Abstract
Centromere protein A (Cenpa for mouse, CENP-A for other species) is a histone H3-like protein that is thought to be involved in the nucleosomal packaging of centromeric DNA. Using gene targeting, we have disrupted the mouse Cenpa gene and demonstrated that the gene is essential. Heterozygous mice are healthy and fertile whereas null mutants fail to survive beyond 6.5 days postconception. Affected embryos show severe mitotic problems, including micronuclei and macronuclei formation, nuclear bridging and blebbing, and chromatin fragmentation and hypercondensation. Immunofluorescence analysis of interphase cells at day 5.5 reveals complete Cenpa depletion, diffuse Cenpb foci, absence of discrete Cenpc signal on centromeres, and dispersion of Cenpb and Cenpc throughout the nucleus. These results suggest that Cenpa is essential for kinetochore targeting of Cenpc and plays an early role in organizing centromeric chromatin at interphase. The evidence is consistent with the proposal of a critical epigenetic function for CENP-A in marking a chromosomal region for centromere formation.
Keywords: kinetochore, epigenetic, gene targeting
The centromere is an essential chromosomal component required for the faithful segregation of chromosomes during mitosis and meiosis. The kinetochore is a DNA-protein complex comprising both constitutive proteins that are present at the centromere throughout the cell cycle and transient proteins that are present at various stages (1). Three of the best-studied constitutive proteins are centromere proteins CENP-A, CENP-B, and CENP-C. CENP-A is a 17-kDa histone H3-like protein involved in centromeric nucleosome formation (ref. 2; described below). CENP-B is an 80-kDa protein that binds a 17-bp motif known as the CENP-B box, which is present in human α-satellite and mouse minor satellite DNA (3, 4). Gene knockout analysis of Cenpb in mice indicates that this protein is not essential (5–7), although a decrease in body weight and testis size accompanied protein deficiency (5). CENP-C is a 140-kDa protein that interacts with chromatin at the inner kinetochore plate (8). In vitro DNA binding studies suggest that CENP-C may bind to DNA (9). CENP-C null mutation results in embryonic lethality at 3.5 days postconception (pc), with a missegregation phenotype and metaphase arrest (10, 11). Metaphase arrest also is observed after microinjection of anti-CENP-C antibodies at interphase (12). CENP-C shares a region of homology with Mif2, a Saccharomyces cerevisiae protein. Mutations in the MIF2 gene result in defective chromosome segregation and delayed progression through mitosis (13). However, CENP-C alone is not sufficient to induce centromeric formation (14).
A number of transient centromere proteins now have been described (1, 15–19). Of particular relevance to the present study involving the use of the gene targeting technique is the inner CENP (INCENP). This protein localizes to the centromere at early mitosis and is present on the metaphase plate at the metaphase-anaphase transition (20). Gene disruption in mice reveals embryonic lethality at 2.5 days pc, accompanied by enlarged nuclei containing an increased number of nucleoli, nuclear bridging, chromosome condensation, and spindle fiber bundling (21).
The role of CENP-A in centromeric function has yet to be elucidated. CENP-A is a histone-H3 like protein that is conserved in mammals (22, 23) and S. cerevisiae (24). The C terminal (residues 48–135) of CENP-A is 62% identical to that of histone H3 and corresponds to the histone fold domain. The histone fold domain consists of three α-helices (HI, HII, and HIII) separated by two β-sheet structures (strand A and strand B) (25) (see Fig. 1). This domain of histone H3 has been shown to be sufficient for nucleosome assembly in vitro (26) and in vivo (27). There is no similarity seen between the N-termini sequences (residues 1–47) of CENP-A and normal histone H3 (2). Although this divergence initially was thought to provide CENP-A with the centromere targeting property, a histone H3 chimeric protein containing the N terminus of CENP-A and the histone H3 histone fold domain failed to localize to the centromere, indicating that the C-terminal end is responsible for centromere targeting (28). CENP-A synthesis appears to be coupled with centromere replication during mid-S to early G2 phase, whereas histone H3 expression peaks early in S phase (28). Expression of CENP-A under the histone H3 promoter fails to localize at the centromere (28). These studies suggest that CENP-A is involved in the packaging of centromeric chromatin and that the protein may provide an early epigenetic marker for centromere formation (29).
Figure 1.
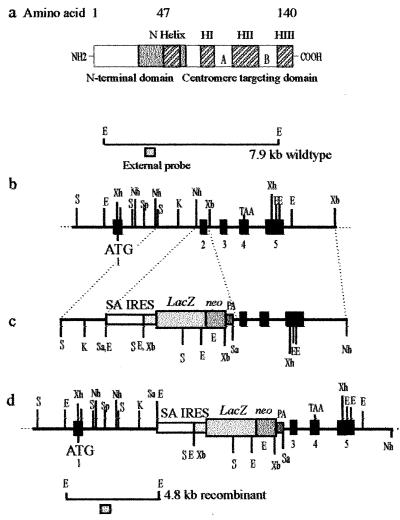
Targeted disruption of the mouse Cenpa gene. (a) The mouse Cenpa protein showing the different subdomains, in particular those at the C terminus that are required for centromere targeting. Our targeting construct (see below) was designed to delete amino acids 29–64 (gray box), which will effectively remove the entire centromere-targeting domain. (b) A restriction map of the Cenpa gene. The exons are denoted by black boxes (23). (c) The gene replacement construct, where the selectable marker cassette consists of a splice-acceptor site (SA), a picornaviral IRES, a lacZ-neomycin-resistance fusion gene, and a simian virus 40 polyadenylation sequence (PA). (d) The Cenpa locus after gene disruption. The positions of external probes used in Southern analysis are shown and the expected size fragments are 7.9-kb wild-type allele and a 4.8-kb targeted allele. ATG and TAA are translation start and stop codons, respectively. Restriction enzymes used were SacI (S), SalI (Sa), EcoRI (E), XbaI (Xb), XhoI (Xh), KpnI (K), NheI (Nh), and SpeI (Sp).
Only limited functional data are available for CENP-A. Microinjection of antibodies raised against the N terminus of CENP-A into HeLa cells within 3 hr of G1/S release resulted in interphase arrest (30). Highly condensed nuclei, granular cytoplasm, and loss of cell division capability were observed. Antibody injection in midinterphase did not disrupt mitosis; however, a mitotic lag was observed possibly because of the antibody interfering with microtubule attachment (31). Studies on CSE4p (chromosome segregation protein), an S. cerevisiae homolog of CENP-A, have demonstrated the protein to be a component of the core centromere (32). Mutation in CSE4p results in missegregation and cell arrest in mitosis; however, the increase in chromosome loss is slight (33). The arrest phenotype is consistent with a specific cell division block that appears to occur after the mitotic spindle has formed but before the onset of anaphase (33). The arrested cells have a 2n DNA content, indicating that DNA replication has taken place before arrest.
To further understand the role of CENP-A in centromere function, we have used the technique of gene targeting by homologous recombination to enable the production of Cenpa null mice. Our analysis of Cenpa null mutants has enabled us to elucidate the involvement of this protein in mitotic cell division and, in particular, its role in kinetochore assembly.
Methods
Construction of Targeting Vectors.
The targeting construct contains 6.4 kb of the Cenpa gene (23) and was used to delete exon 2 (amino acids 29–64) and disrupt the protein through the introduction of the selectable marker cassette. Exon 2 was deleted via the flanking NheI and XbaI sites followed by the insertion of a selectable marker cassette isolated from pGT1.8IRES.βgeo, where IRES is the internal ribosome-entry site (34). This construct, when homologously recombined into the mouse Cenpa locus, will result in a truncated protein lacking the centromere targeting domain (Fig. 1).
Generation of Targeted Embryonic Stem (ES) Cells and Mice.
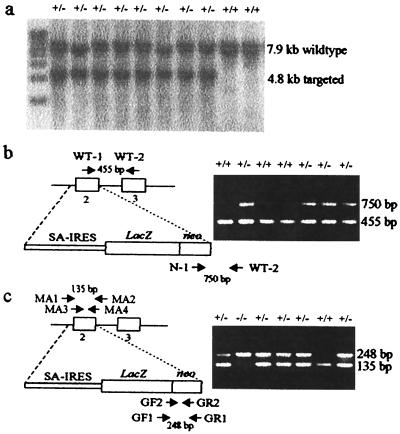
Mouse ES cells (129/1) were electroporated with 40 μg of linearized construct DNA, grown on STO/NeoR feeders, and growth-selected by using G418 (21). Resistant colonies were genotyped by Southern blot (Fig. 2a). Chimeric mice were produced as described (21). Mice were genotyped by PCR. DNA was extracted from mouse tails as described (21). Primers designed by using geneworks were: WT-1 (5′-TCAGACACTGCGCAGAAGAC); WT-2 (5′-GAGCTTAGGAACTGGCATGG); and N-1 (5′-TTCTATCGCCTTCTTGACGAG) (Fig. 2b).
Figure 2.
Southern blotting and PCR genotyping of cell line, tail, and embryo DNA. (a) Southern blot analysis of putative targeted ES cell colonies after EcoRI digestion and probed with an external probe (see Fig. 1 b and d). (b) PCR analysis of mouse tail DNA showing a wild-type product of 455 bp detected by WT-1 and WT-2 primers, and a targeted product of 750 bp detected by N-1 and WT-2 primers. SA, splice-acceptor site. (c) Nested PCR of mouse embryos resulting in a 135-bp wild-type product when using primers MA1, MA2, MA3, and MA4, and a 248-bp targeted product when using primers GF1, GR1, GF2, and GR2.
Genotyping of Preimplantation Embryos.
Preimplantation embryos were obtained from heterozygous mice. Breeding pairs were examined daily for vaginal plugs (an indicator of 0.5 days gestation) and denoted as 0.5 days pc. A nested PCR protocol was designed and used for the amplification of the 2.5-day embryonic DNA. Embryos were flushed and transferred to PCR tubes in 25 μl of dH2O. Nest 1a: denaturation at 95°C for 15 min. Nest 1b: addition of 10× buffer (containing 15 mM MgCl2) (Perkin–Elmer), 0.2 mM dNTP, 250 ng of wild-type primers MA1, MA2, and LacZ-neomycin primers GF1 and GR1, 1 unit of AmpliTaq DNA Polymerase (Perkin–Elmer) in a final volume of 50 μl. Cycle 1: 95°C for 2 min; 55°C for 3 min; 72°C for 90 sec. Cycles 2–30: 95°C for 60 sec; 57°C for 60 sec, and 72°C for 90 sec. Nest 2: Using 1 μl of the Nest 1b product, add 10× buffer, 0.2 mM dNTP, primers MA3, MA4, GR2, GF2, 1 unit of AmpliTaq, in a final volume of 25 μl. Cycle 1: 95°C, 2 min; 58°C, 60 sec; 72°C, 90 sec. Cycles 2–30: 95°C, 1 min; 58°C, 1 min; 72°C, 90 sec. Oligonucleotide primer sequences were: MA1 (5′-TGGAACTGCAGTCTGGGAAC); MA2 (5′-TCTGTCTTCTGCGCAGTGTC); GF1 (5′-AGTATCGGCGGAATTCCAG); GR1 (5′-GATGTTTCGCTTGGTGGTC); MA3 (5′-CCCAAAGCTCAGAGCAAATTC); MA4 (5′-AGTATGTGGCAGCACAGCAG); GR2 (5′-CCTCGTCCTGCAGTTCATGTCTGGTG); and GF2 (5′-CCATTACCAGTTGGTCTGGTG) (Fig. 2c).
Genotyping of Day-8.5 Embryos.
Individual embryos were dissected from their implantation site at 8.5 days gestation, washed twice in PBS, and transferred to a microfuge tube. Mouse tail lysis buffer and proteinase K (1 μg/ml) were added and incubated at 50°C for 4 hr. DNA was extracted twice with phenol-chloroform and once with chloroform followed by ethanol precipitation using 1 μl of glycogen. This precipitate was resuspended in 20 μl of Tris-EDTA from which 5 μl was used for each PCR.
Culturing of Mouse Embryos.
A culturing protocol was introduced to assess embryo development between 3.5 and 8.5 days pc. Embryos were flushed at 3.5 days and cultured in 15-mm diameter dishes (Nunc) with ES media supplemented with leukemia inhibiting factor (AMRAD, Melbourne, Australia) at 37°C, 5% CO2. Embryos were photographed daily and harvested at 8.5 days if normal in appearance or earlier if signs of degeneration were apparent. The embryos were rinsed in PBS and treated with 0.25% trypsin for 3–5 min, resulting in detachment of the trophectoderm cells. Micro-glass pipettes were used to collect the cells. The PCR protocol was the same as that used for the 8.5-day embryos (see Fig. 2c).
Giemsa Staining of Embryos.
Embryos (2.5 and 3.5 days) were placed in M16 media (Sigma) under oil, then transferred to a microwell containing 0.6% trisodium citrate for 4–8 min. Individual embryos were placed on glass slides and fixed in a droplet of methanol/acetic acid (3:1). After two rinses in fixative, the embryos were stained with 10% Giemsa in PBS, pH 6.8 (Gurr), for 10 min, air-dried, and mounted in DPX (BDH) for analysis. To enable morphological analysis of 4.5-, 5.5-, and 6.5-day embryos, embryos were cultured on gelatinized (0.1% gelatin in PBS) coverslips (22 mm × 22 mm) in 35-mm Petri dishes (Nunc). Embryos that failed to attach to the coverslips were harvested and treated in the same manner as the 2.5- and 3.5-day embryos. Embryos were fixed in methanol/acetic acid for 10 min, stained as above, and examined on an Olympus 1×70 microscope/Nikon F-601 camera system.
Mitotic Index Determination.
Cultured embryos were either Giemsa-stained (as above) or fixed in 2% paraformaldehyde and mounted in Vectorshield containing 4′,6-diamidino-2-phenylindole (DAPI). DAPI staining of embryos was used to enable mitotic detection in various focal planes. The number of cells undergoing mitosis was calculated as a percentage of the total cell number using both methods.
Immunofluorescence Analysis of Embryos.
Embryos were cultured on coverslips, rinsed twice in PBS, fixed for 5 min in 2% paraformaldehyde in PBS, permeabilized with 0.1% Triton X-100 in PBS for 2 min, and rinsed an additional two times in PBS (35). Antibodies were applied to coverslips for 1 hr at 37°C. The coverslips were washed three times for 5 min in 1× KB buffer (10 mM Tris⋅HCl/15 mM NaCl/0.1% BSA), the secondary antibody was applied for 1 hr and washing was repeated. The coverslips then were rinsed in PBS and stored in PBS at 4°C. Embryos were mounted in 4′,6-diamidino-2-phenylindole. Image analysis was performed by using an Axioskop fluorescence microscope equipped with a 63× objective (Zeiss), a charge-coupled device camera (Photometrics Image Point, Tucson, AZ) and iplab software (Signal Analytics, Vienna, VA).
Results
Generation of Cenpa Heterozygous Cell Lines and Mice.
The Cenpa-neoR construct (Fig. 1c) was transfected into 129/1 ES cells grown on STO/NeoR feeder cells and placed under G418 selection for 7–10 days. Screening of 48 resistant clones by Southern blot analysis gave 26 positive clones (Fig. 2a), indicating a targeting efficiency of 54%. Microinjection of these heterozygous targeted cell lines into blastocysts resulted in four germ-line chimeras. These mice were crossed with C57BL/6 to produce heterozygous mice (Fig. 2b).
Embryonic Lethality of Cenpa Null Offspring Occurs Postimplantation Between Days 3.5 and 8.5 pc.
The heterozygous mice were phenotypically normal with no obvious impairment of growth or fertility. Intercrossing of heterozygous mice resulted in a total of 186 progeny of which 63 (34%) were +/+ and 123 (66%) were +/−, indicating embryonic lethality of the homozygous mutant state. Embryonic lethality was also evident from the reduced average litter size of 6.0 ± 2.4 for +/− × +/− crosses (n = 23 litters), compared with 9.1 ± 2.6 and 8.6 ± 3.3 for the +/+ × +/+ (n = 31 litters) and +/− × +/+ (n = 13 litters) crosses, respectively.
To determine the point of Cenpa null lethality, embryos at 2.5 and 8.5 days pc were genotyped by nested PCR (Fig. 2c). Matings of +/− × +/− yielded 21 embryos at 2.5 days and 37 embryos at 8.5 days. Genotyping by PCR indicated that null mutants were viable (and healthy looking by Giemsa staining; not shown) at 2.5 days (Table 1). The results indicated that the time of embryo death apparently had occurred before 8.5 days because no homozygous mutants were detected. Examination of the uteri of female mice for the 8.5-day embryos revealed eight (21%; Table 1) abortive implantation sites. These abortive sites were either empty or contained remnants of highly degenerate and resorbing embryos. This analysis indicated that the point of embryonic lethality occurred postimplantation between 3.5 and 8.5 days.
Table 1.
PCR genotyping of embryos at days 2.5 and 8.5 pc, showing the number of embryos and, in brackets, % of total
| Genotype | Day 2.5 | Day 8.5 |
|---|---|---|
| +/+ | 3 (14%) | 11 (30%) |
| +/− | 7 (34%) | 18 (49%) |
| −/− | 4 (19%) | 0 |
| No PCR result | 7 (33%) | 0 |
| Abortive implantation sites | N.R. | 8 (21%) |
| Total no. of embryos | 21 | 37 |
N.R., not relevant because embryos have not implanted at day 2.5.
Morphological Degeneration of Cultured Cenpa Null Embryos at Day 5.5 pc.
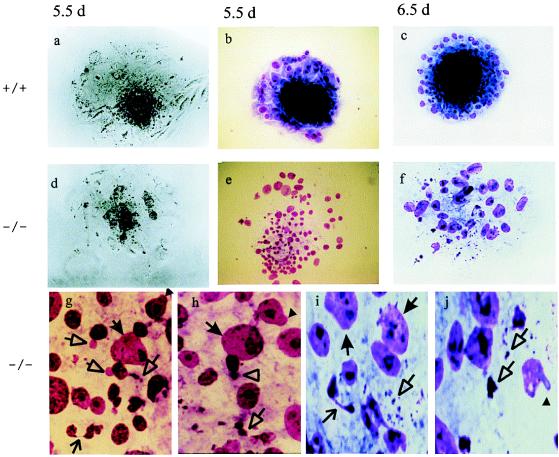
To further investigate postimplantation development, embryos from +/− × +/− crosses were flushed at 3.5 days and cultured individually. The embryos were photographed daily by phase contrast or stained with Giemsa, and the images were captured for more detailed examination. (In the following discussion, the age of cultured embryo continues to refer to the number of days pc; that is, 3.5 days in utero plus days in culture.) At 5.5 days pc (i.e., 2 days in culture), the inner cell mass of a +/+ or +/− embryo was prominent and surrounded by trophectoderm outgrowth after attachment to the coverslip (Fig. 3 a and b) (the inner cell mass subsequently would give rise to the embryo proper whereas the trophectoderm would form the extra-embryonic tissue). In a number of embryos, degeneration of the inner cell mass and trophectoderm cells was apparent (Fig. 3 d and e). Rapid degeneration of these embryos continued and, at 6.5 days, a defined inner cell mass was no longer visible, while the trophectoderm cell number also declined dramatically (Fig. 3f). The remaining embryos maintained a healthy inner cell mass, trophectoderm growth, and morphology from day 6.5 (Fig. 3c) to day 8.5. Healthy embryos were harvested at day 8.5 whereas the degenerating embryos were harvested on day 6.5 for PCR genotyping. The results indicated that all of the day-8.5 embryos were either +/+ or +/−, whereas all of the day-6.5 degenerating embryos showed a 100% coincidence with the −/− genotype (Table 2).
Figure 3.
Phase contrast and Giemsa staining images of day-5.5 and -6.5 embryos. Day-5.5 normal embryo photographed by phase (a) or stained with Giemsa (b). (c) Day-6.5 normal embryo stained with Giemsa. Note the compact, dark inner cell mass and the surrounding trophectoderm outgrowth. Day-5.5 −/− embryo photographed by phase (d) or stained with Giemsa (e). (f) Day-6.5 −/− embryo stained with Giemsa. Note the absence of a defined inner cell mass and the incoherent cells in both the day-5.5 and -6.5 −/− embryos. Magnification for a–f: ×150. (g and h) Close-ups of e and (i and j) close-ups of f, showing micronuclei (empty-triangle arrow), macronuclei (filled-triangle arrow), nuclear bridging (open arrow), nuclear blebbing (filled arrowhead), and highly condensed chromatin bodies (empty arrowhead).
Table 2.
Correlation of genotype with morphology of cultured embryos
| Genotype | Time of harvest, days | Condition of embryo | No. of embryos (% total) |
|---|---|---|---|
| +/+ | 8.5 | Healthy | 9 (26) |
| +/− | 8.5 | Healthy | 19 (54) |
| −/− | 6.5 | Degenerating | 5 (15) |
| No PCR result | 6.5 | Degenerating | 2 (5) |
| Total no. of embryos | 35 (100) |
Embryos were flushed from the uterus at day 3.5. Healthy embryos were harvested at day 8.5 (i.e., 5 days in culture), while degenerating embryos were harvested at day 6.5 (i.e., 3.5 days in culture) for nested PCR analysis.
Chromosomal Missegregation Phenotype.
Close examination of the Giemsa-stained, degenerating cultured Cenpa null embryos at days 5.5 and 6.5 revealed evidence of a severe chromosomal missegregation phenotype. At day 5.5 (Fig. 3 e, g, and h), a substantial number of micronuclei, formed from lagging chromosomes, was observed. Some macronuclei indicative of enlarged genomic content caused by failure of chromosomes to divide properly were apparent. Chromosomal lagging or polar missegregation also affected normal cytokinesis, resulting in nuclear bridging and blebbing of the nuclear membrane. Chromatin fragmentation and hypercondensation were apparent. Similar mitotic problems were observed in the day-6.5 embryos except for an increased degree of severity (Fig. 3 f, i, and j). At 6.5 days, most of the cells were macronucleated, suggesting that chromosome segregation and normal cytokinesis had come to a halt. When the mitotic indices of day-5.5 cultured embryos were determined, a result of 4.7% for normal embryos (n = 23) and 1.1% for Cenpa null embryos (n = 14) was obtained. The chromosomes seen in the null embryos appeared morphologically more condensed and scattered than those of normal embryos, as was previously described for the Cenpc and Incenp null embryos (10, 21) (not shown). Except for an increased background of highly condensed chromatin bodies, no discernible mitotic chromosomes were apparent in the 6.5-day null embryos examined, suggesting cessation of mitosis at this point.
Immunofluorescence Studies.
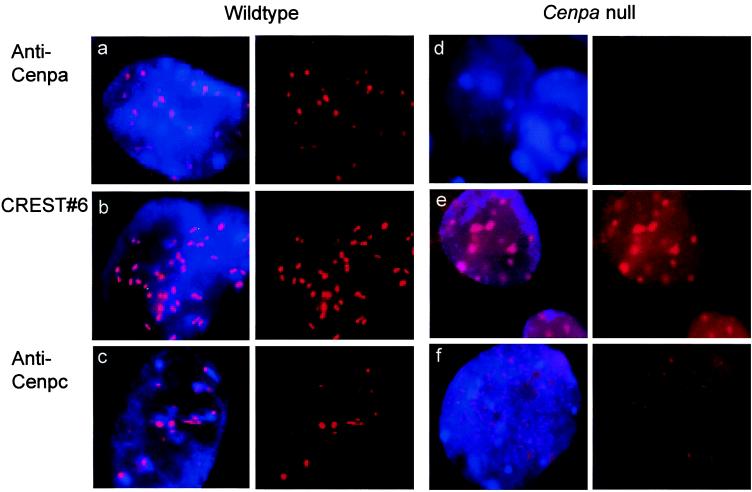
Embryos were analyzed by using antibodies raised against mouse Cenpa (36) or Cenpc (10) or an autoimmune serum (CREST#6) that recognizes human CENP-A and CENP-B (37). Fig. 4 shows typical results obtained on interphase cells in day-5.5 embryos. All three antisera gave strong, discrete signals on the interphase cells of normal embryos (Fig. 4 a–c). In the −/− embryos, with the exception of the occasional cell showing weak residual Cenpa staining, most interphases were negative for this antibody (Fig. 4d), suggesting a complete or near complete depletion of Cenpa (from maternal cytoplasm) by this stage. The CREST#6 antiserum gave positive signals on the −/− embryos (Fig. 4e), but the signals were discernibly different from those seen in the normal embryo in that there was high background staining throughout the nucleus and the enhanced signals on chromatin were unusually diffuse through all focal planes. Because CENP-A was absent from these embryos, the observed CREST#6 signals could be attributed to the staining of CENP-B. When tested with the anti-Cenpc antibody, the prominent and discrete signals seen in normal embryos were undetected in the −/− interphases (Fig. 4f). Instead, these interphase cells showed a profuse speckling of Cenpc signals throughout the entire nucleus.
Figure 4.
Immunofluorescence analysis of day-5.5 embryos. (a–c) Wild-type interphase cells stained with anti-Cenpa, CREST#6 autoimmune serum, and anti-Cenpc, respectively. (d–f) Cenpa null interphase cells stained with anti-Cenpa, CREST#6, and anti-Cenpc, respectively. Although these pictures represented the results taken at one focal plane of a three-dimensional interphase cell nucleus, direct microscopic analysis through all the planes indicated only variation in the total number of observable signals but not the morphology of the signals (e.g., the more diffuse spots in e, and the higher background signals throughout the nuclei in both e and f compared with their respective controls). (Left) Simultaneous staining of chromatin with 4′,6-diamidino-2-phenylindole (blue) and centromere with anticentromere antibody (red). (Right) Split image of Left showing anticentromere antibody staining (red) only.
Discussion
Our gene targeting construct was designed to cause a premature translational termination and deletion of the centromere targeting domain of Cenpa. Evidence that complete gene knockout has been achieved comes from the complete absence of immunofluorescence-detectable Cenpa proteins in morphologically degenerating Cenpa null embryos. The apparently normal phenotype of the Cenpa-targeted heterozygous ES cell line and mice also indicates that our gene disruption strategy does not produce any observable dominant-negative effect.
In previous studies, we reported that targeted gene disruption of the centromere proteins Cenpc and Incenp in mice results in preimplantation embryonic lethality before day 3.5 pc (10, 21). In comparison, expression of a severe phenotype in the Cenpa null embryos appears slightly delayed because these embryos are able to implant into the uterus. Embryonic cell division and development up to this stage presumably are sustained by residual maternal cytoplasmic Cenpa protein (the presence of which has been demonstrated in our immunofluorescence experiments on these early embryos; data not shown). The slightly longer survival time for Cenpa null embryos could reflect a greater stability of the maternal Cenpa mRNA and/or its translated protein compared with those of Cenpc and Incenp. As the maternal Cenpa becomes depleted at day 5.5 pc, mitotic impairment becomes apparent. Chromosomal missegregation results in the formation of a large number of micronuclei (because of lagging chromosomes), macronuclei (because of nonseparated genomes), and nuclear bridging and blebbing (because of tethering chromosomes affecting cytokinesis). The detection of highly condensed, dark Giemsa-staining bodies reflects shrinking chromatin that prompts speculation of apoptotic cell death (38).
Immunofluorescence analysis has provided insight into the cause of mitotic disarray and the role of Cenpa. Depletion of Cenpa in day-5.5 −/− embryos is accompanied by a significant alteration in Cenpb binding in the interphase cells. Instead of the usual discrete and compact signals, Cenpb binding on interphase chromosomes now becomes more diffuse, suggesting that the local chromatin is no longer as highly condensed as that for a normal interphase centromere (compare Fig. 4 e with a–c). This disrupted chromatin structure also appears to affect the normal sequestration of Cenpb from the nuclear pool as evident from the increased Cenpb signal seen throughout the nucleus. At present, it is unclear what the role of Cenpb is because the protein appears to be nonessential for normal mitotic and meiotic cell divisions (5–7).
Anti-Cenpc antibody has been used to further investigate whether a structurally normal interphase kinetochore is formed in Cenpa-deficient cells. Cenpc is a good marker for this because the protein is present only on active centromeres (39, 40) and is functionally essential (10). Our results indicate that depletion of Cenpa leads to an absence of discrete Cenpc binding to interphase centromere. As with Cenpb, sequestration of nuclear Cenpc also becomes impaired, resulting in a diffuse and speckled distribution of the noncentromere-targeted protein throughout the nucleus. The profuse interphase staining seen with both anti-Cenpc and anti-Cenpb antibodies in the −/− embryos also indicates that these proteins are not limiting but are incapable, in the absence of Cenpa, to precipitate kinetochore formation. The observed failure of a structurally normal kinetochore to be properly assembled during interphase provides a suitable explanation for the progressive deterioration of mitotic chromosomal segregation in the Cenpa null embryos, leading ultimately to severe mitotic disarray and embryonic cell death.
Recent data suggest that centromere formation does not strictly depend on DNA sequence and that epigenetic factors may be involved (41–43). Such epigenetic factors are assumed to operate at the higher-order organizational level, directly on chromatin structures. The histone H3-like property of CENP-A means that the protein can directly influence centromere-specific chromatin organization at the nucleosomal level, making this protein a suitable candidate for epigenetic regulation. The observation that CENP-A synthesis appears to be coupled with centromere replication (28) has prompted the proposal that through direct incorporation of the protein at the time of DNA replication, a chromosomal region becomes marked for kinetochore assembly (29). A possible role of replication not withstanding, the results of the present study indicate that CENP-A is required for the kinetochore targeting of functionally critical CENP-C and, by virtue of its nucleosomal nature (expected to play a commanding role in establishing the primary level of chromatin packaging before the seeding of other kinetochore proteins such as CENP-C) probably constitutes one of the earliest events in the interphase kinetochore assembly pathway. These properties are consistent with an epigenetic role of CENP-A in marking a chromosomal region for centromere formation. Future studies aimed at understanding the mechanisms that facilitate the recruitment of CENP-A to a potential centromeric site should provide further important insight.
Acknowledgments
We thank P. Mountford for the IRES-βgeo marker, G. Kay for the 129/1 cell line, E. Robertson for STO/NeoR feeder cells, S. Gazeas, M. Sibson, A. Sylvain, and J. Ladhams for excellent technical assistance, and E. Earle for helpful discussion. This work was supported by the National Health and Medical Research Council of Australia. K.H.A.C. is a Principal Research Fellow of the Council.
Abbreviations
- ES
embryonic stem
- CENP
centromere protein
- INCENP
inner CENP
- pc
postconception
- IRES
internal ribosome-entry site
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Choo K H A. The Centromere. Oxford: Oxford Univ. Press; 1997. [Google Scholar]
- 2.Sullivan K, Hechenberger M, Masri K. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pietras D F, Bennett K L, Siracusa L D, Woodworth-Gutai M, Chapman V M, Gross K W, Kane-Haas C, Hastie N D. Nucleic Acids Res. 1983;11:6965–6983. doi: 10.1093/nar/11.20.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rattner J B. BioEssays. 1991;13:51–56. doi: 10.1002/bies.950130202. [DOI] [PubMed] [Google Scholar]
- 5.Hudson D F, Fowler K, Earle E, Saffery R, Kalitsis P, Trowell H, Hill J, Wreford N, de Kretser D, Cancilla M, et al. J Cell Biol. 1998;141:309–319. doi: 10.1083/jcb.141.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapoor M, De Oca Luna R M, Liu G, Lozano G, Cummings C, Mancini M, Ouspenski I, Brinkley B, May G. Chromosoma. 1998;107:570–576. doi: 10.1007/s004120050343. [DOI] [PubMed] [Google Scholar]
- 7.Perez-Castro A, Shamanski F, Meneses J, Lovato T, Vogel K, Moyzis R, Pedersen R. Dev Biol. 1998;201:135–143. doi: 10.1006/dbio.1998.9005. [DOI] [PubMed] [Google Scholar]
- 8.Saitoh H, Tomkiel J, Cooke C, Ratrie H, Maurer M, Rothfield N F, Earnshaw W C. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- 9.Yang C A, Tomkiel J, Saitoh H, Johnson D H, Earnshaw W C. Mol Cell Biol. 1996;16:3576–3586. doi: 10.1128/mcb.16.7.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalitsis P, Fowler K, Earle E, Hill J, Choo K H A. Proc Natl Acad Sci USA. 1998;95:1136–1141. doi: 10.1073/pnas.95.3.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukagawa T, Brown W R A. Hum Mol Genet. 1997;6:2301–2308. doi: 10.1093/hmg/6.13.2301. [DOI] [PubMed] [Google Scholar]
- 12.Tomkiel J E, Cooke C A, Saitoh H, Bernat R L, Earnshaw W C. J Cell Biol. 1994;125:531–545. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown M, Goetsch L, Hartwell L H. J Cell Biol. 1993;123:387–403. doi: 10.1083/jcb.123.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukagawa T, Pendon C, Morris J, Brown W. EMBO J. 1999;18:4196–4209. doi: 10.1093/emboj/18.15.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craig J M, Earnshaw W C, Vagnarelli P. Exp Cell Res. 1998;246:249–262. doi: 10.1006/excr.1998.4278. [DOI] [PubMed] [Google Scholar]
- 16.Rieder C L, Salmon E D. Trends Cell Biol. 1998;8:310–317. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skibbens R V, Hieter P. Annu Rev Genet. 1998;32:307–337. doi: 10.1146/annurev.genet.32.1.307. [DOI] [PubMed] [Google Scholar]
- 18.Amon A. Curr Opin Genet Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- 19.Dobie K W, Hari K L, Maggert K A, Karpen G H. Curr Opin Genet Dev. 1999;9:206–217. doi: 10.1016/S0959-437X(99)80031-8. [DOI] [PubMed] [Google Scholar]
- 20.Eckley D M, Ainsztein A M, Mackay A M, Goldberg I G, Earnshaw W C. J Cell Biol. 1997;136:1169–1183. doi: 10.1083/jcb.136.6.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cutts S, Fowler K, Kile B, Hii L, O'Dowd R, Hudson D, Saffery R, Kalitsis P, Earle E, Choo K H A. Hum Mol Gen. 1999;8:1145–1155. doi: 10.1093/hmg/8.7.1145. [DOI] [PubMed] [Google Scholar]
- 22.Palmer D K, O'Day K, Trong H L, Charbonneau H, Margolis R L. Proc Natl Acad Sci USA. 1991;88:3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalitsis P, MacDonald A, Newson A, Hudson D, Choo K H A. Genomics. 1998;47:108–114. doi: 10.1006/geno.1997.5109. [DOI] [PubMed] [Google Scholar]
- 24.Basrai M A, Hieter P. BioEssays. 1995;17:669–672. doi: 10.1002/bies.950170802. [DOI] [PubMed] [Google Scholar]
- 25.Arents G, Burlingame R W, Wang B C, Love W E, Moudrianakis E N. Proc Natl Acad Sci USA. 1991;88:10148–52. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Holde K E. Chromatin. New York: Springer; 1989. [Google Scholar]
- 27.Mann R K, Grunstein M. EMBO J. 1992;11:3297–3306. doi: 10.1002/j.1460-2075.1992.tb05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shelby R, Vafa O, Sullivan K. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henikoff S, Csink A K. Trends Genet. 1998;14:200–204. doi: 10.1016/s0168-9525(98)01444-9. [DOI] [PubMed] [Google Scholar]
- 30.Valdivia M M, Figueroa J, Iglesias C, Ortiz M. FEBS Lett. 1998;422:5–9. doi: 10.1016/s0014-5793(97)01583-4. [DOI] [PubMed] [Google Scholar]
- 31.Figueroa J, Saffrich R, Ansorge W, Valdivia M. Chromosoma. 1998;107:397–405. doi: 10.1007/s004120050323. [DOI] [PubMed] [Google Scholar]
- 32.Meluh P B, Yang P, Glowczewski L, Koshland D, Mitchell Smith M. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 33.Stoler S, Keith K, Curnick K, Fitzgerald-Hayes M. Gene Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 34.Mountford P, Zevnik B, Duwel A, Nichols J, Li M, Dani C, Robertson M, Chambers I, Smith A. Proc Natl Acad Sci USA. 1994;91:4303–4307. doi: 10.1073/pnas.91.10.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hendzel M J, Wei Y, Mancini M, Van Hooser A, Ranalli T, Brinkey B, Bazett-Jones D, Allis C D. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 36.Saffery R, Earle E, Irvine D, Kalitsis P, Choo K H A. Chromosome Res. 1999;7:261–265. doi: 10.1023/a:1009222729850. [DOI] [PubMed] [Google Scholar]
- 37.du Sart D, Cancilla M, Earle E, Mao J, Saffery R, Tainton K, Kalitsis P, Martyn J, Barry A, Choo K H A. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- 38.El-Sheraby A M, Hinchliffe J R. J Embryol Exp Morphol. 1974;31:643–654. [PubMed] [Google Scholar]
- 39.Earnshaw W C, Ratrie H, Stetten G. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- 40.Page S L, Earnshaw W C, Choo K H A, Shaffer L G. Hum Mol Genet. 1995;4:289–294. doi: 10.1093/hmg/4.2.289. [DOI] [PubMed] [Google Scholar]
- 41.Karpen G, Allshire R. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- 42.Wiens G, Sorger P. Cell. 1998;93:313–316. doi: 10.1016/s0092-8674(00)81157-5. [DOI] [PubMed] [Google Scholar]
- 43.Barry A, Howman E V, Cancilla M, Saffery R, Choo K H A. Hum Mol Gen. 1999;8:217–227. doi: 10.1093/hmg/8.2.217. [DOI] [PubMed] [Google Scholar]