Abstract
To accelerate the biological annotation of novel genes discovered in sequenced regions of mammalian genomes, we are creating large deletions in the mouse genome targeted to include clusters of such genes. Here we describe the targeted deletion of a 450-kb region on mouse chromosome 11, which, based on computational analysis of the deleted murine sequences and human 5q orthologous sequences, codes for nine putative genes. Mice homozygous for the deletion had a variety of abnormalities, including severe hypertriglyceridemia, hepatic and cardiac enlargement, growth retardation, and premature mortality. Analysis of triglyceride metabolism in these animals demonstrated a several-fold increase in hepatic very-low density lipoprotein triglyceride secretion, the most prevalent mechanism responsible for hypertriglyceridemia in humans. A series of mouse BAC and human YAC transgenes covering different intervals of the 450-kb deleted region were assessed for their ability to complement the deletion induced abnormalities. These studies revealed that OCTN2, a gene recently shown to play a role in carnitine transport, was able to correct the triglyceride abnormalities. The discovery of this previously unappreciated relationship between OCTN2, carnitine, and hepatic triglyceride production is of particular importance because of the clinical consequence of hypertriglyceridemia and the paucity of genes known to modulate triglyceride secretion.
As the DNA sequencing phase of the Human Genome Project progresses, the linking of sequence information to biological function becomes an increasingly significant bottleneck. Though robust strategies have been developed for connecting sequence to endophenotypes such as timing, location, and level of gene expression, the task of linking sequence to phenotypes displayed at the level of the whole organism has proven more demanding. This has proven to be extremely challenging for unicellular organisms and is even more so for higher organisms, such as mammals.
One means for studying the in vivo function of several genes in parallel in mammals has been mutagen-induced or naturally occurring large genomic deletions. These have historically been used to localize genes whose altered dosage results in distinct organismal phenotypes (1), and several examples exist from studies of human contiguous gene syndromes in which single genes have been identified as contributing to distinct features of the syndrome (2–4). Although naturally occurring or mutagen-induced aneuploidy states serve as a powerful means for sifting a large genomic interval for the function of genes contained within the interval, the inability to target the location or size of such genomic alterations has limited their general utility.
Gene targeting in embryonic stem cells, although largely used for studying the function of individual genes in vivo, has recently been coupled with Cre-lox technology, enabling the creation of large chromosomal alterations. This has included the generation of deletions, inversions, translocations, and chromosome loss (5–8). In particular, consecutive insertion of two loxP recombination sites at the borders of a genomic region of interest followed by Cre-induced recombination has been shown to create targeted deletions, inversions, and duplications of up to 3–4 centimorgans in size in embryonic stem cells, which can then be transmitted into the mouse germ line (5). This combination of technologies has provided the means to create and study the effect of altering the dosage of multiple genes contained within any predetermined region of the mouse genome.
In the present study, we have used this approach to examine a 450-kb region of mouse chromosome 11, orthologous to a region of human 5q31. This interval was chosen for analysis because it has been sequenced in both humans and mice and contains a dense clustering of genes largely of unknown function (9, 10) Rather than examining genes one at a time in these studies, we were able to sift through the entire interval for genes whose absence resulted in detectable in vivo phenotypes. Through the analysis of the resulting mice, we were able to demonstrate that a recently discovered gene, OCTN2, is able to modulate hepatic triglyceride secretion, a clinically relevant but poorly understood process.
Methods
Generation of the Genomic Deletion.
A genomic DNA clone corresponding to Irf-1 was isolated from a 129SV genomic library (Stratagene) by using a probe generated by PCR with the following primers: 5′-AGAGCATAGCACTGGCCCTA-3′ and 5′-AGGCCTAGACTGGGGAGAAA- 3′. Vector ploxPNI was made by digesting ploxPneo-1 (11) with HindIII and religating (to remove a loxP site), and then digesting with BamHI and EcoRI, blunting the BamHI end with Klenow, and ligating with a 6.2-kb SmaI-EcoRI fragment from the 3′ end of Irf-1. PloxPNI was linearized with BamHI and was electroporated into R1 ES cells (12), and neo-resistant colonies were selected in the presence of 175 μg/ml of G418 (GIBCO/BRL). This construct deleted exon 10 of Irf-1, which is composed of the last 42 codons and the 3′UTR of the gene. A Csfgm genomic DNA clone was isolated from the 129SV genomic library by using a probe generated by PCR with the following primers: 5′-CCCTCTGAAAACGCTGACTC-3′ and 5′-CCATGACAGCATTGACTCTCA-3′. To create vector ploxPHTC, a 1.8-kb BamHI-SmaI fragment containing the PGK-hygromycin resistance gene was excised from pBS-PGKhyg/loxP/HPRT-D5′ (13) and was cloned adjacent to the HSV-tk gene in pPNT (14) digested with BamHI and EcoRI (blunted with Klenow). The 4.6-kb fragment containing PGK-hygromycin and HSV-tk in a head-to-tail arrangement was excised by BamHI and HindIII and was inserted into ploxPneo-1 digested with BglII and HindIII. The resulting vector, containing the cassette HSV-tk/PGK-hygromycin/loxP, was digested with HindIII and was ligated with a 7.6-kb HindIII genomic fragment located 2 kb downstream of Csfgm. PloxPHTC was linearized with XhoI, was electroporated in ES cells already targeted with the ploxPNI vector, and was grown under selection [180 μg/ml of hygromycin B (Sigma)]. Generation of deletion mice from the targeted ES cells was carried out as described (15). ES clones carrying the 450-kb deletion were injected into blastocysts of C57BL/6J mice. The males were bred to 129/SvEvTacfBR (Taconic Farms) to generate heterozygous mutant F1 mice.
Analytical Procedures.
Mice fed a regular mouse Chow diet were fasted overnight and were killed, and their livers and hearts were removed and homogenized. Lipids were extracted from the cell suspension as described (16) and were dissolved in PBS containing 5% Triton X-100. Quantitation of the triglyceride (without measuring free glycerol) and cholesterol levels were determined by using the commercially available enzymatic kits 337-B (Sigma GPO-Trinder kit) and 236691 (Boehringer Mannheim GmbH), respectively. For plasma lipid analysis, ≈100 μl of blood was collected by tail bleeding from mice fasted overnight. The concentrations of triglycerides (corrected for free glycerol) and total cholesterol in plasma were determined as described. For plasma-free fatty acids determination, 50 μl of blood was drawn in chilled paraoxonized capillary tubes (17). The tubes were placed in ice and immediately were centrifuged at 4°C. Free fatty acids were measured with the NEFA-C kit from Wako Pure Chemical (Osaka) GmbH, according to the manufacturer's recommendation. For fast protein liquid chromatography (FPLC) fractionation, 40 μl of pooled plasma was injected onto a Superose 6 column (3.2 × 30 mm, Smart-system, Amersham Pharmacia) and was eluted at a constant flow rate of 50 μl/min with PBS (pH 7.4, containing 1 mmol/liter EDTA). Fractions of 50 μl were collected and assayed for total cholesterol and triglycerides. Enzyme activities of lipoprotein lipase and hepatic lipase in postheparin plasma obtained from control (n = 5) and del11/del11 (n = 5) mice 5 min after bolus injection of sodium heparin (100 units/kg) into the tail vein were assayed as described (18).
Very-Low Density Lipoprotein (VLDL) Uptake Determination.
VLDL was isolated from pooled plasma of healthy human volunteers by ultracentrifugation at density 1.006 g/ml [40,000 rpm, SW40 rotor (Beckman Instruments, Palo Alto, CA) for 18 h at 4°C]. For VLDL turnover studies, VLDL was radiolabeled with 125I by using IODO-BEADS (Pierce). The specific activity of 125I-VLDL was 76 cpm/ng. Mice were fasted overnight and were injected in the tail vein with labeled VLDL (10 μg of tracer in 200 μl of 0.9% NaCl containing 2 mg/ml BSA). Blood samples of ≈50 μl were withdrawn from the retroorbital plexus into heparinized capillary tubes 2, 10, 20, 45, 90, 180, and 300 min after injection. The plasma content of 125I-labeled apoB was determined after propan-2-ol precipitation and measurement of 125I content of the pellet. The data were modeled by using a biexponential curve from which the fractional catabolic rate was calculated by using the reciprocal area under the curve.
RNase Protection Assay.
Total RNA was extracted from various tissues by using Trizol (GIBCO/BRL) according to the manufacturer's protocol. Radiolabeled antisense riboprobes were generated by using the MAXIscript kit (Ambion) from DNA templates generated by PCR from genomic DNA. RNase protection assays were performed by using the RPAII kit (Ambion) according to the manufacturer's instructions. The protected fragments were separated on a 5% acrylamide denaturing gel, and the dried gel was exposed to x-ray film. A β-actin probe was used as a control.
Results and Discussion
Creation of the Targeted 450-kb Deletion.
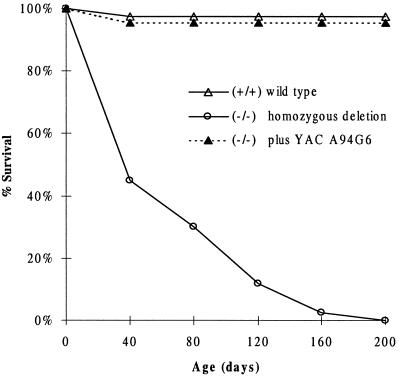
To delete the 450-kb genomic interval between Irf-1 and Csfgm (Fig. 1), two loxP sites bracketing this region were introduced sequentially into embryonic stem (ES) cells. The first loxP site was introduced into the last exon of the Irf-1 gene by using the targeting vector ploxPNI, containing a neomycin resistance (neor) gene. After G418r selection, correctly targeted ES cells were identified by Southern blot analysis. The second loxP site was introduced 2 kb downstream of Csfgm into the already ploxPNI-targeted ES cells via the targeting vector ploxPHTC, which contains tandemly linked bacterial hygromycin resistance (13) and herpes simplex virus thymidine kinase (HSV-tk) (14) genes. Southern blot analysis of hygromycin resistant clones was used to identify those that had undergone the correct targeting events. Twelve of the doubly targeted ES clones were expanded and electroporated with the Cre recombinase-expressing plasmid pBS185 (19). Because of the presence of loxP sites bracketing the hygromycin and HSV-tk markers, ES cells that had undergone Cre recombinase-mediated loxP recombination were identified by selecting for hygromycin sensitivity and 1-(2-deoxy-2-fluoro-β-d-arabinofuransyl)-5-iodouracil (FIAU) resistance. Cells surviving this selection were confirmed to contain the predicted 450-kb cis deletion (del11) by Southern blot analysis (data not shown). The clones heterozygous for the deleted chromosome were injected into blastocysts to generate chimeric mice, and germline transmission of the deletion from two independently targeted ES cell lines was obtained. Analysis of 461 live births from the mating of heterozygous animals revealed the following genotypes: 25% (+/+), 49% (+/del11), and 26% (del11/del11). Although this Mendelian ratio indicates the viability of the homozygous deletion mice, the majority of the del11/del11 mice died by 6 weeks of age, with the oldest surviving to 30 weeks (Fig. 2).
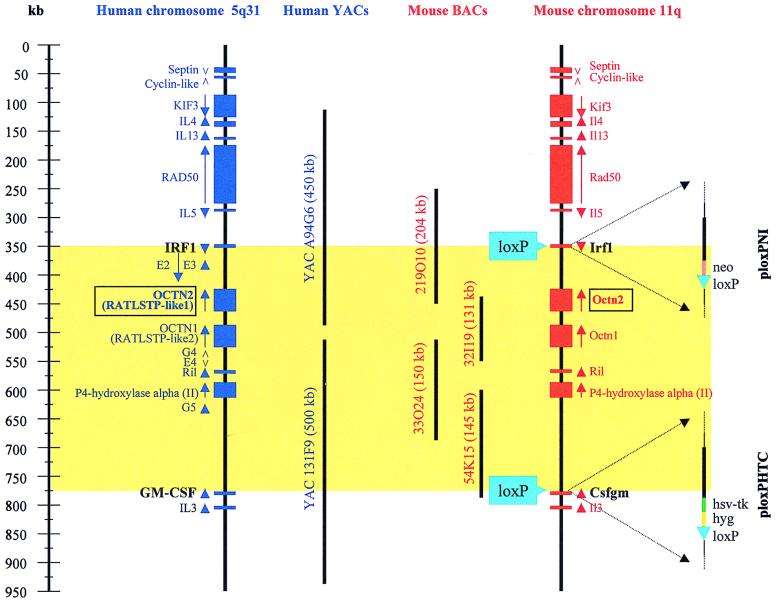
Figure 1.
The genomic organizations of the syntenic regions on mouse chromosome 11 and human chromosome 5q31. Computational analysis of human 5q31 sequences (9) identified putative genes by homology to known proteins (blue rectangles), by exact expressed sequence tag matches (E2, E3, E4, and E5) or by analysis of GRAIL-predicted exons (G4 and G5). For each gene, a vertical arrow indicates the direction of transcription. The genomic locations of the murine genes (red rectangles) in the diagram have been confirmed by content mapping of mouse chromosome 11 clones (YACs and BACs) and genomic DNA of homozygous deletion mice, as well as analyses of mouse chromosome 11 sequences (data not shown). The organization of the loxP targeting vectors, ploxPNI and ploxPHTC, introduced into the mouse genome and used to delete the 450-kb region between Irf-1 and Csfgm (shaded yellow) are shown on the right. Positions of the human YAC (medium-length bars) and mouse BAC (short bars) clones used to generate transgenic mice for the complementation studies are indicated. To create mouse BAC transgenics, BAC DNA was isolated by using an alkaline lysis method and was microinjected at a concentration of ≈1 ng/μl into fertilized FVB hybrid mouse eggs by using standard procedures. Transgenic mice were identified by PCR using primers that amplified vector sequences and genes encoded by the BAC sequences in transgenic homozygous deletion animals. Generation of the human YAC transgenic mice used in this study has been described previously (9).
Figure 2.
Life span of wild-type (n = 114), del11/del11 (n = 120), and del11/del11plus YAC A94G6 (n = 22) mice.
Analysis of del11/del11 Mice.
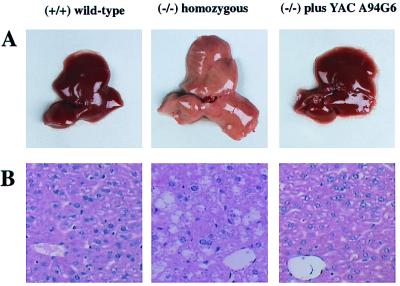
The del11/del11 mice appeared normal at birth but by 1 day of age developed abdominal swelling, the result of dramatic hepatomegaly. Gross examination revealed enlargement of livers and hearts of the del11/del11 mice (Table 1 and Fig. 3A). The body weight of these animals was significantly reduced compared with del11/+ and wild-type littermates (Table 1). Microscopic analysis of liver sections stained with hematoxylin and eosin (Fig. 3B) and Oil Red O (data not shown) demonstrated cytosolic lipid accumulation. Triglyceride content was elevated dramatically in the livers and hearts of the del11/del11 mice whereas total cholesterol level in these organs was only minimally elevated (Table 1). Changes in the lipid content observed in the hearts and livers of the del11/del11 mice were also reflected in their plasma. Free fatty acid and triglyceride levels were markedly increased whereas minimal changes were observed in their plasma cholesterol levels (Table 1). The increased plasma triglycerides, as determined by fast protein liquid chromatography, were mainly confined to the VLDL-sized fractions (results not shown).
Table 1.
Body and tissue weight, lipid levels of the various genotypes of mice: wild-type controls (+/+), +/del11 mice (+/−), del11/del11 mice (−/−), and del11/del11 mice hemizygous for human YAC A94G6 (−/−) (YAC)
| Mice genotype | Body weight, g | Liver
|
Heart
|
Plasma
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Weight, percent of body weight | TG, mg/g | TC, mg/g | Weight, percent of body weight | TG, mg/g | TC, mg/g | TG, mg/dl | TC, mg/dl | FFA, mmol/liter | ||
| +/+ | 15.7 ± 3.2 | 4.7 ± 0.7 | 3.2 ± 1.0 | 2.9 ± 0.7 | 0.5 ± 0.1 | 3.5 ± 0.9 | 1.7 ± 0.2 | 40.6 ± 24.6 | 142.5 ± 22.2 | 0.3 ± 0.1 |
| +/− | 16.1 ± 2.9 | 4.8 ± 0.7 | 5.9 ± 3.5 | 2.4 ± 0.6 | 0.5 ± 0.1 | 4.1 ± 2.4 | 1.7 ± 0.5 | 30.3 ± 6.2 | 145.7 ± 34.3 | 0.3 ± 0.1 |
| −/− | 10.1 ± 1.6* | 10.7 ± 2.4* | 45.6 ± 13.6* | 3.3 ± 0.9 | 1.3 ± 0.2* | 13.6 ± 4.3* | 2.4 ± 0.9 | 407.1 ± 172.5* | 162.9 ± 57.2 | 1.6 ± 0.1* |
| −/− (YAC) | 17.2 ± 2.1 | 5.3 ± 0.3 | 6.4 ± 2.7 | 2.2 ± 0.2 | 0.5 ± 0.1 | 3.1 ± 1.2 | 1.3 ± 0.5 | 56.8 ± 20.4 | 142.4 ± 34.3 | ND |
At 28 days of age, mice fed a regular Chow diet were weighed and killed, and their livers and hearts were removed and weighed. Body and tissue weights are expressed as the mean ± SD of 10 mice per group. A second group of mice at 28 days of age was fasted overnight and killed, the triglycerides (TG) and total cholesterol (TC) levels were measured in their livers, hearts, and plasma, and free fatty acids (FFA) levels were determined in plasma. Values are expressed as the mean ± SD of at least 5 mice per group. ND, not determined.
P < 0.05, significantly different between the homozygous (−/−) deletion mice and wild-type controls (nonparametric Mann–Whitney U test).
Figure 3.
(A) Livers from 28-day-old mice fed a normal mouse Chow diet. (B) The livers shown in A were formalin-fixed and paraffin-embedded, and 4-μm sections were stained with hematoxylin and eosin (H&E). Cytosolic lipid accumulation is apparent in the hepatocytes of the liver from the del11/del11 mice. Original magnification: ×200.
To explore the mechanism underlying the hypertriglyceridemia, an investigation of triglyceride metabolism in mice of the various genotypes was performed. These studies analyzed the three major steps involved in VLDL triglyceride metabolism: lipolysis, hepatic uptake, and hepatic secretion. To determine whether the elevated levels of plasma triglycerides in the del11/del11 mice were attributable to impaired lipolysis, the activity of both hepatic lipase and lipoprotein lipase was measured in postheparin plasma. Similar activities were noted in plasma from del11/del11 and wild-type mice for hepatic lipase (10.6 ± 1.9 vs. 12.3 ± 0.7 μmol/ml/h, P = 0.076) and lipoprotein lipase (14.3 ± 4.8 vs. 18.1 ± 2.7 μmol/ml/h, P = 0.83). The hepatic uptake of human VLDL-apoB was assessed by injecting 125I-labeled VLDL into the tail vein of animals of the different genotypes and determining the rate of VLDL removal. No significant differences were observed in clearance of VLDL particles from the circulation of del11/del11 and wild-type mice as reflected by the calculated VLDL fractional catabolic rates (0.33 ± 0.05 vs. 0.43 ± 0.08 pool apoB/h, respectively, P = 0.1573).
VLDL Metabolism.
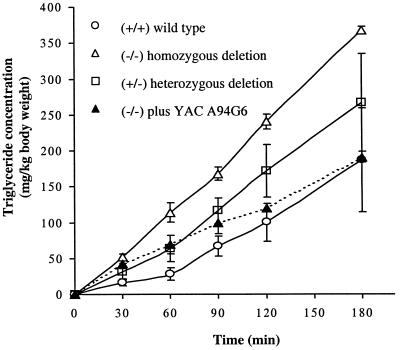
Alterations in hepatic VLDL-triglyceride secretion as a cause of the elevated plasma triglycerides levels were assessed by injecting fasted animals with Triton WR1339 coupled with timed triglyceride measurements (20). Triton WR 1339 inhibits lipolysis and hepatic uptake of VLDL triglycerides, thereby enabling assessment of changes in plasma VLDL triglycerides due to hepatic VLDL triglyceride secretion. After Triton WR 1339 injections, the hepatic VLDL-triglyceride secretion rates were ≈3- and 2-fold elevated in del11/del11 and del11/+ mice, respectively, compared with control animals (Fig. 4). These data indicate increased hepatic triglyceride secretion as a major etiology of the hypertriglyceridemia in mice containing the deleted chromosome.
Figure 4.
Hepatic VLDL-triglyceride secretion rates in the various phenotypes of mice. Triton WR 1339 (500 mg/kg body weight) was injected into four groups of mice (n = 3–4 for each group). Plasma triglyceride levels were determined at 30, 60, 90, 120, and 180 min after injection, were corrected for the triglyceride level at the time of injection (0 min), and were related to the body mass of the animals. Values are given as means ± SD. The hepatic triglyceride secretion rate was calculated from the slope of the curve and was significantly different for both del11/del11 (115.4 ± 1.8 mg/h/kg body weight, P < 0.034) and +/del11 (78.1 ± 16.7 mg/h/kg body weight, P < 0.043) mice compared with wild-type controls (45.5 ± 12.8 mg/h/kg body weight). The hepatic triglyceride secretion rate in the del11/del11 mice that were hemizygous for human YAC A94G6 was not significantly different from wild-type mice (66.8 ± 6.5 mg/h/kg body weight P < 0.157). Statistical analysis was done with the nonparametric Mann–Whitney U test.
Transgene Complementation of del11/del11 Phenotype.
The abnormalities observed in the del11/del11 mice (decreased body weight, enlarged fatty livers and hearts, shortened life span, and increased hepatic triglyceride secretion) are, most likely, the consequence of loss of gene(s) function, and, therefore, transgene complementation was used to identify the individual gene(s) responsible for these traits. Transgenic mice were created by using mouse chromosome 11 BACs—219O10, 32I19, 33O24, and 54K15—and human YACs from the homologous region of human 5q31—A94G6 and 131F9 (Fig. 1). The four overlapping mouse BAC clones together encompass the entire deleted interval whereas the two human YAC clones span most of the deleted region, with the exception of a small gap (20 kb). Lines of mice homozygous for the deletion as well as hemizygous for each of the murine BAC and human YAC transgenes were generated through breeding and then were phenotypically assessed. The four murine BACs and the human YAC 131F9 failed to complement any of the abnormalities noted in the del11/del11 mice. In contrast, del11/del11 mice harboring the human YAC A94G6 were indistinguishable in body weight and liver and heart size (Table 1), life span (Fig. 2), plasma and hepatic triglyceride levels (Table 1), and hepatic VLDL-TG production (Fig. 4) from wild-type controls.
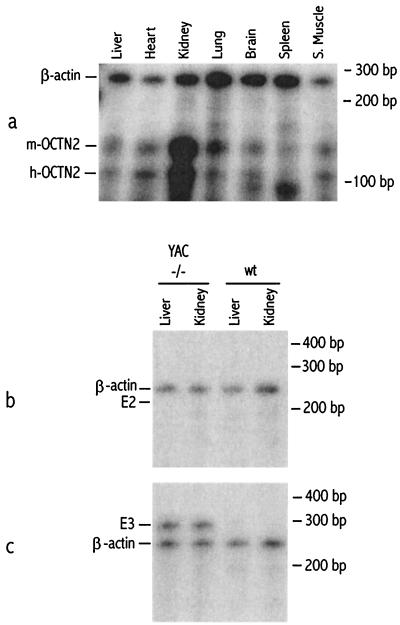
To identify the human gene(s) on YAC A94G6 responsible for complementing the deletion-associated abnormalities, we examined and compared the gene content of human YAC A94G6 and the mouse chromosome 11-deleted interval. A combination of mouse/human sequence comparisons and Southern and Northern blot analyses was used to compare the set of putative genes encoded by sequences on human YAC A94G6 to those present in deletion and wild-type mice. Of the nine putative genes encoded by human YAC A94G6, three, OCTN2, E2, and E3, failed to express because of their absence in the genome of mice homozygous for the chromosome 11 deletion. Of these genes, however, only OCTN2 appears to have a murine homologue, present in wild-type but absent in del11/del11 mice. OCTN2 has recently been identified as an organic cation transporter (21–23). Expression analysis of tissues from the YAC A94G6 transgenic animals, with probes specific for murine and human OCTN2, revealed a similar expression pattern, both genes being expressed at a high level in the kidney and at moderately high levels in the liver, heart, skeletal muscle, brain, and spleen (Fig. 5a). The mouse BACs 219O10 and 32I19 contained parts of, but not the entire, murine OCTN2 gene, and del11/del11 mice containing these BACs as transgenes did not produce a murine OCTN2 transcript (data not shown). The human transcription units designated E2 and E3 present within YAC A94G6 had been originally identified based on exact matches in the human expressed sequence tag database (9). Sequences homologous to these two putative human transcription units have not been reported in the mouse expressed sequence tag database nor detected within the murine genomic sequence that is available for the entire 450-kb deleted interval. Sensitive RNase protection analysis of the A94G6 YAC transgenics failed to detect E2 expression in the several tissues examined (Fig. 5b) whereas a human E3 transcript was easily detected in the liver and kidney (Fig. 5c). Because E3 comprised a small putative ORF (90 bp) and had an expression pattern similar to that of OCTN2, we explored whether it was an untranslated component of the OCTN2 transcript. Reverse transcription–PCR analysis of liver and kidney RNA from the YAC A94G6 transgenics with primers to E3 and the 3′ UTR of the OCTN2 cDNA generated a 0.9-kb product. This result indicates that E3 is not an independent transcription unit, but either a 3′ splice variant or a previously unrecognized 3′ end of the OCTN2 transcript. These reverse transcription–PCR results for E3 coupled with the absence of a murine homologue to E2 or detectable human E2 transcripts in the YAC A94G6 transgenics indicates OCTN2 as the one gene present on human YAC A94G responsible for complementing the deletion associated abnormalities.
Figure 5.
RNase protection analysis of OCTN2, E2, and E3 expression. (a) Total RNA from liver, heart, kidney, lung, brain, spleen, and skeletal muscle from a wild-type mouse hemizygous for YAC A94G6 was hybridized with a probe specific for both murine and human OCTN2. (b) Total RNA from liver and kidney of a del11/del11 mouse hemizygous for YAC A94G6 (YAC −/−) and of a wild-type mouse (wt) was hybridized with a 208-bp probe specific for E2. (c) Total RNA from liver and kidney of a del11/del11 mouse hemizygous for YAC A94G6 (YAC −/−) and of a wild-type mouse (wt) was hybridized with a 280-bp probe specific for E3. A probe for β-actin was used as a control.
OCTN2 and Its Effect on Carnitine and Trigelyceride Metabolism.
OCTN2 has been reported as having the capability of transporting carnitine across the cellular membrane with high specificity (24). Recently, defects in this gene have been implicated as being responsible for primary carnitine deficiency in humans (22, 23) and in the murine model for this condition, the juvenile visceral steatosis (jvs) mouse (25). Based on this information, we investigated whether mice containing the del11 allele suffered from general carnitine deficiency. Total carnitine levels were markedly decreased in plasma of del11/del11 mice compared with wild-type mice (13.1 ± 2.1 vs. 56.0 ± 3.1 nmol/ml, P < 0.021, respectively). In addition, a significant decrease in total carnitine levels was noted in plasma of the del11/+ mice (44.4 ± 1.9 nmol/ml, P < 0.021). Because the phenotypic abnormalities in humans with primary carnitine deficiency and in jvs mice containing an OCTN2 missense mutation are corrected by dietary carnitine supplementation, we investigated whether massive systemic carnitine administration could correct any of the abnormalities in the del11/del11. l-carnitine (1 mg) was injected into del11/del11 mice daily from postnatal days 10–28, which is the dose regimen reported to correct some of the abnormalities in jvs mice (26). This administration of carnitine failed to rescue any of the phenotypic abnormalities noted in the del11/del11 mice.
An unexpected finding of this study was that, despite the complete absence of a contiguous, 450-kb, gene-rich interval, the del11/del11 mice were viable. This represents the largest reported chromosomal deletion in mice or humans associated with viability in the homozygous state (27). Although many of the deletion-associated abnormalities in the del11/del11 mice were corrected by the expression of the human OCTN2-containing transgene, the differences between the jvs and del11/del11 mice, including the failure of carnitine rescue and viability differences, suggest that genes in the deleted interval other than OCTN2 contribute to their phenotype. These mice, with and without the various transgenes, coupled to careful phenotyping, will serve as useful reagents for further deciphering the function of the various genes in the deleted interval. Although the approach used in this study is still labor-intensive, it illustrates a general strategy for screening, in either heterozygous and/or homozygous deletion mice, the properties of several individual genes in parallel contained within a genomic interval of particular interest.
Abnormalities in fatty acid metabolism have long been noted in patients with primary carnitine deficiency [systemic carnitine deficiency; online Mendelian inheritance in man (OMIM) no. 212140] and the jvs mice (28) whereas alterations specifically in the metabolism of triglycerides have not previously been explored. The phenotypic screens and follow-up analyses of the del11/del11 mice, all carried out before identification of OCTN2's physiological role, were unbiased by assumptions of the functions of individual genes within the deleted interval. This facilitated the discovery of a novel and previously unappreciated aspect of the carnitine-deficient phenotype: the relationship between carnitine, hepatic triglyceride production, and OCTN2. The findings of the present study, coupled with reports of carnitine administration significantly reducing plasma triglyceride concentrations in hypertriglyceridemic hemodialysis patients (29, 30) as well as in patients with type IV hyperlipoproteinemia (31), provides genetic evidence in support of a critical role for carnitine transport in the regulation of plasma triglyceride metabolism. The analysis of the del11/del11 as well as the del11/+ mice suggest that reductions in carnitine, whose intracellular concentration appears to be affected by modest decreases in OCTN2 activity, leads to the shunting of fatty acids away from mitochondrial transport into triglyceride storage and secretory pools. In the discovery of OCTN2's ability to modulate hepatic triglyceride secretion and plasma levels, we have identified OCTN2 as a potential intervention target for the management of this cardiovascular risk factor.
Acknowledgments
The authors thank F. Kuipers (Academic Hospital Groningen) for help in quantifying carnitine in plasma and P. Blanche and M. La Belle for help with the lipase assays and 125I-VLDL metabolism experiments. Research was conducted at the E.O. Lawrence Berkeley National Laboratory. The work was supported by National Institute of Health Grants 18574 and 50590 and was performed under Department of Energy Contract DE-AC0376SF00098, University of California.
Abbreviations
- VLDL
very-low density lipoprotein
- ES cell
embryonic stem cell
References
- 1.Holdener-Kenny B, Sharan S K, Magnuson T. BioEssays. 1992;14:831–839. doi: 10.1002/bies.950141208. [DOI] [PubMed] [Google Scholar]
- 2.Frangiskakis J M, Ewart A K, Morris C A, Mervis C B, Bertrand J, Robinson B F, Klein B P, Ensing G J, Everett L A, Green E D, et al. Cell. 1996;86:59–69. doi: 10.1016/s0092-8674(00)80077-x. [DOI] [PubMed] [Google Scholar]
- 3.Yamagishi H, Garg V, Matsuoka R, Thomas T, Srivastava D. Science. 1999;283:1158–1161. doi: 10.1126/science.283.5405.1158. [DOI] [PubMed] [Google Scholar]
- 4.Burch G H, Gong Y, Liu W, Dettman R W, Curry C J, Smith L, Miller W L, Bristow J. Nat Genet. 1997;17:104–108. doi: 10.1038/ng0997-104. [DOI] [PubMed] [Google Scholar]
- 5.Ramirez-Solis R, Liu P, Bradley A. Nature (London) 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 6.Smith A J, De Sousa M A, Kwabi-Addo B, Heppell-Parton A, Impey H, Rabbitts P. Nat Genet. 1995;9:376–385. doi: 10.1038/ng0495-376. [DOI] [PubMed] [Google Scholar]
- 7.Lewandoski M, Martin G R. Nat Genet. 1997;17:223–225. doi: 10.1038/ng1097-223. [DOI] [PubMed] [Google Scholar]
- 8.Herault Y, Rassoulzadegan M, Cuzin F, Duboule D. Nat Genet. 1998;20:381–384. doi: 10.1038/3861. [DOI] [PubMed] [Google Scholar]
- 9.Frazer K L, Ueda Y, Zhu Y, Gifford V R, Garofalo M R, Mohandas N, Martin C H, Palazzolo M J, Cheng J F, Rubin E M. Genome Res. 1997;7:495–512. doi: 10.1101/gr.7.5.495. [DOI] [PubMed] [Google Scholar]
- 10.Symula D J, Frazer K A, Ueda Y, Denefle P, Stevens M E, Wang Z-E, Locksley R, Rubin E M. Nat Genet. 1999;23:241–244. doi: 10.1038/13880. [DOI] [PubMed] [Google Scholar]
- 11.Nagy A, Moens C, Ivanyi E, Pawling J, Gertsenstein M, Hadjantonakis A K, Pirity M, Rossant J. Curr Biol. 1998;8:661–664. doi: 10.1016/s0960-9822(98)70254-4. [DOI] [PubMed] [Google Scholar]
- 12.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith A J, De Sousa M A, Kwabi-Addo B, Heppell-Parton A, Impey H, Rabbitts P. Nat Genet. 1995;9:376–385. doi: 10.1038/ng0495-376. [DOI] [PubMed] [Google Scholar]
- 14.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 15.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embyo: A Laboratory Manual. Plainview, New York: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 16.Bligh E G, Dyer W J. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 17.Zambon A, Hashimoto S I, Brunzell J D. J Lipid Res. 1993;34:1021–1028. [PubMed] [Google Scholar]
- 18.Krauss R M, Levy R, Frederickson D. J Clin Invest. 1974;54:1107–1124. doi: 10.1172/JCI107855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sauer B, Henderson N. New Biol. 1990;2:441–449. [PubMed] [Google Scholar]
- 20.Aalto-Setala K. J Clin Invest. 1992;90:1889–1900. doi: 10.1172/JCI116066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu X, Prasad P D, Leiba F H, Ganapathy V. Biochem Biophys Res Commun. 1998;246:589–595. doi: 10.1006/bbrc.1998.8669. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Ye J, Ganapathy V, Longo N. Proc Natl Acad Sci USA. 1999;96:2365–2360. doi: 10.1073/pnas.96.5.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nezu J, Tamai I, Oku A, Ohashi R, Yabuuchi H, Hashimoto N, Nikaido H, Sai Y, Koizumi A, Shoji Y, et al. Nat Genet. 1999;21:91–94. doi: 10.1038/5030. [DOI] [PubMed] [Google Scholar]
- 24.Tamai I, Ohashi R, Nezu J, Yabuuchi H, Oku A, Shimane M, Sai Y, Tsuji A. J Biol Chem. 1998;273:20378–20382. doi: 10.1074/jbc.273.32.20378. [DOI] [PubMed] [Google Scholar]
- 25.Lu K M, Nishimori H, Nakamura Y, Shima K, Kuwajima M. Biochem Biophys Res Commun. 1998;252:590–594. doi: 10.1006/bbrc.1998.9708. [DOI] [PubMed] [Google Scholar]
- 26.Horiuchi M, Kobayashi K, Tomomura M, Kuwajima M, Imamura Y, Koizumi T, Nikaido H, Hayakawa J, Saheki T. J Biol Chem. 1992;267:5032–5035. [PubMed] [Google Scholar]
- 27.Epstein C J. The Consequences of Chromosome Imbalance: Principles, Mechanisms and Models. Cambridge, U.K.: Cambridge Univ. Press; 1986. [Google Scholar]
- 28.Tomomura M, Tomomura A, Musa D A, Horiuchi M, Takiguchi M, Mori M, Saheki T. J Biochem (Tokyo) 1997;121:172–177. doi: 10.1093/oxfordjournals.jbchem.a021562. [DOI] [PubMed] [Google Scholar]
- 29.Guarnieri G F, Ranieri F, Toigo G, Vasile A, Ciman M, Rozzoli V, Moracchiello M, Campanacci L. Am J Clin Nutr. 1980;33:1489–1492. doi: 10.1093/ajcn/33.7.1489. [DOI] [PubMed] [Google Scholar]
- 30.Elisaf M, Bairaktari E, Katopodis K, Pappas M, Sferopoulos G, Tzallas C, Tsolas O, Siamopoulos K C. Am J Nephrol. 1998;18:416–421. doi: 10.1159/000013386. [DOI] [PubMed] [Google Scholar]
- 31.Maebashi M, Kawamura N, Sato M, Imamura A, Yoshinaga K. Lancet. 1978;ii:805–807. doi: 10.1016/s0140-6736(78)92587-4. [DOI] [PubMed] [Google Scholar]