Abstract
Streptococci and actinomyces that initiate colonization of the tooth surface frequently coaggregate with each other as well as with other oral bacteria. These observations have led to the hypothesis that interbacterial adhesion influences spatiotemporal development of plaque. To assess the role of such interactions in oral biofilm formation in vivo, antibodies directed against bacterial surface components that mediate coaggregation interactions were used as direct immunofluorescent probes in conjunction with laser confocal microscopy to determine the distribution and spatial arrangement of bacteria within intact human plaque formed on retrievable enamel chips. In intrageneric coaggregation, streptococci such as Streptococcus gordonii DL1 recognize receptor polysaccharides (RPS) borne on other streptococci such as Streptococcus oralis 34. To define potentially interactive subsets of streptococci in the developing plaque, an antibody against RPS (anti-RPS) was used together with an antibody against S. gordonii DL1 (anti-DL1). These antibodies reacted primarily with single cells in 4-h-old plaque and with mixed-species microcolonies in 8-h-old plaque. Anti-RPS-reactive bacteria frequently formed microcolonies with anti-DL1-reactive bacteria and with other bacteria distinguished by general nucleic acid stains. In intergeneric coaggregation between streptococci and actinomyces, type 2 fimbriae of actinomyces recognize RPS on the streptococci. Cells reactive with antibody against type 2 fimbriae of Actinomyces naeslundii T14V (anti-type-2) were much less frequent than either subset of streptococci. However, bacteria reactive with anti-type-2 were seen in intimate association with anti-RPS-reactive cells. These results are the first direct demonstration of coaggregation-mediated interactions during initial plaque accumulation in vivo. Further, these results demonstrate the spatiotemporal development and prevalence of mixed-species communities in early dental plaque.
The human oral cavity harbors a complex microbial ecosystem characterized by spatiotemporal variability in species composition. Despite this variability, consensus exists that supra- and subgingival dental plaques develop according to reproducible patterns. Analyses of species composition in supragingival dental plaque have shown that the majority (47 to 90%) of cultivable bacteria are Streptococcus sanguinis (formerly S. sanguis [31]), Streptococcus oralis, and Streptococcus mitis (biovar 1) (24) and that one-third of the remaining bacteria are Actinomyces naeslundii (25). Scanning electron microscopy has shown isolated cells and clusters of cells after 4 h of accumulation, larger “microcolonies” after 8 h, and confluent monolayers after 12 h (22). Transmission electron microscopy has shown that some colonies consisted of gram-negative cells together with gram-positive cells; thus, multispecies colonies were unambiguously identifiable by 24 h (23). These and similar studies together form the basis for present understanding of community evolution in early supragingival dental plaque, and recent analyses of species composition that use molecular approaches (1, 17) support these basic concepts. However, none of these studies have provided information on the spatial organization of bacteria within the plaque; methods have been developed only recently to examine the architecture of natural biofilms (12, 19, 20, 28, 32).
The reproducible sequential appearance of bacterial species during plaque accumulation (16) and also in the development of other biofilm communities (2, 27, 29, 30) has been postulated to depend on interbacterial adhesion. Coaggregation between a number of oral bacteria was first reported by Gibbons and Nygaard (9) and was subsequently widely investigated in the two genera dominant in early plaque development, Streptococcus and Actinomyces. The receptors in many of these lectin-like interactions (mediated by a protein adhesin that recognizes a complementary receptor carbohydrate) are streptococcal cell wall polysaccharides composed of hexa- or heptasaccharide repeating units. These streptococcal receptor polysaccharides (RPS) fall into two types defined by the host-like disaccharide motif that confers receptor specificity (6): Gn RPS has GalNAcβ1→3Gal as the recognition motif, whereas G RPS has Galβ1→3GalNAc as the recognition motif. Within the Gn type, four structures that differ in the remaining saccharide moieties have been identified and named 1Gn, 2Gn, 4Gn, and 5Gn; in the G type, two different structures, named 2G and 3G, are known (7). These different structures are not involved in coaggregation specificity (which is controlled by the disaccharide motif), but the differences do influence antibody reactivity. Coaggregations between streptococci involve recognition of Gn RPS on strains such as S. oralis 34 by protein adhesins on strains such as Streptococcus gordonii DL1 (10). In contrast, coaggregations between streptococci and actinomyces occur through recognition of either Gn or G RPS by type 2 fimbriae of actinomyces (6). A noteworthy biological outcome of interactions set up by coaggregation is exemplified by S. oralis 34 and A. naeslundii T14V. Neither strain reproducibly forms a monoculture biofilm in vitro with saliva as the sole carbon and nitrogen source. Yet, when allowed to interact under the same conditions, they establish a luxuriant interdigitated biofilm (26).
Despite extensive description of coaggregation characteristics compiled by using oral bacterial isolates in vitro, it has been difficult to investigate the occurrence, and thus the significance, of coaggregation in vivo. Interaction between S. oralis and A. naeslundii would be an attractive target for investigation in vivo. However, unambiguous definition of coaggregation-mediated interactions in vivo is not simple for two reasons. First, heterogeneity of coaggregation traits with respect to taxonomy makes firm interpretation of data from plaque difficult. Interactions occur not only between streptococcal species (intrageneric interactions [15]) but also between strains of a single streptococcal species (10), and each streptococcal species is heterogeneous in its coaggregation traits. Thus, simply identifying an organism on the tooth surface as, e.g., S. oralis, does not define the coaggregation interactions in which that organism participates. Rather, identification of these interactions must be based on identification of the coaggregation-mediating components (e.g., RPS and type 2 fimbriae) on each cell, an approach best undertaken through the specificity afforded by antibodies characterized for their reactivity with numerous oral isolates. Intimate association of a cell reactive with antibody against RPS and a cell reactive with antibody against type 2 fimbriae within plaque would be strong evidence for cell-cell recognition in situ. Second, an approach to sampling plaque that maintains spatial relationships must be combined with an approach to detecting spatial relationships that operates at single-cell resolution. A previous study that used appropriate antibodies on plaque scrapings yielded promising results (3). However, bacterial strain specificity of the antibodies was not well characterized, spatial organization of the sample was unlikely to have been reflective of that in situ, and the resolution of the microscopic data was not high. The present study surmounts these difficulties and provides unambiguous evidence for oral bacterial cell-cell recognition in dental plaque in situ.
MATERIALS AND METHODS
Bacterial strains.
Human oral isolates of streptococci (Table 1) have been previously described (10, 13). Most were isolated during the studies of plaque topography and species composition cited in the introduction (21, 25); others were obtained from commercial strain collections (e.g., American Type Culture Collection). Actinomyces spp. strains (Table 2) whose names begin with N were isolated by L. V. H. Moore and W. E. C. Moore, and all strains in Table 2 have been characterized according to the Moores' serotype scheme (11).
TABLE 1.
Indirect immunofluorescence reactions of streptococcal strains with anti-DL1 and anti-RPS
| Species (biovar)a | Strainb | Reactionc with:
|
|
|---|---|---|---|
| Anti- RPS | Anti- DL1 | ||
| S. oralis | SK92, SK111, 9811, 34 | + | − |
| ATCC 10557, SK23, SK143, SK144, C104, 6249 | − | − | |
| S. mitis (1) | SK137 | + | + |
| J22, 15914 | + | − | |
| SK141, SK142, H1d | − | − | |
| S. mitis (2) | SK96, SK148, K103, 903, H127 | − | + |
| I18 | − | − | |
| S. gordonii (1) | 38 | + | + |
| SK6, SK120, SK121 | − | + | |
| S. gordonii (2) | SK184, DL1,e D105, K1, K4, M5, ATCC 10558, G102 | − | + |
| S. gordonii (3) | SK9, SK12 | + | + |
| SK33, SK186 | − | + | |
| S. sanguinis (1) | SK77 | + | + |
| SK75, SK72 | + | − | |
| SK1, SK36, SK37, ATCC 10556, SK76, SK85, SK108 | − | + | |
| S. sanguinis (2) | SK4, SK156, SK157, SK158, SK164 | − | + |
| S. sanguinis (3) | SK163 | + | − |
| SK150, SK159, SK160, SK162 | − | + | |
| S. sanguinis (4) | SK45, SK112 | + | − |
| S. mutans | ATCC33534, NCTC 10449 | − | − |
| S. salivarius | ATCC 9222 | − | + |
| ATCC 25975 | − | − | |
| S. anginosus | SK14, SK47, SK52, SK63, SK66, SK81, SK215, SK216, SK218, SK221 | − | − |
TABLE 2.
Indirect immunofluorescence reactions of actinomyces strains with anti-type-1 and anti-type-2e
| Taxonomic groupa | Strain(s) | Reactionb with:
|
|
|---|---|---|---|
| Anti- type-2 | Anti- type-1 | ||
| A. naeslundii geno- species 1, sero- type I | N28B-15, N34A-24, N35B-3, N37B-2, ATCC 12104 | − | − |
| A. naeslundii geno- species 2 | |||
| Serotype II | N11A16, N12A-2B, ATCC 49339, N11A12, T14V, ATCC 27044 | + | + |
| Serotype III | N33A2B, N37B-13 | + | + |
| N34A-23 | − | + | |
| ATCC 49340 | − | − | |
| Serotype NVc | N28B-1, N29A27 | + | + |
| N32A-8, N37B-9 | − | − | |
| Nonserotypeabled | N38B-10 | + | + |
| N33A3 | + | − | |
| N34A-14 | − | + | |
| Actinomyces sero- | N7B-40, N16B-5, PK 1259, | − | + |
| type WVA 963 | ATCC 49338 | − | − |
| A. odontolyticus | N39A7, N34A-1, N34A19 | − | − |
According to reference 11.
Ten micrograms of primary antibody per ml; +, fluorescence equivalent to antigen strain; −, no fluorescence or extremely dim
Reactive with more than one serotype antibody.
Reactive with no serotype antibody.
All strains coaggregate with S. oralis 34 except those of A. odontolyticus.
Antibodies.
Four rabbit antibodies were used. Antibody against S. gordonii strain DL1 (Challis), prepared by multiple injections of whole bacteria, was absorbed with whole cells of S. oralis 34; this antibody is designated “anti-DL1.” Antibody against 1Gn RPS of S. oralis 34 was purified by elution from an affinity column prepared with the purified polysaccharide. Eight milligrams of purified RPS (7) was subjected to mild periodate oxidation, desalted, and incubated overnight at room temperature with 4 ml of Affi-Gel Hz (Bio-Rad) following recommendations of the manufacturer. The gel was then washed to remove uncoupled material. Six milliliters of diluted antiserum R26 against whole cells of S. oralis 34 (18) was slowly passed through the RPS-derivatized affinity gel column (1.5 ml) in the cold. After extensive washing with phosphate-buffered saline (PBS) to remove unbound material, the bound antibody was eluted with 4 M MgCl2 and dialyzed against PBS. This purified antibody against S. oralis 34 is designated “anti-RPS.” Antibodies R59 against purified type 1 fimbriae and R55 against purified type 2 fimbriae of A. naeslundii T14V have been described previously (4). These antibodies are designated, respectively, “anti-type-1” and “anti-type-2.” Samples of all four immune immunoglobulins G were fluorescently labeled for direct immunofluorescence by using AlexaFluor labeling kits (Molecular Probes, Eugene, Oreg.) following the manufacturer's directions.
Strain specificity of antibodies.
Strains were grown overnight anaerobically in brain heart infusion (Difco), washed in PBS containing 1% bovine serum albumin (PBS-BSA), resuspended in PBS-BSA containing primary antibody (10 μg/ml) for 15 min at room temperature, washed twice in PBS-BSA, resuspended in PBS-BSA containing labeled secondary antibody (Jackson Laboratory) for 15 min, washed twice in PBS-BSA, and then examined using epifluorescence microscopy. Cultures demonstrating fluorescence similar in intensity to the positive controls (A. naeslundii T14V, S. gordonii DL1, and S. oralis 34) were scored positive (+). Cultures with very dim or no fluorescence were scored negative (−).
Enamel chip model.
Details on fabrication of chips and their use in healthy human volunteers have been published previously (28). Briefly, enamel pieces (2 by 2 by 1 mm [length by width by thickness]) were cut from extracted, unerupted human third molars. Chips were cleaned in an ultrasonic bath (1510; Branson; Danbury, Conn.) for 20 min, sterilized with ethylene oxide, and affixed in custom-fabricated acrylic stents using red dental wax. Two bilateral mandibular stents (spanning the posterior buccal surface from the first premolar to first molar), each of which contained three chips, were worn by each volunteer. In certain experiments, visible plaque was first removed and teeth were polished prior to stent insertion (prophylaxis). In certain experiments, a series of 30-s sucrose rinses (20 ml of filter-sterilized 10% sucrose) took place at 90-min intervals beginning immediately after stent insertion. One stent was worn for 4 h and the other for 8 h. No intake of food or liquids (other than water) was allowed except during a lunch period when stents were removed and stored in a humid denture cup at 37°C. Data were gathered from two male volunteers free of periodontal disease who had participated in the entire four-way matrix of protocols (no prophylaxis and no sucrose rinsing; no prophylaxis but sucrose rinsing; prophylaxis but no sucrose rinsing; and prophylaxis and sucrose rinsing).
Staining.
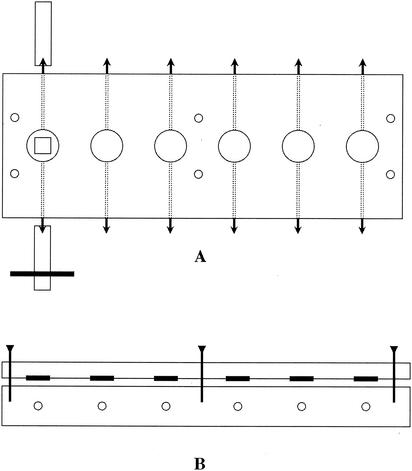
Staining for microscopy began immediately after removal of a stent. The three chips were removed from the stent and were placed in individual wells in a custom-fabricated staining chamber (Fig. 1). The chamber has six 50-μl wells; conduits permit washing and staining procedures to be carried out with the chips completely immersed at all times (i.e., without disturbance by air-liquid interfaces). Each well and conduit are filled with PBS-BSA prior to chip insertion, and then the chamber is sealed with a removable transparent top. Liquids are passed through the wells by injection through the clamped tubing. The total volume of the conduit and well was 150 μl; all injections were 300 μl (i.e., twice the volume of the system). The chips were first rinsed by injection of PBS-BSA, and then each chip was stained by injection of a different mixture of labeled primary antibodies (each antibody at a final concentration of 10 μg/ml) in PBS-BSA that usually included a nucleic acid stain (1 μg of either acridine orange or Syto 59 per ml; Molecular Probes). The three staining mixtures were acridine orange plus anti-RPS plus anti-type-1, anti-DL1 plus anti-RPS plus Syto 59, and anti-type-2 plus anti-RPS plus anti-type-1. After 20 min of reaction time, the stains were washed out by injection of PBS.
FIG. 1.
Diagram of staining chamber. (A) Top view. The six wells are indicated by large circles with the leftmost well containing an enamel chip (square). Conduits drilled through the plastic into each well are indicated by dotted lines. Barbed connectors (black arrowheads) are threaded into each end of the conduits, and tubing is attached; the leftmost conduit shows tubing on each connector, one of which is clamped (black bar). Six screw holes (small circles) provide connections with top plate (not shown). (B) Side view. Three (of six) screws are shown passing through the transparent plastic upper plate. Six O rings on the top plate (black bars) form seals around the periphery of each well; six conduits in the lower plate are indicated by small circles.
Microscopy.
The staining-chamber top was removed, the chamber was placed on the microscope stage, and the chip surfaces were examined with a 63×/0.9 numerical aperture water-immersible lens on a Leica TCS/4D confocal microscope (Leica Microsystems, Exton, Pa.). The microscope was set up by examining the field for bright objects (anti-RPS, anti-DL1, or nucleic acid) and photomultiplier tube (PMT) settings (gain and black level) were adjusted to provide full-range pixel values with the GlowOverUnder LookUpTable. Cells reactive with anti-type-1 were not visible with the oculars; Alexa 647-conjugated anti-type-1 has far-red fluorescence not easily detectable by eye. Alexa 488-conjugated anti-type-2-reactive cells were usually too dim to see over the natural enamel autofluorescence. To visualize cells stained with these antibodies, the laser power and PMT settings were adjusted to high values until examples of positive staining were found, after which the PMT settings and laser power were reduced to the minimum required to image these cells; this laser level and these PMT settings were then used for subsequent samples. All images presented were collected simultaneously into three channels. Channel one collected green fluorescence from one of the following stains: acridine orange, Alexa 488-conjugated anti-type-2, or Alexa 488-conjugated anti-DL1. Channel two was used exclusively to detect Alexa 532-conjugated anti-RPS (red). Channel three (blue) collected fluorescence from either Syto 59 or Alexa 647-conjugated anti-type-1. Images were collected at ×1 magnification (low magnification) and at ×2.5 to ×3 electronic zoom. Image stacks were generally acquired with axial spacing of 0.5 or 0.75 μm. Because the enamel surface was never exactly horizontal with respect to the microscope stage, the number of optical sections is not directly translatable to biofilm thickness. All images presented are maximum projections of the entire confocal image stack. Adobe Photoshop (Adobe Systems Inc., San Jose, Calif.) was used to adjust output levels within the individual channels of the 24-bit RGB color overlay images; no other manipulation of the images presented was performed except as required for image analysis (see below).
Image analysis.
RGB color maximum projections of the image stacks were manually processed in Photoshop to remove debris and enamel fluorescence and were then converted to grayscale. The grayscale images were manually thresholded by using IMAQ ImageBuilder (National Instruments, Austin, Tex.), and particle analysis results were filtered to remove particles that were ≤ 0.77 μm2 in area (area slightly less than that of a 1-μm-diameter coccus).
RESULTS
Antibody reactivity.
Seventy-four oral streptococcal isolates representative of the taxonomic groups present in early plaque and characterized with respect to coaggregation properties (10) and RPS type (7) were tested for reactivity with anti-RPS and anti-DL1. Of the 22 strains known to possess RPS (strain names in boldface in Table 1), 14 reacted with anti-RPS; these strains bear 1Gn, 2Gn, or 2G polysaccharides. The antibody did not label the 4Gn, 5Gn, or 3G polysaccharides borne on the eight remaining RPS-bearing strains. Only 2 of 52 strains that lack RPS (S. gordonii strains SK9 and SK12) gave a positive immunofluorescence reaction and thus represent the sole examples of aberrant anti-RPS reactivity in the 74 streptococcal strains. Anti-DL1 reacted with 40 strains, including all 16 S. gordonii strains and 18 of the 38 S. sanguinis, S. mitis biovar 1, and S. oralis strains thought to be most important in primary colonization (24). Only five strains reacted with both antibodies. Thus, each antibody defines a subset of streptococci: the anti-RPS reactive subset, which contains 63% of the streptococci known to bear RPS, and the anti-DL1-reactive subset, which contains all S. gordonii strains and 77% of S. sanguinis strains.
Actinomyces taxonomy is based to a large degree on serological reactions (11), and fimbriae are an important factor in bacterial antigenicity. It was therefore necessary to characterize anti-type-1 and anti-type-2 against a collection of Actinomyces strains (Table 2) broadly representative of diversity within this genus. Thirteen of 17 A. naeslundii genospecies 2 strains reacted with anti-type-1, and 12 reacted with anti-type-2. None of the A. naeslundii genospecies 1 strains reacted with either antibody, nor did any of the A. odontolyticus strains. None of the Actinomyces serotype WVA 963 strains reacted with anti-type-2, but three of four reacted with anti-type-1. The A. naeslundii genospecies 1 strains and the serotype WVA 963 strains bear type 2 fimbriae that either do not react or react poorly with anti-type-2 produced against strain T14V (5, 14). Only 5 of the 17 genospecies 2 strains that might be expected to react with anti-type-2 raised against T14V fimbriae did not react, and three of those strains are of unclear serology (NV and nonserotypeable strains). Anti-type-2 therefore labels most A. naeslundii genospecies 2 strains that coaggregate with streptococci, but it does not label any strains of genospecies 1 or serotype WVA 963, which have antigenically different type 2 fimbriae. Likewise, anti-type-1 is useful in identifying this fimbrial type on most genospecies 2 A. naeslundii strains. Type 1 fimbriae are known to be important in binding to the salivary pellicle (8).
General features of plaque accumulation.
Each stent contained three chips and thus yielded three replicate plaque samples that were each probed with a different stain combination. These combinations were acridine orange plus anti-RPS plus anti-type-1 (staining all cells, RPS-bearing streptococci, and type-1-fimbria-bearing cells), anti-DL1 plus anti-RPS plus Syto 59 (differentiating between two subsets of streptococci while also staining all cells), and anti-type-2 plus anti-RPS plus anti-type-1 (staining cells bearing either fimbrial type while also revealing RPS-bearing cells). No obvious differences attributable to sucrose rinsing or to prophylaxis were seen between samples from the four-way matrix of treatments. Rather, differences were noted between 4- and 8-h time points.
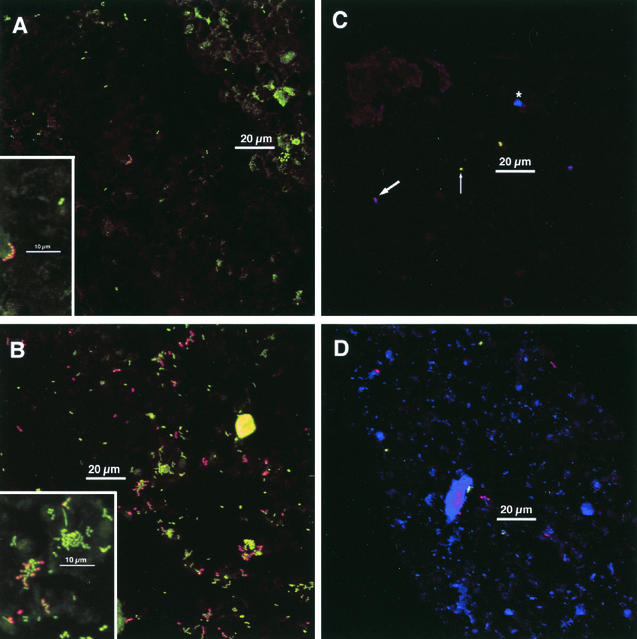
Colonization after 4 h of stent wear was typically rather sparse and was characterized by single cells and small clusters of cells (Fig. 2A and C). However, some fields showed more dense colonization (Fig. 2B and D) with many small clusters of cells (Fig. 2B). Such variation could sometimes be seen on a single chip (Fig. 2C and D). Debris and enamel autofluorescence made unambiguous identification of cells challenging at low magnification. Ideally, it should be demonstrated that the objects identified by fluorescence in these in situ biofilms are in fact bacterial cells and not enamel autofluorescence, debris, or nonspecifically reactive material. Use of a nucleic acid stain together with antibodies allows assurance that the objects stained with antibodies are cellular. Many cells were stained with acridine orange (green) and with anti-RPS (red); the centers of the cells were yellow (green plus red), whereas the edges (where fluorescence of antibody was more prevalent than that of acridine orange) were orange to red. A chain of such cells is seen in Fig. 2A (inset), whereas predominantly single cells exhibiting this staining pattern are seen in Fig. 2B. Syto 59 performed differently as a whole-cell nucleic acid stain from how acridine orange performed. While anti-RPS-reactive cells displayed colocalization of Syto 59 (blue) with anti-RPS (red) and were thus purple, anti-DL1-reactive cells did not and were thus green rather than blue-green (Fig. 2C). This observation may relate to membrane integrity or membrane potential. Syto 59 is used as a viability indicator; cells with an intact membrane and high membrane potential presumably stain more intensely than do those with a damaged membrane or with low membrane potential. Once removed from the mouth, cells on the chips could undergo a reduction in membrane potential. Alternatively, cell walls of anti-DL1-reactive cells may be intrinsically less permeable to Syto 59 than are those of anti-RPS-reactive cells. In summary, 4-h-old plaque is typified by sparse colonization, but a high degree of variability can be seen across even a single chip. Unambiguous interactions between cell types are rare, but fields with heavier colonization already show a trend towards larger cellular aggregates composed of differently stained cell types (Fig. 2B).
FIG. 2.
Typical colonization after 4 h of appliance wear. Insets: electronic zoom of center region. (A and B) Staining with acridine orange (green), anti-RPS (red), and anti-type-1 (blue). (A) Sparse colonization. Chain of anti-RPS reactive cells in center; cluster of acridine orange-stained cells at lower right of scale bar. Most cells are not antibody reactive. No anti-type-1-reactive cells are visible. Marked enamel autofluorescence in upper right. Inset shows homogeneity of anti-RPS reactivity within the chain of cells. Anti-RPS reactive cells are characterized by a yellow center (green plus red) and orange-to-red edges. A pair of antibody-unreactive cells (only acridine orange staining) is visible. Other dim greenish material is debris or enamel autofluorescence. (B) Heavier colonization. Most cells are in clusters. Many anti-RPS-reactive cells. Inset shows anti-RPS-reactive cells together with acridine orange-stained cells within a mixed microcolony. (C and D) Staining with anti-DL1 (green), anti-RPS (red), and Syto 59 (blue). These images show different fields of view from a single chip. (C) Very sparse colonization. Antibody-RPS-reactive cells are purple (red plus blue; thick arrow), whereas anti-DL1-reactive cells are green (limited uptake of Syto 59; thin arrow; see text for details). A cluster of three antibody-unreactive cells (Syto 59-stained; blue; asterisk) is between scale bar and upper edge. (D) Heavier colonization. Several anti-RPS reactive cells (purple) and a few anti-DL1 reactive cells (green) are visible, but most cells are not antibody reactive (blue). Debris or enamel fluorescence (large blue regions) is visible. Maximum projection images of simultaneous-acquisition three-channel confocal stacks. Low-magnification images are shown; dimensions are 158 μm on a side and 25,000-μm2 total area (approximately 1/10 of the total chip area).
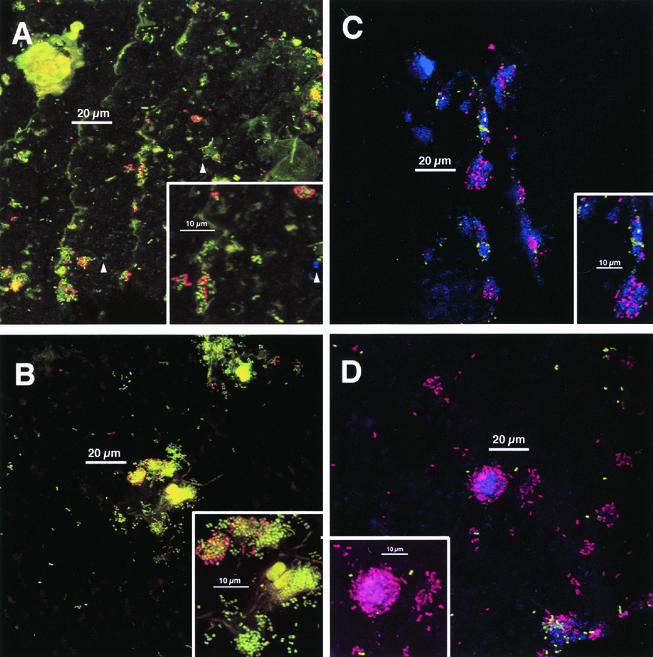
Colonization was heavier and was more consistent between chips and across single chips in 8-h-old plaque than in 4-h-old plaque, and a distinct colonial nature was seen (Fig. 3). Some of the increase in biomass likely occurred through additional colonization resulting in the appearance of new solitary cells as well as the formation of multicellular communities. Thus, although solitary cells were observed, the majority of biomass was found in aggregates, not as single cells as in the 4-h-old plaque. This general description of plaque accumulation is identical to that previously reported by several investigators (e.g., references 21 and 33); however, the present data show that the cell aggregates are typically heterogeneously stained and therefore may consist of more than one cell type. Many of the aggregates were similar in shape to colonies on agar surfaces, and some were up to 10 cell layers thick (e.g., Fig. 3D, inset). At least two staining types were usually seen within the aggregates: antibody-reactive cells were found in direct association with antibody-unreactive (solely acridine orange- or Syto 59-stained) cells. Observation of single colonies containing three staining types (anti-DL1, anti-RPS, and antibody unreactive) was frequent (Fig. 3C and D). Cellular morphology became more diverse (Fig. 3B, inset). These results suggest that the mixed-species microcolonies seen at 8 h were formed by a combination of adherence of planktonic cells to already attached cells (for example, those observed at 4 h) and growth of the cells in the microcolony.
FIG. 3.
Typical colonization after 8 h of appliance wear. Staining as done for Fig. 2. Panels A, B, and D are 8-h plaque samples that correspond directly (i.e., are from the same volunteer in the same experiment) to the 4-h samples shown in Fig. 2. Insets: electronic zoom of center region. (A) Colonial nature of plaque accumulation is apparent. Two anti-type-1-reactive cells are visible (arrowheads). Inset shows heterogeneity of anti-RPS reactivity within colonies and a single anti-type-1 reactive cell (arrowhead). (B) Long, weakly fluorescent rods and large, highly fluorescent cocci (center) are visible. No anti-type-1-reactive cells are visible. Inset confirms heterogeneity of anti-RPS reactivity within colonies. (C) Colonial nature of accumulation and heterogeneity of anti-RPS-reactive cells (purple) as well as of anti-DL1-reactive cells (green) within single colonies is apparent. Inset confirms heterogeneity of antibody reactivity within colonies; three groups of cells occur (anti-DL1 reactive, anti-RPS reactive, and antibody unreactive). (D) Colony composed of anti-RPS-reactive cells, anti-DL1-reactive cells, and antibody-unreactive cells is seen in lower right. Inset shows mixed colony of anti-RPS-reactive cells and Syto 59-stained cells; anti-DL1-reactive cells (green) are in close proximity to anti-RPS-reactive cells as well as in more solitary positions.
Occurrence of actinomyces (anti-type-1- or anti-type-2-reactive) cells.
Anti-type-1 and anti-type-2 frequently failed to outline the entire cell; instead, the antibodies tended to concentrate at the poles of the cell, giving the appearance of two distinct objects (Fig. 4, small inset). The clear bipolar distribution of antifimbrial antibodies seen on cells in vivo was not seen on cultured cells used in antibody strain specificity determinations; antibody distribution on cultured cells was frequently heterogeneous, but bipolar distribution was not apparent (data not shown). Also, in contrast to the rare double labeling of single streptococcal cells by anti-DL1 and anti-RPS, single actinomyces cells that reacted with both actinomyces-directed antibodies were common (Fig. 4, small inset). Although actinomyces cells that reacted with one or with both of the appropriate antibodies were readily discernible, their frequency of occurrence was low on the chips at 4 h and remained low at 8 h (Fig. 4, main image).
FIG. 4.
Antibodies directed against actinomyces fimbriae are heterogeneously distributed on the cell surface, and anti-fimbria-reactive cells are infrequent on chips. Main image and large inset (zoom of central region of main image) show 8-h-old plaque stained with acridine orange (green), anti-RPS (red), and anti-type-1 (blue). Arrow in large inset indicates a cell for which anti-type-1 reactivity (blue) is concentrated at the poles; note nucleic acid stain (green) between the two blue spots. Three other anti-type-1 reactive cells (below scale bar near lower edge) display less heterogeneity. All four of these cells are visible beneath the scale bar in the main image. Arrowheads in main image point to additional anti-type-1-reactive cells that are difficult to discern because of other biomass. The location of these cells was confirmed by examining only the blue channel of the RGB image (representing output of the far-red confocal channel; Alexa 647-conjugated anti-type-1). Small inset is from a different field of view and was stained with anti-type-2 (green), anti-RPS (red), and anti-type-1 (blue). Polar localization of antifimbrial antibodies in the absence of nucleic acid stain results in two pairs of dots (contrast with cell at arrow in large inset); each pair defines a single cell. Each cell is reactive with both antibodies; the leftmost cell has more anti-type-1 reactivity (blue dominates over green) than the cell at right (green dominates over blue).
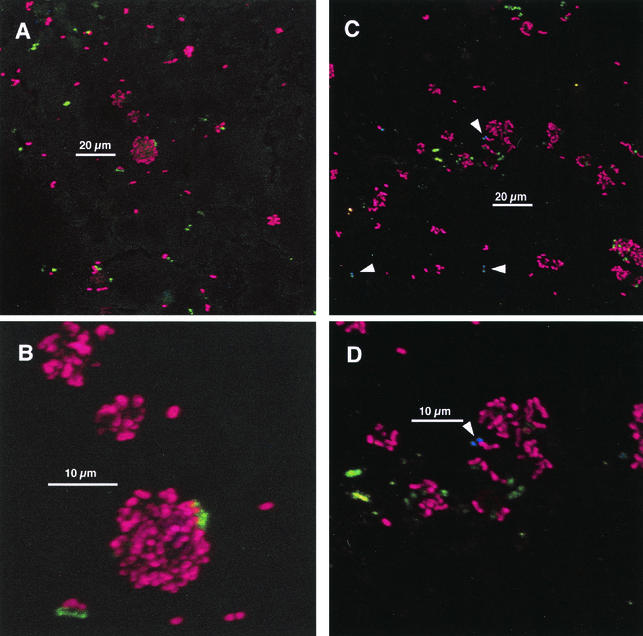
Interactions between coccoid cells.
Three classes of cell-cell interactions between cocci can be distinguished: (i) cells reactive with one of the two streptococcus-directed antibodies in association with cells visible only by nucleic acid stain, (ii) interactions between clusters of cells that react with each antibody but which are not associated with antibody-unreactive cells, and (iii) separate clusters of cells reactive with each antibody that are each in interaction with antibody-unreactive cells. While a single colony that contains solely cells reactive with a single antibody may represent clonal growth, lack of coherence between taxonomy and antibody reactivity (Table 1) precludes this firm conclusion. However, for cases of anti-DL1-reactive cells associated with anti-RPS-reactive cells, the conclusion that the interaction is between distinct cell types is firmer. As shown in Fig. 2 and 3, morphology of cells within colonies was frequently uniform. These colonies could represent interactions between more than one cell type. In some cases, it was unambiguous that the associations comprised at least two cell types (Fig. 5). These data are circumstantial support for a role for coaggregation or coadherence in plaque accumulation. For example, although anti-RPS identifies receptors for intrageneric coaggregation of streptococci (1Gn and 2Gn RPS), it also identifies the 2G that is not involved in intrageneric streptococcal coaggregation. Although it is not possible to positively identify coaggregation as a mechanism in establishment of these interactions, it is clear that cells in juxtaposition can interact.
FIG. 5.
Unambiguous interactions of at least two coccoid genotypes. Staining with anti-DL1 (green), anti-RPS (red), and Syto 59 (blue). (A) Anti-DL1-reactive cells in association with an anti-RPS-reactive cell in 4-h-old plaque. (B) Interaction of anti-DL1-reactive cells, anti-RPS-reactive cells, and antibody-unreactive cells in 8-h-old plaque.
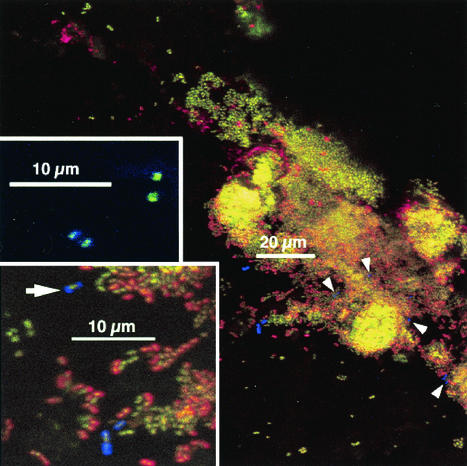
Interaction between anti-RPS-reactive cells and anti-type-2-reactive cells.
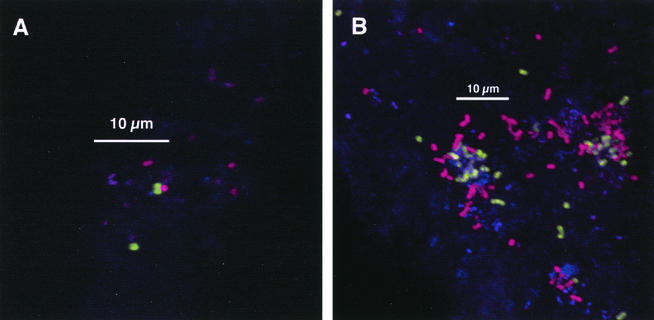
The suite of antibodies used here can unequivocally identify only the coaggregation-coadhesion interaction between RPS-bearing cells (using anti-RPS) and genospecies 2 type-2-fimbria-bearing cells (using anti-type-2). Examples of this interaction are shown in Fig. 6; the receptor required for coaggregation (G or Gn RPS) is identified on one of the cells involved in the interaction, and the respective adhesin (in this case, the fimbriae that bear the adhesin) is identified on the other cell. In some cases, clear juxtaposition of single cells was seen (Fig. 6A and B). In other cases, several anti-type-2-reactive cells were found close to one another and to anti-RPS-reactive cells (Fig. 6C and D) as well as interdigitated in colonies with anti-RPS-reactive cells (Fig. 6C, lower right). Images in Fig. 6 show areas in which actinomyces colonization was higher than that seen in most regions, and similar densities of actinomyces colonization were not found in the absence of anti-RPS-reactive cells. Under all protocol conditions, as noted above, actinomyces were not present in large numbers. These results provide the first unambiguous evidence of a role of coaggregation-mediated cell-cell recognition in plaque development.
FIG. 6.
Interaction between anti-RPS-reactive cells and anti-type-2-reactive cells in 8-h-old plaque. Staining is with anti-type-2 (green), anti-RPS (red), and anti-type-1 (blue). (A and C) Low-magnification views. (B and D) Electronic zoom of central regions for panels A and C. Most actinomyces cells stained primarily with anti-type-2; arrowheads in panels C and D indicate cells for which binding of anti-type-1 predominated.
DISCUSSION
The juxtaposition of anti-type-2-reactive bacteria with anti-RPS-reactive bacteria in undisturbed plaque provides direct evidence that coaggregation and coadherence occurs in plaque and that these cell-cell interactions begin early in oral biofilm community development. Strain specificity of the antibodies shows that anti-type-2 reacts exclusively with genospecies 2 actinomyces (Table 2) and that anti-RPS reacts, with two exceptions (SK9 and SK12), exclusively with RPS-bearing streptococci (Table 1). In fact, the high degree of antibody specificity allowed detection of what may be only a small fraction of such associations: anti-RPS identifies only slightly more than half the streptococcal strains in Table 1 known to bear RPS, and anti-type-2 identifies only one (genospecies 2) of the three actinomyces taxonomic groups known to bear coaggregation-mediating type 2 fimbriae (Table 2). Thus, large numbers of coaggregation-mediated Streptococcus spp.-Actinomyces spp. interactions known to occur in vitro would not be identified with the antibodies used herein, suggesting that the occurrence of such associations in vivo may be higher than identified in the present study.
Many interactions between coccoid cells were detected. Some interactions, as depicted in Fig. 5, unambiguously involved more than one cell type (based on antibody specificity); these could represent intrageneric interactions (e.g., S. oralis 34 with S. gordonii DL1), but they could also represent interactions within a species (e.g., S. sanguinis SK163 with S. sanguinis SK1). Much more complicated are the possibilities arising from interaction of antibody-reactive cells with solely nucleic-acid-stained cells within a single colony (Fig. 3C and D, insets). These data clearly indicate the omnipresence of intimate interactions between different coccoid cell types from the earliest point in plaque community development. The general features of plaque accumulation presented herein fit well with previous descriptions of supragingival plaque accumulation (e.g. references 21 and 33). An extensive species-cataloging study demonstrated that the most prevalent bacteria in 4-h-old plaque were S. sanguinis, S. oralis, and S. mitis biovar 1 (24, 25). Anti-DL1 and anti-RPS label early colonizing strains. Many cells in 4-h plaque were stained by these antibodies (Fig. 2), especially when colonization levels were relatively low. These data support the importance of these species in early plaque while providing a spatial context: the species are frequently in interaction with one another.
Actinomyces, as detected by anti-type-2 and anti-type-1, were infrequent at 4- and 8-h time points, but cells labeled with these antibodies were easier to find at the 8-h time point. The observation also supports the earlier study (25), in which actinomyces were a small percentage of early plaque. Image analysis was performed on the images in Fig. 2 and 3 to estimate the level of colonization and the degree of growth. The estimated cell number was calculated by dividing the area coverage of biological material (total cell area) by 0.78 (area of a 1-μm-diameter coccus). If this number is roughly translated as CFU, then the range of colonization (cell number per chip) in the present study was 2.0 × 102 to 6.8 × 103 per 250,000 μm2 for the 4-h samples and 4.5 × 103 to 1.4 × 104 per 250,000 μm2 for the 8-h samples. Although these numbers are 10- to 100-fold lower than total counts reported in previous studies (25), the increase in cell numbers between 4 and 8 h in the present study and in reference 24 was strikingly similar: 2- to 40-fold. Collectively, the relative amounts of colonization at 4 and 8 h and the relative ratios of actinomyces to streptococci at 4 and 8 h, as well as the predominance of streptococci in early dental plaque, support and extend previous work (21-23, 25).
It is now clear that, from the earliest point, bacteria form mixed-species colonies within dental plaque. Some of these interactions may confer an advantage for the participating organisms. Data have shown that, in vitro, cocultures of A. naeslundii T14V and S. oralis 34 form a luxuriant interdigitated biofilm when grown with saliva as the sole source of carbon and nitrogen, yet monoculture biofilms of these organisms grow poorly (26). The present study would detect such an interaction. The proper cellular arrangement was identified in dental plaque (Fig. 6); however, the physiological outcome of the interaction seen with pure cultures in vitro (luxuriant growth) was not apparent. The time frame over which these chip experiments took place was shorter than that used during the in vitro experiments, and it is possible that the advantage for such an interaction in plaque may first become apparent at time points later than those investigated herein. The study of bacterial interactions begins with the identification of organisms that interact. The approach demonstrated here identifies communities as they develop in undisturbed plaque and thus represents a starting point for spatially resolved isolation and subsequent characterization of oral bacterial interactions that are known to occur in nature.
Acknowledgments
We thank Rosemary Wu for initial work on the enamel chip model system in our laboratory. We also acknowledge the expertise of Divya Mittal and the staff of the NIDCR Clinic.
REFERENCES
- 1.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buswell, C. M., Y. M. Herlihy, P. D. Marsh, C. W. Keevil, and S. A. Leach. 1997. Coaggregation amongst aquatic biofilm bacteria. J. Appl. Microbiol. 83:477-484. [Google Scholar]
- 3.Cisar, J. O., M. J. Brennan, and A. L. Sandberg. 1985. Lectin-specific interaction of Actinomyces fimbriae with oral streptococci, p. 159-163. In S. E. Mergenhagen and B. Rosan (ed.), Molecular basis of oral microbial adhesion. American Society for Microbiology, Washington, D.C.
- 4.Cisar, J. O., S. H. Curl, P. E. Kolenbrander, and A. E. Vatter. 1983. Specific absence of type 2 fimbriae on a coaggregation-defective mutant of Actinomyces viscosus T14V. Infect. Immun. 40:759-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cisar, J. O., V. A. David, S. H. Curl, and A. E. Vatter. 1984. Exclusive presence of lactose-sensitive fimbriae on a typical strain (WVU45) of Actinomyces naeslundii. Infect. Immun. 46:453-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cisar, J. O., A. L. Sandberg, C. Abeygunawardana, G. P. Reddy, and C. A. Bush. 1995. Lectin recognition of host-like saccharide motifs in streptococcal cell-wall polysaccharides. Glycobiology 5:655-662. [DOI] [PubMed] [Google Scholar]
- 7.Cisar, J. O., A. L. Sandberg, G. P. Reddy, C. Abeygunawardana, and C. A. Bush. 1997. Structural and antigenic types of cell wall polysaccharides from viridans group streptococci with receptors for oral actinomyces and streptococcal lectins. Infect. Immun. 65:5035-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibbons, R. J., D. I. Hay, J. O. Cisar, and W. B. Clark. 1988. Adsorbed salivary proline-rich protein 1 and statherin: receptors for type 1 fimbriae of Actinomyces viscosus T14V-J1 on apatitic surfaces. Infect. Immun. 56:2990-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibbons, R. J., and M. Nygaard. 1970. Interbacterial aggregation of plaque bacteria. Arch. Oral Biol. 15:1397-1400. [DOI] [PubMed] [Google Scholar]
- 10.Hsu, S. D., J. O. Cisar, A. L. Sandberg, and M. Kilian. 1994. Adhesive properties of viridans streptoccocal species. Microb. Ecol. Health Dis. 7:125-137. [Google Scholar]
- 11.Johnson, J. L., L. V. H. Moore, B. Kaneko, and W. E. C. Moore. 1990. Actinomyces georgiae sp. nov, Actinomyces gerencseriae sp. nov., designation of two genospecies of Actinomyces naeslundii, and inclusion of Actinomyces naeslundii serotypes II and III and Actinomyces viscosus serotype II in A. naeslundii genospecies 2. Int. J. Syst. Bacteriol. 40:273-286. [DOI] [PubMed] [Google Scholar]
- 12.Juretschko, S., A. Loy, A. Lehner, and M. Wagner. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84-99. [DOI] [PubMed] [Google Scholar]
- 13.Kilian, M., L. Mikkelsen, and J. Henrichsen. 1989. Taxonomic study of viridans streptococci: description of Streptococcus gordonii sp. nov. and emended descriptions of Streptococcus sanguis (White and Niven 1946), Streptococcus oralis (Bridge and Sneath 1982), and Streptococcus mitis (Andrewes and Horder 1906). Int. J. Syst. Bacteriol. 39:471-484. [Google Scholar]
- 14.Klier, C. M., A. G. Roble, and P. E. Kolenbrander. 1998. Actinomyces serovar WVA963 coaggregation-defective mutant strain PK2407 secretes lactose-sensitive adhesin that binds to coaggregation partner Streptococcus oralis 34. Oral Microbiol. Immunol. 13:337-340. [DOI] [PubMed] [Google Scholar]
- 15.Kolenbrander, P. E., R. N. Andersen, and L. V. H. Moore. 1990. Intrageneric coaggregation among strains of human oral bacteria: potential role in primary colonization of the tooth surface. Appl. Environ. Microbiol. 56:3890-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolenbrander, P. E., and J. London. 1993. Adhere today, here tomorrow: oral bacterial adherence. J. Bacteriol. 175:3247-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McIntire, F. C., L. K. Crosby, A. E. Vatter, J. O. Cisar, M. R. McNeil, C. A. Bush, S. S. Tjoa, and P. V. Fennessey. 1988. A polysaccharide from Streptococcus sanguis 34 that inhibits coaggregation of S. sanguis 34 with Actinomyces viscosus T14V. J. Bacteriol. 170:2229-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moter, A., G. Leist, R. Rudolph, K. Schrank, B. K. Choi, M. Wagner, and U. B. Göbel. 1998. Fluorescence in situ hybridization shows spatial distribution of as yet uncultured treponemes in biopsies from digital dermatitis lesions. Microbiology 144:2459-2467. [DOI] [PubMed] [Google Scholar]
- 20.Noiri, Y., L. Li, and S. Ebisu. 2001. The localization of periodontal-disease-associated bacteria in human periodontal pockets. J. Dent. Res. 80:1930-1934. [DOI] [PubMed] [Google Scholar]
- 21.Nyvad, B. 1993. Microbial colonization of human tooth surfaces. APMIS 101:7-45. [PubMed] [Google Scholar]
- 22.Nyvad, B., and O. Fejerskov. 1987. Scanning electron microscopy of early microbial colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:287-296. [DOI] [PubMed] [Google Scholar]
- 23.Nyvad, B., and O. Fejerskov. 1987. Transmission electron microscopy of early microbial colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:297-307. [DOI] [PubMed] [Google Scholar]
- 24.Nyvad, B., and M. Kilian. 1990. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 24:267-272. [DOI] [PubMed] [Google Scholar]
- 25.Nyvad, B., and M. Kilian. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:369-380. [DOI] [PubMed] [Google Scholar]
- 26.Palmer, R. J., Jr., K. Kazmerzak, M. C. Hansen, and P. E. Kolenbrander. 2001. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. J. Bacteriol. 69:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer, R. J., Jr., and D. C. White. 1997. Developmental biology of biofilms: implications for treatment and control. Trends Microbiol. 5:435-440. [DOI] [PubMed] [Google Scholar]
- 28.Palmer, R. J., Jr., R. Wu, S. Gordon, C. Bloomquist, W. F. Liljemark, M. Kilian, and P. E. Kolenbrander. 2001. Retrieval of biofilms from the oral cavity. Methods Enzymol. 337:393-403. [DOI] [PubMed] [Google Scholar]
- 29.Rickard, A. H., S. A. Leach, C. M. Buswell, N. J. High, and P. S. Handley. 2000. Coaggregation between aquatic bacteria is mediated by specific growth-phase-dependent lectin-saccharide interactions. Appl. Environ. Microbiol. 66:431-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rickard, A. H., S. A. Leach, L. S. Hall, C. M. Buswell, N. J. High, and P. S. Handley. 2002. Phylogenetic relationships and coaggregation ability of freshwater biofilm bacteria. Appl. Environ. Microbiol. 68:3644-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trüper, H. G., and L. D. Clari. 1997. Taxonomic note: necessary correction of specific epithets formed as substantives (nouns) “in apposition.” Int. J. Syst. Bacteriol. 47:908-909. [Google Scholar]
- 32.Wecke, J., T. Kersten, K. Madela, A. Moter, U. B. Göbel, A. Friedmann, and J. P. Bernimoulin. 2000. A novel technique for monitoring the development of bacterial biofilms in human periodontal pockets. FEMS Microbiol. Lett. 191:95-101. [DOI] [PubMed] [Google Scholar]
- 33.Zee, K. Y., L. P. Samaranayake, and R. Attström. 1997. Scanning electron microscopy of microbial colonization of ‘rapid’ and ‘slow’ dental-plaque formers in vivo. Arch. Oral Biol. 42:735-742. [DOI] [PubMed] [Google Scholar]