Abstract
Recent experiments have established that Sox9 is required for chondrocyte differentiation. Here, we show that fibroblast growth factors (FGFs) markedly enhance Sox9 expression in mouse primary chondrocytes as well as in C3H10T1/2 cells that express low levels of Sox9. FGFs also strongly increase the activity of a Sox9-dependent chondrocyte-specific enhancer in the gene for collagen type II. Transient transfection experiments using constructs encoding FGF receptors strongly suggested that all FGF receptors, FGFR1–R4, can transduce signals that lead to the increase in Sox9 expression. The increase in Sox9 levels induced by FGF2 was inhibited by a specific mitogen-activated protein kinase kinase (MAPKK)/mitogen-activated protein kinase/ERK kinase (MEK) inhibitor U0126 in primary chondrocytes. In addition, coexpression of a dual-specificity phosphatase, CL100/MKP-1, that is able to dephosphorylate and inactivate mitogen-activated protein kinases (MAPKs) inhibited the FGF2-induced increase in activity of the Sox9-dependent enhancer. Furthermore, coexpression of a constitutively active mutant of MEK1 increased the activity of the Sox9-dependent enhancer in primary chondrocytes and C3H10T1/2 cells, mimicking the effects of FGFs. These results indicate that expression of the gene for the master chondrogenic factor Sox9 is stimulated by FGFs in chondrocytes as well as in undifferentiated mesenchymal cells and strongly suggest that this regulation is mediated by the MAPK pathway. Because Sox9 is essential for chondrocyte differentiation, we propose that FGFs and the MAPK pathway play an important role in chondrogenesis.
Sox9 is a high mobility group DNA-binding domain-containing transcription factor that is expressed in all prechondrocytic and chondrocytic cells during mouse embryonic development; its expression pattern closely parallels that of the gene for type II collagen (Col2a1) (1, 2). Recent work from our laboratory based on mouse embryo chimeras derived from Sox9 homozygous mutant embryonic stem (ES) cells has demonstrated that Sox9 is a master regulatory factor that is required for chondrocyte differentiation (3). Indeed, in these chimeras, Sox9 homozygous mutant cells were blocked from differentiating into chondrocytes; they instead persisted as mesenchymal cells. The mutant cells were unable to express the genes for chondrocyte-specific markers, such as collagen types II, IX, and XI and aggrecan. In addition, no cartilages were formed in teratomas derived from Sox9 homozygous mutant ES cells. Furthermore, heterozygous mutations in and around the SOX9 gene in humans cause campomelic dysplasia (CD), a severe dwarfism syndrome characterized by anomalies in all skeletal elements derived from cartilages (4–6). CD is also often associated with XY sex reversal. In many instances, the disease is thought to be due to haploinsufficiency, i.e., the remaining 50 percent of SOX9 is insufficient to perform the physiological function of SOX9. SOX9 binds to and activates chondrocyte-specific enhancer elements in the Col2a1 and Col11a2 genes, and ectopic expression of SOX9 in transgenic mice activates the endogenous Col2a1 gene, providing evidence that these genes are direct targets of Sox9 (7–9). SOX9 was recently also shown to regulate the gene for a cartilage protein CD-RAP (10).
Sox9 expression starts in mesenchymal chondroprogenitor cells and reaches a high level of expression in differentiated chondrocytes (1, 2, 11). Sox9 expression is completely down-regulated in hypertrophic chondrocytes. In addition, Sox9 is expressed in discrete areas of the urogenital system, brain, and heart (1, 2, 11, 12).
Fibroblast growth factors (FGF) have important roles in various processes of embryonic development, including the earliest stages of limb development. Various FGFs are expressed in the apical ectodermal ridge during development of vertebrate limbs, and these ligands in turn regulate the proliferation of the underlying mesenchyme and outgrowth of the limb bud (13–16). In addition, at least two FGFs, FGF1 and FGF2, are expressed in differentiated chondrocytes (17, 18). FGF2 has been shown to stabilize the phenotype of chondrocytes plated at low density and to inhibit differentiation of chondrocytes into hypertrophic chondrocytes in culture (19, 20). Furthermore, intraarticular administration of FGF2 enhances matrix formation in vivo (21–23). FGFs signal by binding to a four-member family of transmembrane tyrosine kinase receptors encoded by Fgfr1, Fgfr2, Fgfr3, and Fgfr4. During embryonic development, each FGF receptor displays distinct patterns of expression (24–26). FGFR1 is the predominant isoform expressed in the mesenchyme of the developing limbs, and its expression is maintained throughout chondrocyte differentiation, including hypertrophic chondrocytes. Fgfr2 and Fgfr4 are also expressed in cartilage primordia. In contrast, Fgfr3 expression is restricted to the resting and proliferating zones of cartilages in the growth plates (27, 28). It is possible, therefore, that the different receptors play distinct roles during chondrocyte differentiation. In humans, mutations in FGFR3 are associated with several forms of dwarfism, including achondroplasia, thanatophoric dysplasia, and hypochondroplasia (29–33). Animal models for achondroplasia and thanatophoric dysplasia are characterized by a shorter growth plate, in which proliferation of chondrocytes is markedly inhibited (28, 34, 35).
We show here that expression of Sox9 is up-regulated by FGF in primary chondrocytes and in Sox9-expressing mesenchymal cells. We further present evidence that FGF stimulation of Sox9 expression is mediated by the mitogen-activated protein kinase (MAPK) cascade, a signal transduction pathway that is activated by growth factors such as FGF. Our data strongly suggest that FGF and the MAPK pathway play an important role in the regulation of Sox9 expression during chondrocyte differentiation.
Materials and Methods
Reagents.
Recombinant FGF1 was prepared as previously described (36). Recombinant human FGF2 was from Amersham Pharmacia. Recombinant FGF7 was either prepared as described (37) or purchased from PeproTech (Rocky Hill, NJ). Recombinant epidermal growth factor (EGF), insulin, and heparin were from Sigma. Recombinant bone morphogenetic protein (BMP) 2 and transforming growth factor (TGF)-β1 were from Genetics Institute (Cambridge, MA) and Life Technologies (Rockville, MD), respectively. Mitogen-activated protein kinase/ERK kinase (MEK) inhibitor U0126 was from Promega.
Cell Cultures.
Mouse costal chondrocytes were obtained from 1- to 5-day-old mice (38). Prechondrocytic ATDC5 cells were provided by Tadao Atsumi (The Institute of Physical and Chemical Research, Tsukuba, Japan) (39). All other cells were described previously (7, 40, 41). The cells were maintained in DMEM supplemented with penicillin (50 units/ml), streptomycin (50 μg/ml), l-glutamine (2 mM), and 10% FBS.
RNA Preparation and Northern Blot Analysis.
Total cellular RNA was extracted from cultures by using the TRIZOL Reagent (Life Technologies). RNA aliquots of 10 μg were loaded on each lane, fractionated by electrophoresis, and transferred onto nylon filters (Zeta-Probe GT; Bio-Rad). The filters were hybridized with 32P-labeled Sox9 and 18S rRNA probes as described previously (7, 38). Hybridization signals were quantified by scanning densitometry (Intelligent Quantifier; BioImage, Ann Arbor, MI).
Western Blot Analysis.
Total cellular protein was prepared by lysing cells in 20 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 5 μg/ml pepstatin, and 1 mM PMSF. Protein concentrations were determined by the Bradford method. Thirty to forty micrograms of protein was separated by 8% SDS/PAGE and electrophoretically transferred to nitrocellulose filters (PROTRAN; Schleicher & Schuell). The filters were blocked in 5% nonfat dry milk in Tris-buffered saline (pH 7.5) containing 0.1% Tween 20 and then incubated with antibodies K-23 (ERK1 and ERK2) or E-4 (Tyr-204 phosphorylated ERK1 and ERK2) (Santa Cruz Biotechnology) or the Sox9 antibody (8). Filters were then incubated with the second antibody (horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG; Amersham Pharmacia), and the signal was detected by enhanced chemiluminescence (Amersham Pharmacia).
Plasmids.
4x48-p89 Col2a1 luciferase construction and its MA6 mutant were described previously (41, 42). pcDNA1/Neo (Invitrogen) expression vectors encoding FGFR1IIIc, FGFR2IIIc, FGFR3IIIc, and FGFR4 were either previously described or generated by subcloning the cDNAs (43–45). pcDNA3.1/Zeo (Invitrogen) expression plasmids encoding the extracellular and transmembrane domains of FGFR2βIIIb and the kinase domains of FGFR1 or FGFR2 were described previously (46). The expression plasmid encoding the chimeric FGF receptor consisting of the extracellular and transmembrane domains of FGFR2βIIIb and the kinase domain of FGFR3 was generated by ligating the cDNAs at the conserved ApaLI site in the juxtamembrane domain and subcloned into pcDNA3.1/Zeo. pCS2+MT vectors encoding the wild-type FGFR3IIIc, achondroplasia (G380R), thanatophoric dysplasia type I (R248C), and type II (K650E) mutants were generously provided by Michael Naski and David Ornitz (Washington University School of Medicine, St. Louis, MO) (47). pFC-MEK1 encoding a constitutively active mutant of MEK1 (S218/222E, Δ32–51) was from Stratagene. pSG5 expression vector (Stratagene) encoding myc-tagged CL100 was a generous gift from Stephen M. Keyse (Biomedical Research Centre, Ninewells Hospital, Dundee, U.K.) (48). pcDNA3 m-Stat1-His-713M/Flag and pcDNA3 m-Stat1-Tyr-701M/Flag encoding the dominant-negative mutants of Stat1 were kindly provided by Michael J. Walter and Michael J. Holtzman (Washington University School of Medicine) (49).
Transient Transfections.
Cells were transiently transfected with the reporter constructs by using FuGene6 (Roche Molecular Biochemicals). Briefly, 1.7 μl of Fugene6 was mixed with a total of 600–800 ng of plasmid DNA in 50 μl of standard medium. The mixture was preincubated for 15 min and added to preestablished monolayers of 3.0 × 105 cells per 4-cm2 well. Cells were harvested at 24–48 h after transfection. For some of the experiments, cells were incubated with FGF or other agent for the last 18–24 h. Reporter plasmids and pSVβgal (Promega), used as an internal control for transfection efficiency, were cotransfected in a 3:1 ratio. Luciferase and β-galactosidase activities were assayed as described previously (40). Promoter activities were normalized for transfection efficiency and expressed as fold activity relative to the basal activity of p89, a construct that harbors the 89-bp minimal Col2a1 promoter but lacks the 48-bp Col2a1 enhancer.
Results
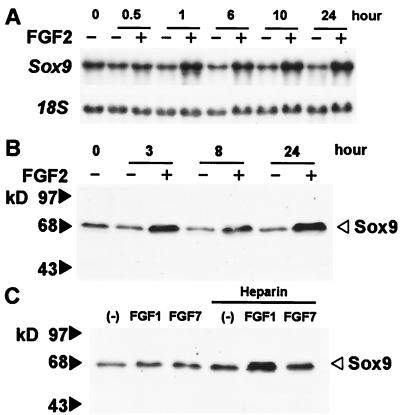
We first examined the effects of FGF2 on Sox9 mRNA levels in mouse primary chondrocytes. FGF2 increased the levels of Sox9 mRNA as early as 30 min after addition of FGF2; the increase lasted for at least 24 h. (Fig. 1A). FGF2 also increased Sox9 protein levels in these cells, which was detected within 90 min (data not shown, and Fig. 1B), and enhanced the levels of Sox9 protein in the prechondrocytic ATDC5 cells (data not shown). An increase in Sox9 mRNA and protein was also observed after treatment of the mesenchymal cell line C3H10T1/2 with FGF2 (data not shown). Both ATDC5 cells and C3H10T1/2 cells express low levels of Sox9.
Figure 1.
FGFs increase Sox9 mRNA and protein levels in chondrocytes. (A) Mouse primary chondrocytes were cultured for the indicated periods of time in the presence or absence of 2 ng/ml FGF2. Total RNA was used for Northern blots to measure Sox9 mRNA levels; an 18S rRNA probe was used as loading control. FGF2 increased Sox9 mRNA about 3-fold at 24 h. (B) Mouse primary chondrocytes were cultured for the indicated periods of time in the presence or absence of 2 ng/ml FGF2. Total cell lysates were examined by Western blot analysis using a Sox9 antibody. FGF2 increased Sox9 protein about 7-fold at 24 h. (C) Mouse primary chondrocytes were treated with 60 ng/ml FGF1, 60 ng/ml FGF7, or vehicle for 19 h in the presence or absence of 200 μg/ml heparin. The figure presents data from one of two experiments that produced very similar results.
We tested other FGFs for their ability to elicit a similar response, specifically FGF1 and FGF7. Heparin is known to potentiate the activity of FGF1 in various cell systems (50–52). FGF1, which binds to all isoforms of FGF receptors, increased the levels of Sox9 protein in the presence of heparin in primary chondrocytes (Fig. 1C). In contrast, FGF7 had little or no effect on Sox9 expression in these cells in the presence or absence of heparin. FGF7 only binds to a receptor isoform FGFR2IIIb, an alternatively spliced isoform of FGFR2 selectively expressed in epithelial cells (50, 53). One possible explanation for the inability of FGF7 to increase Sox9 expression in chondrocytes would be that these cells do not express FGFR2IIIb.
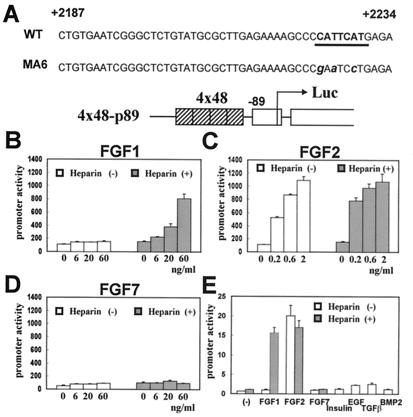
To analyze the mechanisms of the regulation of Sox9 expression by FGFs, we used the activity of a 48-bp Col2a1 enhancer as a functional measurement of Sox9. In the construct that we used, four copies of a 48-bp chondrocyte-specific enhancer of the Col2a1 gene were cloned directly upstream of an 89-bp minimal Col2a1 promoter (Fig. 2A). The activity of this construct is entirely dependent on Sox9, and a mutation in the Sox9 binding site of this enhancer abolishes the activity of this enhancer (7, 42). We first transfected primary chondrocytes with the 4x48-p89 construct and subsequently treated the cells with FGF1, -2, or -7 in the presence or absence of heparin. FGF1 increased the activity of this enhancer in the presence of heparin in a dose-dependent manner (Fig. 2B). FGF2 similarly activated this enhancer, but this activation was not heparin-dependent (Fig. 2C). In contrast, FGF7 did not affect the activity of the enhancer (Fig. 2D). A similar pattern of activation of this enhancer was also observed in C3H10T1/2 cells (Fig. 2E). The activation of the Sox9-dependent enhancer was specific to FGFs, since insulin, EGF, TGF-β, and BMP2 did not affect the enhancer activity. Although BMP2 was reported to enhance Sox9 expression in other systems (54, 55), we did not observe up-regulation of Sox9 mRNA and protein in these cells (data not shown). The increase in the activity of the 48-bp Col2a1 enhancer by FGFs paralleled the increase in Sox9 levels as determined by Western blot analysis. In addition, both in primary chondrocytes and C3H10T1/2 cells, FGF2 was unable to activate an identical construct containing a mutant 48-bp enhancer that abolishes SOX9 binding and fails to be activated by SOX9 (data not shown). Furthermore, FGF2 did not activate the 48-bp Col2a1 enhancer in BALB/3T3 fibroblasts and in the osteoblastic osteosarcoma ROS17/2.8 cells, both of which do not express Sox9 (data not shown). However, FGF2 activated this enhancer in ATDC5 cells and the immortalized mouse chondrocyte line MC615, both of which express Sox9 (data not shown).
Figure 2.
FGFs increase the activity of a Sox9-dependent Col2a1 enhancer construct in chondrocytes and C3H10T1/2 cells. (A) Schematic representation of a 48-bp chondrocyte-specific Col2a1 enhancer construction and its MA6 mutant. The end points of this enhancer relative to the position of the Col2a1 transcription start site, the Sox9 binding site (underlined segment), and the mutations in this site (lowercase letters) are indicated. (B–D) Dose-dependent activation of the enhancer activity by FGFs. Primary chondrocytes were transiently transfected with 4x48-p89 and pSVβgal. FGF1 (B), FGF2 (C), and FGF7 (D) at indicated concentrations were added to the culture medium in the presence or absence of heparin at 100 μg/ml (B and C) or 200 μg/ml (D). (E) FGF-specific activation of the enhancer activity in C3H10T1/2 cells. The cells were transiently transfected with 4x48-p89 and pSVβgal and treated with FGF1 (50 ng/ml), FGF2 (5 ng/ml), FGF7 (10 ng/ml), EGF (10 ng/ml), insulin (50 μg/ml), TGFβ (2 ng/ml), and BMP2 (100 ng/ml). Heparin was added at 100 μg/ml. In all panels, promoter activities are average values ± standard deviations for three independently transfected cultures from one representative experiment.
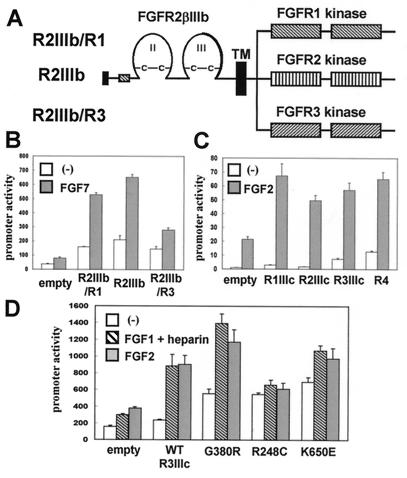
We then used the activity of the 48-bp Col2a1 enhancer to identify FGF receptors that mediated the increase in Sox9 expression produced by FGFs. To bypass the activation of endogenous FGF receptors, we used vectors expressing chimeric FGF receptors consisting of the extracellular and transmembrane domains of FGFR2βIIIb and the kinase domains of FGFR1, FGFR2, or FGFR3 (Fig. 3A). Transfections of these chimeric receptors into primary mouse chondrocytes allowed us to use FGF7 as ligand, since it requires FGFR2IIIb for binding (50). The constructs encoding the chimeric FGF receptors were cotransfected with the 4x48-p89 Col2a1 construct, and the cells treated with FGF7. Since primary chondrocytes and C3H10T1/2 cells show virtually no response to FGF7, activation of the enhancer should be because of signaling from the transfected chimeric FGF receptors. In both cell types, coexpression of chimeric receptors activated the Sox9-dependent enhancer, and this activity was further enhanced by FGF7 (Fig. 3B, and data not shown). Thus, the kinase domains of FGFR1, FGFR2, and FGFR3 were able to transduce signals that activate the Sox9-dependent enhancer.
Figure 3.
Activation of a Sox9-dependent Col2a1 enhancer by coexpression of FGF receptors and FGF treatment. (A) Schematic representation of chimeric FGF receptors encoding an ectodomain of FGFR2βIIIb and kinase domains of FGFR1, FGFR2, and FGFR3. FGFR2βIIIb is an alternatively spliced isoform of FGFR2 that lacks the first Ig-like domain and utilizes IIIb exon for the second half of the third Ig-like domain. (B) Activation in primary chondrocytes of a Sox9-dependent Col2a1 enhancer by coexpression of a chimeric FGF receptor and FGF7 treatment. The cells were transiently transfected with 4x48-p89, pSVβgal, and 10 ng of the FGFR constructs. FGF7 was added at 20 ng/ml. (C) Activation in C3H10T1/2 cells of a Sox9-dependent Col2a1 enhancer by coexpression of FGFR1IIIc, FGFR2IIIc, FGFR3IIIc, and FGFR4 and FGF2 treatment. The cells were transiently transfected with 4x48-p89, pSVβgal, and 30 ng of the FGFR constructs. FGF2 was added at 5 ng/ml. (D) Activation in primary chondrocytes of a Sox9-dependent Col2a1 enhancer by coexpression of wild-type FGFR3IIIc or its achondroplasia (G380R), thanatophoric dysplasia type I (R248C), or type II (K650E) mutants in the presence or absence of FGFs. The cells were transiently transfected with 4x48-p89, pSVβgal, and 100 ng of FGFR3 constructs. FGF1 was added at 60 ng/ml in the presence of 100 μg/ml heparin. FGF2 was added at 2 ng/ml. In all panels, promoter activities are average values ± standard deviations for three independently transfected cultures from one representative experiment.
We also tested whether the wild-type FGF receptors FGFR1IIIc, FGFR2IIIc, FGFR3IIIc, and FGFR4 could further increase the activation of the Col2a1 enhancer element by FGF2. The results in Fig. 3C, which presents an experiment performed in C3H10T1/2 cells, show that coexpression of each of the wild-type FGF receptors increased the activation of the Sox9-dependent enhancer by FGF2 alone. These data strongly suggest that all four FGF receptor tyrosine kinases can transduce signals that lead to the activation of the Sox9-dependent enhancer element and imply that a common pathway downstream of the four FGF receptors is utilized to enhance Sox9 expression.
Because FGFR3IIIc is expressed in resting and proliferating chondrocytes of growth plates in a pattern that overlaps with the expression of Sox9 (2, 27, 28) and because activating mutations of FGFR3 have been implicated in the pathogenesis of chondrodysplasias, we examined whether these FGFR3 mutants activated the Col2a1 enhancer more than the wild-type FGFR3. Coexpression of achondroplasia (G380R), thanatophoric dysplasia type I (R248C), and type II (K650E) mutants of FGFR3IIIc each resulted in an increased activity of the Sox9-dependent enhancer compared with the wild-type receptor in the absence of ligand. Treatment with FGF1 in combination with heparin or FGF2 further increased the activity of the enhancer in cells transfected with the wild-type FGFR3IIIc or its G380R or K650E mutants. These results are consistent with previous observations that achondroplasia and thanatophoric dysplasia mutations are partially activating mutations and that the G380R and K650E mutants, but not the R248C mutant, retain ligand dependency (47). These results also suggest the hypothesis that expression of Sox9 could be regulated by these mutant FGFR3 receptors in achondroplasia and thanatophoric dysplasias.
We then sought to identify the pathway responsible for the FGF regulation of Sox9 expression. Because the MAPK pathway has been shown to be activated by FGFs in various cell types including chondrocytes (56, 57), we first examined the involvement of this pathway using a specific mitogen-activated protein kinase kinase (MAPKK)/MEK inhibitor, U0126 (58, 59). This inhibitor exerts its effects at two discrete steps. U0126 inhibits both the activation of MEK1 and MEK2 by Raf-1 and the activation of MAPKs/extracellular signal-regulated kinase (ERK)1 and ERK2 by MEKs. Because serum is a potent activator of the MAPK cascade, serum concentration was reduced to 1% 24 h prior to the experiment. FGF2 increased Sox9 protein levels in parallel with the phosphorylation of ERK1 and ERK2 in primary chondrocytes (Fig. 4). The MEK inhibitor U0126 markedly inhibited both the effects of FGF2 on the levels of Sox9 protein and the phosphorylation of ERK1 and ERK2. These results support the hypothesis that the increase in Sox9 expression caused by FGF is mediated by the MEK-MAPK pathway. In a parallel experiment, we examined the time course of ERK1 and ERK2 phosphorylation after addition of FGF2. ERK1 and ERK2 were phosphorylated within 5 min after FGF2 treatment in primary chondrocytes, preceding the accumulation of Sox9 mRNA (data not shown). These data are consistent with the notion that FGFs may enhance Sox9 mRNA expression through activation of the MEK-MAPK pathway.
Figure 4.
Inhibition by the MEK inhibitor U0126 of FGF2-induced increase in Sox9 protein and phosphorylation of ERK1 and ERK2 in primary chondrocytes. Confluent primary chondrocytes were cultured in the presence of 1% FCS for 24 h and incubated with or without 20 μM U0126 for 15 min before addition of 2 ng/ml FGF2 (F) or control vehicle (C). Total cell lysates were prepared 3 h later and analyzed by Western blot using antibodies that recognize Sox9, ERK1 and ERK2, and their Tyr 204 phosphorylated forms (p-ERK1, p-ERK2). FGF2 increased Sox9 protein levels 7-fold in the absence of U0126. U0126 inhibited the increase 85%. The figure represents one of two experiments that produced very similar results.
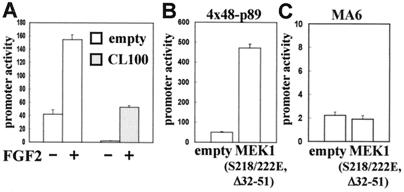
We also examined whether CL100/MAPK phosphatase-1 (MKP-1) can inhibit FGF2 activation of the Sox9-dependent enhancer in primary chondrocytes. CL100/MKP-1 is a dual-specificity protein phosphatase that efficiently dephosphorylates phosphotyrosines and phosphothreonines and inactivates ERK1 and ERK2 (60, 61). Coexpression of CL100/MKP-1 strongly inhibited the activation of the Sox9-dependent enhancer by FGF2 (Fig. 5A). This result further supports the hypothesis that FGF enhances Sox9 expression by means of the MAPK pathway. Interestingly, coexpression of CL100/MKP-1 remarkably reduced the basal activity of the Sox9-dependent enhancer, suggesting that the endogenous level of Sox9 is also regulated by MAPK in these cells.
Figure 5.
Expression of CL100 or constitutively active MEK1 mutant (S218/222E, Δ32–51) modulates the activity of a Sox9-dependent Col2a1 enhancer in primary chondrocytes. (A) Inhibition of FGF effects on the activity of a Sox9-dependent Col2a1 enhancer by CL100. Primary chondrocytes were transiently transfected with 4x48-p89, pSVβgal, and 100 ng of the CL100-expressing or empty pSG5 vector. Cells were incubated in the presence (+) or absence (−) of FGF2 at 2 ng/ml. (B) Primary chondrocytes were transiently transfected with 4x48-p89, pSVβgal, and 30 ng of pFC-MEK1 or its empty vector. (C) Primary chondrocytes were transiently transfected with the MA6 mutant of 4x48-p89, pSVβgal, and 30 ng of pFC-MEK1 or its empty vector. In all panels, promoter activities are average values ± standard deviations for three independently transfected cultures from one representative experiment.
Because FGFR3 activates signal transducer and activator of transcription (STAT)1, we also examined the involvement of STAT1 in the FGF regulation of the Sox9-dependent enhancer (57, 62). Either a dominant-negative mutant of STAT1 or STAT1β, a naturally occurring truncated isoform that inhibits the activity of full-length STAT1, was cotransfected with 4x48-p89 in primary chondrocytes (49). Coexpression of these dominant-negative STAT1 constructs did not inhibit the effects of FGF2 and did not affect the basal activity of the Sox9-dependent enhancer (data not shown). These results strongly suggest that FGF regulation of Sox9 is independent of STAT1.
We then tested whether activation of the MEK-MAPK pathway is sufficient to activate the Sox9-dependent enhancer in chondrocytes. We cotransfected a constitutively active mutant of MEK1 with the 4x48-p89 construct. This mutant MEK1 contains serine-to-glutamic acid substitutions of the two phosphoacceptors at amino acids 218 and 222 in combination with an internal deletion from amino acid 32 to amino acid 51. These mutations result in high constitutive activity of MEK1, and the mutant does not need to be activated by other protein kinases (63, 64). Coexpression of this mutant MEK1 strongly increased the activity of the Sox9-dependent enhancer both in primary chondrocytes and C3H10T1/2 cells, mimicking the effect of FGFs (Fig. 5B, and data not shown). Sox9 very likely mediates this activation, since the constitutively active MEK1 mutant failed to activate the 48-bp Col2a1 enhancer construct containing a mutation that abolishes SOX9 binding (Fig. 5C). Furthermore, the constitutively active MEK1 mutant did not activate the Sox9-dependent enhancer in osteoblastic osteosarcoma ROS17/2.8 cells that do not express Sox9 (data not shown). Altogether, these results strongly suggest that FGFs enhance Sox9 expression by activating the MEK-MAPK pathway.
Discussion
We have shown that FGFs markedly enhanced expression of Sox9 in primary chondrocytes. Sox9 mRNA increased within 30 min in these cells, and this increase was reflected in Sox9 protein levels within 90 min after addition of FGF2. The rapid response of Sox9 expression after FGF2 treatment strongly suggests that FGF signaling directly regulates Sox9 expression. FGF2 also increased Sox9 levels in ATDC5 prechondrocytic cells and in C3H10T1/2 cells, both of which express low levels of Sox9. However, in BALB/3T3 fibroblasts that do not express Sox9, FGF2 was unable to induce Sox9 expression (data not shown). Despite the increase in Sox9, we did not observe an increase in Col2a1 mRNA levels in primary chondrocytes and C3H10T1/2 cells (data not shown), maybe because Col2a1 is already maximally expressed in primary chondrocytes or because an increase in Col2a1 expression in C3H10T1/2 cells might need additional factors. Nevertheless, given that Sox9 is essential for chondrocyte differentiation and for expression of a number of genes in chondrocytes (3), we hypothesize that FGF signaling may have an important role in this process.
To explore the pathway by which FGFs increased Sox9 expression in chondrocytes, we used the activity of a 48-bp Col2a1 enhancer as a functional measurement of Sox9. All four FGF receptors, FGFR1–4, enhanced the activity of the Sox9-dependent enhancer, implying that various FGF ligands can participate in the regulation of Sox9, since multiple ligands can bind to each of the FGF receptors (50). FGFs could increase the levels of Sox9 at various steps in the chondrocyte differentiation pathway, given that FGF receptors are expressed throughout this pathway. Our findings that activation of FGFR3 increases the activity of the Sox9-dependent Col2a1 enhancer suggest the possibility that expression of Sox9 may be regulated by FGFR3 in growth plate chondrocytes. Because we found that achondroplasia and thanatophoric dysplasia mutants of FGFR3 activated the Sox9-dependent enhancer in chondrocytes, it is possible that Sox9 expression in cartilage is increased in these disorders.
Our study also strongly suggests that the increase in Sox9 expression was mediated by the MEK-MAPK pathway, one of the major pathways downstream of FGF receptors. Signals from FGF receptors lead to the activation of MEK, which in turn phosphorylates and activates MAPK/ERK. MAPK then phosphorylates and modulates the activity of a variety of kinases and transcription factors. A specific MEK inhibitor U0126, which abolished the phosphorylation of ERK1 and ERK2, almost completely inhibited the increase in Sox9 levels produced by FGF2. Furthermore, coexpression of a constitutively active MEK1 mutant strongly increased the activity of the Sox9-dependent Col2a1 enhancer both in primary chondrocytes and C3H10T1/2 cells, mimicking the effect of FGFs. In contrast, cotransfection of a dual-specificity phosphatase, CL100/MKP-1, that efficiently dephosphorylates and inactivates ERK1 and ERK2, inhibited the increase in activity of the Sox9-dependent enhancer in primary chondrocytes. Interestingly, the basal chondrocyte-specific activity of the enhancer was strongly inhibited by CL100/MKP-1 in primary chondrocytes, and a similar inhibition of the basal activity was also observed in the rat chondrosarcoma cell line RCS, which stably expresses the chondrocyte phenotype (data not shown). This suggests that endogenous Sox9 levels may be under the control of ERK1 and ERK2 in chondrocytic cells. The activity of the MAPK pathway has been implicated in mesoderm induction in Xenopus and in neuronal differentiation in the PC12 pheochromocytoma cells (65, 66). Because Sox9 is essential for chondrocyte differentiation and expression of chondrocyte-specific genes, we hypothesize that the activity of MAPK plays an important role in chondrocyte differentiation.
In conclusion, our results demonstrate that FGFs markedly enhance Sox9 expression both in chondrocytes and Sox9-expressing mesenchymal and prechondrocytic cells. Our evidence strongly suggests that this FGF-induced increase in Sox9 expression is mediated by the MEK-MAPK pathway. Because Sox9 is essential for chondrocyte differentiation and expression of chondrocyte-specific genes, we hypothesize that FGFs and the activity of the MAPK play an important role in chondrocyte differentiation.
Acknowledgments
We thank Kazuhisa Nakashima and Véronique Lefebvre for helpful discussions, Sandra McKinney for technical assistance, and Pat Aru-baleze for editorial assistance. We also thank James H. Kimura for RCS cells, Bjorn Olsen for MC615 cells, Tadao Atsumi for ATDC5 cells, William T. Butler for ROS 17/2.8 cells, Stephen M. Keyse for the pSG5 vector encoding CL100, Michael J. Walter and Michael J. Holtzman for the dominant-negative STAT1 constructs, Michael Naski and David Ornitz for the mutant FGFR3 constructs, and Genetics Institute for BMP2. This work was funded by National Institutes of Health Grants AR42919 (to B.d.C.) and DK40739 (to W.L.M.).
Abbreviations
- FGF
fibroblast growth factor
- MAPK
mitogen-activated protein kinase
- EGF
epidermal growth factor
- TGF
transforming growth factor
- ERK
extracellular signal-regulated kinase
- MKP
MAPK phosphatase
- STAT
signal transducer and activator of transcription
- MEK
mitogen-activated protein kinase/ERK kinase
- BMP
bone morphogenetic protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ng L J, Wheatley S, Muscat G E, Conway-Campbell J, Bowles J, Wright E, Bell D M, Tam P P, Cheah K S, Koopman P. Dev Biol. 1997;183:108–121. doi: 10.1006/dbio.1996.8487. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Q, Eberspaecher H, Lefebvre V, de Crombrugghe B. Dev Dyn. 1997;209:377–386. doi: 10.1002/(SICI)1097-0177(199708)209:4<377::AID-AJA5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 3.Bi W, Deng J M, Zhang Z, Behringer R R, de Crombrugghe B. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 4.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli F D, Keutel J, Hustert E, et al. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 5.Foster J W, Dominguez-Steglich M A, Guioli S, Kowk G, Weller P A, Stevanovic M, Weissenbach J, Mansour S, Young I D, et al. Nature (London) 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- 6.Südbeck P, Schmitz M L, Baeuerle P A, Scherer G. Nat Genet. 1996;13:230–232. doi: 10.1038/ng0696-230. [DOI] [PubMed] [Google Scholar]
- 7.Lefebvre V, Huang W, Harley V R, Goodfellow P N, de Crombrugghe B. Mol Cell Biol. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bridgewater L C, Lefebvre V, de Crombrugghe B. J Biol Chem. 1998;273:14998–15006. doi: 10.1074/jbc.273.24.14998. [DOI] [PubMed] [Google Scholar]
- 9.Bell D M, Leung K K, Wheatley S C, Ng L J, Zhou S, Ling K W, Sham M H, Koopman P, Tam P P, Cheah K S. Nat Genet. 1997;16:174–178. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- 10.Xie W F, Zhang X, Sakano S, Lefebvre V, Sandell L J. J Bone Miner Res. 1999;14:757–763. doi: 10.1359/jbmr.1999.14.5.757. [DOI] [PubMed] [Google Scholar]
- 11.Wright E, Hargrave M R, Christiansen J, Cooper L, Kun J, Evans T, Gangadharan U, Greenfield A, Koopman P. Nat Genet. 1995;9:15–20. doi: 10.1038/ng0195-15. [DOI] [PubMed] [Google Scholar]
- 12.Kent J, Wheatley S C, Andrews J E, Sinclair A H, Koopman P. Development (Cambridge, UK) 1996;122:2813–2822. doi: 10.1242/dev.122.9.2813. [DOI] [PubMed] [Google Scholar]
- 13.Savage M P, Fallon J F. Dev Dyn. 1995;202:343–353. doi: 10.1002/aja.1002020404. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki H R, Sakamoto H, Yoshida T, Sugimura T, Terada M, Solursh M. Dev Biol. 1992;150:219–222. doi: 10.1016/0012-1606(92)90020-h. [DOI] [PubMed] [Google Scholar]
- 15.Niswander L, Martin G R. Development (Cambridge, UK) 1992;114:755–768. doi: 10.1242/dev.114.3.755. [DOI] [PubMed] [Google Scholar]
- 16.Crossley P H, Martin G R. Development (Cambridge, UK) 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez A M, Buscaglia M, Ong M, Baird A. J Cell Biol. 1990;110:753–765. doi: 10.1083/jcb.110.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jingushi S, Scully S P, Joyce M E, Sugioka Y, Bolander M E. J Orthop Res. 1995;13:761–768. doi: 10.1002/jor.1100130516. [DOI] [PubMed] [Google Scholar]
- 19.Kato Y, Gospodarowicz D. J Cell Biol. 1985;100:477–485. doi: 10.1083/jcb.100.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato Y, Iwamoto M. J Biol Chem. 1990;265:5903–5909. [PubMed] [Google Scholar]
- 21.Jentzsch K D, Wellmitz G, Heder G, Petzold E, Buntrock P, Oehme P. Acta Biol Med Ger. 1980;39:967–971. [PubMed] [Google Scholar]
- 22.Cuevas P, Burgos J, Baird A. Biochem Biophys Res Commun. 1988;156:611–618. doi: 10.1016/s0006-291x(88)80887-8. [DOI] [PubMed] [Google Scholar]
- 23.Otsuka Y, Mizuta H, Takagi K, Iyama K, Yoshitake Y, Nishikawa K, Suzuki F, Hiraki Y. Dev Growth Differ. 1997;39:143–156. doi: 10.1046/j.1440-169x.1997.t01-1-00003.x. [DOI] [PubMed] [Google Scholar]
- 24.Peters K G, Werner S, Chen G, Williams L T. Development (Cambridge, UK) 1992;114:233–243. doi: 10.1242/dev.114.1.233. [DOI] [PubMed] [Google Scholar]
- 25.Peters K, Ornitz D, Werner S, Williams L. Dev Biol. 1993;155:423–430. doi: 10.1006/dbio.1993.1040. [DOI] [PubMed] [Google Scholar]
- 26.Stark K L, McMahon J A, McMahon A P. Development (Cambridge, UK) 1991;113:641–651. doi: 10.1242/dev.113.2.641. [DOI] [PubMed] [Google Scholar]
- 27.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 28.Naski M C, Colvin J S, Coffin J D, Ornitz D M. Development (Cambridge, UK) 1998;125:4977–4988. doi: 10.1242/dev.125.24.4977. [DOI] [PubMed] [Google Scholar]
- 29.Shiang R, Thompson L M, Zhu Y Z, Church D M, Fielder T J, Bocian M, Winokur S T, Wasmuth J J. Cell. 1994;78:335–342. doi: 10.1016/0092-8674(94)90302-6. [DOI] [PubMed] [Google Scholar]
- 30.Rousseau F, Bonaventure J, Legeai-Mallet L, Pelet A, Rozet J M, Maroteaux P, Le Merrer M, Munnich A. Nature (London) 1994;371:252–254. doi: 10.1038/371252a0. [DOI] [PubMed] [Google Scholar]
- 31.Tavormina P L, Shiang R, Thompson L M, Zhu Y Z, Wilkin D J, Lachman R S, Wilcox W R, Rimoin D L, Cohn D H, Wasmuth J J. Nat Genet. 1995;9:321–328. doi: 10.1038/ng0395-321. [DOI] [PubMed] [Google Scholar]
- 32.Rousseau F, Saugier P, Le Merrer M, Munnich A, Delezoide A L, Maroteaux P, Bonaventure J, Narcy F, Sanak M. Nat Genet. 1995;10:11–12. doi: 10.1038/ng0595-11. [DOI] [PubMed] [Google Scholar]
- 33.Bellus G A, McIntosh I, Smith E A, Aylsworth A S, Kaitila I, Horton W A, Greenhaw G A, Hecht J T, Francomano C A. Nat Genet. 1995;10:357–359. doi: 10.1038/ng0795-357. [DOI] [PubMed] [Google Scholar]
- 34.Li C, Chen L, Iwata T, Kitagawa M, Fu X Y, Deng C X. Hum Mol Genet. 1999;8:35–44. doi: 10.1093/hmg/8.1.35. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Spatz M K, Kannan K, Hayk H, Avivi A, Gorivodsky M, Pines M, Yayon A, Lonai P, Givol D. Proc Natl Acad Sci USA. 1999;96:4455–4460. doi: 10.1073/pnas.96.8.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo Y, Gabriel J L, Wang F, Zhan X, Maciag T, Kan M, McKeehan W L. J Biol Chem. 1996;271:26876–26883. doi: 10.1074/jbc.271.43.26876. [DOI] [PubMed] [Google Scholar]
- 37.Jang J H, Wang F, Kan M. In Vitro Cell Dev Biol Anim. 1997;33:819–824. doi: 10.1007/s11626-997-0162-7. [DOI] [PubMed] [Google Scholar]
- 38.Lefebvre V, Garofalo S, Zhou G, Metsaranta M, Vuorio E, de Crombrugghe B. Matrix Biol. 1994;14:329–335. doi: 10.1016/0945-053x(94)90199-6. [DOI] [PubMed] [Google Scholar]
- 39.Atsumi T, Miwa Y, Kimata K, Ikawa Y. Cell Differ Dev. 1990;30:109–116. doi: 10.1016/0922-3371(90)90079-c. [DOI] [PubMed] [Google Scholar]
- 40.Mukhopadhyay K, Lefebvre V, Zhou G, Garofalo S, Kimura J H, de Crombrugghe B. J Biol Chem. 1995;270:27711–27719. doi: 10.1074/jbc.270.46.27711. [DOI] [PubMed] [Google Scholar]
- 41.Lefebvre V, Zhou G, Mukhopadhyay K, Smith C N, Zhang Z, Eberspaecher H, Zhou X, Sinha S, Maity S N, de Crombrugghe B. Mol Cell Biol. 1996;16:4512–4523. doi: 10.1128/mcb.16.8.4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou G, Lefebvre V, Zhang Z, Eberspaecher H, de Crombrugghe B. J Biol Chem. 1998;273:14989–14997. doi: 10.1074/jbc.273.24.14989. [DOI] [PubMed] [Google Scholar]
- 43.Feng S, Wang F, Matsubara A, Kan M, McKeehan W L. Cancer Res. 1997;57:5369–5378. [PubMed] [Google Scholar]
- 44.Wang F, Kan M, McKeehan K, Jang J H, Feng S, McKeehan W L. J Biol Chem. 1997;272:23887–23895. doi: 10.1074/jbc.272.38.23887. [DOI] [PubMed] [Google Scholar]
- 45.Kan M, Wu X, Wang F, McKeehan W L. J Biol Chem. 1999;274:15947–15952. doi: 10.1074/jbc.274.22.15947. [DOI] [PubMed] [Google Scholar]
- 46.Matsubara A, Kan M, Feng S, McKeehan W L. Cancer Res. 1998;58:1509–1514. [PubMed] [Google Scholar]
- 47.Naski M C, Wang Q, Xu J, Ornitz D M. Nat Genet. 1996;13:233–237. doi: 10.1038/ng0696-233. [DOI] [PubMed] [Google Scholar]
- 48.Lewis T, Groom L A, Sneddon A A, Smythe C, Keyse S M. J Cell Sci. 1995;108:2885–2896. doi: 10.1242/jcs.108.8.2885. [DOI] [PubMed] [Google Scholar]
- 49.Walter M J, Look D C, Tidwell R M, Roswit W T, Holtzman M J. J Biol Chem. 1997;272:28582–28589. doi: 10.1074/jbc.272.45.28582. [DOI] [PubMed] [Google Scholar]
- 50.Ornitz D M, Xu J, Colvin J S, McEwen D G, MacArthur C A, Coulier F, Gao G, Goldfarb M. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 51.Gospodarowicz D, Cheng J. J Cell Physiol. 1986;128:475–484. doi: 10.1002/jcp.1041280317. [DOI] [PubMed] [Google Scholar]
- 52.Uhlrich S, Lagente O, Lenfant M, Courtois Y. Biochem Biophys Res Commun. 1986;137:1205–1213. doi: 10.1016/0006-291x(86)90353-0. [DOI] [PubMed] [Google Scholar]
- 53.Miki T, Fleming T P, Bottaro D P, Rubin J S, Ron D, Aaronson S A. Science. 1991;251:72–75. doi: 10.1126/science.1846048. [DOI] [PubMed] [Google Scholar]
- 54.Healy C, Uwanogho D, Sharpe P T. Dev Dyn. 1999;215:69–78. doi: 10.1002/(SICI)1097-0177(199905)215:1<69::AID-DVDY8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 55.Zehentner B K, Dony C, Burtscher H. J Bone Miner Res. 1999;14:1734–1741. doi: 10.1359/jbmr.1999.14.10.1734. [DOI] [PubMed] [Google Scholar]
- 56.Legeai-Mallet L, Benoist-Lasselin C, Delezoide A L, Munnich A, Bonaventure J. J Biol Chem. 1998;273:13007–13014. doi: 10.1074/jbc.273.21.13007. [DOI] [PubMed] [Google Scholar]
- 57.Sahni M, Ambrosetti D C, Mansukhani A, Gertner R, Levy D, Basilico C. Genes Dev. 1999;13:1361–1366. doi: 10.1101/gad.13.11.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeSilva D R, Jones E A, Favata M F, Jaffee B D, Magolda R L, Trzaskos J M, Scherle P A. J Immunol. 1998;160:4175–4181. [PubMed] [Google Scholar]
- 59.Favata M F, Horiuchi K Y, Manos E J, Daulerio A J, Stradley D A, Feeser W S, Van Dyk D E, Pitts W J, Earl R A, Hobbs F, et al. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 60.Alessi D R, Smythe C, Keyse S M. Oncogene. 1993;8:2015–2020. [PubMed] [Google Scholar]
- 61.Sun H, Charles C H, Lau L F, Tonks N K. Cell. 1993;75:487–493. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- 62.Su W C, Kitagawa M, Xue N, Xie B, Garofalo S, Cho J, Deng C, Horton W A, Fu X Y. Nature (London) 1997;386:288–292. doi: 10.1038/386288a0. [DOI] [PubMed] [Google Scholar]
- 63.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukasawa K, Vande Woude G F, Ahn N G. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 64.Coso O A, Chiariello M, Yu J C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 65.Umbhauer M, Marshall C J, Mason C S, Old R W, Smith J C. Nature (London) 1995;376:58–62. doi: 10.1038/376058a0. [DOI] [PubMed] [Google Scholar]
- 66.Fukuda M, Gotoh Y, Tachibana T, Dell K, Hattori S, Yoneda Y, Nishida E. Oncogene. 1995;11:239–244. [PubMed] [Google Scholar]