Abstract
The “Spanish” influenza pandemic killed over 20 million people in 1918 and 1919, making it the worst infectious pandemic in history. Here, we report the complete sequence of the hemagglutinin (HA) gene of the 1918 virus. Influenza RNA for the analysis was isolated from a formalin-fixed, paraffin-embedded lung tissue sample prepared during the autopsy of a victim of the influenza pandemic in 1918. Influenza RNA was also isolated from lung tissue samples from two additional victims of the lethal 1918 influenza: one formalin-fixed, paraffin-embedded sample and one frozen sample obtained by in situ biopsy of the lung of a victim buried in permafrost since 1918. The complete coding sequence of the A/South Carolina/1/18 HA gene was obtained. The HA1 domain sequence was confirmed by using the two additional isolates (A/New York/1/18 and A/Brevig Mission/1/18). The sequences show little variation. Phylogenetic analyses suggest that the 1918 virus HA gene, although more closely related to avian strains than any other mammalian sequence, is mammalian and may have been adapting in humans before 1918.
The influenza pandemic of 1918 was exceptionally severe, killing 20–40 million people worldwide, with unusually high death rates among young, healthy adults (1). A mild wave of influenza in the spring and summer of 1918 was highly contagious but caused few deaths. In late August, a virulent form of the disease emerged and swept the globe in 6 months, killing over 10,000 people per week in some U.S. cities (1) at the height of the pandemic.
Recently, this laboratory reported the isolation of fragments of RNA from the 1918 influenza virus from preserved lung tissue of a victim of the deadly fall wave of the pandemic. Sequence from 5 of the virus’s 10 genes indicated that the strain was of the H1N1 subtype and different from any other sequenced influenza strain. All of the sequenced genes were more similar to those influenza strains infecting mammals than to those that infect birds (2). We now report the complete sequence of the hemagglutinin (HA) gene of the 1918 influenza virus. The full-length sequence was generated by using RNA fragments isolated from the victim reported in March 1997 [A/South Carolina/1/18 (H1N1)] (2). Additional sequence was obtained from two recently identified 1918 victims.
Influenza is a zoonotic disease, affecting many species of birds and mammals. The HA protein is found on the surface of the influenza virus particle and is responsible for binding to receptors on host cells and initiating infection. HA is also the principal target of the host’s immune system. Thus, in order for influenza to spread in a new host, the HA protein must acquire the ability to bind to the new host’s cells. Once in the new host, the HA protein comes under selective pressure for change to evade the host’s immune system (3).
Pandemic influenza results when an influenza strain emerges with an HA protein to which few people have prior immunity (4). It is thought that the source of HA genes that are new to humans is the extensive pool of influenza viruses infecting wild birds. Of the 15 HA subtypes found in birds, only 3 (H1, H2, and H3) are known to have caused pandemics in humans (5). Recently an H5 virus caused illness in a limited number of people in Hong Kong, China (6, 7). How new HA genes emerge and where they adapt from their characteristic avian form to a form that successfully spreads in humans are not understood perfectly. In both 1957 and 1968, the pandemic strains had new HA genes that were very similar to known avian strains (8, 9). We sought to determine whether the HA gene of the 1918 pandemic strain also had avian characteristics.
MATERIALS AND METHODS
Case Selection.
Autopsy cases of 78 victims of the lethal fall wave of the 1918 pandemic were examined for this study. Evidence from 74 victims consisted of formalin-fixed, paraffin-embedded tissues, stained slides, and clinical records from the files of the Armed Forces Institute of Pathology. All the samples were screened by histologic analysis. The majority of individuals died of secondary acute bacterial pneumonia, the most common cause of death in the 1918 pandemic (10); most of the samples taken from these individuals were not analyzed further, because they were extremely unlikely to retain influenza virus (11, 12). However, 13 samples were selected by histologic and clinical criteria for further analysis. These samples were from patients who experienced acute influenza deaths after clinical courses of less than 1 week. In addition to samples taken from patients with early bronchopneumonia, samples from patients with acute massive pulmonary edema and/or hemorrhage were also selected, reflecting the unusual histopathology observed in 1918 (13). Of these 13 samples, 2 were positive for influenza RNA on subsequent molecular genetic analysis.
Case Histories.
The first patient was a 21-year-old male stationed at Ft. Jackson, SC. He was admitted to the camp hospital on September 20, 1918 with influenza and pneumonia. He had a progressive course with cyanosis and died on September 26, 1918. During the autopsy, it was noted that he had a fatal secondary lobar bacterial pneumonia in his left lung, whereas the right lung showed only focal acute bronchiolitis and alveolitis, indicative of primary influenza pneumonia. Formalin-fixed, paraffin-embedded right lung tissue was positive for influenza RNA [A/South Carolina/1/18 (H1N1)] as reported (2).
The second patient was a 30-year-old male stationed at Camp Upton, NY. He was admitted to the camp hospital with influenza on September 23, 1918, had a very rapid clinical course, and died from acute respiratory failure on September 26, 1918. The autopsy showed massive bilateral pulmonary edema and focal acute bronchopneumonia. Formalin-fixed, paraffin-embedded lung tissue was positive for influenza RNA [A/New York/1/18 (H1N1)]. RNA templates larger than 150 nucleotides could not be amplified in these two cases.
An additional 1918 influenza case was found by examining lung tissue from four 1918 influenza victims exhumed from a mass grave in Brevig Mission on the Seward Peninsula of Alaska. Brevig Mission (called Teller Mission in 1918) suffered extremely high mortality during the influenza pandemic in November 1918. Although individual case records were not available, historical records show that influenza spread through the village in about 5 days, killing 72 people, representing about 85% of the adult population (1, 14). Victims were buried in a mass grave in permafrost. In August 1997, four of these victims were exhumed. Frozen lung tissues were biopsied in situ from each, and tissues were placed in formalin, alcohol fixatives, and RNAzol (Tel-Test, Friendswood, TX). Although the histologic analysis was hampered by artifacts of freezing, these tissues showed evidence of acute massive pulmonary hemorrhage and edema. One of the victims, an Inuit female (age unknown) was influenza RNA positive [A/Brevig Mission/1/18 (H1N1)]. In this case, RNA templates greater than 120 nucleotides could not be amplified.
RNA Extraction.
RNA lysates from the paraffin-embedded tissues were produced as described (15). RNA was isolated from the frozen lung tissue by using RNAzol (Tel-Test) according to the manufacturer’s instructions.
Reverse Transcription–PCR (RT-PCR).
RT was carried out at 37°C for 45 min in 20 μl of 1× RT buffer (GIBCO/BRL) containing 300 units of Moloney murine leukemia virus reverse transcriptase, 5 μM random hexamers, 200 nM dNTP, and 10 mM DTT. RT reaction (2 μl) was added to a 20-μl PCR containing 50 mM KCl, 10 mM Tris, 2.5 mM MgCl2, 1 μM each primer, 100 nM dNTP, 1 unit of Amplitaq Gold (Perkin—Elmer), and 2 μCi of 32P-labeled dATP (3,000 Ci/mmol). The entire HA coding sequence of 1,701 nucleotides was amplified in 22 overlapping fragments, such that the sequences matching primers could be confirmed. The primers were designed as degenerate H1 consensus primers by using alignments of human, swine, and avian H1 HA sequences. The primer sequences used for these amplifications are available on request.
PCR conditions were 9 min at 94°C; 40 cycles of 94°C for 30 sec, 50°C for 30 sec, 72°C for 30 sec; and 72°C for 5 min. Part (one-sixth) of the reaction product was separated on a denaturing 7% polyacrylamide gel, dried, and visualized by autoradiography. Bands were excised, electroeluted, and precipitated with ethanol. Part (one-fourth) of the eluted product was added to a 50-μl PCR (50 mM KCl/10 mM Tris/2.5 mM MgCl2/200 nM dNTP/1 μM each primer/1 unit of Amplitaq Gold) and cycled as above. Reaction product (2 μl) was cloned into the PCR 2.1 vector (Invitrogen), according to the manufacturer’s instructions.
Replicate RT-PCRs from independently produced RNA preparations gave identical sequence results. Only one nucleotide change was noted between the formalin-fixed samples and the RNA isolated from the frozen, unfixed sample (see below).
DNA Sequencing.
Direct PCR that used M13 primers was done on white colonies, and the products were sequenced by cycle sequencing as described (16).
Phylogenetic Analyses.
Phylogenetic analyses of the HA gene and its subsequences HA1 and HA2 employed parsimony and neighbor-joining (NJ) methods and used the software paup, version 3.1.1 (Phylogenetic Analysis Using Parsimony; ref. 17) and mega, version 1.1 (Molecular Evolutionary Genetics Analysis; ref. 18), respectively. The optimization method used in paup was acctran. The software package macclade was used to follow HA1 character evolution and accumulated changes in the influenza HA1 sequence family. macclade allows the evolution of single characters to be traced throughout the phylogenetic tree. Taxa (in this case, viral strains) can be moved around within a parsimony tree to evaluate the effect of different tree topologies on branch lengths, tree lengths, and character transformation (19).
NJ analyses routinely employed the proportion of differences between the sequences as the distance measure (p distance). Analyses were performed on 28 complete HA genes, 64 HA1 subsequences, and 30 HA2 subsequences. All analyses were bootstrapped 100 replications.
One parsimony analysis used 11 HA1 sequences and paup’s exhaustive search option, which examines every possible tree produced by the data set. The sequences used (one 1918, three avian, three swine, and three human, plus one H2 as outgroup) were chosen to give a fair representation of the larger data set. Analysis of these sequences generated 34,459,425 trees with an average length of 1,459 steps (range of 1,022–1,553 steps with a standard deviation of 68.9).
Strains Used for Phylogenetic Analyses.
The following sequences were used in this analysis and were obtained from GenBank, the European Molecular Biology Laboratory, and the DNA Data Base in Japan. Strain abbreviations are listed in ref. 2 or as follows: Texas90, A/Texas/22/90 (H1N1); Qingdao91, A/Qingdao/28/91 (H1N1); Singapore90, A/Singapore/6/90 (H1N1); Fukushima88, A/Fukishima/2/88 (H1N1); Canada88, A/Canada/7/88 (H1N1); Czechoslovakia89, A/Czechoslovakia/2/89 (H1N1); Goroka90, A/Goroka/2/90 (H1N1); Fiji88, A/Fiji/2/88 (H1N1); Suita89, A/Suita/1/89 (H1N1); Ohio83, A/Ohio/101/83 (H1N1); Kiev79, A/Kiev/5/79 (H1N1); Arizona90, A/Arizona/1/90 (H1N1); PR34 (MS), A/PR8/34 (Mt. Sinai) (H1N1); Sw/29/37, A/Swine/29/37 (H1N1); SwQuebec90, A/Swine/Quebec/1747/90 (H1N1); SwNebraska92, A/Swine/Nebraska/1/92 (H1N1); Maryland91, A/Maryland/12/91 (H1N1); SwHong Kong74, A/Swine/Hong Kong/1/74 (H1N1); TyGermany90, A/Turkey/Germany/2482/90 (H1N1); DkHong Kong76, A/Duck/Hong Kong/36/76 (H1N1); MallTennessee85, A/Mallard/Tennessee/11464/85 (H1N1); TyMinnesota81, A/Turkey/Minnesota/1661/81 (H1N1); ChHong Kong76, A/Chicken/Hong Kong/14/76 (H1N1); and Japan57, A/Japan/305/57 (H2N2).
RESULTS
Sequence Analysis.
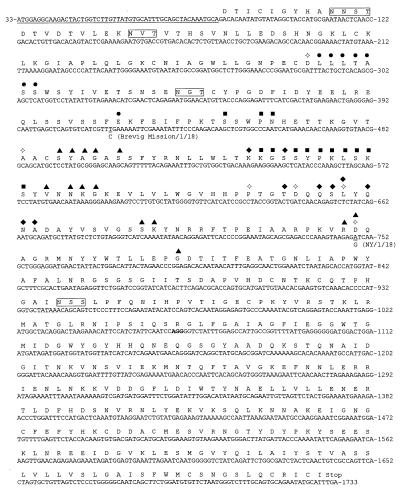
The HA gene sequence of A/South Carolina/1/18 (nucleotides 33–1,733), containing 1,701 nucleotides, and a theoretical translation are shown in Fig. 1. The HA protein is composed of two domains, HA1 and HA2, linked by a basic amino acid. The sequence of HA1 (nucleotides 33–1,064) and part of HA2 (nucleotides 1,065–1,258) were confirmed by using both A/New York/1/18 and A/Brevig Mission/1/18. Only two nucleotide differences were noted among the strains. These changes, at nucleotides 416 and 748, are shown in Fig. 1. There is a T at nucleotide 416 in A/South Carolina/1/18 and A/New York/1/18 and a C in A/Brevig Mission/1/18; however, this change does not alter an amino acid. The change at 748 is nonsynonymous, coding for aspartic acid in A/South Carolina/1/18 and A/Brevig Mission/1/18 and for glycine in A/New York/1/18. These changes were confirmed by independent RT-PCRs.
Figure 1.
Complete coding sequence of the HA gene of the 1918 influenza virus. The sequence for A/South Carolina/1/18 is shown with a theoretical translation of the HA1 and HA2 domains. The numbering of the nucleotide sequence is aligned to PR/8/34 and refers to the sequence of the gene in the sense (mRNA) orientation. The sequence coding for the signal peptide is underlined. The cleavage site (nucleotides 1,062–1,064) between the HA1 and HA2 domains is shown in bold. Sequence differences between this strain and the other 1918 strains are shown as double-underlined nucleotides (nucleotides 416 and 748). The sequences were confirmed by sequencing overlapping RT-PCR products and by replicate RT-PCRs for each case. The GenBank accession number is AF117241 for A/South Carolina/1/18, AF116576 for A/New York/1/18, and AF116575 for A/Brevig Mission/1/18. The theoretical translation of the gene is shown above the nucleotide sequence. Boxed amino acids indicate potential glycosylation sites as predicted by the sequence (26). Receptor-binding sites (open diamonds; ref. 23), Cb antigenic site (open circles), Sa antigenic site (closed squares), Sb antigenic site (closed diamonds), and Ca antigenic site (closed triangles; refs. 28 and 37). Some of the receptor-binding residues are also in known antigenic sites. For these sites, symbols for both are shown.
Phylogenetic Analyses.
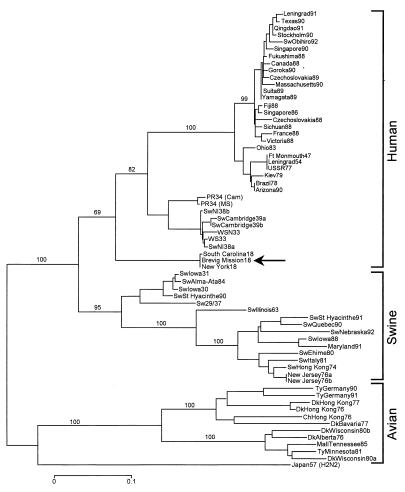
NJ analyses were performed on the complete HA gene and on its two domains, HA1 and HA2; 28 complete H1 sequences plus 1 complete H2 sequence were compared, and three distinct clades—human, swine, and avian—emerged from the analysis. The 1918 sequence was within and near the root of the human clade. A similar result was found with 30 HA2 domain sequences. Analysis of 64 HA1 nucleotide sequences placed 1918 HA1 within and near the root of the swine clade. Parsimony analysis of HA, HA1, and HA2 always placed the 1918 sequences within and near the root of the human clade. None of the NJ or parsimony analyses placed 1918 HA within the avian clade. A subset of 10 HA1 nucleotide sequences (3 avian, 3 swine, 3 human, and a single 1918) were analyzed by paup with the exhaustive search option, which examines every possible tree produced by the data set. The shortest tree was over six standard deviations shorter than the mean tree length and placed 1918 HA1 within the human clade. The 10 shortest trees (1,022–1,035 steps) were examined, and in no case was 1918 HA placed within the avian clade.
Synonymous and nonsynonymous substitutions were analyzed separately. Both NJ and parsimony analysis of synonymous substitutions placed the 1918 sequences within and near the root of the human clade for the complete HA, HA1 (Fig. 2), and HA2. When only nonsynonymous substitutions were examined, NJ analysis placed HA and HA1 within and near the root of the swine clade.
Figure 2.
Phylogenetic tree of the influenza virus HA gene segment, HA1. Sequences were aligned with lasergene software (DNAstar, Madison, WI) and analyzed for phylogenetic relationships by the NJ method with the proportion of sequence differences as the distance measure (0.1 p distance ≈ 22.35 synonymous differences). Synonymous substitutions were analyzed and bootstrap values (100 replications) are given for selected nodes. Human, swine, and avian clades are identified with large brackets. The arrow identifies the position of the 1918 sequences. A distance bar is shown below the tree. Influenza strain abbreviations used in the analyses are described in Materials and Methods and in ref. 2.
Parsimony analysis of the HA protein placed the 1918 sequence within and near the root of the human clade, reflecting the result obtained with the nucleotide sequence. A similar result was obtained by analyzing 55 HA1 protein sequences that had the five most common egg-adaptation sites (20) removed. NJ analysis of the translated HA and HA1 proteins, on the other hand, placed the 1918 sequences within and near the root of the swine clade. This result is consistent with the phylogeny obtained from analyzing only the nonsynonymous substitutions in the gene.
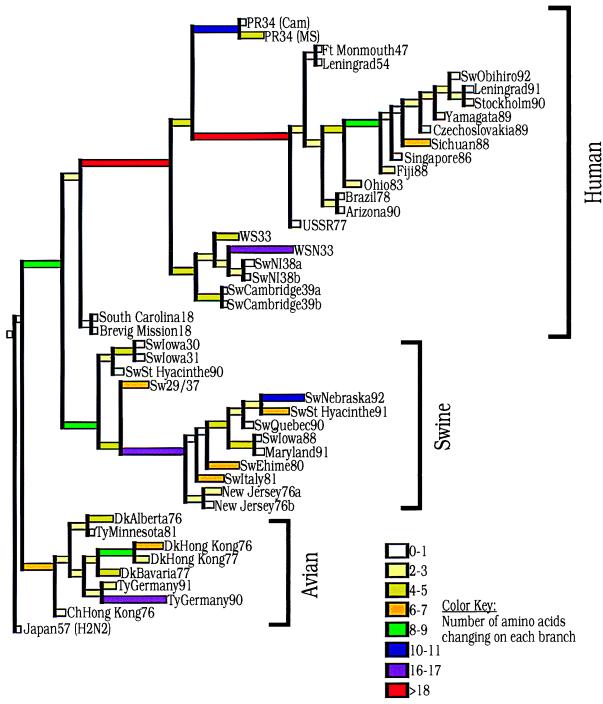
Fig. 3 shows a parsimony tree of 564 steps generated by paup and analyzed by macclade. The total number of changes per branch is shown. The macclade program has reconstructed the sequence of the putative ancestor at each node and then determined the number of changes along the branch leading to the next node. It shows that avian HA1 proteins have changed less over time than human HA1 proteins. Swine HA1 proteins have an intermediate number of changes. For example, the 1918 HA1 has just 4 changes from the putative mammalian ancestral sequence, whereas the closest swine sequence differs by 10 changes from the same ancestral sequence. The 1918 HA1 has 13 changes from the putative avian ancestor, whereas the closest avian sequence differs by 8 changes from that ancestor.
Figure 3.
Amino acid changes in three lineages of the influenza virus HA protein segment, HA1. A phylogenetic tree was derived by using paup (17). The tree was then imported into macclade (19), and the ancestral sequences at each node of the tree were inferred. The tree shows the numbers of unambiguous changes between these sequences, with branch lengths proportional to the number of changes. Colors denote the numbers of changes along each branch and are identified by the color key at the lower right.
With a length of 564 steps, the tree shown in Fig. 3 is the shortest generated by paup. The tree places 1918 within the human clade. However, the tree becomes only three steps longer when 1918 HA1 is considered ancestral to both human and swine lineages. For comparison, supposing that an early human strain like WS33 is ancestral to both human and swine results in a tree of 591 steps; supposing that an early swine strain like Sw/Iowa/30 is ancestral to all mammalian strains results in a tree of 573 steps.
Finally, we performed parsimony analysis by using only the three 1918 cases and nine avian strains to see whether the 1918 cases are placed in the mammalian clade, because all mammalian H1 strains are derived from a strain similar to the pandemic virus. This analysis showed the presence of two clades, avian and 1918, suggesting that the 1918 sequences are indeed phylogenetically distinct from avian strains.
Plotting the total number of amino acid changes from node to node (Fig. 3) vs. the year of isolation for viruses in the human clade before 1954 suggests that the ancestor of the 1918 virus entered humans around 1900. Tracing total numbers of nucleotide changes from node to node of a nucleotide parsimony tree places the 1918 ancestor in humans around 1915.
Receptor-Binding Sites.
Influenza infection requires binding of the HA protein to sialic acid-containing receptors on the host cell surface (21). HAs from influenza strains infecting birds preferentially bind to receptors with sialic acid linked to galactose by the α2,3 linkage (SAα2,3Gal), whereas human-adapted HAs prefer the α2,6 linkage (SAα2,6Gal). A shift from SAα2,3Gal to SAα2,6Gal binding seems to be a critical step in the adaptation of an avian HA to the human host (20, 22, 23). Determining the minimal changes allowing avian H1s to function in humans is complicated by the fact that the earliest H1 strains have been maintained in different laboratory host cells, a practice that selects for mutations in the receptor-binding sites (20). The 1918 sequence, having been derived directly from viral RNA, is free of any possible artifacts of laboratory adaptation.
A subset of amino acids that are unvaried in all avian HAs but vary in mammalian-adapted HAs has been identified (23) and constitutes the minimum receptor-binding site. Changes in these amino acids have been shown to affect receptor-binding specificity (24) and antigenicity (25). Studies in which mammalian strains are adapted to grow in eggs indicate that some of these amino acids are involved in the shift from SAα2,6Gal to SAα2,3Gal binding.
To shift from the avian receptor-binding site to that of swine H1s requires only one amino acid change, E190D (Table 1). The receptor-binding site of one of the 1918 cases (A/New York/1/18) is identical to that of A/Sw/Iowa/31 (a classical swine strain) and A/Sw/Germany/91 (a recent avian introduction into swine). The other two 1918 cases have an additional change from the avian consensus, G225D (Fig. 1; Table 1). Because swine viruses with the same receptor site as Sw/Iowa/31 and Sw/Germany/91 bind both SAα2,3Gal and SAα2,6Gal (20), New York/1/1918 probably also has the capacity to bind both receptors. The change at residue 190 may represent the minimal change needed to allow an avian H1-subtype HA to bind the SAα2,6Gal receptor.
Table 1.
HA receptor-binding residues of selected H1 influenza viruses
| Strain* | HA receptor-binding
residues†
|
|||||
|---|---|---|---|---|---|---|
| 77 | 138 | 186 | 190 | 194 | 225 | |
| Avian strains | D | A | P | E | L | G |
| Sw/Germany/91 | D | A | P | D | L | G |
| 1918 strains | D | A | P | D | L | G (1), D (2)‡ |
| Sw/Iowa/31 | D | A | P | D | L | G |
| Sw/Illinois/63 | E | A | P | D | L | G |
| PR/8/34 | D | A | S | D | I | D |
| Ft. Monmouth/47 | E | S | S | D | I | G |
| USSR/77 | E | S | S | D | I | G |
Selected strains are shown. These six amino acid positions are unvaried in all avian HAs (23). The 1918 strains are South Carolina/1/18, New York/1/18, and Brevig Mission/1/18. Other sequences were obtained from GenBank. Complete strain names are listed in Materials and Methods and/or ref. 2.
Amino acid numbering is aligned to the H3 influenza HA (27).
The 1918 strains differ at this site. New York/1/18 has a glycine residue (G), whereas both South Carolina/1/18 and Brevig Mission/1/18 have an aspartic acid (D) at this site.
Glycosylation Sites.
Some glycosylation sites are necessary for the function of the influenza virus (26), but the accumulation of additional glycosylation sites is a common event in the adaptation of influenza strains to human hosts and seems to be a mechanism for masking antigenic sites. All avian and swine viruses share four glycosylation sites, whereas human strains from the 1930s have two additional sites, and modern human H1N1 viruses have up to five additional sites (27). The 1918 strains, in contrast, have only the four glycosylation sites shared with avian and swine strains (Fig. 1), suggesting that acquisition of additional glycosylation sites is not required for efficient replication in humans.
Antigenic Sites.
On the H1 protein, four antigenic sites, encompassing 42 amino acids, have been identified (refs. 25 and 28; Fig. 1). In human strains, these are among the most variable amino acids in the HA protein. At the Sa site, the 1918 strains match the avian sequences at all 12 amino acids. At the Sb site (9 amino acids), the 1918 strains share the avian consensus at 8 amino acids, but at amino acid 156, the 1918 strains have a serine that is found only in one other H1 strain (A/Sw/Illinois/63). At the Ca site (7 amino acids), the 1918 sequences have one change, N74S, from the avian consensus. This change is found in all swine strains but not in any human strains. At the Cb site (13 amino acids), the 1918 sequences differ from the avian consensus by two amino acids, S139A and T166V. The first change is found in all swine and some early human strains, and the second is found in all swine and human strains. Overall, the antigenic sites resemble the avian consensus closely, suggesting that there may have been little selective antigenic pressure on the HA protein before 1918.
Cleavage Site.
The HA of the recent H5 influenza outbreak in Hong Kong, China contained a cleavage-site mutation characteristic of highly lethal H5 and H7 avian viruses (6, 7). As reported (2), A/South Carolina/1/18 does not contain such a mutation, and the sequencing of the cleavage site in A/Brevig Mission/1/18 and A/New York/1/18 confirms the absence of this mutation (Fig. 1).
DISCUSSION
Previous phylogenetic analyses of H1 sequences suggest that the swine and human lineages diverged from a common, avian-like ancestor around 1905 (29, 30). Other analyses, which propose a higher rate of mutation early in the adaptation of avian influenza strains to mammalian hosts, suggest that the pandemic virus derived from an avian virus immediately before 1918, possibly through a swine intermediary (31, 32).
Of all mammalian H1s, those of the 1918 viruses are most similar to their avian counterparts. However, the 1918 HA gene has accumulated enough nonselected, mammal-associated changes to place it consistently in the mammalian clade phylogenetically (Fig. 2). The 1918 HA1 sequence differs from its closest avian relative by 26 amino acids, whereas the 1957 H2, the 1968 H3, and 1997 H5 HAs had 16, 10, and 3 differences, respectively, with their closest avian relatives (6–9). Phylogenetic analyses of the 1957, 1968, and 1997 HAs place them in avian clades and suggest that all three strains derived from the Eurasian group of avian strains. By contrast, phylogenetic analyses invariably place the 1918 sequence outside the avian clade and suggest that the 1918 sequence is phylogenetically equidistant from the Eurasian and North American avian strains.
Although the 1918 HA always falls within the mammalian group, it will sometimes seem to be more human-like, sometimes more swine-like, depending on the method of analysis. These results may be explained, at least in part, by the widely differing substitution rates in different lineages making up the tree (see Fig. 3). Human strains, because of strong immune selection, have many amino acid substitutions that are not shared with the 1918 strains or swine strains. As a result, phylogenetic analyses based on amino acid sequence or nonsynonymous nucleotide substitutions may tend to group 1918 and swine strains together. Synonymous substitutions, on the other hand, reflect drift unrelated to immune selection and indicate a closer relationship between 1918 strains and subsequent human strains. In all of the phylogenetic analyses, branch lengths near the root of the tree were very short, suggesting that there may be few differences between the 1918 sequences and the common mammalian influenza virus ancestor.
The existing strain to which the 1918 sequences are most closely related is A/Sw/Iowa/30, the oldest classical swine flu strain. It has been known since the 1930s that survivors of the 1918 influenza had antibodies that neutralized classic swine influenza virus (33), and A/Sw/Iowa/30 is very similar to the 1918 strains at the antigenic sites. Because individual swine do not live long enough to exert immune selective pressure on the virus, influenza drifts more slowly in swine than in humans (30). Therefore, one would expect a swine virus isolated in the 1930s to retain more characteristics of the 1918 strain than a 1930s human virus. Based on the calculated rate of amino acid change in swine HA genes (30), one would expect approximately 20 changes between the 1918 strains and A/Swine/Iowa/30. We find 22 changes (of which 4 are in antigenic sites), consistent with the hypothesis that a virus similar to the 1918 strains entered the swine population in 1918 and changed gradually over the subsequent 12 years. By contrast, there are 43 amino acid differences between the 1918 strains and the human virus A/PR/8/34 (Cam), of which 15 are in antigenic sites, a rate of change consistent with a virus under substantial immune pressure.
Historical accounts document a widespread but mild wave of influenza in humans in the spring of 1918. Victims of the spring wave were reported to be immune from severe disease in the fall, suggesting that the spring and fall waves were closely related (10). Swine were first affected in the fall, when severe influenza-like disease outbreaks were noted in swine in the United States, Hungary, and China (34–36). Contemporary veterinarians had not seen this disease in pigs and believed it to be the same disease affecting humans. This sequence of events supports the theory that the 1918 influenza spread from humans to swine and is difficult to reconcile with the theory that separate human and swine H1 viruses were circulating from 1905 to 1918.
Analysis of the 1918 HA sequence permits alternative interpretations as to its origin. The 1918 sequences are phylogenetically distinct from current avian strains. One possibility is that around 1918 there existed an avian strain more similar to the pandemic virus than current avian strains and that its HA entered with little modification into the human population. If avian viruses have not drifted over the past 80 years, such a strain would differ from current avian strains phylogenetically. This hypothesis cannot be tested, because avian H1 influenza-virus isolates from that time do not exist. A second possibility is that the pandemic virus had been adapting in mammals before 1918 and that it had accumulated enough changes to make its HA gene seem more mammalian by many phylogenetic criteria (e.g., parsimony and NJ). Our data and those of others (29) suggest that an entry date into the human population between 1900 and 1915 is reasonable.
The 1918 influenza virus HA gene does not possess the cleavage site mutation seen in virulent avian influenza strains. No other known genetic changes were observed in the 1918 HA sequence that would account for the exceptional virulence of this pandemic virus. What determines the virulence of a particular influenza strain is quite complex and involves host adaptation, transmissibility, tissue tropism, and replication efficiency. The genetic basis for virulence of other influenza strains (for which complete genomic sequence is available) cannot be determined yet, but it is most likely polygenic in nature (4). Further sequence analyses of the remaining 1918 influenza genes are currently in progress, and it is hoped that these studies will shed additional light on the nature of the 1918 influenza virus.
Acknowledgments
We are grateful to the people of Brevig Mission, Alaska, for their generosity and willingness to support this project and to Timothy J. O’Leary for his foresight and ongoing support. This work was supported in part by grants from the American Registry of Pathology and the Department of Veterans Affairs and by the intramural funds of the Armed Forces Institute of Pathology.
ABBREVIATIONS
- HA
hemagglutinin
- NJ
neighbor-joining
- RT-PCR
reverse transcription–PCR
Footnotes
References
- 1.Crosby A. America’s Forgotten Pandemic. Cambridge, U.K.: Cambridge Univ. Press; 1989. [Google Scholar]
- 2.Taubenberger J K, Reid A H, Krafft A E, Bijwaard K E, Fanning T G. Science. 1997;275:1793–1796. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 3.Murphy B, Webster R. In: Fields Virology. Fields B, Knipe D, Howley P, editors. Philadelphia: Lippincott; 1996. pp. 1397–1445. [Google Scholar]
- 4.Kilbourne E. J Am Med Assoc. 1977;237:1225–1228. [Google Scholar]
- 5.Kilbourne, E. D. (1997) J. Infec. Dis.176, Suppl. 1, S29–S31. [DOI] [PubMed]
- 6.Claas E C, Osterhaus A D, van Beek R, De Jong J C, Rimmelzwaan G F, Senne D A, Krauss S, Shortridge K F, Webster R G. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 7.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, et al. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 8.Schafer J R, Kawaoka Y, Bean W J, Suss J, Senne D, Webster R G. Virology. 1993;194:781–788. doi: 10.1006/viro.1993.1319. [DOI] [PubMed] [Google Scholar]
- 9.Bean W, Schell M, Katz J, Kawaoka Y, Naeve C, Gorman O, Webster R. J Virol. 1992;66:1129–1138. doi: 10.1128/jvi.66.2.1129-1138.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shope R. Public Health Rep. 1958;73:165–178. [PMC free article] [PubMed] [Google Scholar]
- 11.Chmel H, Bendinelli M, Friedman H. Pulmonary Infections and Immunity. New York: Plenum; 1994. p. 286. [Google Scholar]
- 12.Braude A I, Davis C E, Fierer J. Infectious Diseases and Medical Microbiology. Philadelphia: Saunders; 1986. p. 783. [Google Scholar]
- 13.Wolbach S B. Johns Hopkins Hosp Bull. 1919;30:104–109. [Google Scholar]
- 14.Fosso C. In: Alaska: Reflections on Land and Spirit. Hedin R, Holthaus G, editors. Tucson, AZ: Univ. of Arizona Press; 1989. pp. 215–222. [Google Scholar]
- 15.Krafft A E, Duncan D W, Bijwaard K E, Taubenberger J K, Lichy J H. Mol Diagn. 1997;2:217–230. doi: 10.1054/MODI00200217. [DOI] [PubMed] [Google Scholar]
- 16.Reid A H, Cunningham R E, Frizzera G, O’Leary T J. Am J Pathol. 1993;142:395–402. [PMC free article] [PubMed] [Google Scholar]
- 17.Swofford D L. paup, Phylogenetic Analysis Using Parsimony. Champaign, IL: Illinois Natural History Survey; 1991. , Version 3.1.1. [Google Scholar]
- 18.Kumar S, Tamura K, Nei M. mega, Molecular Evolutionary Genetics Analysis. University Park, PA: Pennsylvania State Univ.; 1993. , Version 1.01. [Google Scholar]
- 19.Maddison W P, Maddison D R. macclade, Analysis of Phylogeny and Character Evolution. Sunderland, MA: Sinauer Associates; 1992. , Version 3. [Google Scholar]
- 20.Gambaryan A, Tuzikov A, Piskarev V, Yamnikova S, Lvov D, Robertson J, Bovin N, Matrosovich M. Virology. 1997;232:345–350. doi: 10.1006/viro.1997.8572. [DOI] [PubMed] [Google Scholar]
- 21.Weis W, Brown J H, Cusack S, Paulson J C, Skehel J J, Wiley D C. Nature (London) 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 22.Ito T, Suzuki Y, Takada A, Kawamoto A, Otsuki K, Masuda H, Yamada M, Suzuki T, Kida H, Kawaoka Y. J Virol. 1997;71:3357–3362. doi: 10.1128/jvi.71.4.3357-3362.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matrosovich M, Gambaryan A, Teneberg S, Piskarev V, Yamnikova S, Lvov D, Robertson J, Karlsson K. Virology. 1997;233:224–234. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- 24.Rogers G, D’Souza B. Virology. 1989;173:317–322. doi: 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 25.Raymond F, Caton A, Cox N, Kendal A P, Brownlee G G. Virology. 1986;148:275–287. doi: 10.1016/0042-6822(86)90325-9. [DOI] [PubMed] [Google Scholar]
- 26.Schulze, I. T. (1997) J. Infect. Dis.176, Suppl. 1, S24–S28. [DOI] [PubMed]
- 27.Winter G, Fields S, Brownlee G G. Nature (London) 1981;292:72–75. doi: 10.1038/292072a0. [DOI] [PubMed] [Google Scholar]
- 28.Caton A J, Brownlee G G, Yewdell J W, Gerhard W. Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 29.Kanegae Y, Sugita S, Sortridge K, Yoshioka Y, Nerome K. Arch Virol. 1994;134:17–28. doi: 10.1007/BF01379103. [DOI] [PubMed] [Google Scholar]
- 30.Sugita S, Yoshioka Y, Itamura S, Kanegae Y, Oguchi K, Gojobori T, Nerome K, Oya A. J Mol Evol. 1991;32:16–23. doi: 10.1007/BF02099924. [DOI] [PubMed] [Google Scholar]
- 31.Gorman O T, Bean W J, Webster R G. Curr Top Microbiol Immunol. 1992;176:75–97. doi: 10.1007/978-3-642-77011-1_6. [DOI] [PubMed] [Google Scholar]
- 32.Gorman O, Bean W, Kawaoka Y, Donatelli I, Guo Y, Webster R. J Virol. 1991;65:3704–3714. doi: 10.1128/jvi.65.7.3704-3714.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shope R E. J Exp Med. 1936;63:669. doi: 10.1084/jem.63.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beveridge W. Influenza: The Last Great Plague, an Unfinished Story of Discovery. New York: Prodist; 1977. [Google Scholar]
- 35.Chun J. Natl Med J China. 1919;5:34–44. [Google Scholar]
- 36.Koen J S. Am J Vet Med. 1919;14:468–470. [Google Scholar]
- 37.Luoh S-M, McGregor M W, Hinshaw V S. J Virol. 1992;66:1066–1073. doi: 10.1128/jvi.66.2.1066-1073.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]