Abstract
In bacteria, the regulation of gene expression in response to changes in cell density is called quorum sensing. Quorum-sensing bacteria produce, release, and respond to hormone-like molecules (autoinducers) that accumulate in the external environment as the cell population grows. In the marine bacterium Vibrio harveyi two parallel quorum-sensing systems exist, and each is composed of a sensor–autoinducer pair. V. harveyi reporter strains capable of detecting only autoinducer 1 (AI-1) or autoinducer 2 (AI-2) have been constructed and used to show that many species of bacteria, including Escherichia coli MG1655, E. coli O157:H7, Salmonella typhimurium 14028, and S. typhimurium LT2 produce autoinducers similar or identical to the V. harveyi system 2 autoinducer AI-2. However, the domesticated laboratory strain E. coli DH5α does not produce this signal molecule. Here we report the identification and analysis of the gene responsible for AI-2 production in V. harveyi, S. typhimurium, and E. coli. The genes, which we have named luxSV.h., luxSS.t., and luxSE.c. respectively, are highly homologous to one another but not to any other identified gene. E. coli DH5α can be complemented to AI-2 production by the introduction of the luxS gene from V. harveyi or E. coli O157:H7. Analysis of the E. coli DH5α luxSE.c. gene shows that it contains a frameshift mutation resulting in premature truncation of the LuxSE.c. protein. Our results indicate that the luxS genes define a new family of autoinducer-production genes.
Many species of bacteria regulate gene expression in response to increasing cell population density, and, collectively, this phenomenon is termed quorum sensing (1). Quorum-sensing bacteria produce and release acyl-homoserine lactone-signaling molecules (called autoinducers) that accumulate in the environment as the cell density increases. When a threshold stimulatory concentration of autoinducer is achieved, a signal transduction cascade is initiated that ultimately is translated into a change in behavior of the organism (2–5). Vibrio harveyi, a free-living marine bacterium, possesses two quorum-sensing systems that function in parallel to control the density-dependent expression of bioluminescence (lux). Each of the V. harveyi density-sensing systems (signaling system 1 and signaling system 2) is composed of a sensor and a cognate autoinducer. Therefore, system 1 is composed of sensor 1 and it responds to autoinducer 1 (AI-1), and system 2 is composed of sensor 2 and this system detects autoinducer 2 (AI-2) (6). Sensory information from both systems is integrated via a shared regulatory protein to control the output, light emission (7–9). Genetic analysis of V. harveyi has shown that either system 1 or system 2 is sufficient for the density-dependent expression of luminescence.
The V. harveyi system 1 autoinducer (AI-1) has been purified and identified as hydroxybutanoyl-l-homoserine lactone (10), and its synthesis dependents on the genes luxL and luxM (11). Neither the structure nor the biosynthetic gene(s) for the system 2 autoinducer (AI-2) has been reported. The sensor proteins that detect AI-1 and AI-2 are members of the bacterial family of two-component adaptive regulatory proteins, and the mechanism of signal transduction is a phosphorylation/dephosphorylation cascade (11, 12). Reporter strains of V. harveyi have been constructed that respond specifically to only AI-1 or AI-2. These strains were used in the development of a bioassay capable of detecting autoinducers produced by other species of bacteria. It was observed that many species of bacteria produce an AI-2-like activity; however, only very rarely were species identified that produced an AI-1-like substance (13).
Using the V. harveyi bioassay, several strains of Escherichia coli and Salmonella typhimurium were shown to produce an AI-2-like activity. Analysis of autoinducer production in these enteric bacteria showed that the AI-2 activity is produced maximally in midexponential phase in the presence of certain preferred carbon sources. However, unlike other described quorum-sensing systems, in E. coli and S. typhimurium the AI-2 signal is degraded when the bacteria enter stationary phase (14). Several environmental cues were shown to influence the levels of autoinducer production and degradation in E. coli and S. typhimurium. Rapid logarithmic growth, preferred carbon sources, low pH, and/or high osmolarity all resulted in increased autoinducer production, whereas conditions of stationary phase, the lack of a preferred carbon source, neutral pH, and low osmolarity induced degradation of the AI-2 signal. Protein synthesis was required for both induction of signal production and signal degradation in E. coli and S. typhimurium (15). Interestingly, we showed that the domesticated laboratory strain E. coli DH5α is not capable of AI-2 production or degradation (14).
In the present manuscript we report the analysis of a gene responsible for AI-2 production in V. harveyi, E. coli, and S. typhimurium. The gene identified in all three species of bacteria is highly homologous, and we propose that these genes define a new family of proteins involved in autoinducer production. The genes, which we named luxSV.h., luxSE.c., and luxSS.t. have been identified in many species of bacteria by genome sequencing projects, but until now no function has been ascribed to this gene in any organism. The luxS genes do not bear homology to any other gene known to be involved in autoinducer production.
MATERIALS AND METHODS
Bacterial Strains, Media, and Recombinant DNA Techniques.
V. harveyi BB120 is the wild-type strain (13). S. typhimurium strain LT2 was obtained from K. Hughes (University of Washington), S. typhimurium 14028 is ATCC strain 14028 Organism: Salmonella choleraesuis. E. coli O157:H7 is a clinical isolate supplied by Paddy Gibb (University of Calgary). Luria–Bertani medium (LB) contained 10 g bacto tryptone (Difco), 5 g yeast extract (Difco), and 10 g NaCl per liter (16). The recipe for autoinducer bioassay (AB) medium has been reported (17). Where specified, glucose was added from a sterile 20% stock to a final concentration of 0.5%. Antibiotics were used at the following concentrations (mg/liter): ampicillin (Amp), 100; chloramphenicol (Cm), 10; gentamycin (Gn), 100; kanamycin (Kn), 100; and tetracycline (Tet), 10. DNA isolation, restriction analysis, and transformation of E. coli was performed as described by Sambrook et al. (16). Probes for Southern blot analysis were labeled by using the Multiprime DNA labeling system of Amersham. Sequencing was carried out by using an Applied Biosystems sequencing apparatus. The V. harveyi BB120 genomic library was constructed in the cosmid pLAFR2 as described (11). The method for Tn5 mutagenesis of cloned V. harveyi genes and the allelic replacement technique for inserting Tn5 mutated genes into the V. harveyi chromosome have been reported (11).
Autoinducer Assay.
The AI-2 bioassay using the V. harveyi reporter strain BB170 (sensor 1−, sensor 2+) has been reported (14, 15). Cell-free culture fluids from V. harveyi, E. coli, or S. typhimurium strains to be tested for AI-2 activity were prepared as described (14, 15) and assayed at 10% (vol/vol). AI-2 activity is reported as the fold-induction of the reporter strain over background or as the percentage of activity obtained from V. harveyi BB120 (wild type) cell-free culture fluid.
Mutagenesis and Analysis of the AI-2 Production Gene in S. typhimurium LT2.
MudJ insertion mutants of S. typhimurium LT2 were generated by using a phage P22 delivery system as described (18). After growth to midexponential phase in LB containing 0.5% glucose, the S. typhimurium insertion mutants were tested for AI-2 production using the V. harveyi BB170 bioassay. The site of the MudJ insertion that inactivated the AI-2 production function in S. typhimurium was identified by PCR amplification and sequencing of the chromosomal DNA at the insertion junction. A two-step amplification procedure was used (19). In the first PCR, the arbitrary primer 5′-GGCCACGCGTCGACTAGTCANNNNNNNNNNACGCCC-3′ and the MudJ specific primer 5′-GCACTACAGGCTTGCAAGCCC-3′ were used. Next, 1 μl of this PCR was used as the template in a second PCR amplification employing a second arbitrary primer (5′-GGCCACGCGTCGACTAGTCA-3′) and another MudJ specific primer (5′-TCTAATCCCATCAGATCCCG-3′). The PCR product from the second reaction was purified and sequenced.
Cloning and Sequencing of the E. coli MG1655, E. coli O157:H7, and E. coli DH5α AI-2 Production Genes.
The DNA sequence obtained from the S. typhimurium LT2 MudJ screen was used to search the E. coli MG1655 genome sequence to identify the corresponding E. coli region (20). The gene identified from the E. coli genome project had the designation ygaG. Primers that flanked the ygaG gene and incorporated restriction sites were designed and used to amplify the E. coli MG1655, E. coli O157:H7, and E. coli DH5α ygaG genes. The primers used were 5′-GTGAAGCTTGTTTACTGACTAGATC-3′ and 5′-GTGTCTAGAAAAACACGCCTGACAG-3′. The PCR products were purified, digested, and cloned into pUC19. In each case, the PCR products from three independent reactions were cloned and sequenced.
RESULTS
Identification and Cloning of the Gene Responsible for AI-2 Production in V. harveyi.
We recently have reported that unlike many other E. coli strains, E. coli strain DH5α does not produce an AI-2 signal molecule that can be detected by V. harveyi (14). We reasoned, therefore, that we could use E. coli DH5α as a mutant to clone the V. harveyi AI-2 production gene. A library of wild-type V. harveyi BB120 genomic DNA was transformed into E. coli strain DH5α, and the transformants were screened for AI-2 production in the V. harveyi BB170 AI-2 detection bioassay. The library consisted of 2,500 clones, each containing roughly 25 kb of V. harveyi genomic DNA. Five DH5α clones were identified that resulted in upwards of 300-fold stimulation of the reporter strain in the bioassay.
The recombinant cosmid DNA from the five AI-2-producing E. coli DH5α clones was analyzed by restriction analysis and Southern blotting. All five of the cosmids contained an overlapping subset of identical V. harveyi genomic restriction fragments, indicating that we had cloned the same locus several times. One cosmid, called pBB2929, was selected for further analysis. Random mutagenesis using transposon Tn5 was carried out on cosmid pBB2929, and pools of cosmids harboring Tn5 insertions subsequently were transformed into E. coli DH5α. We tested 962 individual E. coli DH5α/pBB2929∷Tn5 strains for the loss of the ability to produce AI-2. Four E. coli DH5α strains harboring Tn5 insertions in pBB2929 were identified that failed to produce AI-2. We mapped the locations of these Tn5 insertions in pBB2929 and found that all four transposon insertions resided in the same 2.6-kb HindIII V. harveyi genomic DNA fragment (Fig. 1A).
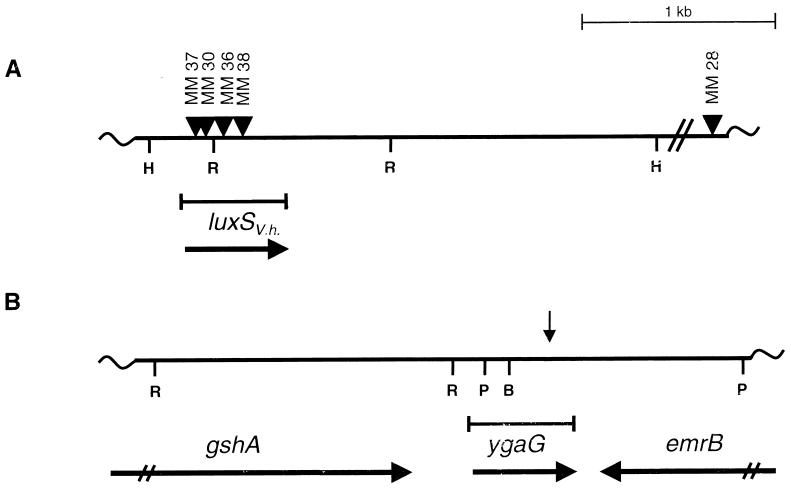
Figure 1.
The luxS and ygaG genes from V. harveyi and E. coli MG1655. (A) A restriction map of the V. harveyi luxSV.h. chromosomal region that was defined by Tn5 insertion. The sites of Tn5 insertions that disrupted the AI-2-production function and one control Tn5 insertion outside of the luxSV.h. locus are shown (triangles). (B) The ygaG region in the E. coli MG1655 chromosome. This ORF is flanked by the emrB and gshA genes. The direction of transcription of each gene is indicated by horizontal arrows. The corresponding position of the MudJ insertion that eliminated AI-2 production in S. typhimurium LT2 is shown by a vertical arrow. H, R, P, and B denote HindIII, EcoRI, PstI, and BamHI restriction sites, respectively.
Cosmid pBB2929 was digested with HindIII, and the eight resulting fragments were subcloned in both orientations into pALTER (Promega). The pALTER subclones were transformed into E. coli DH5α and subsequently tested for AI-2 production. The only strains capable of producing AI-2 contained the 2.6-kb HindIII fragment identified in the Tn5 mutagenesis. This fragment was sequenced and only one ORF could be identified. Its location corresponded to the map positions of the four Tn5 insertions that eliminated AI-2 production. We named the ORF luxSV.h. (Fig. 1A).
Mutagenesis of luxSV.h. in V. harveyi.
We analyzed the effects of luxSV.h. null mutations on AI-2 production in V. harveyi. The four Tn5 insertions that mapped to the luxSV.h. gene and the control Tn5 insertion adjacent to the luxSV.h. locus were transferred to the corresponding locations in the V. harveyi BB120 chromosome to make strains MM37, MM30, MM36, MM38, and MM28, respectively (Fig. 1A). Southern blotting was used to confirm the correct placement of all five Tn5 insertions in the V. harveyi chromosome. The four V. harveyi luxSV.h.∷Tn5 insertion strains were tested for the ability to produce AI-2, and all four strains gave identical results.
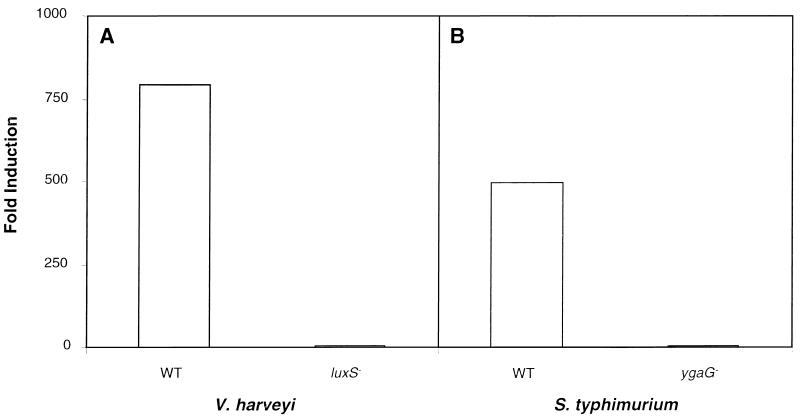
In Fig. 2A, we show the AI-2 production phenotypes of the wild-type control Tn5 insertion strain MM28 and one representative luxSV.h.∷Tn5 insertion strain, MM30. V. harveyi MM28 and MM30 were grown to high cell density, after which cell-free culture fluids were prepared. The culture fluids were assayed for AI-2 activity by the ability to induce luminescence in the AI-2 detector strain BB170. Fig. 2A shows that addition of culture fluids from the control Tn5 insertion strain MM28 induced luminescence in the reporter 780-fold, whereas culture fluid from the luxSV.h.∷Tn5 insertion strain MM30 did not induce the expression of luminescence in the reporter. Therefore, a null mutation in luxSV.h. in V. harveyi eliminates AI-2 production.
Figure 2.
Autoinducer-production phenotypes of V. harveyi and S. typhimurium strains. Cell-free culture fluids from V. harveyi and S. typhimurium strains were prepared and tested for AI-2 activity in the V. harveyi BB170 bioassay. (A) AI-2 production phenotypes of the wild-type V. harveyi strain MM28, which contains a Tn5 insertion outside of luxSV.h. (denoted WT) and the luxSV.h.∷Tn5 mutant strain MM30 (denoted luxS−). (B) AI-2-production phenotypes of wild-type S. typhimurium LT2 (denoted WT) and the ygaG∷MudJ insertion mutant strain CS132 (denoted ygaG−). Activity is reported as fold-induction of luminescence expression of the V. harveyi BB170 reporter strain over that when sterile medium was added.
Identification and Analysis of S. typhimurium Autoinducer-Production Mutants.
To identify the gene responsible for AI-2 production in S. typhimurium, we randomly mutagenized S. typhimurium LT2 using the MudJ transposon (18). Ten thousand S. typhimurium LT2 insertion mutants were assayed for AI-2 production in the V. harveyi BB170 bioassay. One S. typhimurium MudJ insertion mutant (strain CS132) was identified that lacked detectable AI-2 in culture fluids at midexponential phase.
Fig. 2B shows the AI-2 production phenotypes of S. typhimurium strain LT2 and the corresponding MudJ insertion strain CS132. The strains were grown to midexponential phase in LB containing glucose, and cell-free culture fluids were prepared and assayed for AI-2. S. typhimurium LT2 culture fluids induced the reporter strain 500-fold, whereas culture fluids from strain CS132 contained no AI-2 activity. Furthermore, strain CS132 did not produce AI-2 under any of the growth conditions that we have reported previously that induce AI-2 production in S. typhimurium (not shown).
The site of the MudJ insertion in S. typhimurium CS132 was determined by PCR amplification followed by sequencing of the 110 bp of chromosomal DNA adjacent to the transposon. This sequence was used to search the database for DNA homologies. The sequence matched a site (89-/105-bp identity) in the E. coli MG1655 genome that corresponded to an ORF of unknown function denoted ygaG (20). In the chromosome, the E. coli ygaG gene is flanked by the gshA and emrB genes (Fig. 1B). The ygaG gene is transcribed from its own promoter, which is located immediately upstream of the gene, indicating that it is not in an operon with gshA (A. Beeston, M.G.S., and B.L.B., unpublished results). The emrB gene is transcribed in the opposite direction. We PCR-amplified the ygaG region from the chromosomes of E. coli O157:H7 and E. coli MG1655, and the two E. coli ygaG genes were cloned into pUC19.
Complementation of S. typhimurium and E. coli AI-2− Mutants.
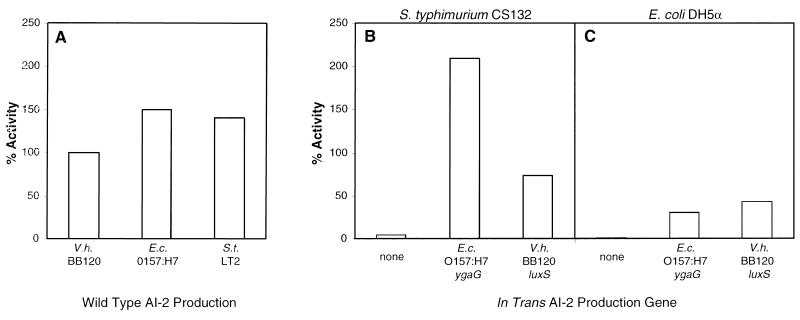
We tested whether the E. coli O157:H7 ygaG gene and the V. harveyi luxSV.h. gene could restore AI-2 production in the AI-2− strains S. typhimurium CS132 and E. coli DH5α. In Fig. 3A, we show the AI-2 activity produced by wild-type V. harveyi BB120, E. coli O157:H7, and S. typhimurium LT2. In this figure, the level of AI-2 activity present in V. harveyi BB120 cell-free culture fluids was normalized to 100%, and the activities in cell-free culture fluids from E. coli and S. typhimurium were compared with that. In this experiment, E. coli O157:H7 produced 1.5 times and S. typhimurium LT2 produced 1.4 times more AI-2 activity than V. harveyi BB120 (i.e., 150% and 141%, respectively).
Figure 3.
Complementation of AI-2 production in S. typhimurium CS132 and E. coli DH5α. Cell-free culture fluids from E. coli and S. typhimurium strains were tested for AI-2 activity in the bioassay. The activity present in these fluids was compared with that produced by wild-type V. harveyi BB120. The level of BB120 activity was normalized to 100%. (A) AI-2 activity in cell-free fluids from wild-type V. harveyi BB120, E. coli O157:H7, and S. typhimurium LT2. (B) Complementation of S. typhimurium CS132 (ygaG∷MudJ). (C) Complementation of E. coli DH5α. In B and C, the in trans AI-2 production genes are the following: vector control (“none”), E. coli O157:H7 ygaG, and V. harveyi BB120 luxSV.h.. E.c., E. coli; V.h., V. harveyi.
Fig. 3 B and C shows the AI-2 complementation results for S. typhimurium CS132 and E. coli DH5α. Fig. 3B demonstrates that introduction of the E. coli O157:H7 ygaG gene into S. typhimurium CS132 restored AI-2 production beyond the level of production of wild-type S. typhimurium (i.e., 209% activity). Comparison of the data in Fig. 3 A and B shows that the E. coli ygaG gene in S. typhimurium resulted in AI-2 production, exceeding that produced in vivo by E. coli O157:H7. Introduction of the V. harveyi luxSV.h. gene into S. typhimurium resulted in AI-2 production at slightly less than the level produced by wild-type V. harveyi BB120 (i.e., 73% of the level of V. harveyi BB120). Fig. 3C shows that introduction of the cloned E. coli O157:H7 and the V. harveyi BB120 AI-2 production genes in E. coli DH5α resulted in AI-2 production. However, expression of E. coli O157:H7 ygaG and V. harveyi BB120 luxSV.h. in E. coli DH5α resulted in only 31% and 43% of the V. harveyi BB120 AI-2 activity, respectively. Fig. 3 B and C shows that the control vectors produced no activity in the complementation experiments.
Analysis of the AI-2 Production Genes from V. harveyi, E. coli, and S. typhimurium.
We sequenced the AI-2 production gene luxSV.h. from V. harveyi BB120 and the ygaG loci from E. coli O157:H7, E. coli MG1655, and E. coli DH5α. The translated protein sequences encoded by the ygaG ORFs are shown in Fig. 4, and they are aligned with the translated LuxS protein sequence from V. harveyi. The underlined amino acids indicate the residues in the E. coli proteins that differ from the V. harveyi LuxS protein. The ygaG loci from E. coli encode proteins that are highly homologous to one another and also to LuxS from V. harveyi. The E. coli MG1655 and the E. coli O157:H7 YgaG proteins are 77% and 76% identical to LuxS from V. harveyi BB120. The DNA sequence we determined for ygaG from E. coli O157:H7 differs at five sites from the reported (and our) sequence for the E. coli MG1655 ygaG gene. Four of the changes are silent; the fifth results in a conservative Ala-to-Val alteration at amino acid residue 103 in the E. coli O157:H7 protein.
Figure 4.
Alignment of LuxS and YgaG protein sequences. The translated protein sequences for the AI-2 production family of proteins are shown. We determined the sequences for the luxSV.h. gene from V. harveyi BB120 and the ygaG genes from E. coli MG1655, E. coli O157:H7, and E. coli DH5α. The S. typhimurium LT2 ygaG partial sequence came from the S. typhimurium database. Amino acid residues that are not identical to the LuxSV.h. protein are underlined. The site of the frameshift mutation in the E. coli DH5α DNA sequence is denoted by ∗. The 20 altered amino acid residues that are translated after the frameshift are enclosed by the box.
Identification of the ygaG locus in E. coli MG1655 and E. coli O157:H7 allowed us to investigate the AI-2 production defect in E. coli DH5α. E. coli DH5α possesses the ygaG gene because we could PCR-amplify this region from the chromosome using the same primers we employed to amplify it from E. coli MG1655 and E. coli O157:H7. Examination of the E. coli DH5α ygaG promoter showed that it is identical to that of E. coli MG1655, indicating that the AI-2 defect in E. coli DH5α is not simply a result of decreased transcription of ygaG (not shown). However, sequence analysis of the E. coli DH5α ygaG coding region showed that a 1-bp G-C deletion and a T-to-A transversion exist at base pairs 222 and 224, respectively. The frameshift mutation resulting from the G/C deletion causes premature truncation of the E. coli DH5α protein. Fig. 4 shows that the truncated E. coli DH5α protein is 111 aa, whereas the E. coli MG1655 and E. coli O157:H7 proteins are 171 residues. Twenty altered amino acids are translated after the frameshift and before termination of the protein. Our complementation results (Fig. 3) demonstrate that the AI-2 production defect in E. coli DH5α is recessive to in trans expression of ygaG, which is consistent with the defect being a result of a null mutation caused by the frameshift in the E. coli DH5α ygaG gene.
We searched the S. typhimurium database by using the sequence we obtained adjacent to the MudJ that inactivated the AI-2-production function in S. typhimurium CS132. A perfect match (110/110 bp) was identified to fragment B_TR7095.85-T7 in the S. typhimurium LT2 genome sequencing database (Genome Sequencing Center, Washington University, St. Louis). However, the S. typhimurium LT2 database ygaG sequence is incomplete (Fig. 4). The translated sequence matches the E. coli and V. harveyi sequences beginning at amino acid residue 8. The translated sequence shows that the S. typhimurium protein is 75% identical to LuxS of V. harveyi. To align the S. typhimurium sequence with the V. harveyi LuxS protein, we corrected three apparent frameshift errors in the database sequence. Considering that only crude, unannotated sequence data are currently available for S. typhimurium, we predict that the S. typhimurium protein contains seven more amino acids and that the frameshift mutations are sequencing errors. We were unsuccessful at PCR-amplifying either the S. typhimurium 14028 or the S. typhimurium LT2 ygaG gene using the primers designed for E. coli, so we do not yet have the complete sequence of the S. typhimurium gene.
DISCUSSION
Our results indicate that the genes identified and analyzed in this report encode a novel family of proteins responsible for autoinducer production. We have designated the members of this family of autoinducer-production genes as luxSE.c., luxSS.t., and luxSV.h. for E. coli, S. typhimurium, and V. harveyi, respectively.
Mutagenesis of luxS in V. harveyi, S. typhimurium, and E. coli eliminates AI-2 production in all three species of bacteria (Fig. 2 and DH5α results). S. typhimurium could be complemented to full AI-2 production by the introduction of either the E. coli O157:H7 luxSE.c. gene or the V. harveyi BB120 luxSV.h. gene (Fig. 3). These results indicate that both the E. coli and V. harveyi LuxS proteins can function with S. typhimurium cellular components to produce AI-2. E. coli DH5α was only partially complemented to AI-2 production by the introduction of either the E. coli O157:H7 luxSE.c. or the V. harveyi BB120 luxSV.h. gene (Fig. 3). Because in trans expression of luxS genes in E. coli DH5α did not completely restore autoinducer production, we hypothesize that other biochemical or physiological factors may contribute to signal production. We already know that the regulation of AI-2 production differs between pathogenic and nonpathogenic strains. For example, E. coli O157:H7 strains produce AI-2 at 30° and 37°C with or without glucose whereas E. coli K-12 strains do not produce AI-2 in the absence of a preferred carbon source. Also, all of the E. coli O157 strains that we have tested produce greater AI-2 activity than nonpathogenic E. coli strains. Likewise, pathogenic S. typhimurium 14028 produces significantly more AI-2 than does S. typhimurium LT2 (K. Knapp, M.G.S., and B.L.B., unpublished results). We have begun investigating these differences in regulation of autoinducer production and whether they impact virulence in these bacteria.
Our sequence analysis shows that the LuxS proteins are highly similar (Fig. 4), and our complementation data suggest that the proteins can function across species (Fig. 3). These results indicate that the enzymatic activity carried out by the LuxS proteins and any other cellular machinery that contributes to AI-2 synthesis must be conserved. We could not identify any amino acid sequence motif in the LuxS proteins that is indicative of a particular function. Therefore, we hypothesize that the LuxS proteins catalyze one specific enzymatic step in biosynthesis of the autoinducer. The remainder of the steps involved in AI-2 biosynthesis could be a consequence of normal intermediary metabolic processes.
As we have reported, we do not yet know the structure of AI-2 from V. harveyi, E. coli, or S. typhimurium. Furthermore, the AI-2s cannot be purified by conventional techniques used for the isolation of acyl-homoserine lactone autoinducers (14, 15). These results have led us to suspect that the AI-2s are not acyl-homoserine lactones. The luxS genes identified here bear no homology to other genes known to be involved in production of HSL autoinducers [luxI-like (1), luxLM-ainS-like (11, 21)], further indicating that the AI-2 class of autoinducers is novel.
Database analysis of finished and unfinished bacterial genomes revealed that many other species of bacteria possess a gene homologous to luxS from V. harveyi, S. typhimurium, and E. coli. The species of bacteria identified and the percent homology/identity (H/I) to the LuxS protein of V. harveyi are as follows: Haemophilus influenzae (88/72), Helicobacter pylori (62/40), Bacillus subtilis (58/38), Borrelia burgdorferi (52/32), Neisseria meningitidis (89/80), Neisseria gonorrhoeae (89/80), Yersinia pestis (85/77), Campylobacter jejuni (85/74), Vibrio cholerae (95/90), Deinococcus radiodurans (65/45), Mycobacterium tuberculosis (59/41), Enterococcus faecalis (60/44), Streptococcus pneumoniae (57/36), and Streptococcus pyrogenes (57/36). In an earlier report (13), a few of these species were tested for AI-2 production. We showed that V. cholerae and Y. enterocolitica but not B. subtilis produced AI-2 activity. We predict that B. subtilis does produce AI-2 but we have not uncovered the environmental conditions that induce its synthesis. We further predict that all of the species identified in the database analysis produce an AI-2-like molecule.
We have suggested that the cues influencing autoinducer production and degradation in E. coli and S. typhimurium indicate that AI-2 is important for regulating the transition from a nonpathogenic existence outside a host to a pathogenic existence inside a host (14, 15). In support of this hypothesis, conditioned medium from E. coli O157:H7 that had been enriched for AI-2 has been shown to induce the expression of the Type III secretion system, a known virulence target. This result indicates further that AI-2 plays a role in pathogenicity (J. Kaper, personal communication). We have constructed luxS null mutants in S. typhimurium 14028 and E. coli O157:H7, and, using animal models, we currently are testing whether these mutant strains have virulence defects.
Acknowledgments
This work was supported by a National Science Foundation grant (B.B.) and grants from the Alberta Heritage Foundation for Medical Research and The Medical Research Council of Canada (M.S.).
ABBREVIATIONS
- AI-1 and AI-2
autoinducer 1 and 2, respectively
Note Added in Proof
Recently a full-length Salmonella luxS gene sequence became available from the Salmonella typhi sequencing group at The Sanger Centre. The sequence can be obtained from the following web site: http://www.sanger.ac.uk/Projects/S_typhi/blast_server.shtml. The S. typhi protein is the same length as the V. harveyi and E. coli LuxS proteins and it differs from the E. coli MG1655 LuxS protein at 11 positions.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF120098).
References
- 1.Fuqua W C, Winans S C, Greenberg E P. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engebrecht J, Nealson K, Silverman M. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 3.Bainton N J, Bycroft B W, Chhabra S, Stead P, Gledhill L, Hill P J, Rees C E D, Winson M K, Salmond G P C, Stewart G S A B, Williams P. Gene. 1992;116:87–91. doi: 10.1016/0378-1119(92)90633-z. [DOI] [PubMed] [Google Scholar]
- 4.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 5.Piper K R, Beck von Bodman S, Farrand S K. Nature (London) 1993;362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- 6.Bassler B L, Silverman M R. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 431–445. [Google Scholar]
- 7.Bassler B L, Wright M, Silverman M R. Mol Microbiol. 1994;12:403–412. doi: 10.1111/j.1365-2958.1994.tb01029.x. [DOI] [PubMed] [Google Scholar]
- 8.Freeman J A, Bassler B L. Mol Microbiol. 1999;31:665–668. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- 9.Freeman, J. A. & Bassler, B. L. (1999) J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 10.Cao J, Meighen E A. J Biol Chem. 1989;264:21670–21676. [PubMed] [Google Scholar]
- 11.Bassler B L, Wright M, Showalter R E, Silverman M R. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 12.Bassler B L, Wright M, Silverman M R. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 13.Bassler B L, Greenberg E P, Stevens A M. J Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surette M G, Bassler B L. Proc Natl Acad Sci USA. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Surette M G, Bassler B L. Mol Microbiol. 1999;31:585–596. doi: 10.1046/j.1365-2958.1999.01199.x. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.Greenberg E P, Hastings J W, Ulitzur S. Arch Microbiol. 1979;120:87–91. [Google Scholar]
- 18.Maloy S R, Stewart V J, Taylor R K. Genetic Analysis of Pathogenic Bacteria: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. [Google Scholar]
- 19.Caetano-Annoles G. Methods Appl. 1993;3:85–92. [Google Scholar]
- 20.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 21.Gilson L, Kuo A, Dunlap P V. J Bacteriol. 1995;177:6946–6951. doi: 10.1128/jb.177.23.6946-6951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]