Abstract
mariner family transposons are widespread among eukaryotic organisms. These transposons are apparently horizontally transmitted among diverse eukaryotes and can also transpose in vitro in the absence of added cofactors. Here we show that transposons derived from the mariner element Himar1 can efficiently transpose in bacteria in vivo. We have developed simple transposition systems by using minitransposons, made up of short inverted repeats flanking antibiotic resistance markers. These elements can efficiently transpose after expression of transposase from an appropriate bacterial promoter. We found that transposition of mariner-based elements in Escherichia coli produces diverse insertion mutations in either a targeted plasmid or a chromosomal gene. With Himar1-derived transposons we were able to isolate phage-resistant mutants of both E. coli and Mycobacterium smegmatis. mariner-based transposons will provide valuable tools for mutagenesis and genetic manipulation of bacteria that currently lack well developed genetic systems.
The ability to create random DNA insertions in bacterial chromosomes has been a very powerful technique for discovering new genes, discovering new functions of known genes, and studying protein functions (1). A number of well characterized, naturally occurring mobile DNA elements have been used to create insertions in bacterial chromosomes. Several transposons have been described that function in Gram-negative bacteria. However, these transposons do not appear functional in many other bacterial species. For example, Tn10, which has been used widely in Escherichia coli, appears to integrate site specifically in Mycobacterium smegmatis (2). In fact, in mycobacteria, the only transposons that have been found to produce insertion mutations in diverse sites are derived from other mycobacteria (3–6). These transposons do not function in all mycobacterial species and currently lack many of the features that have been introduced into other transposons, such as separation of transposase function from the transposon and introduction of cloned genes to create reporter constructs. Transposon mutagenesis may also be complicated by the presence of indigenous transposons that might inhibit transposition, serve as sites for homologous recombination, or transpose in response to the introduction of related elements creating unmarked mutations. A transposon that avoids these problems and functions in any bacterial species would provide a universal tool for genetic studies of several species including many important pathogens.
The genomes of diverse eukaryotic organisms have been found to contain members of the mariner/Tc1 superfamily of transposable elements (7–9). Elements of this superfamily share certain amino acid identities, have similar overall organization, and have similar “cut-and-paste” mechanisms of transposition (10–12). Comparisons of sequences of mariner/Tc1 elements from insects strongly suggest that there has been recent horizontal transmission (10) and, by implication, that a single transposon is capable of function in diverse eukaryotic hosts. This surprising observation suggested that it might be possible to reconstitute transposition in vitro. In vitro transposition has been observed by using two transposases of the mariner/Tc1 superfamily in the absence of any added host cofactors (11, 12), a property that has been used to make insertion mutations in naturally competent bacteria (13). In fact, both mariner-like and Tc1-like elements have been used to transfer markers between insects (14–16), and the mariner element Mos1, originally derived from Drosophila mauritania, has been shown to transpose in Leishmania major (17).
We reasoned that, with appropriate signals for transposase expression, a transposon derived from a Himar1, a mariner family element isolated from the horn fly Haematobia irritans, would transpose in bacteria. We found that transposition occurs in both E. coli and in mycobacteria. Transposition appears to have little site specificity beyond the known requirement for the dinucleotide TA. We have characterized inactivating mutations in E. coli genes and have also identified a transposon insertion that apparently activates the transcription of a downstream gene. mariner-based transposons should be broadly useful as genetic tools, particularly in bacterial species lacking indigenous transposable elements.
MATERIALS AND METHODS
Plasmids and Strains.
E. coli strains DH5αλpir, SM10λpir (18), and BW20767 (19) and M. smegmatis strain mc2155 (20) were maintained by standard methods. Plasmids pPR23 (21) and pBMML2S were maintained in E. coli. Bacteriophages, including a virulent λ phage and mycobacteriophage D29, were maintained by standard methods. Cloning and transformation was performed with standard molecular biology protocols (22).
Construction of E. coli Transposons.
Himar1-based minitransposons were constructed by cloning the Ecl136I fragment containing the kanamycin resistance gene from plasmid pBSL80 (23) into the unique SmaI site in a pMinimariner insertion into an ampicillin resistance gene (24) to create pEMKan. In magellan3, the kanamycin allele was replaced with the kanamycin resistance allele from plasmid TyK (25) by cleavage of pTyK with BamHI and PstI, filling in to create blunt ends, adding MluI linkers, and cloning into MluI-digested pEMKan. The Himar1 transposase was cloned from pET13a/mariner (11) by digestion with PstI, filling in to create blunt ends, and digestion with XhoI. The fragment containing the transposase was cloned into pBC KS+ (Stratagene) which had been digested with XhoI and Ecl136I to create pBCMar. After digestion of pBCMar with BglII and a fill-in reaction to create blunt ends, the ScaI–XmnI fragment from pEMKan was inserted to create pMEnt. To make a suicide plasmid, pMEnt was cleaved with SapI, the ends were filled in, and then digested with BsaAI, and the fragment containing the transposon and transposase was cloned into the EcoRV site of pGP704 (26) to create pFD1.
Construction of Mycobacterial Transposon.
To create a mycobacterial promoter (27), the primers 5′-GCTCTAGACCGTCCAGTCTGGCAGGCCGGAACATCGGTCAGCAGATAGGCTTTACCAGTAAGAAGGAG-3′ and 5′-CGAATTCCATATGTATATCTCCTTCTTACTGGTAA-3′ were annealed, extended with Taq polymerase, digested with XbaI and NdeI, and ligated into pET13a/mariner, which had also been digested with XbaI and NdeI to create pMM. This was digested with XbaI, and an XbaI–SpeI fragment from pEMKan was cloned to create pMME. A fragment of DNA containing oriR6Kγ was added to the transposon by PCR, amplifying a fragment of plasmid pWM41 (19) with primers 5′-AGATCTCAAACTGGAACAACACTCAACCC-3′ and 5′-TTAATTAACCCCGAAAAGTGCCACCTGACG-3′ and cloning the product into the SmaI site of pMME to create pMyr6K. This was digested with XbaI and SpeI, and the fragment containing the transposon and transposase was cloned into XbaI-digested pPR23 to make pMycoMar.
Identification of Insertions in a Target Plasmid.
pMML2S, a large plasmid that contains an RP4 origin of transfer, was introduced by electroporation into SM10λpir (pMEnt), allowed to recover without antibiotics for 1 h, and then plated on plates containing ampicillin and kanamycin. After overnight incubation, cells were mated with TOP10 by cross-streaking and allowed to conjugate for ≈6 h. Cells were then harvested from plates and selected on plates containing streptomycin (to select against donor cells) and kanamycin. Surviving colonies were pooled and plasmid was prepared.
The PCR was performed with a primer that hybridizes within the ampicillin resistance gene (5′-CGGGAGGGCTTACCATCTGGC-3′) and a 6-carboxyfluorescein-labeled version of MarOUT (5′-CGGGGACTTATCAGCCAACC-3′), which hybridizes to the inverted repeat of the Himar1-derived transposons. PCR products were analyzed on an ABI377 sequencer with GeneScan software (Applied Biosystems) according to the manufacturer’s instructions to obtain a read length of 500 bases.
Selection of λ Phage-Resistant Transposon Mutants.
Plasmid pFD1 was transferred by conjugation from BW20767 (pFD1) into TOP10 by conjugation. Cells were harvested from mating plates and replated on selection plates containing kanamycin and streptomycin that had been top spread with a virulent λ. Surviving cells were restreaked on Maconkey-maltose plates and scored for the ability to metabolize maltose. Strains that were able to metabolize maltose were analyzed for insertions in the lamB gene by PCR with a primer that hybridizes within the lamB gene (5′-GCGGTGAACAACAGTGTTTCCAGAC-3′) and MarOUT. PCR products were analyzed on a 2% agarose gel.
Transposon Mutagenesis of M. smegmatis.
Plasmid pMycoMar was introduced into M. smegmatis mc2155 by electroporation applying conditions described (21). After overnight recovery in 7H9 broth at 30°C, cells were plated on Luria-Bertani medium plates containing kanamycin at either 30°C (to determine the total number of transformants) or 39°C (to select for insertion mutants). Chromosomal DNA was prepared from individual mutants as described (28). Transposon insertions were cloned by digesting chromosomal DNA with either BssHII or BamHI, ligating, and transforming DH5αλpir.
RESULTS
In Vivo Transposition in E. coli.
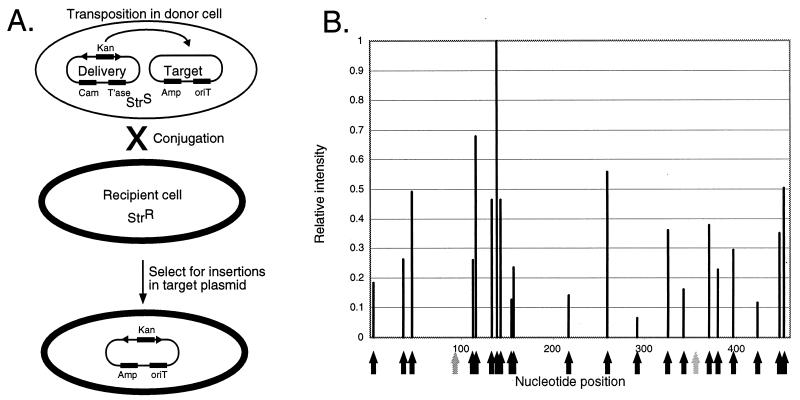
To test whether Himar1 transposase could direct transposition in a prokaryotic cell we devised a system to detect transposition between a delivery plasmid and a second target plasmid. We first constructed a vector that contained the transposase, under the transcriptional control of the lac promoter, and the minitransposon magellan3, which contains a gene encoding kanamycin resistance flanked by Himar1-inverted repeats. The resulting delivery plasmid, pMEnt, was introduced by electroporation into a streptomycin-sensitive strain, SM10λpir (18), which expresses the RP4 genes required for mobilization of a plasmid containing an appropriate origin of transfer. A target plasmid, pBMML2S, which contains a transfer origin, was transformed into the same strain, resulting in cells that contain both plasmids (Fig. 1A). After overnight growth, the target plasmid was transferred into a streptomycin-resistant E. coli strain by conjugation. The transconjugants were selected for growth on kanamycin and streptomycin.
Figure 1.
Location of magellan3 insertions in a plasmid. Transposition from a delivery plasmid to a target plasmid occurred in a donor cell (A) and target plasmid was transferred to a recipient cell by conjugation. The location of insertions in the target plasmid was mapped by PCR and a DNA sequencer that also quantitated fluorescence intensity (B). Arrows indicate the position of TA dinucleotides. Positions with insertions are marked with black arrows and those without insertions are marked with gray arrows.
With this protocol we obtained ≈105 kanamycin- and streptomycin-resistant colonies. To determine the sites of integration of the transposon, we pooled ≈104 colonies and isolated plasmid DNA. The pooled plasmids were then analyzed by PCR footprinting with a primer that hybridizes within the ampicillin resistance gene and a second fluorescently labeled primer, MarOUT, which hybridizes to the inverted repeat of magellan3. The products were analyzed with a sequencing gel, allowing us to determine the exact sites of insertion within the pool. Because there was no selection for ampicillin, we expected that several sites within the ampicillin resistance gene would be available for transposition. As can be seen in Fig. 1B, within the 500 bp analyzed we found insertions in 21 of 23 possible TA dinucleotide insertion sites. No insertions were identified in other dinucleotides. This result demonstrates that the transposon maintains the same site specificity seen in eukaryotes and in vitro. Beyond the requirement for this dinucleotide, however, we could find no consensus sequence for insertion. There was considerable variation in the fluorescence intensity of products at various sites. This could represent site preference for the transposon, variability of growth rate among the mutants, or, perhaps, preferential amplification of certain insertions in the PCR.
Transposition into the E. coli Chromosome.
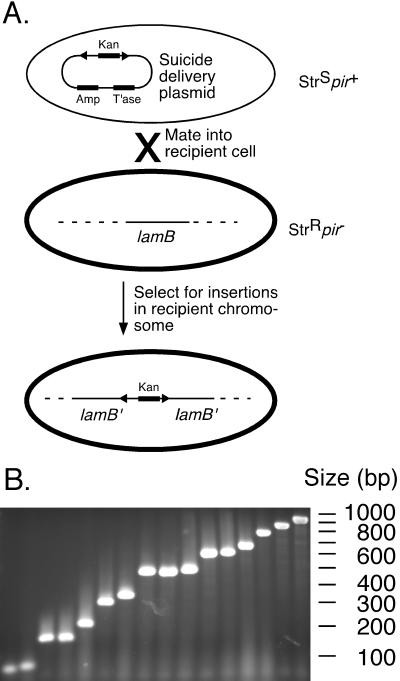
To create transposon insertions in the E. coli chromosome, we constructed a suicide delivery system (Fig. 2A). A Himar1-derived minitransposon that carried a gene encoding resistance to kanamycin and the Himar1 transposase gene under the transcriptional control of the lac promoter were cloned onto plasmid pGP704 (26). This plasmid contains both an origin of transfer allowing mobilization and the R6Kγ origin of replication and, therefore, depends on the presence of the pir gene for replication. When transferred to a pir− host by conjugation, the plasmid fails to replicate and the transposon marker is lost unless transposition occurs. With this system we are able to obtain large libraries of transposon mutants (105–107) in E. coli and other Gram-negative bacteria.
Figure 2.
(A) magellan3 insertions in the E. coli lamB gene. A library of magellan3 insertions in E. coli was constructed by conjugating a suicide vector encoding the mariner transposase and magellan3 into a recipient strain. Mutants that were resistant to lysis with a virulent λ phage were selected. To map those insertions that were within the lamB gene, individual colonies were selected and PCR was performed with a primer that hybridized within the lamB gene and a primer that hybridized to the inverted repeat of magellan3. (B) PCR products were analyzed on an agarose gel.
To test whether such a library contains a diversity of insertions, we selected for bacteria that were resistant to infection with a virulent mutant of λ phage. λ phage uses the LamB protein, a maltodextrin transporter, as its receptor. Mutants with insertions in either the lamB gene (that retains the ability to metabolize maltose [mal+]) or the regulatory gene malT (that cannot metabolize maltose [mal−]) fail to express the receptor and are, therefore, resistant to λ infection. To obtain such resistant cells, we plated a library of E. coli chromosomal transposon insertions (≈5 × 106 colonies) onto plates that were top-spread with virulent λ phage. Resistant mutants were obtained at a frequency of ≈1/1,000. Of 59 randomly chosen mutants, 37 were mal+ (63%) and 22 (37%) were mal−. These mutations could represent either several copies of identical insertions or insertions in diverse sites. To map the mutations in the lamB gene, we performed PCR with a primer that hybridized 125 bases from the 5′ end of the lamB gene and the MarOUT primer on 32 mal+ mutants. Single PCR products were obtained from all but three of the mutants. These three probably represent insertions in the 5′ end of the gene or in the lamB promoter (i.e., outside the specific interval selected for PCR analysis). The 29 PCR products varied in size (16 representatives are shown in Fig. 2B), representing insertions distributed throughout most of the lamB gene.
Transposition in Mycobacteria.
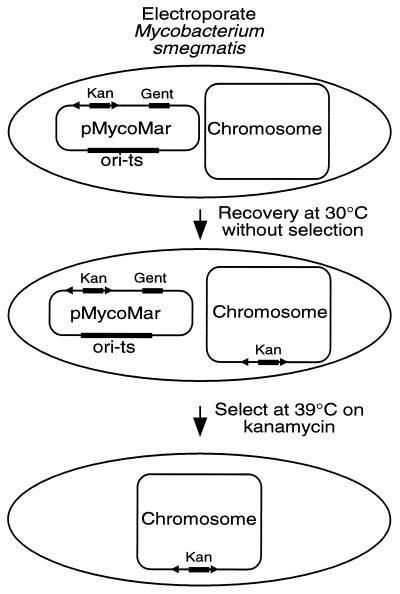
Our mariner transposition results in E. coli suggested that expression of the transposase and efficient delivery of the DNA encoding the transposon are sufficient conditions for transposon mutagenesis in any bacterial species. To determine whether mariner transposition could be observed in unrelated bacteria, we developed a mariner transposition system for mycobacteria. First, we constructed a new transposon, magellan4, which is identical to magellan3 except for the replacement of the kanamycin resistance gene from Tn903 with the kanamycin resistance gene originally derived from Tn5 and the addition of a 662-bp fragment containing oriR6Kγ. Initial attempts to clone this minitransposon into a vector that contained the Himar1 transposase under the transcriptional control of a mycobacterial heat shock promoter were unsuccessful. Because this promoter is also highly active in E. coli, it may be that overexpression of the transposase in the presence of the transposon is toxic. To avoid this problem, we constructed an artificial promoter with a sequence that has been found to produce high-level transcription in mycobacteria but not in E. coli (promoter T6 described by refs. 29 and 30). The T6 promoter/transposase and magellan4 were cloned into the temperature-sensitive mycobacterial replicon pPR23 (21) to create plasmid pMycoMar (Fig. 3).
Figure 3.
mariner transposition system for mycobacteria. Plasmid pMycoMar, which encodes the Himar1 transposase and the magellan4 minitransposon, was introduced into M. smegmatis by electroporation. After overnight recovery at 30°C without selection, transformants were plated on medium containing kanamycin and incubated at either 30°C (to determine the total number of transformants) or 39°C (to isolate insertion mutants).
To produce insertion mutants, pMycoMar was introduced into M. smegmatis mc2155 (20) by electroporation. Cells were grown overnight in broth at 30°C to allow transposition and then were plated on kanamycin-containing medium at either 30°C (allowing plasmid replication) or 39°C (which is not permissive for plasmid replication). Plates incubated at 39°C contained ≈1,000-fold fewer colonies than plates incubated at 30°C. All colonies that survived at 39°C were sensitive to gentamicin, an antibiotic resistance encoded by pMycoMar but not within magellan4, and had, therefore, lost the plasmid. With this method we obtained ≈4,000 independent kanamycin-resistant mutants. Southern blot analysis of 16 randomly chosen colonies showed that each contained a single transposon insertion that produced a unique restriction fragment pattern (data not shown).
Analysis of 13 arbitrarily chosen insertion mutants indicated that mariner transposition in mycobacteria is random. Cloning and sequencing of the insertion junctions revealed that 10 insertions occurred within likely ORFs, most of which were most similar to Mycobacterium tuberculosis genes (the only mycobacterial species for which a complete genome sequence is currently available; Table 1). Two insertions occurred in DNA sequences without significant homology to known genes. As found previously, all insertions occurred at TA dinucleotides. Beyond that sequence specificity, however, there was no clear consensus for insertion in this GC-rich organism.
Table 1.
Sequences of magellan4 insertions in M. smegmatis
| Sequence at junction | Similar ORF | Organism | P value |
|---|---|---|---|
| TACGTGGTGGTGGGCA | Rv1393c | M. tuberculosis | 2.50E − 15 |
| TATTCGAGCTGCAGCG | UvrC | M. tuberculosis | 1.70E − 43 |
| TAGGCCGCGACAAGCA | None | ||
| TACCTCGCAGCACAAG | Rv0560c | M. tuberculosis | 7.60E − 04 |
| TACGTGCTGTCCGACG | FadD8 | M. tuberculosis | 6.70E − 04 |
| TAGTCGATGCGCTCGG | Adjacent to TcmP | Streptomyces glaucescens | 2.40E − 25 |
| TAGGAGTCGGGCCGGT | Transcriptional regulator (AL031317) | Streptomyces coelicolor | 2.70E − 16 |
| TACGTGCCGCGCGGCA | Rv1482c | M. tuberculosis | 1.50E − 44 |
| TAGGTCATGAGCTCGT | AdhA | M. tuberculosis | 1.50E − 78 |
| TATCCCCACGGCATTC | Amidase | Synechocystis sp. | 1.50E − 29 |
| TACATCACGCGCCGGG | Benzene 1,2-dioxygenase α subunit | E. coli | 4.60E − 13 |
| TAGAGCACCGAGCCGG | None | ||
| TATCGCCGCGGGCGCC | Multiphosphoryl transfer protein | Rhodobacter capsulatus | 5.90E − 16 |
The duplicated TA dinucleotides are indicated by bold letters. Homologous open reading frames were identified and significance (P value) was determined by searching the GenBank database by using the blast program (38).
Selection of a Targeted Phenotype in Mycobacteria.
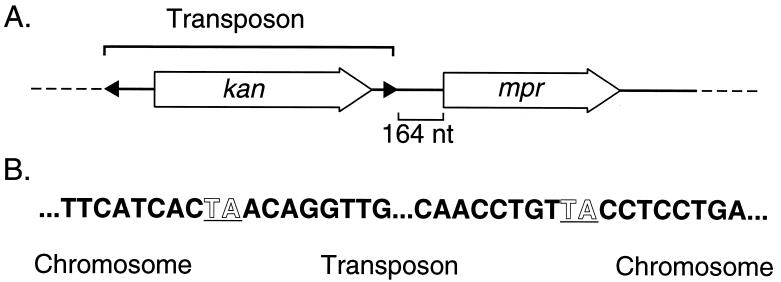
If mariner produces random insertions and is of general utility in mycobacteria, it should be possible to select for a specific phenotype conferred by a transposon insertion. We selected a pool of M. smegmatis transposon insertion mutants for resistance to lysis by the virulent mycobacteriophage D29 (32). We found a single mutant, DR1, from a pool of ≈4,000 mutants, that was highly resistant to D29-mediated lysis. Cloning and sequencing of the insertion junction revealed that the insertion was located 164 bp upstream of the 5′ end of the mpr gene (Fig. 4). This gene was previously shown to mediate resistance to phages L5 and D29 when overexpressed (27). The transposon insertion in strain DR1 is oriented such that the mpr gene is in the same orientation as the magellan4 kanamycin resistance gene. We previously found that insertions in this orientation within Haemophilus influenzae operons allow transcription of downstream genes (unpublished results), suggesting that the magellan4 insertion in strain DR1 probably mediates resistance by overexpression of the mpr gene via the kanamycin resistance gene’s promoter.
Figure 4.
Map of magellan4 insertion in a D29 phage-resistant mutant. (A) The location of the insertion with the orientation of the kanamycin resistance gene (kan) and the chromosomal mpr gene. The inverted repeats of the magellan4 minitransposon are indicated by filled arrowheads. (B) The sequence of the transposon-chromosome junctions. The duplicated TA dinucleotide is indicated with outlined letters.
DISCUSSION
mariner transposons have a remarkable lack of host specificity. They have previously been shown to transpose in distantly related insects (14–16), even more distantly related protozoa (17), and, most recently, vertebrate cells (33–36). Here we show that an insect-derived mariner is capable of transposing efficiently across domain boundaries, with activity in quite distantly related Gram-positive and Gram-negative bacteria. Although some elements can transpose in a variety of bacteria (37), most transposons are restricted in their host range. The determinants of host restriction are largely unknown. Some transposons with well characterized mechanisms interact with specific host proteins that are known to modulate transpositional activity (38). The presence or absence of such host factors may be partly responsible for host restriction.
An additional factor that may limit transposon host range is site specificity because many transposons either integrate site specifically or have a marked site preference (1). Site specificity can be mediated by transposase binding to the target sequence, interaction with host- or transposon-encoded accessory proteins, DNA supercoiling, or chromosomal structure. The Himar1 transposon has previously been shown to have little site specificity in vitro (11, 24). We tested for insertional specificity within the ampicillin resistance gene of a high copy number plasmid and found insertions at almost every TA dinucleotide within the analyzed sequence despite the fact the insertion frequency may be lower in this actively transcribed region than in nontranscribed DNA (1). Similarly, we prepared an E. coli mutant library in the presence of maltose, thus ensuring transcription of the lamB gene, and still saw diverse insertions within the gene. There does appear to be variation in the frequency of insertions into potential sites. This variation may result from transposase specificity or reduced fitness of some mutants. For example, protein fusions in or truncations of some portions of LamB might result in toxicity and, therefore, decreased growth rate.
Transposition of our minitransposon also appears to be random in mycobacteria. There were no recognizable sequence determinants among the 14 insertion junctions sequenced, apart from the required TA dinucleotide. This differs from the mycobacterial transposon Tn53677. Insertion of this transposon results in an 8-bp target duplication. Although there is no consensus sequence for this target, reported sequence targets have been relatively A-T rich (2). Interestingly, when we selected for a specific phenotype in M. smegmatis, we found an insertion that probably results in overexpression of a gene whose overexpression has previously been shown to yield resistance to D29 phage (27). This suggests that there are likely to be few insertions that will produce phage resistance. This is particularly striking because there are only two TA dinucleotides in the 300 bp 5′ of the mpr gene and, thus, a small target for transposition.
In experiments not shown, we found that we can create transposon insertion libraries in a variety of Gram-negative bacteria and in Mycobacterium fortuitum and Mycobacterium bovis BCG with Himar1-derived minitransposons. These transposons are particularly valuable because they are small and use short (31 bp) inverted repeats that have allowed us to make both transcriptional and translational reporter constructs (not shown). It seems likely that this transposon will be active in any bacterial strain for which expression signals are known and there is a system for introducing DNA. mariner transposons are likely to provide generally useful mutagenesis tools for a large number of organisms that were previously genetically intractable.
Acknowledgments
We thank the Microbiology Core Sequencing Facility (Harvard Medical School) for DNA sequencing, Dana Boyd, Jonathan Blum, Graham Hatfull, William Jacobs, Jr., Lalita Ramakrishnan, and Barry Wanner for plasmids and strains and Roberto Kolter for review of the manuscript. This work was supported by National Institutes of Health Grants AI02137 (to E.J.R.), AI33586 (for D.J.L.), AI37901 (to R.N.H.), and AI26289 (to J.J.M). B.J.A. was supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation Fellowship, DRG-1371. E.J.R. was a physician postdoctoral fellow of the Howard Hughes Medical Institute.
References
- 1.Craig N L. Annu Rev Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 2.Bardarov S, Kriakov J, Carriere C, Yu S, Vaamonde C, McAdam R A, Bloom B R, Hatfull G F, Jacobs W R., Jr Proc Natl Acad Sci USA. 1997;94:10961–10966. doi: 10.1073/pnas.94.20.10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin C, Timm J, Rauzier J, Gomez-Lus R, Davies J, Gicquel B. Nature (London) 1990;345:739–743. doi: 10.1038/345739a0. [DOI] [PubMed] [Google Scholar]
- 4.Cirillo J D, Barletta R G, Bloom B R, Jacobs W R., Jr J Bacteriol. 1991;173:7772–7780. doi: 10.1128/jb.173.24.7772-7780.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guilhot C, Otal I, Van Rompaey I, Martin C, Gicquel B. J Bacteriol. 1994;176:535–539. doi: 10.1128/jb.176.2.535-539.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McAdam R A, Weisbrod T R, Martin J, Scuderi J D, Brown A M, Cirillo J D, Bloom B R, Jacobs W R., Jr Infect Immun. 1995;63:1004–1012. doi: 10.1128/iai.63.3.1004-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson H M, Lampe D J. Annu Rev Entomol. 1995;40:333–357. doi: 10.1146/annurev.en.40.010195.002001. [DOI] [PubMed] [Google Scholar]
- 8.Robertson H M, Soto-Adames F N, Walden K K O, Avancini R M P, Lampe D J. In: Horizontal Gene Transfer. Syvanen M, Kado C, editors. London: Chapman & Hall; 1997. pp. 268–284. [Google Scholar]
- 9.Hartl D L, Lohe A R, Lozovskaya E R. Annu Rev Genet. 1997;31:337–358. doi: 10.1146/annurev.genet.31.1.337. [DOI] [PubMed] [Google Scholar]
- 10.Robertson H M, Lampe D J. Mol Biol Evol. 1995;12:850–862. doi: 10.1093/oxfordjournals.molbev.a040262. [DOI] [PubMed] [Google Scholar]
- 11.Lampe D J, Churchill M E, Robertson H M. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 12.Vos J C, De Baere I, Plasterk R H. Genes Dev. 1996;10:755–761. doi: 10.1101/gad.10.6.755. [DOI] [PubMed] [Google Scholar]
- 13.Akerley B J, Rubin E J, Camilli A, Lampe D J, Robertson H M, Mekalanos J J. Proc Natl Acad Sci USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lidholm D A, Lohe A R, Hartl D L. Genetics. 1993;134:859–868. doi: 10.1093/genetics/134.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lozovskaya E R, Nurminsky D I, Hartl D L, Sullivan D T. Genetics. 1996;142:173–177. doi: 10.1093/genetics/142.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loukeris T G, Livadaras I, Arca B, Zabalou S, Savakis C. Science. 1995;270:2002–2005. doi: 10.1126/science.270.5244.2002. [DOI] [PubMed] [Google Scholar]
- 17.Gueiros-Filho F J, Beverley S M. Science. 1997;276:1716–1719. doi: 10.1126/science.276.5319.1716. [DOI] [PubMed] [Google Scholar]
- 18.Simon R, Priefer U, Pühler A. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 19.Metcalf W W, Jiang W, Daniels L L, Kim S K, Haldimann A, Wanner B L. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 20.Snapper S B, Melton R E, Mustafa S, Kieser T, Jacobs W R., Jr Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 21.Pelicic V, Jackson M, Reyrat J M, Jacobs W R, Jr, Gicquel B, Guilhot C. Proc Natl Acad Sci USA. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Alexeyev M F, Shokolenko I N, Croughan T P. Gene. 1995;160:63–67. doi: 10.1016/0378-1119(95)00108-i. [DOI] [PubMed] [Google Scholar]
- 24.Lampe D J, Grant T E, Robertson H M. Genetics. 1998;149:179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garraway L A, Tosi L R, Wang Y, Moore J B, Dobson D E, Beverley S M. Gene. 1997;198:27–35. doi: 10.1016/s0378-1119(97)00288-6. [DOI] [PubMed] [Google Scholar]
- 26.Miller V L, Mekalanos J J. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barsom E K, Hatfull G F. Mol Microbiol. 1996;21:159–170. doi: 10.1046/j.1365-2958.1996.6291342.x. [DOI] [PubMed] [Google Scholar]
- 28.van Soolingen D, Hermans P W, de Haas P E, Soll D R, van Embden J D. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das Gupta S K, Bashyam M D, Tyagi A K. J Bacteriol. 1993;175:5186–5192. doi: 10.1128/jb.175.16.5186-5192.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bashyam M D, Kaushal D, Dasgupta S K, Tyagi A K. J Bacteriol. 1996;178:4847–4853. doi: 10.1128/jb.178.16.4847-4853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ford M E, Sarkis G J, Belanger A E, Hendrix R W, Hatfull G F. J Mol Biol. 1998;279:143–164. doi: 10.1006/jmbi.1997.1610. [DOI] [PubMed] [Google Scholar]
- 32.Ivics Z, Hackett P B, Plasterk R H, Izsvak Z. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 33.Fadool J M, Hartl D L, Dowling J E. Proc Natl Acad Sci USA. 1998;95:5182–5186. doi: 10.1073/pnas.95.9.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raz E, van Luenen H G, Schaerringer B, Plasterk R H A, Driever W. Curr Biol. 1998;8:82–88. doi: 10.1016/s0960-9822(98)70038-7. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Sankar U, Lampe D J, Robertson H M, Graham F L. Nucleic Acids Res. 1998;26:3687–3693. doi: 10.1093/nar/26.16.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott J R, Churchward G G. Annu Rev Microbiol. 1995;49:367–397. doi: 10.1146/annurev.mi.49.100195.002055. [DOI] [PubMed] [Google Scholar]
- 37.Mizuuchi K. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- 38.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]