Abstract
Although the chemical structure and physical properties of peptidoglycan have been elucidated for some time, the precise three-dimensional organization of murein has remained elusive. Earlier published computer simulations of the bacterial murein architecture modeled peptidoglycan strands in either a regular (D. Pink, J. Moeller, B. Quinn, M. Jericho, and T. Beveridge, J. Bacteriol. 182: 5925-5930, 2000) or an irregular (A. Koch, J. Theor. Biol. 204: 533-541, 2000) parallel orientation with respect to the plasma membrane. However, after integrating published experimental data on glycan chain length distribution and the degree of peptide side chain cross-linking into this computer simulation, we now report that the proposed planar network of murein appears largely dysfunctional. In contrast, a scaffold model of murein architecture, which assumes that glycan strands extend perpendicularly to the plasma membrane, was found to accommodate published experimental evidence and yield a viable stress-bearing matrix. Moreover, this model is in accordance with the well-established principle of murein assembly in vivo, i.e., sequential attachment of strands to the preexisting structure. For the first time, the phenomenon of division plane alternation in dividing bacteria can be reconciled with a computer model of the molecular architecture of murein.
The biological role, chemical structure, physical properties, and principles of biogenesis of cell walls of both gram-positive and gram-negative bacteria have been extensively reviewed since the mid-1960s (1, 8, 17-19, 26, 29, 33, 44, 51, 53, 56, 57). The major structural component of all types of bacterial walls is murein, the terms murein and peptidoglycan being synonymous. Although its composition and fine chemical structure vary in different bacteria (52), the general principle of its structural organization holds constant. The material, regardless of the bacterial cell morphology and the wall thickness, is invariably composed of peptidoglycan strands cross-linked via peptide bridges. Other polymers that are associated with the sacculus in different bacteria (teichoic acids, lipoteichoic acids, polysaccharides, proteins, and lipoproteins) are not essential to the mechanical firmness of murein and, therefore, are not further discussed here.
To date, the crucial question of how the three-dimensional organization of murein can be visualized remains unanswered. Since there is no methodology that allows investigation of the architecture of murein in intact cells, researchers have had to deduce the tertiary structure on the basis of indirect evidence, the unambiguous interpretation of which is often difficult.
Currently, three models of the architecture of murein are discussed in the literature. (i) The predominant concept considers the peptidoglycan strands to run regularly and in parallel with the plasma membrane, furnishing a planar network (8, 29). (ii) An analogous model assumes an irregular orientation of peptidoglycan strands in the planar network (33, 34). (iii) The scaffold model proposes that peptidoglycan strands are oriented perpendicular to the plasma membrane (12, 13). The radial orientation of glycan strands within bacterial walls was hypothetically contemplated but rejected by Keleman and Rogers (32). It is important to note that the planar network models consider the thin gram-negative cell walls to consist of one major stress-bearing layer in combination with newly synthesized (innermost) and almost degraded (outermost) loosely cross-linked layers (22), whereas the thick gram-positive cell walls are thought to be composed of several layers linked to each other (18). In these models, therefore, a network layer is the principal structural element of the cell wall architecture. In contrast, the scaffold concept envisages the cell wall as a solid, gel-like matrix (12, 13). Thus, the stress-bearing zone of the murein tertiary structure might be viewed from either a planar or a vertically oriented perspective and currently available experimental data do not readily allow discrimination between these models.
Recently, computer simulation approaches were used to address this problem (35, 49). However, different investigators made different starting assumptions and came to different conclusions. Importantly, the degree of cross-linking, which is known to vary in gram-negative walls from approximately 25 to 50% (7, 26, 43, 49; see also the Appendix), was assumed to be either maximal or was completely disregarded in these studies.
Since, in principle, such studies permit objective comparison of different models of murein architecture, we used a computer simulation approach, too. Our specific aims were (i) to determine the effect of naturally occurring, low-degree cross-linking on the planar murein architecture and (ii) to model peptidoglycan strands as running perpendicularly to the plasma membrane. Our calculations show that a moderate degree of murein cross-linking yields a planar organization of peptidoglycan that appears dysfunctional, suggesting that the published model may need to be modified. In contrast, a scaffold model of murein, with a perpendicular orientation of glycan strands, fits the observed structural data (degree of cross-linking, chain length distribution, and pore size) and the general principle of murein assembly in vivo.
Basic principles.
Peptidoglycan chains consist of alternating β-MurNAc(1→4)GlcNAc disaccharide units linked via β-(1→4) glycosidic bonds (18). Peptide side chains, which are attached to the carboxyl group of muramic acid residues, are primarily composed of five amino acids (l-Ala-d-Glu-X-d-Ala-d-Ala, where X stands either for l-Lys or diaminopimelic acid). Certain variations in both peptide and glycan structures exist (52), but they are not important for computer simulations as reported below.
X-ray diffraction (6) and computer simulation (36, 38) studies showed that single peptidoglycan molecules adopt a fourfold helical conformation with the symmetry order N = +4, each turn of the helix being made of four disaccharide units with four peptide side chains oriented outward and perpendicular to each other, as shown in Fig. 1.
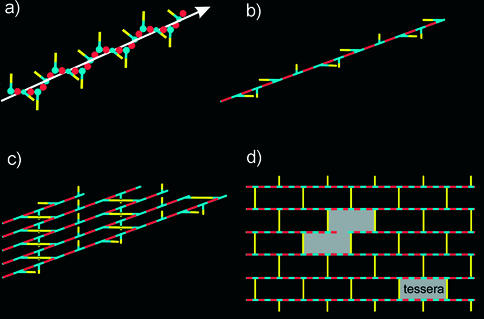
FIG. 1.
Schematic representations of peptidoglycan strand conformation and of a planar murein network. (a) The chain of MurNAc (blue items) and GlcNAc (red items) forms an extended helix. Peptide side chains (yellow bars) project from the helix in a clockwise manner and are perpendicular to each other. (b) Further simplification: the glycan chain is represented as a straight line. (c) Fragment of a murein network composed of long peptidoglycan chains cross-linked to each other via peptide bridges (yellow bars) in a horizontal orientation, whereas vertical peptides remain free. The ratio of bridged to free peptides is 1:1; therefore, the maximal degree of cross-linking is 50%. (d) The most simplified representation of the murein network: the top view includes solely glycan chains and peptide bridges. One tessera and the sample aggregate of two tesserae are shown in gray. The cleavage of long chains (i.e., the existence of short chains) and peptide bridges (i.e., a low degree of cross-linking) will result in the formation of a loose network. Therefore, both the chain length distribution and the degree of cross-linking are critical parameters for the planar murein network.
As convincingly demonstrated by direct analysis with a high-performance liquid chromatography (HPLC)-mass spectrometry technique, glycan strands in murein of different strains of Escherichia coli (27, 31, 45) and in that of Staphylococcus aureus (4) are surprisingly short. At least for these two bacterial species, the question of the orientation of these short helices within the cell wall has not been solved and is difficult to answer a priori. Generally, strands could run either parallel with or perpendicular to the plasma membrane to build up a planar network (regular or irregular) or a vertically oriented matrix, respectively. According to the predominant concept, glycan chains are arranged in parallel with the plasma membrane, are aligned in a head-to-tail manner, and are oriented perpendicularly to the cell axis (8, 9, 29). Taken together, these parameters define the regular murein network, which can be visualized as presented in Fig. 1d.
It is important to note that the degree of cross-linking in gram-negative cell walls is maximal when the ratio of bridged to free peptides is 1:1. Following the proposal of Koch (34), the smallest hole in the murein network is called a tessera and the network is composed exclusively of tesserae if murein is maximally cross-linked (Fig. 1c and d). However, experimental evidence indicates that the degree of cross-linking can be considerably lower, resulting in an averaged ratio of linked to free peptides of about 1:2 (26, 49). Thus, only 33% of all peptides could be involved in a bridge formation, and this means that each third peptide bridge within the planar network may be disconnected, yielding a fabric composed of 33% single and 67% connected tesserae.
Furthermore, glycan strands and peptide bridges of rod-like bacteria are subjected to different surface tension forces, which depend on the local curvature within the sacculus (3). As a result, glycan strands might produce an irregular network. The modified version of the irregular model describes the murein network as a multifaceted mosaic composed of dissimilar small patches with a regular architecture but connected irregularly to each other (34), although, to date, a graphic representation of this model has not been published.
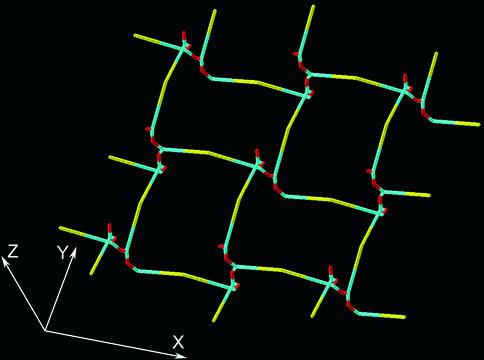
An alternative to a planar orientation is the vertical orientation of glycan strands (12, 13), forming a scaffold-like structure. In principle, the murein architecture viewed by the scaffold model is related to that proposed previously by the periplasmic gel concept (28). The latter considers murein in gram-negative bacteria as a gel-like matrix, which is strongly cross-linked in proximity to one membrane and loosely cross-linked in proximity to another one. To achieve this, glycan chains were proposed to extend beyond the horizontal layer, thereby changing their orientation. Accordingly, the new model envisages peptidoglycan strands as being oriented in perpendicular to the plasma membrane in a zone of high cross-linking. As a consequence, the peptide side chains are arranged parallel to the membrane and are oriented in four distinct directions. Each pair of peptides belonging to two adjacent strands may be bridged if they are located at the same level. Therefore, it was proposed that each strand is able to cross-link with four others (Fig. 2). As is evident from Fig. 2, even short strands can be effectively cross-linked to give a three-dimensional porous matrix. Theoretically, the shortest glycan chain is an octasaccharide that comprises four peptide side chains to be incorporated into the stress-bearing zone of the murein matrix.
FIG. 2.
The principle of murein fabric organization according to the scaffold model. The vertical helices are octasaccharide fragments of glycan chains composed of N-acetylglucosamine (red items) and muramic acid (blue items) residues. Each strand is oriented along the z axis and is able to cross-link with four others to produce a porous matrix. Yellow sticks represent peptide bridges.
Simulation of the architecture of murein. (i) Assumptions.
In agreement with the general principle of murein biogenesis in vivo, the assembly process in silico should proceed via sequential attachment of the growing nascent peptidoglycan strands to the existing murein fabric (15, 16, 22, 55). Other investigators have noted that a computer simulation of a planar murein network assembled by this mechanism is rather problematic, if it is possible at all (49). Nevertheless, when the infinitely long glycan chains in a hypothetical network were selectively cut, planar murein architecture with a maximal degree of cross-linking could indeed be created (49). When applying this cutting technique to the modeling of the planar murein architecture with a low degree of cross-linking, we considered peptide bridges to be distributed not regularly (as in the case of maximal cross-linking) but statistically.
Unlike in the planar murein model, the simulation of the scaffold murein architecture and its assembly via sequential attachment of glycan strands to an existing structure were considered possible because they do not depend on the particular length of the counterparts. Rather, all assumptions relevant to modeling are dictated by both the aim of the simulation and the available parameters. The major constraint was that the assembled murein matrix should exhibit the property of a stress-bearing gel with an appropriate pore size while, at the same time, conforming to experimentally determined parameters such as (i) the glycan chain length distribution pattern and (ii) the degree of cross-linking. To achieve this goal, the following four major assumptions were made. First, strands are oriented perpendicular to the plasma membrane. Second, since the dynamic process of murein biogenesis simultaneously comprises the assembly and turnover processes (47, 48), it was assumed that the vertically oriented murein is composed of two zones, i.e., a functional stress-bearing zone (made of assembled and mature murein) that is located close to the plasma membrane and a zone of murein degradation that is close to the outer membrane in gram-negative bacteria or represents the outermost and mostly worn-out layer in gram-positive walls (23, 24). The degradation zone contains murein remnants composed of very short strands attached to the chains protruding from the functional zone. Third, as both the synthesis of strands and concomitant murein assembly proceed in association with the plasma membrane (55), the assumption was made that glycan chain ends terminated with MurNAc residues point to the plasma membrane. Fourth, as the degrees of cross-linking in gram-positive and gram-negative walls differ significantly, we assumed that cross-linking is close to 100% in the vicinity of the plasma membrane and diminishes either gradually (gram-positive walls) or abruptly (gram-negative walls) toward the degradation zone. This allows the strands in the walls of gram-negative bacteria to make a narrow and highly cross-linked internal structure that is able to function as a stress-bearing zone in the whole murein structure. It bears mentioning that, in agreement with other authors (4, 36, 49), we modeled the glycan chains within murein as rather short and did not consider chains longer than 30 disaccharide units. The computational techniques used in this study are described in the Appendix.
(ii) Simulation of a planar murein network with maximal cross-linking.
By applying the strand-cutting technique, we obtained results similar to those reported by Pink et al. (49). The distribution of slits in the planar network was also very similar. The cutting, scheme as programmed by Pink et al. (49), resulted in strands composed exclusively of even numbers of disaccharide units, which contradicted the fact that both even- and odd-numbered strands are present in bacterial murein. Therefore, an extension of the proposed algorithm was elaborated in order to allow the presence of odd-numbered strands. After the simulation was performed (49), our extended algorithm moved along the simulated glycan strands and, when arriving at a previously fixed cutting point in accordance with the method (49), randomly decided whether this cut should be shifted to elongate the previous strand by one disaccharide or not (a shift within the same tessera and aggregate). The procedure then moved on to the next cut. Obviously, the shape of aggregates and the total number of disaccharide units were not changed by this procedure but the length distribution profile became substantially different. Since odd-numbered strands also emerged, the fraction of molecules of each strand was now about 50% of the values found for the corresponding even-numbered glycan strands (data not shown).
(iii) Comparison of the simulated glycan chain length distribution with observed data.
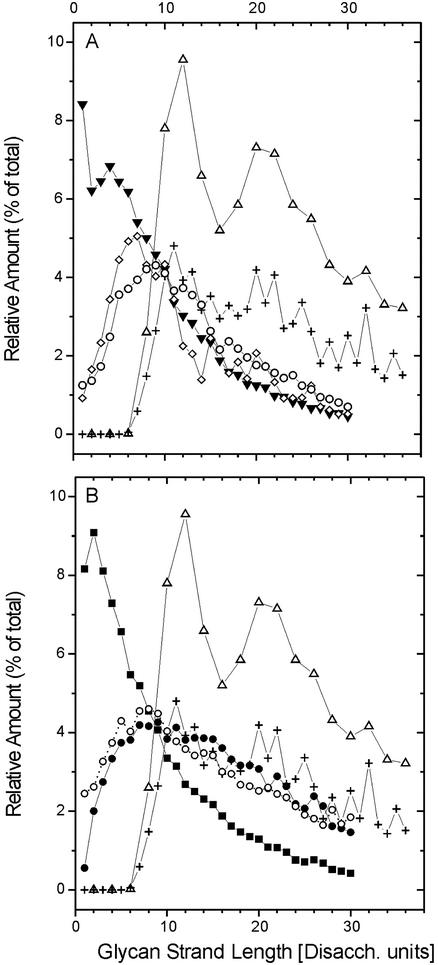
Observed glycan length distribution profiles have generally been published in the form of percent relative amount plotted versus strand length, in which the length was defined by the number of disaccharide units (31, 45) (Fig. 3). We extended the data pool by adding the results of Harz et al. (27), which had originally been presented as HPLC elution profiles and were recalculated to give relative amounts (Appendix). To compare the length distribution profiles obtained by simulation with published experimental data, we transformed our data to reflect relative amounts as shown in Fig. 3. For the two simulated profiles of glycans (with exclusively even numbers and with even and odd numbers, respectively), the amounts for the lengths L = 1 through L = 30 are 71.6 and 72.3% of the total amount. The observed values are 65.5% (31), 70% (27), and 90% (45).
FIG. 3.
Relative amount of glycan strands of length L plotted against glycan strand length L. Amounts indicated by Δ and + in panels A and B were obtained in this study by computer simulation for strands with an even number of disaccharides (Disacch.) or odd and even numbers of disaccharides, respectively, on the basis of data derived from reference 49. Experimental data obtained from E. coli in panel A: ⋄, strain ftsZ84 grown at 42°C (31); ○, strain EH3247 (27); ▾, strain MC4100 (45). Experimental data obtained from E. coli strain P678-54 (45) in panel B: •, cylindrical murein; ○, spherical murein; ▪, whole-cell murein.
Figure 3 shows the profile simulated with strands containing exclusively an even number of disaccharides. This profile does not seem to fit the experimental data at all. In contrast, the simulated profile including odd-numbered glycan chains is much more similar to the observed length distribution, especially in the region with L ≥ 10. However, both simulations differ significantly from the observed profiles in the region with L < 9. Whereas the simulation based on the planar murein model does not generate short glycan chains (up to L = 6), experimental investigations of the E. coli murein have reported up to 25% such glycan strands (45).
(iv) Simulation of a planar murein network with a moderate degree of cross-linking.
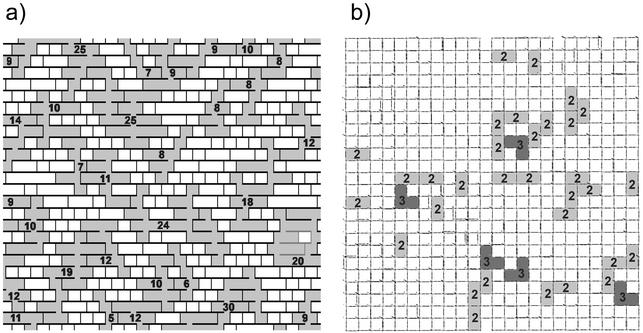
Pink et al. (49) assumed maximal cross-linking of strands, i.e., that 50% of the peptide stems are cross-linked. To investigate the effect of a moderate degree of cross-linking, our computer algorithm randomly selected peptide bridges in the previously obtained slit-containing network and applied virtual cuts to them until only 33% of the peptides in the final planar network were cross-linked. If only peptide bridges are disconnected and glycan strands of infinite length remain intact, then long horizontal slits are produced between the strands to make the network altogether loose. No proper length distribution is generated under these conditions. However, when the cutting of peptide bridges was accompanied by the generation of a glycan length distribution by using the cutting strand procedure (49), an almost disrupted network full of holes was obtained, as shown in Fig. 6a.
FIG. 6.
Comparison of the planar and scaffold models under a condition of 33% cross-linking (top-view projections). (a) Fragment of the planar murein network made of glycan strands arranged in 25 rows. The large (more than four tesserae) holes are depicted in gray, and the number of connected tesserae is given for each such hole. The chains forming a piece of network that lost its connection to the fabric are shown in gray, in contrast to the black chains that represent the network itself. Note that tesserae are shown as squares, in accordance with Fig. 5c and the dimensions given in the text. (b) The low cross-linked murein matrix simulated in accordance with the scaffold model. Average degrees of cross-linking: ∼33% (overall), ∼94% (within the internal zone), and ∼7% (in the degradation zone). The number of connected channels is given for each enlarged hole.
(v) Simulation of the murein architecture in accordance with the scaffold model.
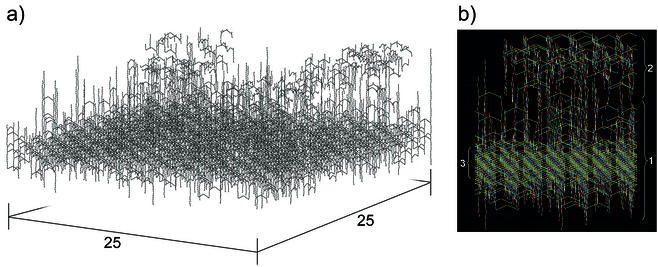
To build up a vertically oriented model of murein, a square piece (matrix) of murein with dimensions of 25 by 25 strands was virtually cleaved from the whole murein fabric. To mend the punctured fabric, randomly selected peptidoglycan strands were sequentially inserted into the square cavity (empty box). The box possessed 625 positions at which to insert randomly selected strands and was bordered at the bottom and at the top by the inner and outer membranes (the latter being absent from gram-positive envelopes), respectively. Importantly, the box was flanked along the perimeter by the existing murein fabric to allow incoming strands to cross-link with it and to bring the simulated process of matrix assembly into agreement with the murein assembly process in vivo (15, 16, 22, 55). The height of the box, i.e., the thickness of the matrix, was estimated to be about 30 disaccharide units, which allowed it to hold all of the strands that were present in the strand pool (see Appendix). All strands were vertically inserted into the box, so that their GlcNAc termini pointed to the outer membrane. It is evident that each disaccharide-peptide unit occupies a definite spatial position at a given distance from the plasma membrane. These distances may be called levels, and the total number of levels within the box was about 30, corresponding to the longest chain composed of 30 disaccharide units. It is important to note that all peptide stems became oriented in parallel with the plasma membrane and each peptide occupied its particular level.
The simulation of the murein architecture was started with the assembly of the functional zone composed of both the assembled and mature murein, which had not yet undergone the process of degradation and thus was free of very short glycan chains. For this purpose, we took the chains from 4 to 30 disaccharide units and inserted them randomly into the box to occupy all 625 positions on levels close to the plasma membrane. Following the logic of murein assembly in vivo, a certain number of strands were modeled as still attached to the membrane, whereas the rest of them were pictured as already detached from the membrane. Thus, some strands (those that were leveled by virtue of being attached to the membrane) could become tightly interwoven with other strands (those recently detached from the membrane) to obtain a maximally cross-linked zone close to the membrane, as shown in Fig. 4. It needs to be stressed that all of the strands in the functional zone participated in the formation of peptide cross-links, thereby ensuring the mechanical stability of the murein structure. The functional zone was free of very short chains; meanwhile, long strands protruded into the degradation zone.
FIG. 4.
Fragment of the murein matrix simulated in accordance with the scaffold model. (a) Panoramic view of the simulated murein matrix composed of 25 × 25 = 625 positions. The whole matrix contained 625 strands in the functional zone (average length, 11.6 disaccharides) and 470 strands in the degradation zone (average length, 2.9 disaccharides). A narrow and highly cross-linked stress-bearing zone is apparent in the lower part of the matrix. (b) Side view of a fragment of the matrix. The functional zone (zone 1) is composed of peptidoglycan strands comprising 4 to 30 disaccharide-peptide units; anhydromuramic acid residues are directed to the inner membrane. The degradation zone (zone 2) contains recently degraded strands in proximity to the outer membrane. The functional zone, which comprises both assembled and mature murein, includes a compact and highly cross-linked narrow internal zone (zone 3) with a height of 4 disaccharides; this internal zone functions as a stress-bearing structure of the murein fabric. To ensure a clear distinction between different structural units, the color coding of the fragments is the same as in Fig. 1 and 2. Note that peptide bridges are shown while free peptide side chains are omitted.
After the functional zone was completed, the remaining short strands were randomly distributed around the long ends of strands protruding from the functional zone and around each other, with the constraint that they should be connected to the whole matrix by at least one peptide bridge. We emphasize that both the functional and degradation zones represent two moieties of one entire entity. The distributions of selected strands within both zones were generated so that their sum was equal to their total distribution, and strands were spread between two zones, with the transition of one into another according to a Boltzmann function centered at a value of 5.5 (Appendix). As a consequence, all strands containing one, two, or three disaccharides were in the degradation zone. The longer the strand was, the higher was the probability that it would be located in the functional zone. The number of strands for each zone was normalized so that it obligatorily corresponded to 625 strands in the functional zone, which gave 470 strands in the degradation zone. The average length of glycan chains was 7.94 disaccharides, with 2.9 disaccharides in the degradation zone and 11.6 disaccharides in the functional zone.
Synchronously with filling of the matrix with strands, all possible cross-links were constructed between adjacent peptides possessing a suitable orientation. This yielded a very high rate of cross-linking (>90%) close to the membrane and 5 to 50% cross-linking beyond it, gradually decreasing while moving further out from the membrane. The picture thus obtained resembled the situation with gram-positive walls, where the degree of cross-linkage could be very high (18, 26) and the outermost part of the wall (an analogue of our degradation zone) is fibrous and almost worn out (23, 24). To model the fact that not all of the peptides in gram-negative walls become cross-linked, some of the bridges (mainly those located beyond the innermost zone of high cross-linking) were cleaved randomly to achieve a desired cross-linking value, which we varied over a range of 30 to 50%. It should be emphasized that our passage from the model of the gram-positive wall to that of the gram-negative wall merely represents a mathematical operation that is not related to particular biosynthetic pathways of murein bridging in vivo. If, in the modeling process, we had gone the other way and performed selective bridging between the inserted strands until the necessary degree of cross-linking was achieved, the result would have been the same.
Figure 4a shows a computer-modeled panorama of the generated E. coli W7 murein matrix with a degree of cross-linking of about 48% (20). Figure 4b depicts a side view of the portion of the generated murein matrix, whereas Fig. 6b shows the top-down view of a matrix with an overall degree of cross-linking of only 33%, about 93% being within the functional zone and about 7% being within the degradation zone. Thus, the scaffold model yields a stable and functional stress-bearing matrix even at low degrees of cross-linking.
Comparative analysis of murein permeability as viewed by the planar and scaffold models.
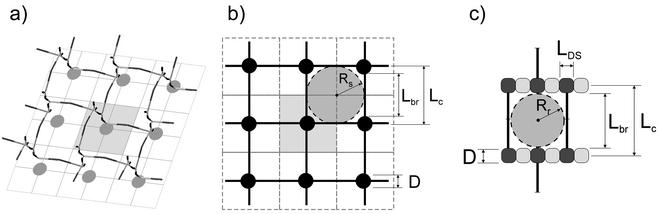
There are two aspects of the problem: (i) what is both the chemical and physical organization of the murein pores in the two models, and (ii) how do these models correspond to experimentally obtained parameters. First, in the planar model, the chemical structure of the pore is accomplished by two oligosaccharide fragments (each consisting of four disaccharide units) and by two peptide bridges, both being the fragments of two strands running parallel to each other. Physically, the pore represents an almost square hole known as a tessera. It is important to note that the oligosaccharide sides of the pore are about 4 nm long (6, 36, 38) and are unable to either expand or shrink, the variable elements of the pore being the peptide bridges. The geometric parameters of the planar pore are depicted in Fig. 5c. Moreover, now it is possible to estimate an average surface area per one disaccharide-peptide unit, which is equal to Lds × Lbr; both parameters are known (see below).
FIG. 5.
Geometric parameters of the structural units and pores in the scaffold and planar murein models. (a) Schematic representation of the spatial murein matrix (fragment composed of nine strands) and its projection onto the membrane plane. The gray circles stand for projections of the glycan chains, and the gray square area illustrates a surface portion occupied by one chain. It is important to note that each strand occupies the same surface area as one disaccharide unit does. (b) Projection of a scaffold fragment on the membrane plane. The black circles stand for projections of glycan chains; the gray square corresponds to the area occupied by one chain. D stands for the glycan chain diameter, approximately 1.1 nm; Lbr is the length of a peptide bridge, approximately 4.1 nm; Lc is the distance between strand axes, approximately 5.2 nm. A big gray circle with a radius (Rs) of about 2.6 nm represents a cross section of the channel pore expanded by the turgor pressure in the stretched sacculus in vivo. (c) Top view of the tessera in a planar model. The light and dark gray boxes stand for disaccharide units, and LDS stands for the length of one disaccharide unit, about 1 nm; the other designations are the same as those described above. A big gray circle with a radius (Rr) of about 2 nm represents the pore formed by the tessera within the relaxed sacculus.
In the scaffold model, the situation is different and the pore is transformed into a helical channel, the latter being as long as the height of the murein matrix. Unlike the planar model, the channel is made up of four cross-linked strands and also possesses a square configuration (Fig. 5a and b). The principal difference between the planar tessera and the helical channel is that four stress-bearing elements in the channel are made of elastic peptide bridges, the perpendicular glycan chains being free of tensile stress. The latter condition is physically important because it allows the channel to regulate its width in a wide range. Experimental data indicate that the mean pore radius (Rr) in the relaxed sacculus of E. coli is 2.06 nm and that for B. subtilis is 2.12 nm (10); the former is 17% less than a stretched radius (Rs) in vivo (10), which may therefore achieve a length of about 2.48 nm. Mathematically, the pore of a definite radius can be envisaged as a circle inscribed either into the tessera or into the cross section of the channel, as depicted in Fig. 5b and c. In other words, the diameter of the circle corresponds to that of a hypothetical spherical molecule that is just able to pass through the channel or the pore. The area occupied by either a relaxed tessera or a relaxed channel could maximally be about 4.12 × 4.12 nm = 17 nm2, whereas the area covered by the stretched tessera or stretched channel is expected to be equal to 4.96 × 4.96 nm = 24.6 nm2. The channel cross section predicted by the scaffold model fits the parameters both for relaxed and stretched sacculi (Fig. 5b). In contrast, the tessera cross section predicted by the planar model is 2Rr × 4LDS = 16.5 nm2 (relaxed) and 2Rs × 4LDS = 19.8 nm2 (stretched). These data show that the cross section of a planar tessera fits the experimental data only for the relaxed tessera (Fig. 5c) but not for the stretched tessera presumably prevalent in vivo.
Discussion.
This study was performed because previously published computer simulations were able to provide a tertiary model of murein only if short glycan strands (known to be present in the bacterial cell wall) were disregarded and a maximal degree of cross-linking of peptide side chains (known not to be the rule in gram-negative bacteria) was assumed (49). We reasoned that an adequate representation of the murein architecture by computer simulation would have to be able to accommodate all of the known experimental data and still yield a functional, stress-bearing fabric.
By using the same technique as Pink et al. (49), we were able to reproduce the proposed slit-type murein monolayer architecture of gram-negative bacteria with a distribution of glycan chain lengths almost identical to that simulated by those authors. However, in contrast to the fact that glycan strands comprise statistically equal numbers of strands with even and odd numbers of disaccharide units, the cutting scheme described by Pink et al. (49) resulted exclusively in the formation of even-numbered glycan chains. To include odd-numbered chains, an extension of the proposed algorithm was elaborated that yielded a simulated chain length distribution more closely related to experimental observations (Fig. 3) than the original procedure.
Nevertheless, some discrepancy remained regarding the existence of short glycan chains, which could make up up to 25% of the total murein content in different bacterial strains. These chains were absent in the chain length distribution generated by the previously published protocols (49) (Fig. 3). In fact, when the original chain-cutting algorithm allowed the formation of short chains, slits immediately became transformed into a variety of different holes, some of which were large (49).
To explore the effect a lower degree of cross-linking might have on the integrity of the planar murein network, our computer algorithm randomly selected peptide bridges in the previously generated slit-type network and performed virtual cuts until only 33% of all peptide stems remained cross-linked. This value was selected because it corresponds to networks with a ratio of bridged and free peptides of 1:2, a situation quite common in gram-negative cell walls (7, 26, 43, 49). This statistical cutting of bridges resulted in an almost disrupted network full of holes (Fig. 6a). It appears that such a structure is not able to function as a stress-bearing layer.
In the published computer simulations of the planar murein network, the cutting approach that was used contradicts the general biological principle of sequential chain attachment (15, 16, 22, 55). A functional, i.e., stable, network could only be created under the condition of a maximal degree of cross-linking. In order to prevent the network from disassembling, an additional condition was introduced (49), namely, that the ends of the aligned glycan chains stay in closest proximity to each other. However, the principles of chemical kinetics dictate that the ends of two adjacent chains would repulse each other (the collision state) if they were kept at a distance of covalent bonds as proposed (49). Under the biological conditions of murein assembly, the repulsive forces would keep the chain ends apart at a larger distance. Therefore, gaps equal in size to one or two tesserae would have to be expected between each pair of ends of the aligned glycan chains. Moreover, separation of aligned glycan chain ends at a distance larger than that of a covalent bond would inevitably result in a decreased degree of cross-linking, with concomitant formation of holes larger than the narrow slits. This problem presents a critical challenge for the concept of the planar murein architecture, because the number of glycan chain ends in the murein network of gram-negative bacteria is very high because glycan chains are short. It has not been our aim to discredit in any way the currently predominant planar model of murein architecture. In fact, as described above, we made a considerable effort to salvage it by recalculating some of its parameters (e.g., for even and odd chain lengths). From our data, however, it appears mandatory to introduce modifications into the current model (e.g., to introduce functional and stress-bearing zones) in order to make the model more compatible with existing data and provide a fully stable matrix.
We decided to explore a completely different approach, namely, to entertain the possibility that peptidoglycan strands run perpendicular to the plasma membrane. To this end, a vertically oriented, scaffold-like matrix was generated by a computer algorithm and randomly packed with peptidoglycan strands of 1 to 30 disaccharide units. This model is the first that accommodates very short glycan strands without disrupting the murein matrix (Fig. 4). It is worth mentioning that, as proposed earlier (28), the major difference in the structural organization of gram-positive and gram-negative walls is rather a quantitative one, both the amount of murein and the degree of cross-linking being the characteristic parameters. Our model supports this conclusion and allows the ready simulation of both types of walls. Indeed, the vertical orientation of glycan strands is perfectly compatible with the high degree of cross-linking characteristic of gram-positive walls (18, 26); the highly cross-linked functional zone automatically becomes thicker.
In order to simulate gram-negative walls, it is sufficient to statistically disconnect a part of the remote bridges in the highly cross-linked murein to be consistent with the experimental data published for E. coli (20). Thus, in the assembled scaffold model of E. coli murein (presented in Fig. 4), the thin stress-bearing zone of the fabric is composed of strands that are almost completely cross-linked in the area close to the membrane. The strands within this thin stress-bearing area are tightly packed and are parallel to each other because this type of regular orientation ensures maximal cross-linking. The ends of long chains are either free or partially cross-linked with the remaining material within the degradation zone. It is possible that these loose ends, together with short strands from the degradation zone, are not arranged in an orderly fashion and adopt a random-coil conformation, allowing this area within the murein matrix to be compressed. Thus, even long strands, measuring about 14 nm in gram-negative bacteria (25, 46), can be arranged within the periplasmic space. Evidently, the vertical orientation of long chains in the walls of gram-positive bacteria is retained because all of the strands are highly cross-linked, explaining why these walls are as thick as 30 nm and more (18, 19, 23).
Although the scaffold model of the architecture of murein fits the available structural data (degree of cross-linking and chain length distribution), the direct proof of this concept is still elusive. Models are most useful when they are able to explain biological phenomena. In this respect, the alternation of the division plane has been difficult to reconcile with the planar model but can be readily explained by the scaffold model.
In rod-like bacteria, such as E. coli (41) and B. subtilis (42), which grow by elongation and divide into equal daughter cells, the division plane is oriented perpendicular to the long axis of the cells. However, it was recently discovered that spheroid cells of E. coli are able to alternate the division plane in either two (2, 3) or three (11, 63) dimensions, each division plane being perpendicular to the preceding one. Each division plane represents a particular type of spherical murein architecture. In E. coli cells, the spherical murein is viewed as a set of progressively smaller concentric rings that are attached by peptide bridges to each other and to the cylindrical portion of the cell (48). Thus, when the rod-like bacterium exhibits the typical pattern of division, the process seems to proceed in agreement with the planar murein architecture. However, the concept runs directly into conflict with the fact that spheroid E. coli cells, like those of some gram-negative cocci (58) and many gram-positive spheroid bacteria, e.g., S. aureus (19, 54, 61), are able to alternate the division plane. Indeed, this biological fact implies that after each division, the cell either has to change the entire pattern of murein assembly (9) or produce progeny with two or three different types of murein architecture within one cell. The “segregated sectors” on the murein surface of dividing spheroid E. coli cells were recently discovered (11), and the question of whether such sectors of one cell really do possess a different architecture arises. Importantly, the scaffold model readily accommodates the biological feature of alternating division planes because the vertical orientation of glycan strands fits all three of the possible dimensions that the division plane can adopt. We recognize that the idea of an irregular planar network (32, 34) is also in agreement with the phenomenon of division plane alternation. For comparison and to check whether it would fit experimental data on murein permeability (10) equally well as the scaffold model, it would be of advantage if the proponents of the irregular model could present a picture of the irregular architecture.
We are not the first to conclude that gram-negative walls are not monolayer peptidoglycan networks. The latter concept has previously been challenged by morphological observations with the use of advanced electron microscopy techniques (23, 28, 39) and by biochemical data (21, 50). These observations were ignored because they seemed to disagree with the assumed surface density of murein. However, data available in the literature on the total number of disaccharide-peptide units on the cell surface are not at all consistent, especially when they are given as the average area (ADSp) per disaccharide-peptide unit: 0.61 (60), 0.77 (30), 1.3 (5), and 2.5 (59) nm2.
The distance between the strands, i.e., the length of the bridge, is obviously a decisive parameter for these calculations. There are several lines of evidence suggesting that previous calculations supporting the concept of murein in gram-negative bacteria as mostly monolayered may have been incorrect. Thus, it was demonstrated that the extended length of one peptide side chain is 2.2 nm (36), which makes the length of the peptide bridge greater than 4 nm. Further, a recent X-ray structural analysis with a resolution of 1.2 Å showed that two peptide chains in the active site of the transpeptidase interact in their fully extended forms (40). This means that the length of the bridge in murein must be greater than 4 nm and that the distance between the centers of adjacent glycan strands (Lc) must be greater than 5 nm because the diameter of the glycan chain is 1.1 nm (5, 13). These values differ substantially from those obtained by calculation with a hypothetical peptide bridge length of 1 nm (59) that led to the conclusion that the number of layers in murein from E. coli is a little more than one (59).
At first glance, therefore, the scaffold model appears to necessitate much more murein material to cover the surface of the bacterial cell. However, this is not the case and the spatial distribution of the existing murein can easily be accomplished. It is important to remember that the perpendicular orientation of the glycan strands implies that only one (terminal!) disaccharide-peptide unit is projected onto the plane of the plasma membrane, as shown on Fig. 5b, and all other structural units of the chain will be projected at the very same place. The distance Lc between the centers of these projected disaccharide-peptide units in the murein scaffold model would be equal to the diameter of the glycan strand (1.1 nm; see references 5 and 13) plus the length of one peptide bridge (at least 4.1 nm). Therefore, one terminal disaccharide-peptide unit within the sacculus occupies an area of about 5.2 by 5.2 nm = 27 nm2 (Fig. 5b), providing sufficient coverage of the entire bacterial surface. Indeed, if the average measured area of one disaccharide-peptide unit on the basis of atomic parameters is SDSp and that determined in experiments based on chemical analysis and electron microscopy data (59) is ADSp, the SDSp/ADSp ratio shows either the number of murein layers in the planar model or the strand length in the scaffold model, the strand length being measured in disaccharide units. Thus, for the scaffold model, this ratio is about 11 disaccharide-peptide units, a value that is in accord with the average glycan chain length used in our simulation.
In conclusion, the proposed scaffold model of murein architecture fulfills the requirements of a stress-bearing structure, allowing it to be compact and stretched (37), highly porous, and elastic (14, 37, 62). A sponge-like fabric is able to retain large amounts of water, thus exhibiting the properties of a gel that is able to shrink and swell (28, 62). These properties of murein provide a clue to the structural organization of the periplasm in gram-negative bacteria. It seems that the model better explains the low mobility of proteins within the periplasm and is in agreement with the idea of periplasm compartmentalization (46); the latter seems to be obligatory for the processes of trans-envelope trafficking of different substances, polymers being the most important. Indeed, in the course of the cell cycle of gram-negative bacteria, all of the components of the outer membrane e.g., β-barrel proteins, lipopolysaccharides, and phospholipids, as well as the capsular polysaccharides and secreted proteins, traverse the murein matrix of the bacterial cell wall. The same is true of the gram-positive bacteria, where the thick and highly cross-linked wall is penetrated by the secreted and surface-associated proteins and is pierced with the teichoic acids, capsular polysaccharides being secreted through the wall as well. It will be important to determine how the proposed scaffold-like murein architecture functions to ensure the success of these processes and whether specialized pores are needed for the translocation of bulky molecules across the peptidoglycan in both gram-positive and gram-negative cells.
There is a shortage of methods by which to precisely determine, by direct structural analysis, the tertiary structure of murein. To fill this gap, both the planar and scaffold models of murein make valuable contributions to deal with the problem conceptually because they reflect theoretically alternative views of the tertiary structure of murein. To its advantage, the scaffold model is in accordance with the biological feature of alternating division planes and the principle of murein assembly in vivo, i.e., by sequential attachment of newly synthesized peptidoglycan strands to the pre-existing material. Moreover, the scaffold model shows that the degree of cross-linking within the matrix could vary over a wide range without considerable network breakdown, even in the presence of short glycan strands.
Acknowledgments
We acknowledge R. Bartels for technical assistance and I. Bouchain for helping to prepare some of the figures. B.A.D. and F.V.T. gratefully acknowledge the Research Center Borstel for providing a research fellowship.
APPENDIX
To explore the planar architecture of the murein network, we used the strand-cutting approach (49) and started from a maximally cross-linked simulated network containing 3,200 single tesserae. By successively cutting the strands at previously described positions (49), we also generated randomly distributed slits in the form of aggregates of five tesserae (size s = 5). Altogether, 25% of the network was randomly covered by 160 generated aggregates. To distribute the slits within the network, we also used the Monte Carlo technique. The aggregates were randomly moved on the plane of the network subject to the effective repulsion strength, V, between pairs of aggregates (49), where
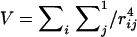 |
(1) |
In equation 1, rij is the distance between tesserae i and j, which belong to two “interacting” aggregates.
One of the curves in Fig. 3a was generated on the basis of the experimental data of Harz et al. (27), which were published as HPLC profiles. We scanned these and obtained the data by averaging the integrated peak areas as determined by UV absorption and radioactivity.
Most experimental chain length distribution data were published as percentages of the relative amount. Therefore, to enable a direct comparison of our results, we transformed the number of strands obtained by computer simulation (49) to this scale. Thus, the relative amount of glycan strands with length L, as shown in Fig. 3, was calculated by
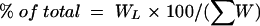 |
(2) |
In equation 2, WL is the relative mass of strands with length L and is defined by: WL = NL × NDS; here, NL is the number of strands with chain length L and NDS is the number of disaccharide units.
To build up a scaffold model of murein, we generated a matrix of 25 by 25 = 625 positions to insert randomly the strands statistically selected from the pool with a known chain length distribution pattern. The chain length distribution pattern of incoming strands corresponded strictly to the experimental data published for a particular E. coli strain (27). All of the strands were inserted into the matrix vertically, so that their anhydro-MurNAc termini pointed to the inner membrane. The thickness of the matrix was selected to be equal to 30 disaccharides, however, being dependent on the generation seed, it varied from 28 to 34 disaccharides. Each disaccharide-peptide unit within the matrix was obligatorily located at a definite height and corresponded to one particular peptide level. Further, each position in the matrix was occupied by one long or middle strand in the functional zone and could also be occupied by additional short strands in the degradation zone. As a result, the total number of strands inserted into the matrix exceeded 625.
The distribution of strands in the functional zone (probability Pf as a function of strand length i) and in the degradation zone (probability Pd as a function of strand length i) was the following:
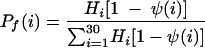 |
(3) |
and
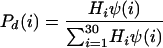 |
(4) |
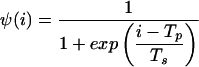 |
Hi is the probability of finding a strand of length i in the experimental distribution (27) (normalized molar fraction), Tp is the position factor for the transition function, selected as 5.5, and Ts is the smoothness factor for the transition function, selected as 0.5 (4.7 and 0.25, respectively, for the simulation shown in Fig. 4a).
All possible cross-links were applied between adjacent peptides (which occupy the same level and possess a suitable orientation) to give hyper cross-linking within the functional zone, decreasing while approaching the outer membrane.
For clarity and convenience, the degree of cross-linking was estimated in this paper as a ratio of bridged peptides to all of the peptides in the murein.
REFERENCES
- 1.Baddiley, J. 1968. The Leeuwenhoek lecture, 1967. Teichoic acids and the molecular structure of bacterial walls. Proc. R. Soc. Lond. B Biol. Sci. 170:331-348. [DOI] [PubMed] [Google Scholar]
- 2.Begg, K. J., B. G. Spratt, and W. D. Donachie. 1986. Interaction between membrane proteins PBP3 and RodA is required for normal cell shape and division in Escherichia coli. J. Bacteriol. 167:1004-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begg, K. J., and W. D. Donachie. 1998. Division planes alternate in spherical cells of Escherichia coli. J. Bacteriol. 180:2564-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boneca, I. G., Z. H. Huang, D. A. Gage, and A. Tomasz. 2000. Characterization of Staphylococcus aureus cell wall glycan strands: evidence for a new β-N-acetylglucosaminidase activity. J. Biol. Chem. 275:9910-9918. [DOI] [PubMed] [Google Scholar]
- 5.Braun, V., H. Gnirke, U. Henning, and K. Rehn. 1973. Model for the structure of the shape-maintaining layer of the Escherichia coli cell envelope. J. Bacteriol. 114:1264-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burge, R. E., A. G. Fowler, and D. A. Reaveley. 1977. Structure of the peptidoglycan of bacterial cell walls. J. Mol. Biol. 177:927-953. [DOI] [PubMed] [Google Scholar]
- 7.Caparrós, M., A. G. Pisabarro, and M. A. de Pedro. 1992. Effect of d-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. J. Bacteriol. 174:5549-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper, S. 1991. Synthesis of the cell surface during the division cycle of rod-shaped, gram-negative bacteria. Microbiol. Rev. 55:649-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper, S. 1997. Division pattern of a round mutant of Escherichia coli. J. Bacteriol. 179:5582-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demchick, P., and A. L. Koch. 1996. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J. Bacteriol. 178:768-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Pedro, M. A., W. D. Donachie, J.-V. Höltje, and H. Schwarz. 2001. Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic protein RodA and penicillin-binding protein 2. J. Bacteriol. 183:4115-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dmitriev, B. A., S. Ehlers, and E. T. Rietschel. 1999. Layered murein revisited: a fundamentally new concept of bacterial wall structure, biogenesis and function. Med. Microbiol. Immunol. 187:173-181. [DOI] [PubMed] [Google Scholar]
- 13.Dmitriev, B. A., S. Ehlers, E. T. Rietschel, and P. J. Brennan. 2000. Molecular mechanics of the mycobacterial cell wall: from horizontal layers to vertical scaffolds. Int. J. Med. Microbiol. 290:251-258. [DOI] [PubMed] [Google Scholar]
- 14.Doyle, R. J., and R. E. Marquis. 1994. Elastic, flexible peptidoglycan and bacterial cell wall properties. Trends Microbiol. 2:57-60. [DOI] [PubMed] [Google Scholar]
- 15.Gally, D. L., and A. R. Archibald. 1993. Cell wall assembly in Staphylococcus aureus: proposed absence of a secondary cross-linking reaction. J. Gen. Microbiol. 139:1907-1913. [DOI] [PubMed] [Google Scholar]
- 16.Gally, D. L., I. C. Hancock, C. R. Harwood, and A. R. Archibald. 1991. Cell wall assembly in Bacillus megaterium: incorporation of new peptidoglycan by a monomer addition process. J. Bacteriol. 173:2548-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghuysen, J.-M. 1968. Use of bacteriolytic enzymes in determination of wall structure and their role in cell metabolism. Bacteriol. Rev. 32:425-464. [PMC free article] [PubMed] [Google Scholar]
- 18.Ghuysen, J.-M., and R. Hakenbeck (ed.). 1994. Bacterial cell wall. Elsevier, Amsterdam, The Netherlands.
- 19.Giesbrecht, P., T. Kersten, H. Maidhof, and J. Wecke. 1998. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol. Mol. Biol. Rev. 62:1371-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glauner, B. 1988. Separation and quantification of muropeptides with high performance liquid chromatography. Anal. Biochem. 182:451-464. [DOI] [PubMed] [Google Scholar]
- 21.Glauner, B., J.-V. Höltje, and U. Schwarz. 1988. The composition of murein of Escherichia coli. J. Biol. Chem. 263:10088-10095. [PubMed] [Google Scholar]
- 22.Glauner, B., and J.-V. Höltje. 1990. Growth pattern of the murein sacculus of Escherichia coli. J. Biol. Chem. 265:18988-18996. [PubMed] [Google Scholar]
- 23.Graham, L. L. 1992. Freeze-substitution studies of bacteria. Electron Microsc. Rev. 5:77-103. [DOI] [PubMed] [Google Scholar]
- 24.Graham, L. L., and T. J. Beveridge. 1994. Structural differentiation of the Bacillus subtilis 168 cell wall. J. Bacteriol. 176:1413-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham, L. L., T. J. Beveridge, and N. Nanninga. 1991. Periplasmic space and the concept of the periplasm. Trends Biochem. Sci. 16:328-329. [DOI] [PubMed] [Google Scholar]
- 26.Hammond, S. M., P. A. Lambert, and A. N. Rycroft. 1984. The bacterial cell surface. Croom Helm, London, England.
- 27.Harz, H., K. Burgdorf, and J.-V. Höltje. 1990. Isolation and separation of the glycan strands from murein of Escherichia coli by reversed-phase HPLC. Anal. Biochem. 190:120-128. [DOI] [PubMed] [Google Scholar]
- 28.Hobot, J. A., E. Carlemalm, W. Villinger, and E. Kellenberger. 1984. Periplasmic gel: new concept resulting from the reinvestigation of the bacterial cell envelope ultrastructure by new methods. J. Bacteriol. 160:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Höltje, J.-V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Höltje, J.-V., and B. Glauner. 1990. Structure and metabolism of Escherichia coli sacculus. Res. Microbiol. 141:75-89. [DOI] [PubMed] [Google Scholar]
- 31.Ishidate, K., A. Ursinus, J.-V. Höltje, and L. Rothfield. 1998. Analysis of the length distribution of murein glycan strands in ftsZ and ftsI mutants of Escherichia coli. FEMS Microbiol. Lett. 168:71-75. [DOI] [PubMed] [Google Scholar]
- 32.Keleman, M. V., and H. J. Rogers. 1971. Three-dimensional molecular models of bacterial cell wall muropeptides (peptidoglycans). Proc. Natl. Acad. Sci. USA 68:992-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koch, A. L. 1988. Biophysics of bacterial walls viewed as stress-bearing fabric. Microbiol. Rev. 52:337-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koch, A. L. 1998. Orientation of the peptidoglycan chains in the sacculus of Escherichia coli. Res. Microbiol. 149:689-701. [DOI] [PubMed] [Google Scholar]
- 35.Koch, A. L. 2000. Length distribution of the peptidoglycan chains in the sacculus of Escherichia coli. J. Theor. Biol. 204:533-541. [DOI] [PubMed] [Google Scholar]
- 36.Koch, A. L. 2000. Simulation of the conformation of the murein fabric: the oligoglycan, penta-muropeptide, and cross-linked nona-peptide. Arch. Microbiol. 174:429-439. [DOI] [PubMed] [Google Scholar]
- 37.Koch, A. L., and S. Woeste. 1992. Elasticity of the sacculus of Escherichia coli. J. Bacteriol. 174:4811-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Labischinski, H., and H. Maidhof. 1994. Bacterial peptidoglycan: overview and evolving concepts, p. 23-28. In J.-M. Ghuysen and R. Hackenbeck (ed.) Bacterial cell wall. Elsevier, Amsterdam, The Netherlands.
- 39.Leduc, M., C. Frehel, E. Siegel, and J. van Heijenoort. 1989. Multilayered distribution of peptidoglycan in the periplasmic space of Escherichia coli. J. Gen. Microbiol. 135:1243-1254. [DOI] [PubMed] [Google Scholar]
- 40.Lee, W., M. A. McDonough, L. Kotra, Z. H. Li, N. R. Salvagi, Y. Takeda, J. A. Kelly, and S. Mobashery. 2001. A 1.2-Å snapshot of the final step of bacterial wall biosynthesis. Proc. Natl. Acad. Sci. USA 98:1427-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marr, A. G., R. J. Harvey, and W. C. Trentini. 1966. Growth and cell division of Escherichia coli. J. Bacteriol. 91:2388-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendelson, N. H., and J. N. Reeve. 1973. Growth of the Bacillus subtilis cell surface. Nature (London) New Biol. 243:62-64. [PubMed] [Google Scholar]
- 43.Mengin-Lecreulx, D., D. Blanot, and J. van Heijenoort. 1994. Replacement of diaminopimelic acid by cystathionine or lanthionine in the peptidoglycan of Escherichia coli. J. Bacteriol. 176:4321-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nanninga, N. 1998. Morphogenesis of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:110-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Obermann, W., and J.-V. Höltje. 1994. Alterations of murein structure and of penicillin-binding proteins in minicells from Escherichia coli. Microbiology 140:79-87. [DOI] [PubMed] [Google Scholar]
- 46.Oliver, D. B. 1996. Periplasm, p. 88-103. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C.
- 47.Park, J. T. 1995. Why does Escherichia coli recycle its cell wall peptides? Mol. Microbiol. 17:421-426. [DOI] [PubMed] [Google Scholar]
- 48.Park, J. T. 1996. The murein sacculus, p. 48-57. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C.
- 49.Pink, D., J. Moeller, B. Quinn, M. Jericho, and T. Beveridge. 2000. On the architecture of the gram-negative bacterial murein sacculus. J. Bacteriol. 182:5925-5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prats, R., and M. A. de Pedro. 1989. Normal growth and division of Escherichia coli with a reduced amount of murein. J. Bacteriol. 171:3740-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salton, M. R. J. 1964. The bacterial cell wall. Elsevier Publishers, London, United Kingdom.
- 52.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strominger, J. L. 1970. Penicillin-sensitive enzymic reactions in bacterial wall synthesis. Harvey Lect. 64:179-213. [PubMed] [Google Scholar]
- 54.Tzagoloff, H., and R. Novick. 1977. Geometry of cell division in Staphylococcus aureus. J. Bacteriol. 129:343-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Heijenoort, J. 1996. Murein synthesis, p. 1025-1034. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology 2nd ed., vol.1. American Society for Microbiology, Washington, D.C.
- 56.Vollmer, W., and J.-V. Höltje. 2001. Morphogenesis of Escherichia coli. Curr. Opin. Microbiol. 4:625-633. [DOI] [PubMed] [Google Scholar]
- 57.Weidel, W., and H. Pelzer. 1964. Bag-shaped macromolecules—a new outlook on bacterial cell walls. Adv. Enzymol. 26:193-232. [DOI] [PubMed] [Google Scholar]
- 58.Westling-Hägström, B., T. Elmros, S. Normark, and B. Windblad. 1977. Growth pattern and cell division in Neisseria gonorrhoeae. J. Bacteriol. 29:333-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wientjes, F. B., C. L. Woldringh, and N. Nanninga. 1991. Amount of peptidoglycan in cell walls of gram-negative bacteria. J. Bacteriol. 173:7684-7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wientjes, F. B., E. Pas, P. E. M. Taschner, and C. L. Woldringh. 1985. Kinetics of uptake and incorporation of meso-diaminopimelic acid in different Escherichia coli strains. J. Bacteriol. 164:331-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamada, S., M. Sugai, H. Komatsusawa, S. Nakashima, T. Oshida, A. Matsumoto, and H. Suginaka. 1996. An autolysin ring associated with cell separation of Staphylococcus aureus. J. Bacteriol. 178:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao, X., M. Jericho, D. Pink, and T. Beveridge. 1999. Thickness and elasticity of gram-negative murein sacculi measured by atomic force microscopy. J. Bacteriol. 181:6865-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zaritsky, A., C. L. Woldringh, I. Fishov, N. O. E. Vischer, and M. Einav. 1999. Varying division planes of secondary constrictions in spheroid Escherichia coli cells. Microbiology 145:1015-1022. [DOI] [PubMed] [Google Scholar]