Abstract
A 314-bp DNA element called Heartbreaker-hm1 (Hbr-hm1) was previously identified in the 3′ untranslated region of a mutant allele of the maize disease resistance gene HM1. This element has structural features of miniature inverted-repeat transposable elements (MITEs) and is a member of a large family of approximately 4,000 copies in the maize genome. Unlike previously described MITEs, most members of the Hbr family display over 90% sequence identity. This, coupled with the insertion of an Hbr element into an allele of the HM1 gene, suggested that this family might have spread recently throughout the genome. Consistent with this view is the finding that Hbr insertion sites are remarkably polymorphic. Ten of ten loci containing Hbr elements were found to be polymorphic for the presence or absence of Hbr among a collection of maize inbred lines and teosinte strains. Despite the fact that over 80% of the maize genome contain moderate to highly repetitive DNA, we find that randomly chosen Hbr elements are predominantly in single or low copy regions. Furthermore, when used to query both the public and private databases of plant genes, over 50% of the sequences flanking these Hbr elements resulted in significant “hits.” Taken together, these data indicate that the presence or absence of Hbr elements is a significant contributory factor to the high level of polymorphism associated with maize genic regions.
Recent studies have shown that different classes of transposable elements (TEs) are associated with the genes of mammals and flowering plants. Two classes of retroelements, long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs) predominate in mammalian genes. As many as 17 LINE-1 elements are within the 65-kb mouse B-globin complex (1), and the 57-kb human hypoxanthine phosphoribosyltransferase gene contains 49 copies of the SINE Alu (2). Although over 40,000 SINEs have been identified in tobacco (3), this element class does not appear to be prevalent in most plant genomes. For example, only two SINEs have been found in the hundreds of grass genes sequenced to date (4).
Rather than containing large numbers of SINEs or LINEs like mammalian genes, many genes from flowering plants, especially those from the grass tribe, harbor miniature inverted-repeat transposable elements (MITEs). The MITE family Tourist appears to be restricted to the grasses (5, 6), whereas the Stowaway family has been associated with the genes of both monocotyledonous and dicotyledonous plants (7, 8). Other MITE families include Alien from bell pepper (9), Emigrant from Arabidopsis (10), and Bigfoot from Medicago (11). Many MITE families have been described in rice (ref. 12; and R. Wing, personal communication). MITE families have also been identified in nonplant species including Caenorhabditis elegans (8), humans (13, 14), the yellow fever mosquito Aedes aegypti (15), and zebrafish (16).
Among the elements analyzed, there is sequence similarity within a MITE family but not between families. However, MITEs as a class share many features, including short length (125–500 bp), terminal inverted repeats (TIR; 10–15 bp), subterminal repeats, target site preference, and high copy number (17). Although they resemble previously characterized nonautonomous DNA elements such as Ds1, their target-site preference and high copy number serve to distinguish MITEs as a group.
Despite the prevalence of MITEs in plant genomes, little is known about their biology. This largely reflects the fact that most MITEs have been identified through database searches (8, 12). It is not currently known, for example, whether their association with genes reflects a true target-site preference or whether this is merely an artifact of identifying elements by searching the gene-rich databases. In addition, database studies do not permit an analysis of how MITEs move.
It has been suggested that MITEs are important tools of evolution because the regulatory regions of some genes were derived from element sequences (6–8, 12, 18). However, since most MITEs isolated to date are present at a particular locus in all or most members of a species, it is difficult to assess their evolutionary impact. Such a determination requires a comparison between alleles that differ solely by the presence or absence of the element. For this to occur, one needs to study element families that were recently active or are still active. In this way, allelic diversity could reflect recent transposition events. Similarly, the study of active MITE families is essential for an understanding of how this element class has attained copy numbers that are one to two orders of magnitude higher than most other DNA elements.
To begin to understand the dynamic aspects of MITE biology, we have characterized a family of MITEs that is distinguished by high sequence identity. This feature is one hallmark of TEs that are either still active or have been active in the recent past. Through our isolation of members of the Heartbreaker (Hbr) family and the analysis of randomly isolated insertion sites from the maize genome, we have determined that insertion sites are highly polymorphic and that they exist preferentially in low copy (genic) regions.
Materials and Methods
Plant Material and Genomic DNA Extraction.
Maize strain GH94-1062 (containing Hbr-hm1) was provided by Guri Johal (University of Missouri, Columbia). All maize inbred lines and sorghum used in this study were obtained from the U.S. Department of Agriculture/Agricultural Research Service Plant Introduction Station at Ames, Iowa. Rice strain IR36 was obtained from Gary Kochert (University of Georgia, Athens, GA), Coix was from Lane Arthur (University of Georgia), and teosintes and Tripsacum were from Sylvestre Marillonnet (University of Georgia).
Plant DNA isolation was as described previously by McCouch et al. (19) using 5 g of leaf tissue.
DNA Blot Analysis.
DNA blot hybridization was performed as described by Zhao and Kochert (20). Blots contained 10 μg of genomic DNA from maize, teosinte, Tripsacum, and Coix, and 3 μg from rice and sorghum digested with HindIII. Probes were labeled with Klenow fragment (BRL) in the presence of [32P]dNTPs. DNA blots were washed either with 2× SSC [1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate (pH 7)]/0.1% SDS at 60°C for 1 hr (moderate stringency), or with 0.1× SSC/0.5% SDS at 67°C for 1 hr (high stringency).
PCR.
PCR was carried out as described by Bureau and Wessler (6), except that different annealing temperatures were used for different pairs of primers. Sequences of primers used in this study are available on request.
Genomic Library Construction and Screening.
DNA from GH94-1062 was first partially digested with BamHI and BglII (neither enzyme has a site in the Hbr-hm1 element) and then partially digested with Sau3A1 (which does not digest Hbr-hm1). DNA fragments of 0.6–1.5 kb were recovered from an agarose gel, purified (Qiagen, Chatsworth, CA), and cloned into the lambda ZAP vector predigested with BamHI (Stratagene). Recombinant phage were screened with labeled Hbr-hm1 DNA. Hybridization was carried out as described by Zhao and Kochert (20), and the filters were washed with either the high- or low-stringency conditions described above.
DNA Sequence Analysis.
Sequences flanking Hbr elements were compared with the 341,073 expressed sequence tag (EST) sequences residing in Pioneer Hi-Bred's (PHI) most comprehensive database (CORNSEQ) and to the 42,756 maize ESTs located in the GenBank updated on November 19, 1999, by using the blastn 2.0 algorithm to search for similarities (21). All matches included in Table 1 have a score >200 and P(N) < 0.001. PHI sequences with similarity to an Hbr-flanking sequence were compared at the nucleotide level to the GenBank public database by using blastn 2.0. If a sequence residing in GenBank matched a PHI EST sequence with a score of at least 150 with P(N) < 0.01, the gene name was associated with the PHI EST sequence and reported in Table 1 under the column “Match”. If additional PHI ESTs were found to have significantly similar nucleotide segments to an Hbr-flanking sequence, the associated GenBank names were included in the Table. A classification of “Unknown” represents an EST with no known function. All templates were sequenced by the Molecular Genetics Instrumentation Facility (University of Georgia).
Table 1.
Database search results using Hbr flanking sequences as queries
| Query | Match | No. of hybridizing bands |
|---|---|---|
| Hbr06flk | None | ND |
| Hbr07flk | None | 1 |
| Hbr10flk | zm tbp1 gene; | |
| 1 unknown zm gene | >10 | |
| Hbr11flk | Ds elements; | |
| 1 unknown zm gene | 2 | |
| Hbr12flk | None | 1 |
| Hbr14flk | None | 3 to 4 |
| Hbr15flk | 1 unknown zm gene | 7 to 8 |
| Hbr21flk | 3 unknown zm genes | ND |
| Hbr22flk | 1 unknown zm gene | 2 to 4 |
| Hbr23flk | None | 1 |
| Hbr24flk | None | 4 to 5 |
| Hbr25flk | None | ND |
| Hbr27flk | 2 unknown zm genes | >10 |
| Hbr29flk | 1 unknown zm gene | Smear |
| Hbr30flk | zm 22KD α-zein gene | |
| 1 unknown zm gene | ND | |
| Hbr32flk | 2 unknown zm genes | 2 to 4 |
| Hbr34flk | None | 1 to 2 |
| Hbr35flk | rice 3 Δ 1-pyr-5-carb syn. | ND |
| Hbr36flk | 1 unknown zm gene | 1 to 2 |
| Hbr38flk | None | 10 |
| Hbr39flk | 4 unknown zm genes | ND |
| Hbr40flk | None | ND |
| Hbr42flk | 1 unknown zm gene | 1 |
| Hbr43flk | None | >10 |
| Hbr45flk | 1 unknown zm gene | 1 |
| Hbr46flk | 2 unknown zm genes | >10 |
| Hbr47flk | 1 unknown zm gene; | |
| zm Tn Bg | ND | |
| Hbr48flk | Arabidopsis Ser/Thr kinase | |
| 1 unknown zm gene | 1 | |
| Hbr50flk | None | 1 |
| Hbr51flk | None | ND |
| Hbr54flk | zm 22KD α-zein gene | ND |
| Hbr56flk | None | ND |
| Hbr58flk | Arabidopsis genomic clone | ND |
| Hbr61flk | 1 unknown zm gene | ND |
| Hbr62flk | None | >10 |
| Hbr65flk | 1 unknown zm gene | 1 |
| Hbr68flk | zm tbp1 gene; | |
| 1 unknown zm gene | ND |
ND, not determined; zm, Zea mays.
Results
Detection of Hbr-hm1-Related Sequences in Selected Grasses.
The first Hbr element was discovered as a 314-bp insertion in the 3′ untranslated region of a mutant allele (hm1-1062) of the maize HM1 gene (22). This element, designated Hbr-hm1, is flanked by a 3-bp direct repeat (DIR) TTA and has a 14-bp TIR that is 80% similar to the TIR of the first discovered MITE family Tourist (5). The internal sequence of Hbr-hm1 is not related to any previously characterized transposable element (data not shown).
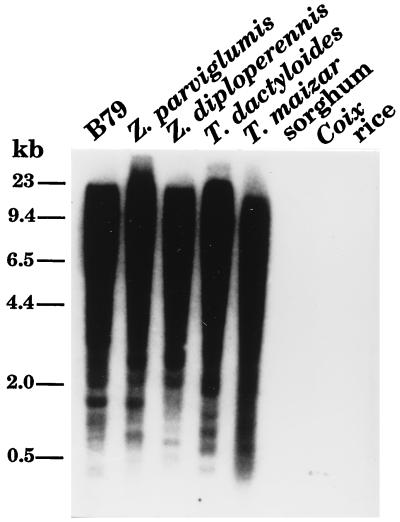
To determine if there were additional copies of Hbr-hm1 in the genome, this element was used to probe a DNA blot containing genomic DNA from selected grasses. Hybridization to hundreds or perhaps thousands of bands was evident at moderate stringency in maize, teosinte, and Tripsacum (Fig. 1). No hybridization signal was detected from the genomic DNA of other grasses, such as Coix, sorghum, or rice.
Figure 1.
DNA blot of genomic DNA from selected grasses probed with Hbr-hm1. Molecular markers (in kb) are shown on the left. B79 is a maize inbred line.
Isolation of Hbr Elements from a Maize Genomic Library.
To obtain the sequences of Hbr elements and information about their insertion sites, several hundred Hbr-containing phage were retrieved from a small insert genomic library (see Materials and Methods). The copy number of Hbr elements was determined by probing the equivalent of 20% of the maize genome (5 × 105 plaques, with about 5 × 105 kb of genomic DNA) (23) with Hbr-hm1. At moderate stringency, 786 plaques hybridized with the probe, while 657 plaques were detected with conditions of high stringency (see Materials and Methods) (data not shown). Based on these values, the copy number of Hbr elements per haploid maize genome was estimated to range from 3,000 (high stringency: 657/0.2 = 3,285) to 4,000 (moderate stringency: 786/0.2 = 3,930) copies.
As a first step toward comparing element sequences and insertion sites, 30 positive clones were chosen at random (from the moderate-stringency condition screen) for sequencing. Twenty-seven inserts contained Hbr elements with >82% identity with Hbr-hm1. Inserts from the remaining three clones contained partial Hbr sequences. Twenty-four of the 27 “complete” elements were 312 to 316 bp and displayed >90% sequence identity (Fig. 2). The remaining three elements share 77–89% sequence identity with other Hbr elements and with each other (data not shown). When each Hbr element was compared with a consensus Hbr sequence derived from the above 27 elements, 14 were found to be >95% identical, and 10 were found to be 90–94% identical. That these elements were chosen at random from the moderate-stringency screen suggests that most of the 3,000–4,000 Hbr elements in the maize genome are highly conserved in both length and sequence.
Figure 2.
Sequence alignment of five genomic Hbr elements and Hbr-hm1 (Hbrhm). The sequences were aligned with pileup program of the Wisconsin GCG computer package (version 8.01) with a gap penalty of 3.0 and gap length penalty of 0.1, and visualized by using boxshade (GCG). Long arrows are over the TIRs, and short arrows indicate the flanking direct repeats.
Analysis of Genomic Polymorphism Associated with Hbr Elements.
The high degree of sequence conservation among Hbr family members coupled with the narrow species distribution of this family (Fig. 1) suggests that Hbr elements have amplified relatively recently. One expectation of the recent spread of Hbr elements is that a significant fraction of Hbr-containing loci should be polymorphic among maize inbred lines and teosinte strains.
The extent of Hbr polymorphism was investigated by comparing 10 of the Hbr-containing loci isolated from GH94-1062 with the corresponding loci in 5 teosinte accessions and 8 maize inbred lines. The availability of the sequences of the Hbr elements and flanking regions of these loci facilitated the design of primers that were used in conjunction with a PCR assay to amplify orthologous loci in these strains.
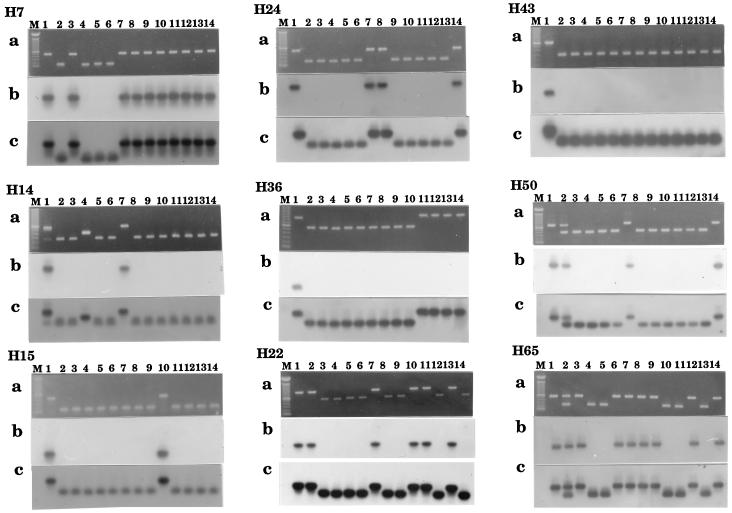
Polymorphism was detected at each locus (Fig. 3, loci are designated H7, H14, etc., and correspond with the number of the resident Hbr element) when the products were visualized on agarose gels (Fig. 3a). In almost all of the cases examined, the fragments detected differed in size by an amount that coincided with the length of the Hbr element, suggesting that the element was absent from the smaller fragments. To confirm this finding, all products were transferred to membranes and probed with Hbr-hm1 (Fig. 3b). In all but one instance (see below), the smaller products did not hybridize with the Hbr-hm1 probe. Similarly, in all but one instance (see below), the larger products hybridized with the element. All lanes hybridized with probes derived from the sequences flanking the corresponding Hbr elements (Fig. 3c), verifying that the orthologous locus had been amplified in each reaction.
Figure 3.
Polymorphism associated with Hbr-containing loci. (a) Agarose gel of the PCR products visualized with ethidium bromide. (b) Autoradiograph of PCR products probed with Hbr-hm1. (c) Autoradiograph of PCR products probed with the flanking genomic sequence of the corresponding Hbr element. M, 100-bp DNA ladder; 1, GH94-1062; 2, Zea parviglumis; 3, Zea mexicana; 4, Zea huehuetenangensis; 5, Zea diploperennis; 6, Zea luxurians; lanes 7–14 are the following maize inbred lines: 7, B73; 8, B79; 9, A554; 10, AC88; 11, 1022; 12, 4722; 13, W23; 14, C13. Cropping of photographs did not remove any visible bands.
Different Elements at Two Loci.
A large PCR product that does not hybridize with the Hbr-hm1 element was detected at the H36 locus in four maize inbred lines (Fig. 3, H36, lanes 11–14). This product was cloned and found to contain an insertion 25 bp from the Hbr-insertion site in GH94-1062. Database searches identified a sequence with 91% identity from the internal region of a previously described Ds2 element (24). This insertion has no identifiable TIRs or DIRs (data not shown).
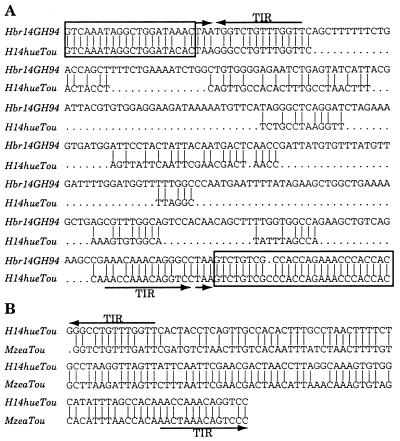
The PCR product from the H14 locus of the teosinte Zea huehuetenangensis is smaller than the Hbr-containing locus but larger than the putative empty sites in most of the other strains (Fig. 3, H14, lane 4). Upon cloning and sequencing, H14 DNA from this strain was found to contain a 128-bp insertion at precisely the same site as the Hbr element in GH94-1062. Furthermore, although this 128-bp sequence shares no internal sequence homology with the Hbr family, it contains a TIR that is virtually identical with the Hbr TIR and a DIR, TAA, that is identical to the DIR flanking Hbr14 in GH94-1062 (Fig. 4A). When used as a query to search GenBank, this element, designated H14hue, showed significant sequence similarity with the previously described Tourist elements (Fig. 4B) (5, 6).
Figure 4.
Another element at the H14 locus in Z. huehuetenangensis. (A) DNA sequence alignment of the elements at the H14 locus in GH94-1062 and Z. huehuetenangensis. Boxed regions represent the genomic sequences flanking Hbr14 in GH94-1062 and H14hueTou in Z. huehuetenangensis. Dots are gaps introduced during sequence comparison. (B) Sequence comparison of MzeaTou and H14hueTou. MzeaTou is a member of the maize Tourist family. Short and long arrows are as in Fig. 2.
Sequences of Empty Sites.
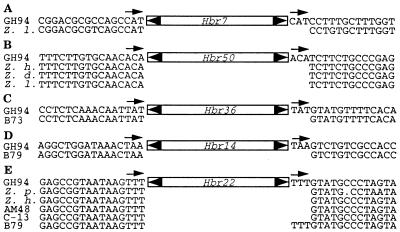
Although it is not known whether MITEs are DNA or RNA (retro) elements, the available evidence favors the view that they are nonautonomous DNA elements (25). As such, two explanations could account for the polymorphism detected at all Hbr-containing loci. The resident Hbr element may have inserted long ago but has excised in strains where it is now absent. Alternatively, an Hbr element has inserted recently and is only present at that locus in some strains. To distinguish these possibilities, we determined the sequence of several empty sites to see if they contained extra nucleotides (transposon footprints) at the putative excision site. Such transposon footprints almost always accompany the excision of plant DNA elements (26).
The sequences of several empty sites at five loci were determined (Fig. 5). These loci were chosen because, in GH94-1062, they contained Hbr elements flanked by a perfect 3-bp DIR (Fig. 5, note arrows). For all empty sites except one, there were no footprints, suggesting that Hbr had never inserted into these loci. It is also a formal possibility that Hbr excision does not generate footprints. The sole exception was an apparent footprint of TTT at the H22 locus in the maize inbred B79 (Fig. 5E). The sequence of four additional empty sites at this locus did not contain transposon footprints. Taken together, these data suggest that most of the polymorphisms at Hbr-containing loci is likely to be due to new insertions rather than frequent excision events.
Figure 5.
Sequences of five polymorphic loci isolated from maize and teosinte. The loci shown are named for the resident element: (A) H7. (B) H50. (C) H36. (D) H14. (E) H22. Short arrows represent DIRs, and filled arrowheads are the Hbr TIRs. GH94, GH94-1062; Z. l., Z. luxurians; Z. h., Z. huehuetenangensis; Z. d., Z. diploperennis; Z. p., Z. parviglumis.
Hbr Inserts Preferentially into Genic Regions.
Previous studies have demonstrated that MITEs are associated with hundreds of plant genes (5–7). However, the question of whether MITEs have a preference for insertion sites in genic regions could not be addressed because elements in these studies were identified following searches of gene-rich databases. In contrast, the Hbr elements isolated in this study provide an opportunity to address this question in an unbiased manner because they where chosen at random from a genomic library.
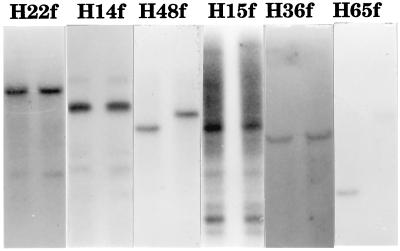
Two methodologies were used to characterize the Hbr insertion sites. First, target-site copy number was estimated by probing DNA blots containing maize genomic DNA with labeled DNA isolated from the regions flanking 24 of the sequenced Hbr-containing fragments from GH94-1062. Twelve probes hybridized with one to two bands, five probes detected three to ten bands, and seven probes detected multiple (more than ten) bands or smear hybridization signals. Data from six of the flanking regions is shown in Fig. 6. Given that up to 80% of the maize genome has been estimated to be middle to highly repetitive DNA (27), these results indicate a preference for insertion into low copy regions.
Figure 6.
DNA blot of maize genomic DNA probed with Hbr-flanking sequences. Each probe was named for the corresponding Hbr element at that locus, followed by “f” for flanking sequence. Genomic DNA is from GH 94-1062 (Left) and W23 (Right).
The second methodology involved the use of 37 Hbr-flanking sequences as queries to search the Pioneer EST/gene database (Pioneer Cornseq) at the nucleotide level. Twenty were found to match at least one of the maize EST/gene sequences at a significance level of P(N) < .01 and a score of at least 200 (Table 1). The average score was 365 (range = 202–653), and the average P(N) was 2.0E-04 (range = 7.90E-20 − 0.0035). Of these 20, only 4 query sequences (Hbr10/Hbr68 and Hbr54/Hbr65) yielded redundant results; however, their sequences were not 100% homologous. Therefore, approximately 54% (20/37) of the Hbr elements analyzed to date have inserted in or near sequences that show significant similarity to known genes or expressed regions of plant genomes.
Discussion
The Hbr element, first identified in the maize Hbr-hm1 allele, is the founding member of a family of MITEs. Like previously characterized MITEs, Hbr elements are short with striking conservation of length (≈314 bp), short TIRs (14 bp), high copy number (≈4,000 copies), and preference for insertion into 3-bp targets that are rich in A and T residues (17).
A Strong Preference for Genic Targets.
Another feature of MITEs that has been suggested but not proven by prior studies is a preference for insertion into low copy (genic) regions. This conclusion is based on database searches that detect MITEs in frequent association with plant genes (5–7, 12). However, because databases contain predominantly gene sequences, these results do not rule out the possibility that MITEs are equally represented in the vast repetitive domains that comprise the majority of most higher plant genomes.
In this study, 37 Hbr-containing loci were selected at random from a maize genomic library. The results of two independent methods demonstrate that a majority of these loci are from low copy regions that are enriched for genes. First, when sequences flanking Hbr elements were used to query the public and private databases, over 50% detected plant ESTs and gene sequences with significant identity. Second, 17 of 24 flanking regions detected 10 bands or fewer on DNA blots when they were used to probe maize genomic DNA (Fig. 6). Since the majority of the maize genome is derived from middle and highly repetitive DNA (27), this result indicates that Hbr elements have a preference for insertion into low copy target sites.
If MITEs prefer to insert into genic regions, then they should be underrepresented in the vast expanses of retrotransposon domains that make up the majority of intergenic DNA. Although few intergenic regions have been sequenced in maize, one striking finding was the complete absence of MITEs in over 180 kb of DNA surrounding Adh1 (27). In contrast, several MITEs were identified in or near the Adh1 gene itself (5).
MITEs Are Probably DNA Elements.
The preference of Hbr elements in particular and possibly MITEs in general for genic regions has important consequences for understanding MITE biology. First, it provides additional evidence that MITEs are nonautonomous DNA elements. Plant DNA elements, including Ac/Ds and Mutator also have a preference for insertion into the genic regions of maize (28–30). Thus, this feature can be added to a growing list of features providing circumstantial evidence that MITEs are DNA and not RNA (retro) elements (17). Perhaps the most direct evidence comes from our recent finding that a short (360 bp) element previously identified as a deletion derivative of a 5.2-kb DNA element called PIF (31) is actually the founding member of a very large MITE family called mPIF (for miniature PIF; Q.Z., W. Eggelston, and S.W., unpublished data).
High Copy Numbers Despite Genic Preference.
There is one striking difference between MITEs and previously studied plant DNA elements that may have important evolutionary consequences. DNA element families like Ac/Ds, Spm/En, and Mutator have copy numbers in the hundreds, usually <100 (32). In contrast, the copy numbers of MITE families are one to two orders of magnitude higher. They are present in thousands, sometimes tens of thousands of copies per haploid genome (5, 11, 15, 16). Such high copy numbers pose two problems: (i) How is the characteristic high copy number attained, and (ii) What are the features of MITEs that permit them to thrive despite a preference for genic targets?
To answer these questions, it will be necessary to study active MITE families. Although MITEs have been identified in virtually all flowering plant genomes and in the genomes of several animals, to date none have been reported to be active. Direct evidence for the excision or insertion of Hbr elements is also not available. However, this family has several of the hallmarks that characterize active TE families, like the maize elements Ac/Ds and Spm/En. First, family members have high sequence identity. That most of the Hbr elements isolated from a screen that employed moderate stringency wash conditions have >90% sequence identity indicates that the majority of Hbr elements in the genome are highly conserved. Insertion sites of active TE family members should also be polymorphic as a result of either excision or new insertions. Ten of ten Hbr loci examined were polymorphic for the presence or absence of Hbr in at least a subset of maize inbred or teosinte lines. The fact that the vast majority of the empty sites lacked TE footprints suggests either that most Hbr insertions are new insertions or that excision of Hbr elements does not usually generate footprints.
One transposition mechanism that would not generate footprints is gap repair, which is responsible for the movement of Drosophila P elements (33). A gap repair mechanism has also been proposed to explain the ability of Mutator elements to almost double their copy number in a single generation (34). Such a mechanism could explain how MITEs attain their high copy numbers. Interestingly, the presence of a different MITE family member at the Hbr14 insertion site (Fig. 4) could also be explained if Hbr elements move by a gap repair mechanism.
Because active transposable element families are usually restricted to a few populations within a species (35), it is not surprising that direct evidence for transposition of the Hbr family has not been obtained, given the limited number of loci and strains that have been examined. In this study, the PCR polymorphism assay did not detect somatic excision at 10 Hbr-containing loci in several maize and teosinte strains (Fig. 3). However, with almost 4,000 Hbr elements in the genome, it is likely that only a small fraction are still active and we have simply not sampled enough elements. In this regard, the detection of MITE activity may present an experimental challenge. Unlike the more traditional element families, MITE activity cannot be monitored by phenotypes such as striped leaves or spotted kernels or high frequencies of germinal reversion because MITEs are in normal genes and excision would probably not alter expression. Also, as discussed above, excision may not be accompanied by the loss of the element from the locus. Rather, to study MITE activity it will be necessary to survey hundreds of family members at one time to detect rare insertion or excision events. The recently described technique called transposon display is such an assay (36) that has been modified to analyze MITE family members in the maize genome (A. Casa and S.R.W., unpublished observations). Use of this technique, in conjunction with MITE families like Hbr, permits routine visualization of hundreds of elements in an equal number of strains and crosses.
MITEs Contribute Significantly to Allelic Diversity in Maize.
Although MITEs are frequently associated with normal plant genes, their evolutionary impact has been difficult to assess because many of the previously identified elements are found at the same locus in most members of the species. In contrast, the data presented in this study indicate that Hbr-containing loci are highly polymorphic and, more importantly, this polymorphism is preferentially associated with maize genic regions. Furthermore, preliminary studies have identified three additional MITE families with 5,000–12,000 copies per haploid genome [B2 (5), mPIF (Q.Z. and S.R.W., unpublished data) and Hb2 (37)], that, like Hbr, have polymorphic insertion sites in genic regions of maize (Q.Z., N. Jiang, and S.R.W., unpublished data). Given the high copy numbers of the four families, these data indicate that the presence or absence of MITEs is a significant contributory factor to the high level of polymorphism shown previously to be characteristic of maize genes (38, 39).
The high level of polymorphism resulting from the presence or absence of MITEs differs dramatically from the situation found in most animal genomes, particularly in primates. Alu elements are frequently associated with human genes. However, it has been estimated that most of the transposition of this retroposon family occurred over 30 million years ago (40). Unlike Hbr elements, the vast majority of Alu insertion sites are conserved in all members of the species and in related species. For this reason, maize and perhaps other plant species may provide unique opportunities to assess the impact of TEs as tools of evolution. After all, there is now an abundance of material available to determine whether the allelic diversity created by MITE insertions leads to differences in gene expression or gene products. Similarly, with the availability of genome-wide methods to visualize MITEs (A. Casa and S.R.W., unpublished observations), it may be possible to identify MITE-containing alleles that have been subject to human (maize) or natural (teosinte) selection.
Acknowledgments
We thank Drs. Guri Johal and Steve Briggs for providing strains and clones containing the Hbr-hm1 element and Dr. John Grey for furnishing unpublished information. This study was supported by a grant from the National Institutes of Health (to S.R.W.).
Abbreviations
- TE
transposable element
- MITE
miniature inverted-repeat transposable element
- LINE
long interspersed nuclear element
- SINE
short interspersed nuclear element
- TIR
terminal inverted repeat
- DIR
direct repeat
- EST
expressed sequence tag
Footnotes
References
- 1.Hutchison C A, Hardies S C, Loeb D D, Shehee W R, Edgell M H. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 593–617. [Google Scholar]
- 2.Edwards A, Voss H, Rice P, Civitello A, Stegemann J, Schwager C, Zimmerman J, Erfle H, Caskey C T, Ansorge W. Genomics. 1990;6:593–608. doi: 10.1016/0888-7543(90)90493-e. [DOI] [PubMed] [Google Scholar]
- 3.Yoshioka Y, Matsumoto S, Kojima S, Ohshima K, Okada N, Machida Y. Proc Natl Acad Sci USA. 1993;90:6562–6566. doi: 10.1073/pnas.90.14.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mochizuki K, Umeda M, Ohtsubo H, Ohtsubo E. Jpn J Genet. 1992;67:155–166. doi: 10.1266/jjg.67.155. [DOI] [PubMed] [Google Scholar]
- 5.Bureau T E, Wessler S R. Plant Cell. 1992;4:1283–1294. doi: 10.1105/tpc.4.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bureau T E, Wessler S R. Proc Natl Acad Sci USA. 1994a;91:1411–1415. doi: 10.1073/pnas.91.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bureau T E, Wessler S R. Plant Cell. 1994b;6:907–916. doi: 10.1105/tpc.6.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oosumi T, Garlick B, Belknap W R. Proc Natl Acad Sci USA. 1995b;92:8886–8890. doi: 10.1073/pnas.92.19.8886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pozueta-Romero J, Houlne G, Schantz R. Gene. 1996;171:147–153. doi: 10.1016/0378-1119(96)00007-8. [DOI] [PubMed] [Google Scholar]
- 10.Casacuberta E, Casacuberta J M, Puigdomenech P, Monfort A. Plant J. 1998;16:79–85. doi: 10.1046/j.1365-313x.1998.00267.x. [DOI] [PubMed] [Google Scholar]
- 11.Charrier B, Foucher F, Kondorosi E, d'Aubenton-Carafa Y, Thermes C, Kondorosi A, Ratet P. Plant J. 1999;18:1–11. doi: 10.1111/j.1365-313x.1999.00469.x. [DOI] [PubMed] [Google Scholar]
- 12.Bureau T E, Ronald P C, Wessler S R. Proc Natl Acad Sci USA. 1996;93:8524–8529. doi: 10.1073/pnas.93.16.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oosumi T, Belknap W R, Garlick B. Nature (London) 1995;378:672. doi: 10.1038/378672a0. [DOI] [PubMed] [Google Scholar]
- 14.Smit A F A, Riggs A D. Proc Natl Acad Sci USA. 1996;93:1443–1448. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tu Z. Proc Natl Acad Sci USA. 1997;94:7476–7480. [Google Scholar]
- 16.Izsvak Z, Ivics Z, Shimoda N, Mohn D, Okamoto H, Hackett P B. J Mol Evol. 1999;48:13–21. doi: 10.1007/pl00006440. [DOI] [PubMed] [Google Scholar]
- 17.Wessler S R, Bureau T E, White S E. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 18.Esen A, Bandaranayake H. Genome. 1998;41:597–604. doi: 10.1139/g98-061. [DOI] [PubMed] [Google Scholar]
- 19.McCouch S R, Kochert G, Yu Z H, Khush G S, Coffman W R, Tanksley S D. Theor Appl Genet. 1988;76:815–829. doi: 10.1007/BF00273666. [DOI] [PubMed] [Google Scholar]
- 20.Zhao X, Kochert G. Mol Gen Genet. 1992;231:353–359. doi: 10.1007/BF00292702. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Johal G S, Briggs S P. Science. 1992;258:985–987. doi: 10.1126/science.1359642. [DOI] [PubMed] [Google Scholar]
- 23.Arumuganathan K, Earle E D. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- 24.Giroux M J, Clancy M, Baier J, Ingham L, McCarty D, Hannah L C. Proc Natl Acad Sci USA. 1994;91:12150–12154. doi: 10.1073/pnas.91.25.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wessler S R. Physiol Plant. 1998;103:581–586. [Google Scholar]
- 26.Sutton W D, Gerlach W L, Schwartz D, Peacock W J. Science. 1983;223:1265–1268. doi: 10.1126/science.223.4642.1265. [DOI] [PubMed] [Google Scholar]
- 27.SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 28.Capel J, Montero L M, Martinez-Zapater J M, Salinas J. Nucleic Acids Res. 1993;21:2369–2373. doi: 10.1093/nar/21.10.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carels N, Barakat A, Bernardi G. Proc Natl Acad Sci USA. 1995;92:11057–11060. doi: 10.1073/pnas.92.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cresse A D, Hulbert S H, Brown W E, Lucas J R, Bennetzen J L. Genetics. 1995;140:315–324. doi: 10.1093/genetics/140.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker E L, Eggleston W B, Demopulos D, Kermicle J, Dellaporta S L. Genetics. 1997;146:681–693. doi: 10.1093/genetics/146.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunze R, Saedler H, Lonnig W E. Adv Bot Res. 1997;27:331–470. [Google Scholar]
- 33.Engels W R, Johnson-Schlitz D M, Eggleston W B, Sved J. Cell. 1990;62:515–525. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- 34.Alleman M, Freeling M. Genetics. 1986;112:107–119. doi: 10.1093/genetics/112.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capy P, Anxolabehere D, Langin T. Trends Genet. 1994;10:7–12. doi: 10.1016/0168-9525(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 36.Van den Broeck D, Maes T, Sauer M, Zethof J, De Keukeleire P, D'Hauw M, Van Montgagu M, Gerats T. Plant J. 1998;13:121–129. doi: 10.1046/j.1365-313X.1998.00004.x. [DOI] [PubMed] [Google Scholar]
- 37.Spell M, Baran G, Wessler S R. Mol Gen Genet. 1988;211:364–366. doi: 10.1007/BF00330617. [DOI] [PubMed] [Google Scholar]
- 38.Johns M A, Strommer J N, Freeling M. Genetics. 1983;105:733–743. doi: 10.1093/genetics/105.3.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wessler S R, Varagona M. Proc Natl Acad Sci USA. 1985;82:4177–4181. doi: 10.1073/pnas.82.12.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Britten R J. Proc Natl Acad Sci USA. 1994;91:6148–6150. doi: 10.1073/pnas.91.13.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]