scholar]