Abstract
The marine bacterium Microbulbifer degradans strain 2-40 produces at least 10 enzyme systems for degrading insoluble complex polysaccharides (ICP). The draft sequence of the 2-40 genome allowed a genome-wide analysis of the chitinolytic system of strain 2-40. The chitinolytic system includes three secreted chitin depolymerases (ChiA, ChiB, and ChiC), a secreted chitin-binding protein (CbpA), periplasmic chitooligosaccharide-modifying enzymes, putative sugar transporters, and a cluster of genes encoding cytoplasmic proteins involved in N-acetyl-d-glucosamine (GlcNAc) metabolism. Each chitin depolymerase was detected in culture supernatants of chitin-grown strain 2-40 and was active against chitin and glycol chitin. The chitin depolymerases also had a specific pattern of activity toward the chitin analogs 4-methylumbelliferyl-β-d-N,N′-diacetylchitobioside (MUF-diNAG) and 4-methylumbelliferyl-β-d-N,N′,N"-triacetylchitotrioside (MUF-triNAG). The depolymerases were modular in nature and contained glycosyl hydrolase family 18 domains, chitin-binding domains, and polycystic kidney disease domains. ChiA and ChiB each possessed polyserine linkers of up to 32 consecutive serine residues. In addition, ChiB and CbpA contained glutamic acid-rich domains. At 1,271 amino acids, ChiB is the largest bacterial chitinase reported to date. A chitodextrinase (CdxA) with activity against chitooligosaccharides (degree of polymerization of 5 to 7) was identified. The activities of two apparent periplasmic (HexA and HexB) N-acetyl-β-d-glucosaminidases and one cytoplasmic (HexC) N-acetyl-β-d-glucosaminidase were demonstrated. Genes involved in GlcNAc metabolism, similar to those of the Escherichia coli K-12 NAG utilization operon, were identified. NagA from strain 2-40, a GlcNAc deacetylase, was shown to complement a nagA mutation in E. coli K-12. Except for the GlcNAc utilization cluster, genes for all other components of the chitinolytic system were dispersed throughout the genome. Further examination of this system may provide additional insight into the mechanisms by which marine bacteria degrade chitin and provide a basis for future research on the ICP-degrading systems of strain 2-40.
Microbulbifer degradans strain 2-40 is a marine γ-proteobacterium that was isolated from decaying Spartina alterniflora, a salt marsh cord grass, in the Chesapeake Bay watershed (2, 14). Consistent with its isolation from decaying plant matter, 2-40 can degrade cellulose, pectin, and xylan, which are common components of the cell walls of higher plants. Strain 2-40 is also able to depolymerize algal cell wall components, such as agar, agarose, and laminarin, as well as protein, starch, pullulan, and alginic acid. Chitin, a component of crustacean and insect exoskeletons, yeast and fungal cell walls, and diatoms, is also depolymerized by 2-40 enzymes. In addition to degrading this plethora of polymers, 2-40 can utilize each of the polysaccharides as the sole carbon source. Therefore, strain 2-40 is not only an excellent model of microbial degradation of insoluble complex polysaccharides (ICP) but can also be used as a paradigm for complete metabolism of these ICP (11, 51).
Chitin is a homopolymer composed of repeating units of N-acetyl-d-glucosamine (GlcNAc) and is found in various forms throughout the marine environment. It is usually at least 90% acetylated and is often in a complex with proteins and other carbohydrates (30). The microcrystalline structure of chitin varies between antiparallel sheets (alpha chitin), parallel sheets (beta chitin), and a mixture of both (gamma chitin) (30). Alpha chitin is found in the calyces of hydrozoa, mollusks, plankton, and as a component of the cuticles of arthropods. Beta chitin, a less stable and more degradable form of chitin, is found in mollusks, squid pen, diatoms, and insect exoskeletons and cocoons and is the major component of fungal cell walls (30). Chitin is resistant to chemical degradation and is difficult to digest enzymatically because of the multiple steps required to expose and cleave the polymer. Because chitin resists chemical and physical breakdown, microorganisms must play a major role in its degradation (15, 52). Surprisingly, almost no free chitin is found in marine sediments (52), demonstrating the efficiency of these microbial systems. Therefore, chitin represents an abundant source of carbon and nitrogen to microorganisms in the marine environment.
The degradation and metabolism of chitin by marine microorganisms appear to involve the synergistic action of multiple proteins, including several extracellular chitin depolymerases, noncatalytic chitin-binding proteins, chitodextrinases, and periplasmic and cytoplasmic N-acetylglucosaminidases (chitobiases and N-acetylhexosaminidases) (22, 23, 44). These proteins typically include conserved modules that function as catalytic domains or chitin-binding domains and may also contain domains of unknown function such as fibronectin type III and/or polycystic kidney disease (PKD) domains (30). Many of the genes for these enzymes have been cloned individually from chitin-degrading organisms.
The mechanism by which M. degradans strain 2-40 degrades and metabolizes chitin was investigated using biochemical, genetic, and genomic analyses. The results indicate that strain 2-40 expresses extracellular, periplasmic, and cytoplasmic systems for the depolymerization, transport, and metabolism of chitin-derived products. The proteins of these systems contain domains similar to catalytic and binding regions of other microbial chitinases and in some cases polyserine- and hydroxyamino acid-rich linkers of unknown function.
MATERIALS AND METHODS
Growth of bacterial strains.
M. degradans strain 2-40 was grown in minimal medium containing (per liter): 2.3% Instant Ocean (Aquarium Systems, Mentor, Ohio), 0.5% ammonium chloride, 0.2% glucose, and 50 mM Tris HCl, pH 7.6. Other carbon sources were added to a final concentration of 0.1%. Agar was added to a final concentration of 1.5% to prepare solid media. All cultures were incubated at 25°C. Escherichia coli EC300, DH5αE, and Tuner strains were grown in Luria-Bertani (LB) broth or agar supplemented with the appropriate antibiotics and incubated at 37°C.
Cloning and molecular biology protocols.
All DNA manipulations and standard molecular biology protocols were performed as described by Sambrook et al. (38).
Materials and reagents.
Individual genes were amplified using ProofPro DNA polymerase (Continental Lab Products, San Diego, Calif.). Restriction enzymes, T4 DNA ligase, amylose resin, and the pMal-2pX expression vector were obtained from New England Biolabs (Beverly, Mass.). Ni-nitrilotriacetic acid (Ni-NTA) agarose and the pETBlue-2 expression vector were obtained from Novagen (Madison, Wis.). The pCC1 fosmid vector with the copy control feature was obtained from Epicentre Technologies (Madison, Wis.). All other reagents and substrates were obtained from Sigma-Aldrich (St. Louis, Mo.) unless otherwise noted. Centrifugal concentrators were obtained from Millipore (Bedford, Mass.).
Construction of an M. degradans strain 2-40 genomic library.
Strain 2-40 chromosomal DNA was isolated and prepared for ligation into pCC1 according to the manufacturer's protocol. Sau3A fragments of 30 to 40 kb were isolated using gel extraction and ligated into BamHI-digested pCC1. The vector was packaged into phage and used to infect E. coli EC300. Transductants were selected using chloramphenicol (30 μg/ml).
Screening of the M. degradans strain 2-40 genomic library for chitinase activity.
E. coli transductants were initially screened for chitin depolymerase activity by plating the library on LB agar supplemented with 0.1% chitin or 0.08% chitin azure and incubating for 5 days at 37°C. Chitin depolymerase activity was identified by zones of clearing around bacterial colonies.
Alternatively, the chitin analogs 4-methylumbelliferyl-β-d-N,N′-diacetylchitobioside (MUF-diNAG) and 4-methylumbelliferyl-β-d-N,N′,N"-triacetylchitotrioside (MUF-triNAG) were used to screen transductants for chitinase activity. Single transductants were grown in 100 μl of LB broth supplemented with chloramphenicol (30 μg/ml). Cultures were incubated with gentle shaking at 25°C for 12 h. A MUF analog was added to a final concentration of 1.5 μM and incubated with shaking at 25°C for an additional 24 h. Cleavage of the analog was visualized using long-wavelength UV light.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and zymogram analysis.
Concentrated culture supernatants of M. degradans strain 2-40 were prepared from 50-ml cultures grown at 25°C for 50 h in minimal medium without glucose and supplemented with 0.1% chitin. All subsequent steps were performed at 4°C. Cultures were centrifuged at 10,000 × g for 20 min and then sterilized by filtration through a 0.22-μm-pore-size filter. The filter-sterilized supernatant was then concentrated 100-fold using a centrifugal concentrator with a 10-kDa cutoff filter (Millipore).
Proteins in concentrated culture supernatants were fractionated by SDS-PAGE with a stacking gel by the method of Laemmli (26) in an 8% acrylamide separating gel with a final concentration of 0.01% glycol chitin. Gels were then incubated in refolding buffer (50 mM Tris, 1 mM EDTA, 5 mM 2-mercaptoethanol [pH 7.5]) at 4°C for 24 h. Gels were washed for 1 h in 100 mM sodium phosphate buffer (pH 7) at 25°C and then incubated in 100 mM sodium phosphate buffer (pH 7) for 16 h at 37°C. Gels were rinsed and washed in developing buffer (0.5 M Tris, 0.01% Calcofluor [pH 7.5]) for 5 min and then rinsed with distilled water for 2 h with frequent changes of wash water. Zones of chitin depolymerase activity appeared as dark bands when viewed under UV light (45).
Protein expression and purification.
Genes of interest were amplified using PCR and tailed primers (Table 1). Each gene was digested with the appropriate restriction enzyme, ligated into the pETBlue-2 or pMal-2pX expression vector as indicated, and transformed into E. coli Tuner (Novagen) or E. coli DH5αE (Invitrogen) cells. A 50-ml culture of transformants carrying the clone of interest was grown at 37°C to a optical density at 600 nm of 0.5 to 0.6, induced with isopropyl-β-d-thiogalactopyranoside (IPTG), and grown for an additional 3 h at 37°C. Cells were harvested and resuspended in lysis buffer, and clarified lysates were prepared as recommended by the manufacturer of the affinity resin. pETBlue-2 His tag fusions were bound to Ni-NTA agarose, and pMal-2pX maltose-binding protein (MBP) fusions were purified using amylose resin. Fusion proteins were eluted with imidazole and maltose solutions, respectively. Fractions of interest were concentrated using centrifugal concentrators with 10-kDa cutoff filters, aliquoted, and stored at −80°C.
TABLE 1.
Primers used in this study
| Primera | Sequence (5′ to 3′)b |
|---|---|
| ChiA-F | TGCTCTAGAGCATTCGCAGCAACCAAT |
| ChiA-R | AACTGCAGCGGTAAGCTGTGAACCTT |
| ChiB-F | GGAATTCGCAGTGGCTGCACTTAG |
| ChiB-R | TGCTCTAGACTGCTTTTCGTTGCCGAA |
| ChiC-F | GGAATTCATGAACCCTATAGCTAAACT |
| ChiC-R | TGCTCTAGAGCCATTATCAATCGCTCTAA |
| HexA-F | CGGGATCCATGAAACTAAGATTATTACC |
| HexA-R | TGCTCTAGATTTAACGATTATTTCATCTAA |
| HexB-F | CGGGATCCATGGCGTTATTTAGCAAGTAT |
| HexB-R | TGCTCTAGACTCAACAGGTAACGCACG |
| CdxA-F | CGCGGATCCGATGAAAAATAAGCACTGC |
| CdxA-R | AGGCGCGCCAACGTACATACCAAGTCCC |
| NagA-F | GCTCTAGACCCACACCTAAACAAGGT |
| NagA-R | ACCGTGATATCGCTAGTAGCTAGCAGCTA |
Forward (-F) and reverse (-R) primers are indicated.
Restriction sites are underlined.
Chitinase activity assays using chitin analogs and chitooligosaccharides.
To determine the specific activities of chitin depolymerases against the chitin analogs MUF-diNAG and MUF-triNAG, 900 μl of a 50 μM solution of each chitin analog was added to 100 μl of purified enzyme and incubated at 37°C for 30 min. The fluorescence of the reaction (excitation wavelength, 365 nm; emitted wavelength, 460 nm) was determined using a Hoefer TKO 100 fluorometer and compared to a standard curve prepared with 4′-methylumbelliferone. One unit of activity is defined as l μmol of 4′-methylumbelliferone released per mg of purified enzyme per min. Products of chitooligosaccharide-modifying enzyme reactions were identified by thin-layer chromatography (47). Enzyme reaction mixtures contained 0.45 μmol of chitooligosaccharide substrate in 10 mM Tris HCl at pH 7.5. After 1 h at 30°C, reactions were stopped by boiling for 10 min. Degradation products were fractionated on silica gel plates. Plates were developed in 2-propanol-ethanol-distilled water at a ratio of 5:2:1 for 1 h. The plate was air dried and sprayed with 10% sulfuric acid in ethanol. The plate was dried and baked at 120°C for 20 min. Chitooligosaccharide spots appeared brown and were compared to standards composed of chitooligosaccharides of known sizes.
DNA and protein sequence manipulations and analyses.
Protein modules and domains were identified using the Simple Modular Architecture Tool (SMART) and pFAM database (www.smart.embl-heidelberg.de) (27, 39). Similarity searches were performed using the BLAST algorithm at the National Center for Biotechnology Information (NCBI) server (www.ncbi.nih.nlm.gov) (1). Type II secretion signals were identified using the iPSORT program (www.hypothesiscreator.net/iPSORT) (3) and the SignalP version 1.1 program (www.cbs.dtu.dk/services/SignalP) (31). Multiple-sequence alignments were performed using the ClustalW program (20) (www.searchlauncher.bcm.tmc.edu). Estimated protein molecular masses were calculated using the Peptide Mass Tool at the ExPASy server of the Swiss Institute of Bioinformatics (www.us.expasy.org).
Complementation of a nagA mutant.
The M. degradans strain 2-40 nagA gene was amplified using PCR and tailed primers with 2-40 genomic DNA as the template. The amplified DNA and pBluescript SK+ (Ampr) were digested with the appropriate restriction enzymes and ligated using T4 DNA ligase to create pNagA. E. coli K-12 strain IBPC531 (nagA::cm) (36) was transformed with pNagA and plated on GlcNAc-containing minimal medium, which contains M63 minimal salts, 0.2% GlcNAc, ampicillin (50 μg/ml), and chloramphenicol (30 μg/ml).
RESULTS AND DISCUSSION
M. degradans produces multiple chitinases in the presence of chitin.
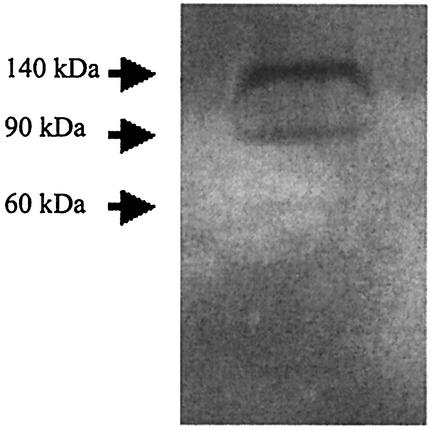
To determine the number of chitin depolymerases produced by M. degradans strain 2-40, concentrated culture supernatants were screened for chitinase activity using zymograms. Late-log-phase cultures grown with alpha chitin as the sole carbon source were harvested, and the culture supernatants were concentrated and subjected to SDS-PAGE with glycol chitin incorporated into the separating gel (45). After renaturation and incubation, zones of chitin depolymerization were visualized using Calcofluor. Three chitin depolymerases were observed in culture filtrates of strain 2-40 with apparent masses of 140, 90, and 60 kDa (Fig. 1).
FIG. 1.
M. degradans strain 2-40 produces three chitin depolymerases in the presence of alpha chitin. Strain 2-40 was grown in the presence of alpha chitin as the sole carbon source. Culture supernatants were collected, concentrated 100-fold, and fractionated in an 8% polyacrylamide gel containing 0.01% glycol chitin in the separating gel. Gels were incubated in refolding buffer overnight at 4°C and then incubated at 37°C for 16 h. Chitin depolymerase activity was visualized by staining the gel with Calcofluor.
Biochemical analysis of an M. degradans strain 2-40 genomic library.
To identify the genes encoding the chitin depolymerases of strain 2-40, a genomic library of 2-40 carrying 40-kb fragments was prepared using the Epifos fosmid library system (Epicentre Technologies). E. coli EC300 transformants carrying the 2-40 library were assayed for chitinase activity on agar plates containing collodial chitin or chitin azure. Although a sufficient number of clones were surveyed for threefold coverage of the genome, only one clone was found to express chitinase activity on these plates as indicated by a 2-mm zone of clearing. Lysates of this transformant produced a single apparent chitin depolymerase that migrated in zymograms with a mass of 140 kDa (data not shown).
Because the expected 90- and 60-kDa chitin depolymerases were not detected in the previous screen, a more sensitive screen that employed the chitin analogs MUF-diNAG and MUF-triNAG was devised. These fluorogenic substrates are degraded by some chitin depolymerases, releasing the fluorescent product methylumbelliferone (49). Six clones in the strain 2-40 library were capable of cleaving either substrate analog. One of these clones was also able to degrade colloidal chitin and was subsequently shown to carry the same genomic fragment as the clone identified above. We hypothesized that the other clones identified in this screen carried genes for chitodextrinases and/or N-acetylglucosaminidases.
The genomic sequence of M. degradans contains genes typical of a chitinolytic system.
A draft sequence of the M. degradans strain 2-40 genome was obtained in conjunction with the United States Department of Energy Joint Genome Institute (http://www.jgi.doe.gov). To identify the genes for the chitin depolymerases and other chitin-processing enzymes, the genomic sequence of strain 2-40 was initially surveyed using the sequences of chitinases from the closely related bacterium Alteromonas (46) and later with additional chitinolytic enzymes and catalytic or binding domains from other bacterial species. A wide variety of carbohydrate-binding modules and glycosyl hydrolase catalytic domains have been identified and classified by protein sequence; a comprehensive list of these domains can be found at http://afmb.cnrs-mrs.fr/. Open reading frames (ORFs) were identified in the M. degradans strain 2-40 genomic sequence whose products shared sequence similarity to conserved domains of known chitin depolymerases, chitiodextrinases, chitin-binding proteins, and N-acetylglucosaminidases.
Identification of three major chitin depolymerases in the M. degradans genome.
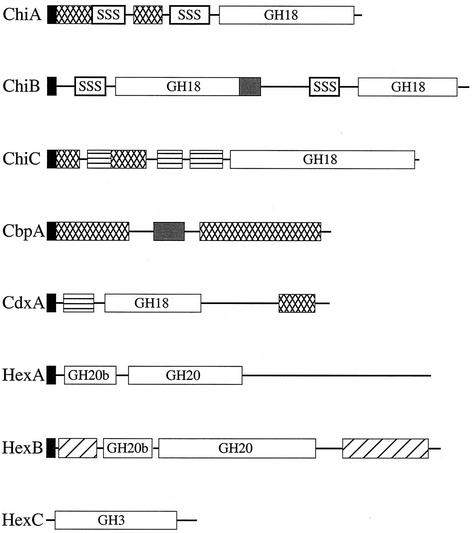
Three of the proteins identified in the above sequence searches exhibited similarity to known microbial chitin depolymerases. Each putative chitin depolymerase of M. degradans strain 2-40 carried the following: (i) an apparent type II N-terminal secretion signal as calculated by the iPSORT and SignalP version 1.1 programs, (ii) a glycosyl hydrolase family 18 (GH18) catalytic domain, and (iii) accessory domains of unknown function, such as PKD domains (Fig. 2). Two proteins also carried Cbd3 (type three chitin-binding domain) motifs. The arrangement of these domains in strain 2-40 chitin depolymerases was distinct from their homologs in other species, and the sequences of regions outside the conserved motifs had little similarity to the sequences of known proteins.
FIG. 2.
M. degradans strain 2-40 chitinases contain polyserine linkers and glutamic acid-rich domains, as well as conserved modules found in other microbial chitinases. Protein sequence analysis revealed the presence of the following elements: putative type II secretion signals; GH18, GH20, and GH3 domains; chitin- and carbohydrate-binding domains; PKD domains; and regions of amino acid repeats. Type II secretion signal (black boxes), polyserine linker (SSS), chitin-binding domain (cross-hatched boxes), glutamic acid-rich domain (grey shaded boxes), PKD domain (boxes with horizontal lines), and soluble sugar-binding domain (hatched boxes) are indicated.
The first chitin depolymerase identified in the M. degradans strain 2-40 genome was ChiA, a 543-amino-acid (aa) protein with a calculated mass of 57.0 kDa (Table 2). ChiA carried two Cbd3 motifs and a GH18 domain. The first Cbd3 consisted of 46 residues and was most similar to a Cbd3 of ChiA from Pseudoalteromonas sp. strain S91 (60% identity and 75% similarity [60% I, 75% S]) (44), while the sequence of the second 47-aa domain was similar to the Cbd3 sequence from ChiA of Vibrio cholerae (44% I, 57% S) (9). The 299-aa GH18 domain exhibited the highest identity with the GH18 domain of ChiA from V. cholerae (65% I, 77% S) (9) (Fig. 2).
TABLE 2.
Relevant characteristics of components of the chitinolytic system of M. degradans strain 2-40
| Protein | Putative localizationa | No. of amino acids | Estimated MWb | GC contentc | Hydropathy of N terminusd | Repetitive AA domain(s)e | GenBank accession no. |
|---|---|---|---|---|---|---|---|
| ChiA | Extracellular | 543 | 57.0 | 49.7 | 1.573 | Ser | BK001043 |
| ChiB | Extracellular | 1,271 | 136.1 | 46.9 | 2.167 | Ser, Glu | BK001042 |
| ChiC | Extracellular | 792 | 87.1 | 54.2 | 1.000 | BK001044 | |
| CbpA | Extracellular | 449 | 47.6 | 49.7 | 1.427 | Glu | BK001045 |
| CdxA | Periplasm | 1,088 | 115.6 | 52.7 | 1.547 | AY233270 | |
| HexA | Periplasm | 795 | 88.5 | 46.0 | 1.527 | BK001046 | |
| HexB | Periplasm | 889 | 98.4 | 47.2 | 0.993 | BK001047 | |
| HexC | Cytoplasm | 345 | 37.4 | 46.9 | 0.487 | BK001048 |
Putative localization of proteins was based on the presence of a signal sequence, known localization of similar proteins, and zymogram analysis of culture supernatants.
Estimated molecular weights (MW) were calculated using the Peptide Mass Tool at the Swiss Institute of Bioinformatics.
The GC content of each gene was calculated using DNasis software.
The hydropathy of each N terminus was calculated using the iPSORT program.
Repetitive amino acid (AA) domains are indicated as follows: Ser, polyserine domains; Glu, glutamic acid-rich domains.
A second chitin depolymerase, ChiB, was a 1,271-aa protein with a calculated mass of 136.1 kDa (Table 2). ChiB consisted of two discrete GH18 domains but had no apparent chitin- or carbohydrate-binding domains (Fig. 2). The first GH18 domain was 384 aa long and was most similar to the active site of ChiB from Serratia marcescens strain BJL200 (55% I, 69% S) (50), while the second was most similar to a chitinase from Vibrio sp. strain 5SM-1 (49% I, 66% S) and was 394 aa long (Fig. 2). This is the largest bacterial chitinase described to date. The apparent presence of two discrete GH18 catalytic modules is novel, but the significance of this feature has not yet been determined.
A third deduced chitin depolymerase, ChiC, was a 792-aa protein with a calculated molecular mass of 87.1 kDa (Table 2). ChiC had two Cbd3 domains; the first, a 46-aa domain, was most similar to the Cbd3 of ChiB from Vibrio harveyi (57% I, 63% S) (S. Ni Chadhain and D. Kirchman, unpublished data), while the second, consisting of 49 aa, was most similar to the Cbd3 of ChiA from V. cholerae (57% I, 75% S) (9). ChiC also contained three PKD-like domains. ChiC had a 350-aa C-terminal GH18 catalytic domain with strong similarity to ChiC from Streptomyces peucetius (61% I, 71% S) (48) (Fig. 2).
Unusual domains in the chitin depolymerases.
Several unusual domains of repetitive amino acid sequences were identified in ChiA, ChiB, and CbpA (discussed later). Two polyserine domains were present in ChiA. The first, a 46-aa polyserine domain (93% serine), was located between the Cbd3 domains of ChiA. The next was located between the second Cbd3 domain and the catalytic site and consisted of 36 residues, 31 of which were serine (Fig. 2). Several polyserine domains were also present in the deduced ChiB sequence. The first began 15 residues after the predicted cleavage site of the secretion signal and was 28 aa long. This was followed, after a spacer of 12 aa, by a 109-aa serine-rich region that was 70% serine. A third polyserine region in ChiB consisted of a 51-aa region beginning at residue 788 that contained 39 serines (Fig. 2). All six codons for serine were utilized to encode these domains, and no obvious pattern of codon usage was detected.
Polyserine linkers have been hypothesized to be regions of high flexibility that may be required for full substrate accessibility (7). This type of polyserine motif has not previously been reported in a chitinase, and there are only a few examples of similar domains in prokaryotic proteins. Cellulases, xylanases, and pectate lyases (e.g., GenBank accession numbers CAA38389, CAA383802, and AAG29353) with serine-rich domains (though smaller and less rich in serine than the 2-40 polyserine domains) have been reported for Cellvibrio japonicus (formerly Pseudomonas cellulosa) (17). Xylella fastidiosa, a gram-negative plant pathogen, encodes a putative 1,4-β-cellobiosidase with a single serine-rich domain (41), though no other proteins from this organism are reported to have such motifs. Streptococcus cristatus encodes a cell wall-associated protein, SrpA, that contains serine-rich domains (GenBank accession number AAF34780.1). While the function of these domains is unknown, the polyserine linker of a xylanase from Cellvibrio japonicus (formerly Pseudomonas cellulosa) was shown to be required for full enzymatic function (7, 16, 17).
In addition to the polyserine domains described above, an acidic domain was also detected in ChiB. A Glu-Thr-rich region was present in the region separating the two deduced catalytic domains of ChiB and consisted of Thr-Glu followed by (Glu-Thr)10. Regions rich in hydroxyamino acids or proline have been identified as linker sequences in a number of microbial ICP-degrading enzymes (40). Linkers of this type have been reported in cellulases from Cellulomonas fimi and are hypothesized to be glycosylation sites (13). Therefore, it is possible that similar domains in M. degradans strain 2-40 are glycosylated, though this has not been demonstrated empirically.
ChiA, ChiB, and ChiC hydrolyze chitin and chitin analogs.
The predicted sizes of ChiA, ChiB, and ChiC were nearly identical to the sizes of the chitin depolymerases identified in culture filtrates of M. degradans strain 2-40. To confirm their identification as chitin depolymerases, chiA, chiB, and chiC were individually amplified by PCR using strain 2-40 genomic DNA as the template and cloned into the pMal-2pX expression vector to create amino-terminal MBP fusions. Each of the expressed proteins was then purified from E. coli DH5αE transformants and assayed for chitin depolymerase activity using glycol chitin zymograms. Chitin depolymerase activity was detected at 180, 130, and 100 kDa for transformants expressing the ChiB, ChiC, and ChiA fusion constructs, respectively (data not shown). After correction for the 40-kDa mass of the fusion tag, these activities were in good agreement with the activities identified in strain 2-40 culture filtrates and the deduced masses of each gene product.
The presence of multiple chitin depolymerases containing different domains and with different modular organizations suggested that each might have distinct biochemical properties. The chitin analogs MUF-diNAG and MUF-triNAG have been used to distinguish the biochemical properties of chitin-degrading enzymes (49), though specific function (i.e., endo- or exochitinase activity) cannot be determined by activity against analogs alone. Each chitin depolymerase had a specific pattern of activity against the chitin analogs (Table 3). ChiA was not active against either MUF analog, while ChiB degraded only MUF-diNAG. ChiC was active against both MUF-diNAG and MUF-triNAG but cleaved MUF-triNAG much more rapidly than it cleaved MUF-diNAG. Different substrate specificities support the conclusion that each chitin depolymerase has a specific biochemical activity and could have a distinct role in the depolymerization of various forms of chitin.
TABLE 3.
Biochemical activities of extracellular chitin depolymerases of M. degradans strain 2-40
| Enzyme | Depolymerizationa of substrate:
|
Enzyme activityb (U/mg) on substrate:
|
||
|---|---|---|---|---|
| Chitin and chitin azure | Glycol chitin | MUF-diNAG | MUF-triNAG | |
| ChiA | + | + | 0 | 0 |
| ChiB | + | + | 7.5 | 0 |
| ChiC | + | + | 12.5 | 31.5 |
Depolymerization (+) of chitin and chitin azure as indicated by the appearance of a 2-mm zone of clearing around colonies expressing these enzymes. Depolymerization (+) of glycol chitin was determined using zymogram analysis and purified protein preparations as described in the legend to Fig. 1.
E. coli expressing each enzyme and purified enzyme preparations were analyzed with MUF-diNAG and MUF-triNAG. One unit of activity is defined as the number of micromoles of 4′-methylumbelliferone released per minute per milligram of purified enzyme. Activities were assayed as described in Materials and Methods.
Identification of periplasmic enzymes involved in chitin metabolism by M. degradans.
Chitooligosaccharides produced by the activity of extracellular chitin depolymerases are imported into the periplasm via an outer membrane chitoporin in some organisms and hydrolyzed to chitobiose and GlcNAc (21). Consistent with this model, apparent N-acetyl-d-glucosaminidases and/or chitodextrinases had been detected in the MUF analog screens of the M. degradans strain 2-40 genomic library described above. During the survey of the 2-40 genome for genes encoding GH18 domains, a putative chitodextrinase was identified. This putative chitodextrinase, termed CdxA, was a 1,088-aa protein, with a calculated molecular mass of 115.6 kDa, and carried a typical type II-dependent secretion signal, two PKD domains, a 403-aa GH18 catalytic site, and a 41-aa Cbd3 chitin-binding domain (Fig. 2). The GH18 domain was most similar to that of chitodextrinase ChiD from Alteromonas sp. strain O-7 (72% I, 83% S) (47). The Cbd3 domain was most similar to the Cbd3 in Pseudoalteromonas sp. strain S91 ChiA (64% I, 71% S) (44). The presence of an apparent Cbd3 domain in a chitodextrinase is unusual, and its function is unknown. A PCR screen using primers unique to this gene revealed that two of the six clones capable of cleaving the MUF derivatives identified in the genomic library screen carried this gene.
Two predicted periplasmic N-acetylglucosaminidases were identified in the M. degradans strain 2-40 genome (Fig. 2). Each had an N-terminal type II-dependent secretion signal. One of these, HexA, was a 795-aa protein with a predicted molecular mass of 88.5 kDa (Table 2). HexA carried a GH20b domain (glycosyl hydrolase family 20 catalytic domain 2) that was most similar to the active site of the Alteromonas sp. strain O-7 N-acetylhexosaminidase (32% I, 57% S) (47) and a 348-aa GH20 domain related to the active site of the Pseudoalteromonas sp. strain S91 N-acetylglucosaminidase (58% I, 73% S) (44) (Fig. 2). The other, HexB, was an 889-aa protein with a predicted mass of 98.4 kDa that contained a putative carbohydrate-binding domain, a GH20b domain found in the N-acetylhexosaminidase B of Alteromonas sp. strain O-7 (36% I, 55% S) (47), and a 406-aa GH20 domain identified as the active site of the N-acetylhexosaminidase of Vibrio vulnificus (55% I, 70% S) (42) (Table 2). HexB also contained an N-acetylhexosaminidase-like C-terminal domain related to the N-acetyl-β-d-glucosaminidase from Enterobacter sp. strain G-1 (44% I, 55% S) (28) (Fig. 2). The overall similarity of HexA and HexB to other N-acetylglucosaminidases and retention of key catalytic domains are consistent with their proposed activity. In addition, a PCR screen using primers specific to hexA and hexB demonstrated that the remaining three genomic clones from the original genomic library with activity against MUF derivatives contained one of these genes.
CdxA, HexA, and HexB hydrolyze chitin analogs and chitooligosaccharides.
To evaluate the deduced functions of these putative chitooligosaccharide-processing enzymes, cdxA, hexA, and hexB were amplified from M. degradans strain 2-40 genomic DNA using PCR and cloned into the pETBlue-2 or pMal-2pX expression vector to create His tag or MBP fusions, respectively, for purification. Each of the fusions was purified on Ni-NTA columns (for His tag constructs) or amylose columns (for MBP constructs) and assayed for activity against colloidal chitin, chitin azure, glycol chitin, chitooligosaccharides, chitobiose, GlcNAc, and chitin analogs MUF-diNAG and MUF-triNAG. Consistent with its identification as a chitodextrinase, CdxA was active against MUF-diNAG, MUF-triNAG, and chitooligosaccharides (Table 4) but did not degrade chitobiose, chitin, chitin azure, or glycol chitin. HexA and HexB hydrolyzed MUF-diNAG, MUF-triNAG, and chitobiose, but not chitooligosaccharides, colloidal chitin, chitin azure, or glycol chitin. These biochemical data, together with the deduced structural features of each protein, are consistent with the proposed functions of CdxA as a chitodextrinase and HexA and HexB as N-acetylhexosamidases.
TABLE 4.
Biochemical activities of periplasmic chitooligosaccharide-degrading enzymes of M. degradans strain 2-40
| Enzyme | Degradationa of substrate:
|
|||
|---|---|---|---|---|
| MUF-diNAG | MUF-triNAG | Chitooligosaccharides | Chitobiose | |
| CdxA | + | + | + | − |
| HexA | + | + | − | + |
| HexB | + | + | − | + |
Crude enzyme preparations of these enzymes degraded (+) the analogs MUF-diNAG and MUF-triNAG, resulting in a fluorescing reaction mixture. Chitooligosaccharides (degree of polymerization of 5 to 7) and chitobiose were added to crude enzyme preparations followed by thin-layer chromatographic analysis of the reaction mixture. Symbols: +, degradation indicated by the disappearance of the thin-layer chromatographic spot corresponding to each chitooligosaccharide and the appearance of degradation products; −, no degradation.
Cytoplasmic components of the M. degradans chitinase system.
The combined activity of the M. degradans strain 2-40 chitin depolymerases and the chitooligosaccharide- and chitobiose-processing enzymes (CdxA, HexA, and HexB) would degrade chitin to GlcNAc and chitobiose, which in some bacteria are imported into the cytoplasm via phosphotransferase system (PTS)-dependent transporters (4, 25, 37). Imported chitobiose can be cleaved by cytoplasmic N-acetylglucosaminidases to yield GlcNAc and GlcNAc-6-phosphate (GlcNAc-6-P); GlcNAc can also be imported directly (22). In E. coli, cytoplasmic GlcNAc is metabolized by the products of the nagBACD operon and the divergently expressed nagE gene (33, 36). NagC is a negative-acting regulatory protein that binds to the promoter region between nagB and nagE (35), whereas NagE is part of an inner membrane GlcNAc transporter, NagA is a cytoplasmic GlcNAc deacetylase, and NagB is a cytoplasmic glucosamine-6-P isomerase (36). The function of NagD remains unknown (35, 36).
An apparent cytoplasmic N-acetylglucosaminidase, HexC, was identified in the M. degradans strain 2-40 genome as a 345-aa protein with a predicted mass of 37.4 kDa that lacked an apparent N-terminal secretion signal but had a GH3N domain (glycosyl hydrolase family 3 N-terminal domain) similar to that of Pseudomonas aeruginosa N-acetylglucosaminidase (50% I, 65% S) (43) (Table 2). HexC likely degrades cytoplasmic chitobiose; this activity could have a role in the regulation of genes activated by the presence of chitobiose and would also release GlcNAc for use as an energy source.
Other cytoplasmic components of the chitinolytic system include homologs to NagA, NagB, and NagC (but not NagD). Genes for these proteins were identified in the M. degradans strain 2-40 genome as an apparent nagCBA operon. The predicted NagC from strain 2-40 (NagC2-40) was a member of the LacI family of transcriptional regulators and was most similar to LacI of X. fastidiosa strain 9a5c (47% I, 65% S) (41). Like NagC proteins from other bacteria (34), NagC2-40 contained an amino-terminal helix-turn-helix motif typical of this family of transcriptional regulators (19). The deduced NagA2-40 contained a putative NAGA (N-acetylglucosamine-6-P deacetylase) domain and was overall most similar to the Caulobacter crescentus N-acetylglucosamine-6-P deacetylase (55% I, 70% S) (32). Expression of NagA2-40 in E. coli K-12 strain IBPC531, a mutant lacking a functional nagA gene, restored growth of IBPC531 on GlcNAc-containing minimal medium, indicating complementation. NagB2-40 included two predicted SIS (sugar isomerase) domains (5) and exhibited substantial similarity to the glucosamine-fructose-6-P aminotransferase of C. crescentus (56% I, 69% S) (32).
A homolog to the E. coli NagE protein was also identified in the M. degradans strain 2-40 genome that was expressed from an apparent nagF-hexA-nagE-ORF4 operon located on the reverse strand directly downstream from the nagCBA operon. NagE2-40 is postulated to contain an N-terminal secretion signal and 11 transmembrane domains. It also contains a 399-aa domain with similarity to a sugar transporter. Overall, it was similar to a deduced Glu/Gal transporter from X. fastidiosa strain 9a5c (43% I, 61% S) (41). The sequence similarity of NagF to the GlcNAc kinase of Brucella melitensis (32% I, 48% S) (10) predicts that nagF encodes a GlcNAc kinase. The product of ORF4 has no predicted function at this time. The orientation of these operons differs from that in E. coli K-12. In E. coli K-12, the corresponding operons are divergently expressed from a shared promoter region. In strain 2-40, the GlcNAc utilization operons are also clustered but are convergently expressed from separate promoters.
There is some evidence that M. degradans strain 2-40 utilizes a PTS to transport GlcNAc to the cytoplasm. Elements of a PTS, such as putative genes for PTS enzymes I and HPr, were identified in the genomic sequence of strain 2-40. It has recently been suggested that sensitivity to streptozotocin may be an indicator of PTS activity (37). When streptozotocin is taken up and phosphorylated by a cell, it produces the strong alkylating agent diazomethane that interferes with DNA synthesis (37). Strain 2-40 was sensitive to streptozotocin (20 μg/ml) when grown in 2216 marine medium and minimal medium containing GlcNAc as the sole carbon source, suggesting that 2-40 may express a functional PTS. However, PTS are rare among strict aerobes (37). Because streptozotocin is a GlcNAc derivative, it may be that 2-40 imports streptozotocin (and therefore GlcNAc) via a non-PTS transporter (NagE) and then phosphorylates it in the cytoplasm by the activity of NagF. Such a pathway could explain non-PTS-dependent sensitivity to streptozotocin.
Other proteins associated with chitin degradation.
Additional proteins that may be involved in chitin degradation were identified in the M. degradans strain 2-40 genome. A putative chitin-binding protein, CbpA, consisted of two carbohydrate-binding domains but had no apparent catalytic domain (Fig. 2 and Table 2). The first chitin-binding domain consisted of 220 aa and was most similar to the chitin-binding module of P. aeruginosa CbpD (32% I, 45% S) (43). The second was a 95-aa type 2 carbohydrate-binding module with similarity to the CBM2 of a rhamnogalacturonan lyase from Cellvibrio japonicus (formerly Pseudomonas cellulosa) (64% I, 74% S) (8). Similar chitin-binding proteins have been reported in a number of marine microorganisms, though their role in chitin degradation is poorly understood (12, 44, 47). It has been hypothesized that chitin-binding proteins may keep a bacterium in close proximity to the chitin polymer to facilitate efficient degradation, though there is no direct evidence that these proteins bind both the cell and chitin simultaneously. A Glu-Pro-rich domain consisting of (Glu-Pro)7 was located between the carbohydrate-binding modules of CbpA.
Two genes encoding components of an apparent diacetylchitobiose transport system were present in the M. degradans strain 2-40 genome. The first ORF had some similarity to a diacetylchitobiose phosphorylase from Vibrio furnissii (33% I, 49% S) (24), and the second ORF, 111 bases downstream from the first, encoded a predicted protein with 11 putative transmembrane domains and a putative N-terminal secretion signal and overall was most similar to a Na+/galactoside permease from Salmonella enterica serovar Typhimurium LT2 (36% I, 55% S) (29).
M. degradans strain 2-40 also encodes several putative polysaccharide deacetylases and secreted proteases. Fungi often deacetylate chitin to form chitosan, which is subsequently degraded. Deacetylases, however, are also utilized in xylan degradation; therefore, it was not possible to determine which, if any, of these proteins are components of the chitinolytic system of strain 2-40 at this time. In some bacterial species, specific proteases are secreted in the presence of chitin, likely because chitin is often complexed with large amounts of protein (30). In the case of strain 2-40, none of the putative secreted proteases has an obvious chitin-binding domain, as was observed in Alteromonas sp. strain O-7 (46), though this does not preclude them from activity toward chitin.
Genetic organization of the M. degradans chitinase system.
It was expected that the chitinase-related genes of M. degradans strain 2-40 would be found in operons or gene clusters. Several marine microorganisms, such as Alteromonas and Pseudoalteromonas, contain clustered chitinase genes that express, for example, a chitin-binding protein and several chitin depolymerase genes (44, 47). Interestingly, none of the chitinase-related genes of strain 2-40, with the exception of the NAG operons, appeared to be part of an operon or chitinase cluster. Instead, the genes for major chitin-processing enzymes (e.g., chiA, chiB, chiC, hexB, cdxA, and cbpA) were dispersed throughout the genome of strain 2-40 and were separated by a minimum of 50 kb. To our knowledge, the only other organisms with such dispersed chitin-degrading systems are Streptomyces coelicolor A3(2) (2, 6) and V. cholerae (18). There was no significant difference between the GC content of each of these genes and the overall GC content of the genome (44.6%) (Table 2). The regions upstream of each gene were analyzed for conserved regulatory sequences, but no common sequences were apparent.
Concluding remarks.
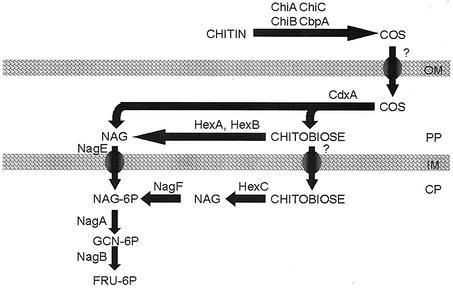
On the basis of biochemical data and sequence analysis, a model for chitin degradation by M. degradans strain 2-40 is proposed (Fig. 3). Consistent with its ability to utilize chitin as the sole carbon source, strain 2-40 encodes the proteins likely to mediate the complete depolymerization of chitin, the transport and depolymerization of chitooligosaccharides, and the transport, modification, and metabolism of GlcNAc. Several extracellular chitin depolymerases were identified, and their activities were demonstrated. A postulated extracellular chitin-binding protein that could function in the in situ degradation of chitin was detected. In V. furnissii, chitooligosaccharides are produced from the action of chitin depolymerases and taken up via an outer membrane chitoporin; attempts to detect a homolog to a chitoporin in the genome of strain 2-40 were unsuccessful. Once transported into the periplasm, chitooligosaccharides appear to be converted into chitobiose and GlcNAc via the activity of CdxA (a chitodextrinase) and HexA and HexB (N-acetylglucosaminidases). Transport of periplasmic GlcNAc into the cytoplasm probably involves the 2-40 NagE homolog, as observed in E. coli (35). Due to the apparent cytoplasmic localization of HexC (also an N-acetylglucosaminidase), a transport system for chitobiose may also be produced by strain 2-40. Cytoplasmic GlcNAc appears to be phosphorylated, either during transport or by the activity of the apparent GlcNAc kinase, NagF. NagA, which has been demonstrated to function as a GlcNAc deacetylase in E. coli, likely converts GlcNAc-6-P to glucosamine-6-P. NagB is hypothesized to then isomerize glucosamine-6-P to form fructose-6-P, which subsequently enters glycolysis, as in E. coli (36).
FIG. 3.
Predicted pathways for degradation, transport, and metabolism of chitin by M. degradans strain 2-40. Chitin in the environment is depolymerized to chitooligosaccharides (COS) by secreted ChiA, ChiB, and ChiC. Secreted CbpA may act as an anchor between the polymer and the bacterium. Once degraded, chitooligosaccharides are transported to the periplasm and degraded to GlcNAc and chitobiose by CdxA. Chitobiose is further degraded to GlcNAc by HexA and HexB. GlcNAc and some chitobiose are likely to be transported to the cytoplasm, perhaps via specific PTS transporters. Once in the cytoplasm, chitobiose may serve as a signaling molecule or can be degraded by HexC to form GlcNAc. GlcNAc may be shunted to cell wall biogenesis or modified and isomerized to form fructose-6-P, which is then metabolized via the glycolytic pathway. Abbreviations: OM, outer membrane; PP, periplasm; IM, inner membrane; CP, cytoplasm.
While the chitinolytic system of M. degradans strain 2-40 has some similarity to other microbial chitinase systems, the 2-40 system is novel in several ways. The major extracellular and periplasmic components of the system have domain arrangements distinct from other enzymes. For example, while GH18-, Cbd3-, and PKD-like domains were present in the chitin depolymerases of strain 2-40 (as in other chitinases), the positions of these domains were distinct from all published homologs. This indicates that these enzymes are not orthologs of known microbial chitinases. Polyserine linkers (a motif that has not been reported for other chitinases) were identified in ChiA and ChiB. ChiB represents one of the most intriguing chitinases heretofore reported. At 1,271 aa, it is the largest bacterial chitinase known. In addition to three polyserine domains, ChiB contains a Glu-rich domain. ChiB also contains two complete GH18 catalytic domains, another feature not previously reported in a microbial chitinase. None of the genes encoding major chitin-processing enzymes in strain 2-40 (with the exception of hexA) were clustered with other chitinase-related genes, leading to questions as to how these genes are regulated and how they were acquired.
The genomic sequence of M. degradans strain 2-40 not only provides the opportunity to identify components of the chitinolytic system based on sequence similarity for subsequent biochemical analysis but also allows investigation into what other novel proteins and regulatory factors may be involved in chitin degradation in the marine environment. Experiments are under way to identify other components of the chitinase system and to evaluate the possible synergistic activity of other carbohydrate-metabolizing systems with the chitinolytic system of strain 2-40.
Acknowledgments
This work was supported in part by grants from the Maryland Sea Grant College (SA7528051E) and the National Science Foundation (DEB0109869).
We thank the United States Department of Energy Joint Genome Institute (USDOE/JGI) for sequencing the genome of strain 2-40, Oak Ridge National Laboratories (ONRL) for annotating the draft genomic sequence of 2-40, and J. Bretz for valuable comments on the manuscript. We are grateful to J. Plumbridge (IBPC, Paris, France) for generously providing E. coli K-12 strain IBPC531.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrykovich, G., and I. Marx. 1988. Isolation of a new polysaccharide digesting bacterium from a salt marsh. Appl. Microbiol. Biotechnol. 54:1061-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannai, H., Y. Tamada, O. Maruyama, K. Nakai, and S. Miyano. 2002. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18:298-305. [DOI] [PubMed] [Google Scholar]
- 4.Bassler, B., P. Gibbons, C. Yu, and S. Roseman. 1991. Chemotaxis to chitin oligosaccharides by Vibrio furnissii. J. Biol. Chem. 266:24268-24275. [PubMed] [Google Scholar]
- 5.Bateman, A. 1999. The SIS domain: a phosphosugar-binding domain. Trends Biochem. Sci. 24:94-95. [DOI] [PubMed] [Google Scholar]
- 6.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 7.Black, G. W., J. E. Rixon, J. H. Clarke, G. P. Hazlewood, M. K. Theodorou, P. Morris, and H. J. Gilbert. 1996. Evidence that linker sequences and cellulose-binding domains enhance the activity of hemicellulases against complex substrates. Biochem. J. 319:515-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, I. E., M. H. Mallen, S. J. Charnock, G. J. Davies, and G. W. Black. 2001. Pectate lyase 10A from Pseudomonas cellulosa is a modular enzyme containing a family 2a carbohydrate-binding module. Biochem. J. 355:155-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connell, T., D. Metzger, J. Lynch, and J. Folster. 1998. Endochitinase is transported to the extracellular milieu by the eps-encoded general secretory pathway of Vibrio cholerae. J. Bacteriol. 180:5591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, et al. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ensor, L., S. Stosz, and R. Weiner. 1999. Expression of multiple complex polysaccharide-degrading enzyme systems by marine bacterium strain 2-40. J. Ind. Microbiol. Biotechnol. 21:123-126. [DOI] [PubMed] [Google Scholar]
- 12.Folders, J., J. Algra, M. Roelofs, L. Loon, J. Tommassen, and W. Bitter. 2001. Characterization of Pseudomonas aeruginosa chitinase, a gradually secreted protein. J. Bacteriol. 183:7044-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilkes, N. R., R. A. Warren, R. C. Miller, Jr., and D. G. Kilburn. 1988. Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J. Biol. Chem. 263:10401-10407. [PubMed] [Google Scholar]
- 14.Gonzalez, J., and R. Weiner. 2000. Phylogenetic characterization of a marine bacterium strain 2-40, a degrader of complex polysaccharides. Int. J. Syst. Bacteriol. 8:831-834. [DOI] [PubMed] [Google Scholar]
- 15.Gooday, G. 1994. Physiology of microbial degradation of chitin and chitosan, p. 279-312. In C. Ratledge (ed.), Biochemistry of microbial degradation. Kluwer, Dordrecht, The Netherlands.
- 16.Hall, J., G. W. Black, L. M. Ferreira, S. J. Millward-Sadler, B. R. Ali, G. P. Hazlewood, and H. J. Gilbert. 1995. The non-catalytic cellulose-binding domain of a novel cellulase from Pseudomonas fluorescens subsp. cellulosa is important for the efficient hydrolysis of Avicel. Biochem. J. 309:749-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall, J., G. P. Hazlewood, N. S. Huskisson, A. J. Durrant, and H. J. Gilbert. 1989. Conserved serine-rich sequences in xylanase and cellulase from Pseudomonas fluorescens subspecies cellulosa: internal signal sequence and unusual protein processing. Mol. Microbiol. 3:1211-1219. [DOI] [PubMed] [Google Scholar]
- 18.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, et al. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henikoff, S., G. Haughn, J. Calvo, and J. Wallace. 1988. A large family of bacterial activator proteins. Proc. Natl. Acad. Sci. USA 85:6602-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 21.Keyhani, N., X. Li, and S. Roseman. 2000. Identification and molecular cloning of a chitoporin. J. Biol. Chem. 275:33068-33076. [DOI] [PubMed] [Google Scholar]
- 22.Keyhani, N., and S. Roseman. 1996. The chitin catabolic cascade in the marine bacterium Vibrio furnissii. Molecular cloning, isolation, and characterization of a periplasmic chitodextrinase. J. Biol. Chem. 271:33414-33424. [DOI] [PubMed] [Google Scholar]
- 23.Keyhani, N., and S. Roseman. 1996. The chitin catabolic cascade in the marine bacterium Vibrio furnissii. Molecular cloning, isolation, and characterization of a periplasmic beta-N-acetylglucosaminidase. J. Biol. Chem. 271:33425-33432. [DOI] [PubMed] [Google Scholar]
- 24.Keyhani, N., L. Wang, Y. Lee, and S. Roseman. 1996. Characterization of an N,N′-diacetylchitobiose transport system. J. Biol. Chem. 271:33409-33413. [DOI] [PubMed] [Google Scholar]
- 25.Keyhani, N., L. Wang, Y. Lee, and S. Roseman. 2000. The chitin disaccharide, N,N′-diacetylchitobiose, is catabolized by E. coli and is transported/phosphorylated by the phosphoenolpyruvate:glycose phosphotransferase system. J. Biol. Chem. 275:33084-33090. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Letunic, I., L. Goodstadt, N. J. Dickens, T. Doerks, J. Schultz, R. Mott, F. Ciccarelli, R. R. Copley, C. P. Ponting, and P. Bork. 2002. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30:242-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuo, Y., M. Kurita, J. K. Park, K. Tanaka, T. Nakagawa, M. Kawamukai, and H. Matsuda. 1999. Purification, characterization and gene analysis of N-acetylglucosaminidase from Enterobacter sp. G-1. Biosci. Biotechnol. Biochem. 63:1261-1268. [DOI] [PubMed] [Google Scholar]
- 29.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, et al. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 30.Muzzarelli, R. 1999. Native, industrial, and fossil chitins, p. 1-5. In R. A. Muzzarelli and P. Jooaes (ed.), Chitin and chitinases. Birkhauser, Basel, Switzerland. [DOI] [PubMed]
- 31.Nielsen, H., J. Englebrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 32.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, et al. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peri, K. G., H. Goldie, and E. B. Waygood. 1990. Cloning and characterization of the N-acetylglucosamine operon of Escherichia coli. Biochem. Cell Biol. 68:123-137. [DOI] [PubMed] [Google Scholar]
- 34.Plumbridge, J. 2001. Regulation of PTS gene expression by the homologous transcriptional regulators, Mlc and NagC, in Escherichia coli (or how two similar repressors can behave differently). J. Mol. Microbiol. Biotechnol. 3:371-380. [PubMed] [Google Scholar]
- 35.Plumbridge, J., and A. Kolb. 1991. CAP and Nag repressor binding to the regulatory regions of the nagE-B and manX genes of Escherichia coli. J. Mol. Biol. 217:661-679. [DOI] [PubMed] [Google Scholar]
- 36.Plumbridge, J. A. 1989. Sequence of the nagBACD operon in Escherichia coli K12 and pattern of transcription within the nag regulon. Mol. Microbiol. 3:505-515. [DOI] [PubMed] [Google Scholar]
- 37.Riemann, L., and F. Azam. 2002. Widespread N-acetyl-d-glucosamine uptake among pelagic marine bacteria and its ecological implications. Appl. Environ. Microbiol. 68:5554-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Schultz, J., R. R. Copley, T. Doerks, C. P. Ponting, and P. Bork. 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28:231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen, H., M. Schmuck, I. Pilz, N. R. Gilkes, D. G. Kilburn, R. C. Miller, Jr., and R. A. Warren. 1991. Deletion of the linker connecting the catalytic and cellulose-binding domains of endoglucanase A (CenA) of Cellulomonas fimi alters its conformation and catalytic activity. J. Biol. Chem. 266:11335-11340. [PubMed] [Google Scholar]
- 41.Simpson, A. J., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, R. Alvarenga, L. M. Alves, J. E. Araya, G. S. Baia, C. S. Baptista, M. H. Barros, E. D. Bonaccorsi, S. Bordin, J. M. Bove, et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 42.Somerville, C. C., and R. R. Colwell. 1993. Sequence analysis of the beta-N-acetylhexosaminidase gene of Vibrio vulnificus: evidence for a common evolutionary origin of hexosaminidases. Proc. Natl. Acad. Sci. USA 90:6751-6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 44.Techkarnjanaruk, S., and A. Goodman. 1999. Multiple genes involved in chitin degradation from the marine bacterium Pseudoaltermonas sp. strain S91. Microbiology 145:925-934. [DOI] [PubMed] [Google Scholar]
- 45.Trudel, J., and A. Asselin. 1990. Detection of chitin deacetylase activity after polyacrylamide gel electrophoresis. Anal. Biochem. 189:249-253. [DOI] [PubMed] [Google Scholar]
- 46.Tsujibo, H., H. Orikoshi, K. Shiotani, M. Hayashi, J. Umeda, K. Miyamoto, C. Imada, Y. Okami, and Y. Inamori. 1998. Characterization of chitinase C from a marine bacterium, Alteromonas sp. strain O-7, and its corresponding gene and domain structure. Appl. Environ. Microbiol. 64:472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsujibo, H., H. Orikoshi, N. Baba, M. Miyahara, K. Miyamoto, M. Yasuda, and Y. Inamori. 2002. Identification and characterization of the gene cluster involved in chitin degradation in a marine bacterium, Alteromonas sp. strain O-7. Appl. Environ. Microbiol. 68:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vetrivel, K. S., S. K. Pandian, U. Chaudhary, and K. Dharmalingam. 2001. Purification, cloning, and DNA sequence analysis of a chitinase from an overproducing mutant of Streptomyces peucetius defective in daunorubicin biosynthesis. Can. J. Microbiol. 47:179-187. [PubMed] [Google Scholar]
- 49.Wang, L., N. Keyhani, S. Roseman, and Y. Lee. 1997. 4-Methylumbelliferyl glycosides of N-acetyl 4-thiochito-oligosaccharides as fluorogenic substrates for chitodextrinase from Vibrio furnissii. Glycobiology 7:855-860. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe, T., K. Kimura, T. Sumiya, N. Nikaidou, K. Suzuki, M. Suzuki, M. Taiyoji, S. Ferrer, and M. Regue. 1997. Genetic analysis of the chitinase system of Serratia marcescens 2170. J. Bacteriol. 179:7111-7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiner, R., L. Taylor, N. Ekborg, and L. Whitehead. 2001. Degradosomes: potential importance in the ocean's carbon cycle and in aquaculture and algal culture, p. 259-268. In N. Seaxena (ed.), Recent advances in marine science and technology, 2000. Pacon International, New York, N.Y.
- 52.Zobell, C., and S. Rittenberg. 1937. The occurrence and characteristics of chitinoclastic bacteria in the sea. J. Bacteriol. 35:275-287. [DOI] [PMC free article] [PubMed] [Google Scholar]