Abstract
In eukaryotes the complex processes of development, differentiation, and proliferation require carefully orchestrated changes in cellular morphology. Single-celled eukaryotes provide tractable models for the elucidation of signaling pathways involved in morphogenesis. Here we describe a pathway regulating cell polarization and separation in the human pathogenic fungus Cryptococcus neoformans. An insertional mutagenesis screen identified roles for the ARF1, CAP60, NDH1, KIC1, CBK1, SOG2, and TAO3 genes in establishing normal colony morphology. ARF1 and CAP60 are also required for capsule production, a virulence factor, and ARF1 confers resistance to the antifungal fluconazole. KIC1, CBK1, SOG2, and TAO3 are homologues of genes conserved in other eukaryotes; in Saccharomyces cerevisiae they constitute components of the RAM (regulation of Ace2p activity and cellular morphogenesis) signaling pathway. A targeted deletion of a fifth component of RAM (MOB2) conferred identical phenotypes to kic1, cbk1, sog2, or tao3 mutations. Characterization of these genes in C. neoformans revealed unique features of the RAM pathway in this organism. Loss of any of these genes caused constitutive hyperpolarization instead of the loss of polarity seen in S. cerevisiae. Furthermore, sensitivity to the drugs FK506 and cyclosporin A demonstrates that the RAM pathway acts in parallel with the protein phosphatase calcineurin in C. neoformans but not in S. cerevisiae. These results indicate that conserved signaling pathways serve both similar and divergent cellular roles in morphogenesis in these divergent organisms.
INTRODUCTION
Regulation of morphogenesis is a central feature of eukaryotic life. Organisms require coordinated signaling pathways to control polarized growth, asymmetric cell division, and specialized tissue development. For example, establishment and maintenance of proper cell polarity is critical to the function of neurons and epithelial cells (Wiggin et al., 2005). Elucidating the mechanisms that generate cellular asymmetry has benefited from the study of this phenomena in phylogenetically diverse models including the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, the nematode Caenorhabditis elegans, the fruit fly Drosophila melanogaster, and cultured mammalian cells (Drubin and Nelson, 1996; Zallen et al., 2000; Emoto et al., 2004).
Morphogenesis also plays a critical role in the virulence of many pathogens including viruses, bacteria, and dimorphic fungi such as the human pathogen Candida albicans and the plant pathogen Ustilago maydis (Bölker, 2001; Liu, 2002). For example, the influenza virus has multiple morphological forms and altered virion morphology has been correlated with virulence (Mitnaul et al., 1996). The sensitivity of Staphylococcus bacteria to penicillin has been attributed in part to changes in cellular morphology (Giesbrecht et al., 1998). In C. albicans, conversion between the yeast and the hyphal form is associated with increased virulence (Lo et al., 1997; Saville et al., 2003; Noverr and Huffnagle, 2004). Similarly, signaling pathways that control morphology switches in U. maydis have been studied in order to provide insight into the stages of its pathogenesis on plants (Bölker, 2001).
Here, we utilize the model fungal pathogen, Cryptococcus neoformans, to identify novel genetic requirements for normal morphogenesis and polarized growth in this eukaryote. C. neoformans, a basidiomycete pathogen of world-wide distribution, is a major cause of life-threatening infections among immunocompromised persons, though infections in immunocompetent people also occur (Casadevall and Perfect, 1998; Hoang et al., 2004). Concurrent with the rise in number of immunocompromised populations over the last several decades due to AIDS, cancer chemotherapy, and organ transplantation immunosuppression, the frequency of Cryptococcus infections has continued to mount (Mitchell and Perfect, 1995). Like S. cerevisiae, C. neoformans can undergo several regulated changes in morphology in response to environmental conditions or the presence of appropriate mating partners. During growth on rich medium under standard conditions, C. neoformans adopts a budding yeast morphology very similar to its ascomycete relatives, forming smooth round colonies on agar. During mating, a filamentous dikaryon is produced that terminates in a specialized structure (the basidium) where meiosis occurs to produce four chains of spores. In addition, pseudohyphal forms of C. neoformans are occasionally isolated from the wild or as phenotypic switch mutants (Fries et al., 1999, Neilson et al., 1978) and these forms may be better adapted to survival under some conditions. Visible morphological defects of yeast colonies, then, might indicate abnormal growth patterns or altered cell wall structure. These in turn could reflect underlying temperature and/or drug susceptibilities, or deficiencies in the production of the polysaccharide capsule, a key virulence factor of this pathogen (Casadevall and Perfect, 1998). To explore the role of morphology in the virulence and drug susceptibility of C. neoformans, we performed a functional genomics screen to identify mutations which cause morphological defects.
Using Agrobacterium-mediated insertional mutagenesis, more than 50,000 mutants were screened for changes in colony morphology. This type of mutagenesis has been applied previously in C. neoformans to identify novel genes related to virulence (Idnurm et al., 2004; Walton et al., 2005). Through this approach, the genes ARF1, CAP60, NDH1, KIC1, CBK1, SOG2, and TAO3 were found to be required for normal colony morphology of C. neoformans. Furthermore, the roles of each in capsule production, growth at high temperature, susceptibility to antifungal agents, and cell division were characterized. Interestingly, KIC1, CBK1, SOG2, and TAO3 are homologues of the genes in S. cerevisiae that constitute part of the RAM (regulation of Ace2p activity and cellular morphogenesis) signaling pathway (Nelson et al., 2003). Here this pathway is shown to play both conserved and divergent roles during the growth of these two fungi.
MATERIALS AND METHODS
Strains and Media
C. neoformans serotype A strains KN99a and KN99α were used as wild-type strains (Nielsen et al., 2003). Agrobacterium tumefaciens strains LBA4404 or EHA105 containing binary vectors able to confer drug resistance were used for transformation of C. neoformans KN99α. Morphology was analyzed on standard yeast extract, peptone, and dextrose (YPD) medium. Strains exhibiting an altered colony morphology that were identified from insertional mutagenesis are listed in Table 1. Drug susceptibility was assessed on YPD media with the addition of rapamycin (100 ng/ml), caspofungin (10 μg/ml), fluconazole (10 μg/ml), FK506 (1 μg/ml), or cyclosporin A (CsA) (100 μg/ml). C. neoformans and S. cerevisiae strains are also listed in Table 2.
Table 1.
Gene identification in insertion mutants affected in morphology
| Strain | Linkage | Disrupted gene-allele | GenBank accession |
|---|---|---|---|
| ST20S | Yes (14) | ndh1 | CNA07650 |
| ST20D | No (13) | ||
| ST119 | No (3) | cbk1-2a | CNG00360 |
| ST218 | Yes (10) | arf1 | CNI03960 |
| ST219 | No (17) | ||
| ST224 | No (9) | ||
| ST239 | No (8) | ||
| ST258 | Yes (14) | kic1-1 | CNA03940 |
| ST306 | Yes (26) | cap60 | CNA05830 |
| ST308 | No (6) | ||
| SR22 | No (8) | kic1-2a | CNA03940 |
| MS1 | Yes (8) | cbk1-1 | CNG00360 |
| RM2 | Yes (31) | tao3-1 | CNB01240 |
| AI109 | No (10) | sog2-2a | CNB04070 |
| DR3 | No (6) | tao3-2a | CNB01240 |
| DR8 | Yes (12) | cbk1-3 | CNG00360 |
| DR17 | Yes (12) | kic1-3 | CNA03940 |
| DR24 | No (17) | tao3-3a | CNB01240 |
| AI131 | Yes (28) | sog2-1 | CNB04070 |
| AI133 | No (3) | cbk1-4a | CNG00360 |
| AI134 | ND | tao3-4a | CNB01240 |
| AI135 | Yes (23) | kic1-4 | CNA03940 |
| AI136 | Yes (10) | tao3-5 | CNB01240 |
| FS1 | Yes (9) | kic1-5 | CNA03940 |
All strains are mating type α except for AI109, which is mating type a. STXXX refers to a unique 40-base pair signature tag adjacent to the NAT resistance cassette. Values in parentheses are number of progeny analyzed. ND, not determined.
a Although these mutations are unlinked to the NAT insertion, the mutated gene has been identified through crossing with mutants of known genotype, followed by progeny analysis.
Table 2.
Strain list
| Strain | Parent | Genotype | Reference |
|---|---|---|---|
| C. neoformans | |||
| KN99α | MATα | Nielsen et al. (2003) | |
| KN99a | MATa | Nielsen et al. (2003) | |
| KK8 | KN99a | MATacna1Δ::NEO | Kojima et al. (2006) |
| FJW8 | KN99α | MATα kic1Δ::NAT | This study |
| FJW9 | KN99α | MATα cbk1Δ::NAT | This study |
| FJW10 | KN99α | MATα mob2Δ::NAT | This study |
| FJW11 | FJW9 | MATα cbk1Δ::NAT CBK1-NEO | This study |
| FJW12 | FJW10 | MATα mob2Δ::NAT MOB2-NEO | This study |
| FJW13 | KN99α | MATα PH3-CBK1-DsRED-NAT | This study |
| FJW14 | KN99α | MATα PH3-KIC1-DsRED-NAT | This study |
| FJW15 | DR24 | MATα tao3 PH3-KIC1-DsRED-NAT | This study |
| FJW16 | FJW8 | MATα kic1Δ::NAT PH3-KIC1-DsRED-NAT | This study |
| FJW17 | FJW9 | MATα cbk1Δ::NAT PH3-CBK1-DsRED-NAT | This study |
| ST258a | KN99a × ST258 | MATakic1::T-DNA (NAT) | This study |
| SR22a | KN99a × SR22 | MATakic1 | This study |
| MS1a | KN99a × MS1 | MATacbk1::T-DNA (NAT) | This study |
| RM2a | KN99a × RM2 | MATatao3::T-DNA (NAT) | This study |
| DR3a | KN99a × DR3 | MATatao3 | This study |
| DR8a | KN99a × DR8 | MATacbk1::T-DNA (NAT) | This study |
| DR17a | KN99a × DR17 | MATakic1::T-DNA (NAT) | This study |
| DR24a | KN99a × DR24 | MATatao3 | This study |
| AI131a | KN99a × AI131 | MATasog2::T-DNA (NAT) | This study |
| AI135a | KN99a × AI135 | MATakic1::T-DNA (NAT) | This study |
| AI136a | KN99a × AI134 | MATatao3::T-DNA (NAT) | This study |
| FJW8a | KN99a × FJW8 | MATakic1Δ::NAT | This study |
| FJW9a | KN99a × FJW9 | MATacbk1Δ::NAT | This study |
| FJW10a | KN99a × FJW10 | MATamob2Δ::NAT | This study |
| AI139 | FJW9 × ST20S | MATacbk1Δ::NAT ndh1::T-DNA (NAT) | This study |
| S. cerevisiae | |||
| PJ69-4a | MATatrp1-901 leu2-3112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al. (1996) | |
| Y1630 | W303 | MATassd1-d2 ura3 leu2 his3 trp1 ade2 CAN1–100 | |
| BY4743 | S288C | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 MET15/met15Δ0 LYS2/lys2Δ0 ura3Δ0/ura3Δ0 | Giaever et al. (2002) |
| FLY757 | S288C | MATacbk1Δ::KANMX ura3-52 leu2-3112 trp1Δ1 his3Δ200 ssd1-d | Nelson et al. (2003) |
| FLY1466 | S288C | MATasog2Δ::KANMX ura3-52 leu2-3112 trp1Δ1 his3Δ200 ssd1-d | Nelson et al. (2003) |
| FLY1001 | W303 | MATaCBK1-13myc::HIS3 mob2Δ::KanR ura3-1 leu2-3112 his3-11,15 ade2-1 can1-100 lys2 trp1-1 ssd1-d | Nelson et al. (2003) |
| FLY1002 | W303 | MATaCBK1-13myc::HIS3 hym1Δ::URA3 ura3-1 leu2-3112 his3-11,15 ade2-1 can1-100 trp1-1 ssd1-d | Nelson et al. (2003) |
| FLY1003 | W303 | MATaCBK1-13myc::HIS3 kic1Δ::KANMX ura3-1 leu2-3112 his3-11,15 ade2-1 can1-100 lys2 trp1-1 ssd1-d | Nelson et al. (2003) |
| FLY1004 | W303 | MATaCBK1-13myc::HIS3 tao3Δ::KANMX ura3-1 leu2-3112 his3-11,15 ade2-1 can1-100 lys2 trp1-1 ssd1-d | Nelson et al. (2003) |
Insertional Mutagenesis and Genotypic Analysis
Insertional mutagenesis was performed using Agrobacterium-mediated T-DNA delivery of constructs bearing cassettes for drug resistance markers as described previously in a screen for melanin mutants (Idnurm et al., 2004; Walton et al., 2005). To determine if the altered colony morphology phenotypes were linked to the insertion of the nourseothricin resistance cassette, mutant strains were crossed with the congenic strain KN99a on V8 juice agar media (pH 5) in the dark to induce mating. After 2 to 4 wk, basidiospores were isolated using a micromanipulator, and the progeny of each cross were analyzed for nourseothricin drug resistance (by growth on YPD with 100 μg/ml nourseothricin) and morphology. Regions flanking the T-DNA insertion were determined by inverse PCR, and the sequence was compared with the TIGR serotype D and Broad Institute/Duke University serotype A C. neoformans genome databases.
Targeted Deletion and Complementation of the MOB2, CBK1, and KIC1 Genes
A mob2 gene disruption allele was produced using overlap PCR (Fraser et al., 2003). Primers (listed in Table 3) were used to amplify regions with ∼1 kb homology on either side of the MOB2 gene and the nourseothricin acetyltransferase (NAT) marker was amplified from plasmid pAI3 by PCR. Equimolar amounts of the three PCR products were amplified using the primers JOHE15969 and JOHE15972 to generate the disruption allele with a NAT marker flanked on either side by DNA homologous to the genomic region just upstream and downstream of the MOB2 gene. The disruption allele, conferring nourseothricin resistance, was used to transform strain KN99α with a biolistic apparatus (Bio-Rad Model PDS-1000/He Biolistic Particle Delivery System, Richmond, CA; Toffaletti et al., 1993). Transformants were analyzed by PCR and Southern hybridization to confirm deletion of the MOB2 gene. The same procedure was used to generate targeted deletions of the coding regions of the KIC1 and CBK1 genes to confirm that the hyperpolarized phenotype was the result of loss of gene function and not the production of truncated or otherwise aberrant proteins.
Table 3.
Primer list
| Primer name | Sequence (5′–3′) |
|---|---|
| Inverse PCR | |
| JOHE8956 | AACAGTTGCGCAGCCTGAATG |
| JOHE8957 | AGAGGCGGTTTGCGTATTGG |
| Deletion of MOB2 | |
| JOHE15969 | TTGGGAGGTTGGTGAAGG |
| JOHE15970 | GCTTATGTGAGTCCTCCCCTGACGAAGTGTATGGAG |
| JOHE15971 | TCGTTTCTACATCTCTTCTGCATAGCTCTGCATTGG |
| JOHE15972 | CTACTTCAGACGTGAGCC |
| Deletion of KIC1 | |
| JOHE15337 | GCTGAGACAGTCTTTGTGATACATG |
| JOHE14611 | GCTTATGTGAGTCCTCCCCCATCCAGTAAGGTGTGC |
| JOHE14306 | TCGTTTCTACATCTCTTCAACAAGGATCGACCAGCC |
| JOHE15338 | GTCGAAGAGCTTGTCCAAAGTTGCTG |
| Deletion of CBK1 | |
| JOHE15898 | AGGTATCATTCTGCCAAG |
| JOHE15899 | GCTTATGTGAGTCCTCCCTCCTCGCAAGCACACTCG |
| JOHE15900 | TCGTTTCTACATCTCTTC GTAGAACTGAACATGAGC |
| JOHE15901 | CTTCCGAGCTAATCACTC |
| Complementation of MOB2 | |
| JOHE15999 | GGATCCTCCTCTGTGCCATCGAGC |
| JOHE16000 | GGATCCTCCAGTTCTCTTACTGAC |
| Complementation of CBK1 | |
| JOHE15902 | GGATCCCTGCCACTGGTAATATCG |
| JOHE15903 | GGATCCTCTAACGGTCGAACGGTC |
| KIC1-DsRED fusion | |
| JOHE15626 | GGATCCATGGCCGCCTCGGAAGGGCG |
| JOHE15908 | CCATGGCAGCATTCTGGGAGTCTACCT |
| CBK1-DsRED fusion | |
| JOHE15904 | GGATCCATGGCGTATCGCCCAATC |
| JOHE15905 | CCATGGCCAGCATCTCGTATCGTCG |
| CBK1 cDNA | |
| JOHE15904 | GGATCCATGGCGTATCGCCCAATC |
| JOHE15906 | GGATCCCAGCATCTCGTATCGTCG |
| KIC1 cDNA | |
| JOHE15626 | GGATCCATGGCCGCCTCGGAAGGGCG |
| JOHE15907 | GGATCCAGCATTCTGGGAGTCTACCT |
| MOB2 cDNA | |
| JOHE15973 | GGATCCATGGCAGGCTTCCTCAAC |
| JOHE15975 | GGATCCTGACTCCAAAATGCCCAT |
To complement the mob2Δ mutation, the MOB2 coding region and ∼1 kb of upstream sequence was amplified and subcloned into plasmid pPZP-NEO1, which contains the selectable neomycin resistance marker (Walton et al., 2005). Agrobacterium strain EHA105 was transformed with this plasmid, and the mob2Δ strain was transformed by coincubation with that EHA105 strain. Transformants were selected on medium containing neomycin (200 μg/ml) and cefotaxime (100 μg/ml).
Light and Fluorescence Microscopy
To visualize the capsule, strains were grown for 4 d in liquid iron-deplete medium and then stained with India ink; stained cells were analyzed with a light microscope. A Zeiss Axioskop 2 Plus fluorescent microscope (Thornwood, NY) with an attached AxioCam MRM digital camera was used for microscopy. For calcofluor white and sytox green staining, cells were grown to midlog phase in liquid YPD medium. Calcofluor white (0.005% wt/vol) was added to the cells for 15 min before fixing. Cells were fixed with ice-cold 70% ethanol for 20 min, then washed in phosphate-buffered saline (PBS), permeabilized with 1% Triton-X100 (in PBS) for 5 min, and washed again and resuspended in PBS. One microliter of this suspension was mixed on a slide with 1 μl of sytox green mixture (1 μl/ml sytox green [Molecular Probes, Eugene, OR] in Vectashield). For actin staining, cells were grown to midlog and fixed with 37% EM grade formaldehyde for 20 min and then washed and permeabilized as above. To 200 μl cell suspension was added 10 μl rhodamine-conjugated phalloidin (10 μl/ml; Molecular Probes), and this mix was incubated for one h before washing and resuspension in PBS. One μl of the suspension was added to 1 μl Vectashield (10 mg/ml; Vector Laboratories, Burlingame, CA; Nichols et al., 2004).
Subcellular Localization with Protein-DsRED Fusions
To localize the RAM proteins, the coding regions of the genes were subcloned with the open reading frame adjacent to the N-terminal end of the red fluorescent protein, DsRed from the coral genus Discosoma (Invitrogen, Carlsbad, CA). Primers used for amplification are listed in Table 3. Expression of the construct was controlled by the C. neoformans histone H3 promotor. Strains KN99α or tao3, cbk1, and kic1 mutant strains were transformed with the plasmid containing the NAT-selectable marker and the RAM gene-DsRed fusion using Agrobacterium-mediated transformation. Transformants selected from several independent plasmid constructs were selected and analyzed for fluorescence.
Protein Interactions
The cDNAs for the CBK1, KIC1, and MOB2 genes were cloned into plasmids for yeast two-hybrid assays. cDNAs were amplified using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) with the polyT oligo nucleotide primer, followed by PCR with gene specific primers (Table 3) on mRNA isolated from total RNA using PolyATtract mRNA Isolation System IV (Promega, Madison, WI) according to the manufacturer’s protocol. Products were cloned into plasmids pGAD.c1 and pGBD.c1 (James et al., 1996). The S. cerevisiae reporter strain PJ69–4a was cotransformed with plasmids using the lithium acetate/heat shock method (James et al., 1996). Transformants harboring both plasmids were selected on medium lacking leucine and tryptophan. Interactions were assessed by growth in the absence of adenine or histidine (+5 mM 3-aminotriazole) and by β-galactosidase assays (Cardenas et al., 1994).
Complementation of S. cerevisiae RAM Mutants with C. neoformans Genes
S. cerevisiae RAM mutants were the generous gift of Dr. Francis Luca (Nelson et al., 2003). These strains were transformed with plasmid constructs containing the corresponding C. neoformans homologues that were functional in the yeast two-hybrid assays. Strains harboring the pGAD.c1 or pGBD.c1 plasmid with the appropriate C. neoformans homolog were selected on media lacking either leucine or tryptophan, respectively. Several transformants from each independent plasmid construct were analyzed with a light microscope for complementation of the morphology defect and on medium containing 50 μg/ml calcofluor white.
RESULTS
Identification of Morphology Mutants
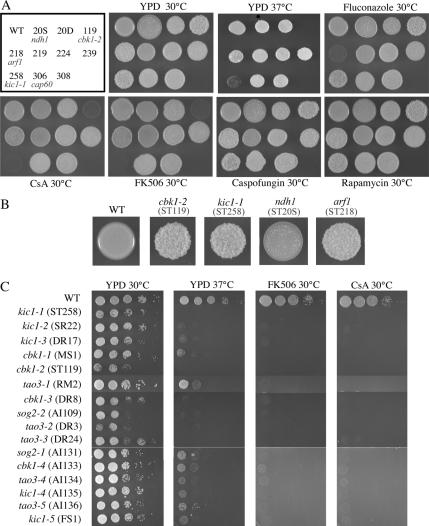
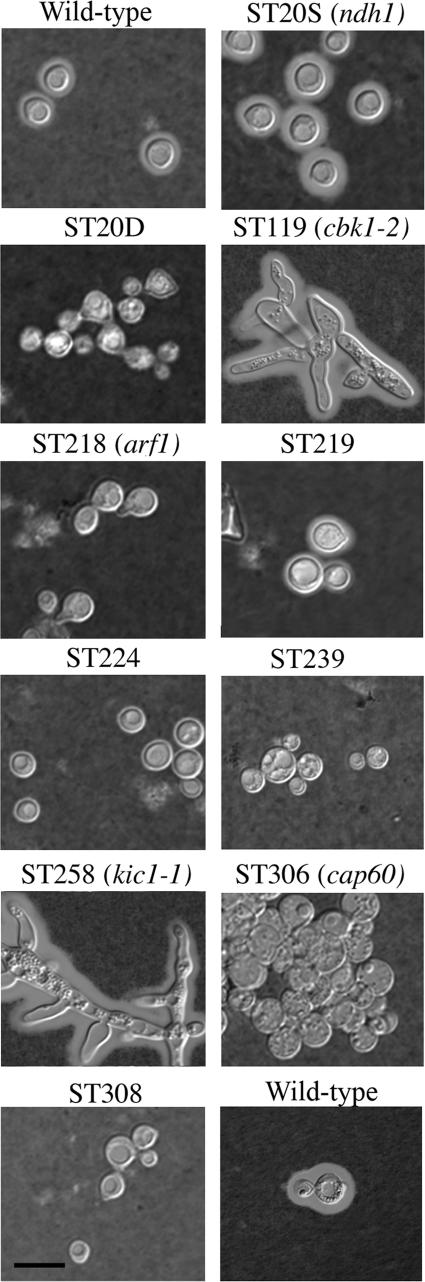
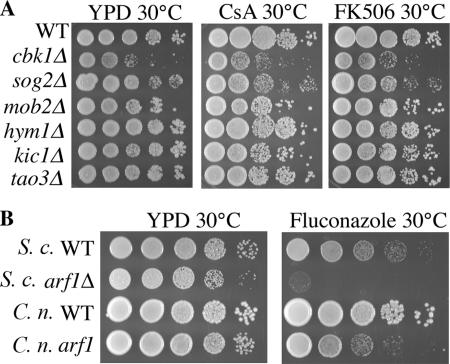
Agrobacterium-mediated insertional mutagenesis is a powerful tool for discovery of novel gene functions in fungi, especially in C. neoformans (Idnurm et al., 2004; McClelland et al., 2005; Michielse et al., 2005; Walton et al., 2005). C. neoformans wild-type strain KN99α was previously transformed with A. tumefaciens cells harboring a Ti-plasmid containing a NAT cassette and molecular signature tag to isolate a collection of more than 12,000 mutant strains (Walton et al., 2005). These strains were screened for abnormal colony morphology as an indicator of capsule production or cell wall defects. Mutants with a dry or crinkled colony surface compared with the smooth, round surface of a wild-type yeast colony were selected for further analysis. In total, 10 mutants with altered colony morphology were isolated from this collection (∼0.1%; Figure 1A). The strains were named according to the signature tag adjacent to the NAT cassette, with letters assigned following the signature tag number if multiple mutants were isolated from that background (i.e., ST20D and ST20S; Table 1). Two mutants, ST119 (cbk1-2) and ST258 (kic1-1), had a unique, crinkled colony morphology and a hyperpolarized, unseparated cellular morphology (Figures 1 and 5), a morphological defect so remarkable that at first they were considered to be contaminants. We therefore screened every subsequent Agrobacterium transformation event relating to other research in the laboratory for additional mutants with this hyperpolarized phenotype. An estimated 50,000 more Agrobacterium-insertional mutants were screened for this hyperpolarized phenotype and 14 additional mutant strains were identified (Figure 1C).
Figure 1.
Colony morphology and drug susceptibility of mutant strains. (A) Ten strains with altered colony morphology were identified in a screen of 12,000 insertional mutants. Dilutions of these strains were plated onto YPD rich medium at 30 or 37°C or YPD at 30°C with fluconazole (10 μg/ml), cyclosporin A (CsA; 100 μg/ml), FK506 (1 μg/ml), caspofungin (10 μg/ml), or rapamycin (100 ng/ml) and grown for 48 h. The arf1 (ST218) mutant shows increased susceptibility to fluconazole. Strains cbk1-2 (ST119) and kic1-1 (ST258) show increased susceptibility to FK506 and CsA. (B) Enlarged photographs of wild-type and mutant growth patches. (C) Additional mutants, n = 50,000, were screened to identify 14 new mutants that share the hyperpolarization phenotype of isolates cbk1-2 (ST119) and kic1-1 (ST258). Tenfold serial dilutions of these strains were plated onto YPD or YPD with FK506 and CsA at 30 or 37°C and grown for 48 h.
Figure 5.
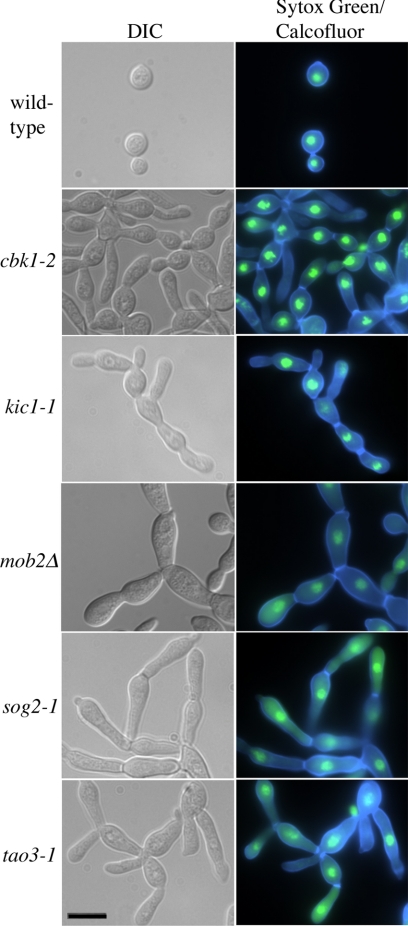
Visualization of the cell walls and nuclei of hyperpolarized mutant strains. Wild-type and mutant cells were grown to midlog phase in liquid YPD medium and stained with calcofluor white and sytox green to visualize cell walls/septa and nuclei, respectively. Scale bar, 10 μm.
Gene Identification in Mutants
To determine if the morphology phenotypes are linked to the insertion of the T-DNA, which confers resistance to nourseothricin, mutant strains were crossed with a congenic strain of opposite mating type, KN99a, and meiotic progeny were analyzed. Basidiospores were isolated from each mating reaction, germinated, and evaluated for cosegregation of the phenotypes and marker. Eight to 31 progeny were scored to confirm linkage (Table 1). Linkage was established for 12 of the 24 mutants, and the number that were linked (50%) was comparable to that previously observed in a similar screen for melanin-deficient C. neoformans mutants (Walton et al., 2005).
In linked mutants, disruption of the C. neoformans genome by T-DNA insertion is the cause of the mutant phenotype rather than a spontaneous unlinked mutation; therefore, the mutated gene causing that phenotype can be identified by sequencing the flanking genomic DNA on either side of the inserted T-DNA. Inverse PCR was performed on each linked mutant, and the sequence of each product was compared with the C. neoformans genome databases at The Institute for Genome Research, the Broad Institute, and Duke University (Loftus, 2005). The putative gene sequences were compared using nucleotide BLAST searches against the NCBI GenBank database to identity homology with genes from other organisms to infer presumptive functions. The results of gene identification in the linked morphology mutants are listed in Table 1.
Two strains bore mutations in previously identified genes of C. neoformans. In the acapsular strain ST306, the T-DNA has inserted into the CAP60 gene that is required for capsule production (Chang and Kwon-Chung, 1998). Acapsular mutant ST218 contains a T-DNA insertion into the ARF1 gene, whose protein product is an ADP-ribosylating factor involved in vesicle formation (Orci et al., 1986; Serafini et al., 1991; Lodge et al., 1994). Mutant ST20S contains a T-DNA insertion into an uncharacterized gene, NDH1, encoding a predicted NADH dehydrogenase.
Several mutants contained disruptions in genes homologous to those in the S. cerevisiae RAM (regulation of Ace2p activity and cellular morphogenesis) signaling pathway. In mutants ST258, DR17 and AI135, the T-DNA has disrupted the KIC1 gene, a homolog of the S. cerevisiae KIC1 gene that encodes a serine/threonine kinase similar to the Ste20-like kinases. Two other linked hyperpolarized mutants, MS1 and DR8, each contain a disruption in the C. neoformans homolog of S. cerevisiae CBK1, which encodes the second serine-threonine kinase in the RAM pathway (Nelson et al., 2003). Mutants RM2 and AI136 are linked and contain insertions in the TAO3 gene. Mutant AI131 contains a T-DNA insertion in the closest match to the S. cerevisiae SOG2 gene.
The other unlinked hyperpolarized mutant strains were crossed with five representative mutant strains (cbk1 [MS1], kic1 [ST258], mob2Δ [FJW10], tao3 [RM2], and sog2 [AI131]) and each other in order to identify allelic mutations based on the segregation of haploid meiotic progeny. In a cross of two strains harboring mutations in the same gene, no meiotic progeny should be found with a wild-type phenotype. Alternatively, when two strains harboring mutations in different genes are crossed, both mutant (∼75%) and wild-type (∼25%) meiotic progeny are produced. C. neoformans RAM mutants were less robust in mating than other strains; mating reactions were regularly conducted for 4 to 6 wk before basidiospore dissection by micromanipulation. Nevertheless filaments and viable basidiospores were produced that could be dissected for genetic analysis. Through this analysis, it was determined that strains ST258, SR22, DR17, AI135, and FS1 all contain mutations in the KIC1 gene, whereas strains MS1, ST119, AI133, and DR8 contain mutations in the CBK1 gene. Strains RM2, DR3, DR24, AI134, and AI136 all contain mutations in the TAO3 gene. Strain AI109 harbors a RAM mutation that is allelic with the sog2 mutation in strain AI131. Thus, by this approach we provide additional evidence that the NAT insertion in ST258, MS1, AI131, and RM2 in KIC1, CBK1, SOG2, and TAO3 confer the phenotype and identify 12 additional mutations in the RAM genes.
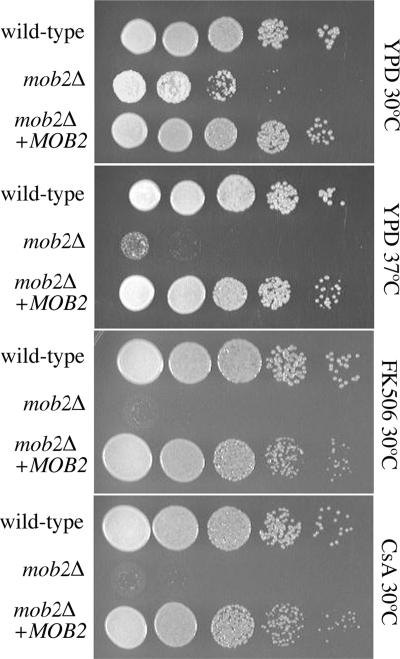
Because Mob2 is the conserved binding partner required for Cbk1 kinase activity characterized in both S. cerevisiae and S. pombe (Weiss et al., 2002) and no insertional mutant in this gene was identified in our screen, a targeted mob2Δ deletion strain was generated. This strain had identical phenotypes to the other C. neoformans RAM mutants and was complemented by reintroduction of the wild-type MOB2 gene (Figure 2). One reason this gene may not have been found in the genomic insertional mutant screen is its relatively small size, with a coding sequence of only 964 nucleotides compared with the KIC1, CBK1, SOG2, and TAO3 genes which are 3171, 2237, 3846, and 7622 nucleotides, respectively. Furthermore, although 16 independent RAM mutants were identified with the same hyperpolarized phenotype, we wanted to confirm that this defect resulted specifically from the loss of a RAM gene and was not an unusual effect of the mutagenesis process or production of aberrant protein versions. Thus targeted deletions of the complete open reading frames of the KIC1 and CBK1 genes were generated in addition to the targeted MOB2 deletion strain. These two disruption strains appeared identical to the insertion mutants, confirming that the hyperpolarization defect is a true loss of function phenotype of the C. neoformans RAM mutants.
Figure 2.
Deletion of C. neoformans MOB2. The predicted MOB2 homolog was replaced by a nourseothricin resistance cassette (mob2Δ) and displays the same phenotypes as other RAM mutants. These phenotypes were complemented by reintroduction of a wild-type copy of the MOB2 gene (mob2Δ + MOB2).
Capsule Production Is Abolished in Five Mutants with Altered Colony Morphology
Capsule production is a key virulence factor in C. neoformans, and an altered colony morphology may serve as an indication of altered production of capsular polysaccharide. The collection of morphology mutants was analyzed for levels of encapsulation by microscopy of cultures grown in low iron medium and stained with India ink. Five acapsular strains, including ST306 (cap60) and ST218 (arf1), were identified (Figure 3). The CAP60 gene was previously shown to be required for capsule production (Chang and Kwon-Chung, 1998); however, the requirement of the ARF1 gene for capsule synthesis is a new discovery and implicates proper vesicle transport in synthesizing and transporting appropriately modified capsular polysaccharides to the cell surface. Although loss of any of the RAM genes results in severe morphological defects at the colony and cellular levels, these mutants produce normal levels of capsule (Figure 3).
Figure 3.
Capsule production in altered morphology mutant strains. Strains were grown for 4 d at 30°C in liquid low-iron medium, and then exclusion of India ink was examined. Strain ST224 shows greatly diminished capsule production, but is not abolished. Scale bar, 10 μm.
arf1 and RAM Mutants Exhibit Increased Drug Susceptibilities
Altered morphology may indicate abnormal cell wall composition. The hyperpolarized RAM mutants could not grow at 37°C; growth at human body temperature is required for virulence in C. neoformans (Figure 1). Neither this temperature sensitivity, nor the altered colony morphology of any of the mutants could be rescued by the addition of sorbitol, an osmotic stabilizer that can rescue the growth of cell wall mutants (unpublished data). We also screened these mutants for sensitivity to several drugs, including the immunosuppressive agents rapamycin, FK506, and cyclosporin A (CsA), and the antifungal agents fluconazole and caspofungin (Figure 1A). None of the mutants showed increased susceptibility to rapamycin or caspofungin, but ST118 (arf1) is hypersensitive to fluconazole. The S. cerevisiae arf1 mutant is also hypersensitive to fluconazole, a phenotype of that mutant which had not, to our knowledge, been previously reported (Figure 4B). Additionally, S. cerevisiae ARF2/arf2 and ARF3/arf3 heterozygous mutants are hypersensitive to fluconazole (Giaever et al., 2004). This link between ADP-ribosylation factors and azole sensitivity warrants further study given that the azoles are the most commonly used antifungal drugs.
Figure 4.
Drug susceptibility of S. cerevisiae mutants. (A) Serial dilutions of S. cerevisiae RAM mutants were plated onto YPD alone or with cyclosporin A (CsA; 100 μg/ml) or FK506 (1 μg/ml) at 30°C for 48 h. (B) Serial dilutions of S. cerevisiae and C. neoformans wild-type and arf1 mutant strains were plated onto YPD or YPD with fluconazole (10 μg/ml).
All of the hyperpolarized RAM mutants are hypersensitive to the immunosuppressive agents FK506 and CsA. On media containing FK506 (1 μg/ml) or CsA (100 μg/ml), the RAM mutants did not grow, whereas the wild-type strain, KN99α, and the other morphology mutants grew as well as on YPD-rich medium (Figure 1). We tested survival of the mutants over time when grown at high temperature or in FK506 supplemented media. Liquid cultures of each RAM mutant were grown in YPD at 30°C, YPD at 37°C, or YPD with 1 μg/ml FK506 for 0.5, 1, 4, or 24 h. They were then plated on solid YPD medium at 30°C, and colony forming units were counted after 3 d. All mutants grown for a period of time up to 24 h at 37°C survived but did not replicate during that time. This implies that the RAM genes are not required for viability at 37°C, but rather are required for proliferation at that temperature. Growth in YPD with FK506, on the other hand, completely killed the mutant strains after as little as 0.5 h; no cells were viable when plated onto rich medium. Thus the mechanism of action of FK506 in a RAM mutant background is fungicidal, whereas the mechanism of temperature sensitivity is fungistatic.
Synthetic Lethality between RAM and Calcineurin Pathways
The increased sensitivity of RAM mutants to drugs that target the calcineurin pathway suggested that cells compromised for the two pathways are likely to be inviable. To test this, we crossed a RAM mutant (cbk1::NAT) with a calcineurin mutant (cna1::NEO) and investigated the possible synthetic lethality between the RAM pathway and calcineurin pathways. From the 75 basidiospores that germinated (175 total), 28 were wild-type (NATs NEOs), 23 were cbk1 (NATr NEOs), 23 were cna1 (NATs NEOr), and only 1 was a candidate cbk1 cna1 double mutant (NATr NEOr). The cellular morphology of this rare NATr NEOr strain was that of the cbk1 mutant and by Southern blot analysis the strain was heterozygous at the CNA1 locus (CNA1/cna1::NEO). Aneuploid progeny are occasionally obtained from C. neoformans crosses, explaining this unusual segregant. The proportion of strains obtained from the cross is highly skewed from the predicted 1:1:1:1 ratio (χ2 test p < 0.001). The CBK1 and CNA1 genes are located on separate large contigs in the serotype A genome sequencing project so there is no chance that this recombination distortion is due to physical linkage. We also determined the mating type in the 75 progeny and observed that a and α alleles segregate independently of the other markers, further demonstrating that the population exhibits normal recombination. This data, together with the observation of cell death in RAM mutants in the presence of FK506, indicates that the RAM pathway and calcineurin pathways function in parallel and their simultaneous inhibition leads to inviability.
FK506 and CsA bind to FKBP12 and cyclophilin A, respectively, to inhibit calcineurin activity, and thereby exert their antifungal and immunosuppressive activities (Rusnak and Mertz, 2000). The synthetic lethality of calcineurin inhibition and RAM gene mutation in C. neoformans suggests that these two pathways share an overlapping essential function, indicating a novel connection between calcineurin and RAM signaling. Interestingly, however, S. cerevisiae RAM mutants are not hypersensitive to FK506 or CsA (Figure 4A), providing further evidence of functional divergence of this pathway in these two species.
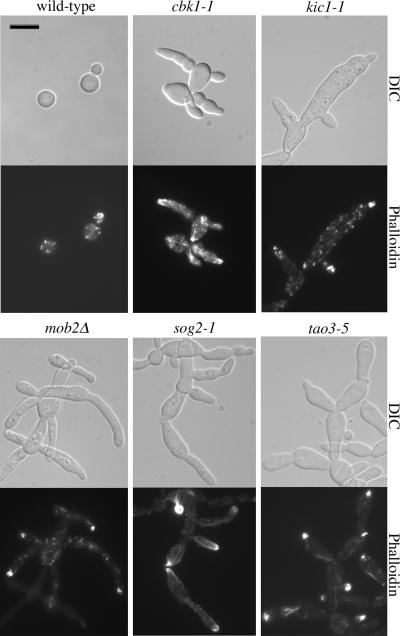
Hyperpolarization Mutants Exhibit Defective Cytokinesis But Normal Nuclear Partitioning
In S. cerevisiae, the RAM mutants are abnormally round and fail to separate, indicating a loss of polarity and an inability to complete cytokinesis (Nelson et al., 2003). Fluorescence microscopy of C. neoformans RAM mutants reveals a similar lack of detachment between mother and daughter cells, but instead of a loss of polarity, these mutant cells are strikingly hyperpolarized and result in cell aggregates reminiscent of pseudohyphae (Figure 5). Staining nuclei and cell walls with sytox green and calcofluor white, respectively, showed proper nuclear segregation and the formation of septa between dividing cells, indicating that the loss of the RAM genes leads to an inability to complete the last stages of cytokinesis that lead to full cell separation. Similarly, in U. maydis mutation of DON3, a gene encoding a product in the same family of serine/threonine kinases as that encoded by KIC1, does not inhibit primary septum formation, but leads to the loss of initiation of the secondary septum, which normally allows the daughter cell to separate from the mother (Weinzierl et al., 2002). The hyperelongation of C. neoformans RAM mutants, however, is unique as the homologous S. cerevisiae, S. pombe, and U. maydis mutants lose polarization (Verde et al., 1998; Durrenberger and Kronstad, 1999; Nelson et al., 2003).
Polarized growth involves changes in actin structure and positioning; we therefore hypothesized that the hyperpolarization defect of the RAM mutants may be a result of perturbation of actin localization. Actin localization in wild-type C. neoformans strain KN99α appeared evenly distributed throughout the cell, visualized as punctate patches and occasional filaments when stained with rhodamine-conjugated phalloidin. RAM mutants, on the other hand, show increased amounts of actin at the bud tips (Figure 6). Despite differing polarization defects in RAM mutants of S. pombe and C. neoformans, both mutant phenotypes appear to be correlated with actin mislocalization, suggesting that the RAM proteins may be conserved in regulating fundamental cellular phenomena including actin localization. The reason that deletion of the homologous genes confers opposing polarization phenotypes in S. pombe or S. cerevisiae and C. neoformans is unclear.
Figure 6.
Actin localization in hyperpolarized mutant strains. Wild-type and mutant cells were grown to midlog phase in liquid YPD medium and stained with rhodamine-conjugated phalloidin. Actin, visualized as punctate dots and occasional filaments, appears evenly distributed throughout the wild-type cells. Cells of the RAM mutants, however, often accumulate an increased amount of actin at the bud tips compared with wild-type, suggesting a role for Kic1 and Cbk1 in actin localization in C. neoformans. Scale bar, 10 μm.
C. neoformans Cbk1 and Kic1 Localize to Punctate Structures and the Septum during Cell Division
To delineate functional differences between S. cerevisiae and C. neoformans RAM mutants, we analyzed the subcellular localization of the Cbk1 and Kic1 proteins tagged with fluorescent DsRED protein in the wild-type KN99α background. To confirm that these fusion proteins were functional in vivo, we transformed kic1 and cbk1 deletion mutants with the plasmids carrying the corresponding DsRED fusion protein. Transformants bearing an integrated transgene were analyzed and showed phenotypes equivalent to wild-type cells, thus confirming that the DsRED fusion proteins complement the deletion of CBK1 or KIC1. As an additional control, the kic1+KIC1-DsRED transformants were crossed to wild-type KN99a, and meiotic progeny were isolated by dissection. The isolation of mutant meiotic progeny further confirmed that the DsRED fusion protein could complement mutant strains.
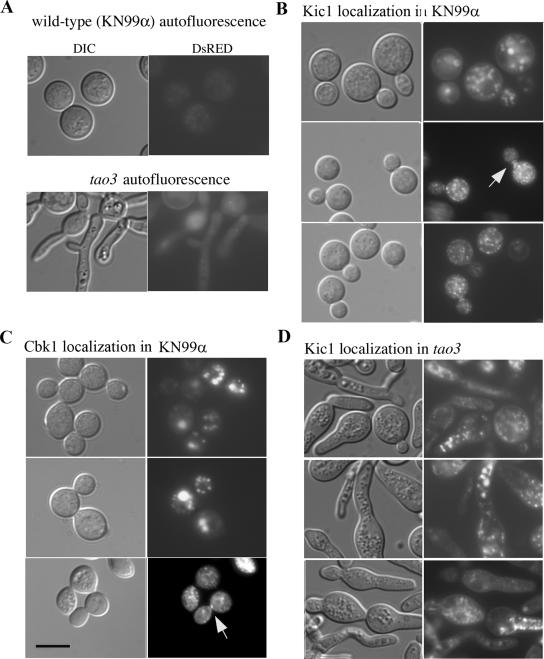
At present, there is no system available to synchronize C. neoformans cell cultures, hampering localization studies. In the majority of cells, Cbk1 is localized diffusely throughout the cytoplasm and to punctate spots resembling vesicles (Figure 7C). The biological significance of the localization of Cbk1 to potential vesicles is unknown. However, in D. melanogaster, Trc and Fry also localize to punctate spots, which are suggested to be vesicles; furthermore, the Trc/Fry pathway genetically interacts with Dral, the D. melanogaster homolog of Ral, which is involved in vesicle trafficking (Feig, 2003; He et al., 2005). Importantly, in actively budding cells of C. neoformans, Cbk1 localizes briefly to the septum, corroborating its specific role in the separation of mother-daughter cells (Figure 7C). Septal localization was seen in ∼2% of budding cells (4/188). S. cerevisiae RAM proteins, including Cbk1, also localize temporally in the cell cycle to the septum (Nelson et al., 2003).
Figure 7.
Subcellular localization of Cbk1 and Kic1. Wild-type or tao3 (strain DR24) cells were transformed with a construct containing Cbk1 or Kic1 tagged with the fluorescent DsRed protein. (A) Autofluorescence of untransformed strains. (B) In wild-type background, Kic1 shows punctate cytoplasmic localization in the majority of cells and temporal localization to the septa in ∼1% (3/236) of dividing cells. (C) Similarly, Cbk1 localizes diffusely to the cytoplasm and to punctate spots resembling vesicles in the majority of cells. Notably, Cbk1 localizes to the septa in actively budding cells (lower panel) in ∼2% of budding cells (4/188). (D) Kic1 localization in the tao3–1 mutant DR24 differs from localization in a wild-type strain. Localization of Kic1 to the septa was never observed (0/>400), suggesting a role for Tao3 in the localization of Kic1. Scale bar, 10 μm.
We also observed the subcellular localization of Kic1 tagged with the fluorescent DsRED protein in the KN99α background. In a pattern similar to Cbk1, the Kic1 protein localizes diffusely throughout the cytoplasm in the majority of the cells; however at a brief interval during cell division, the protein localizes to the septum where we hypothesize it mediates the final stages of cytokinesis in conjunction with other RAM proteins (Figure 7B). Septal localization was seen in ∼1% of budding cells (3/236). We also analyzed the localization of Kic1 in a tao3 background (strain DR24), and were no longer able to detect localization of Kic1 to the septa (0/>400), suggesting a role for Tao3 in the localization of Kic1 (Figure 7D). Mislocalization of Kic1 in the tao3 mutant may be partially responsible for the morphology or cell separation defect.
Cbk1, Kic1, and Mob2 Physically Interact
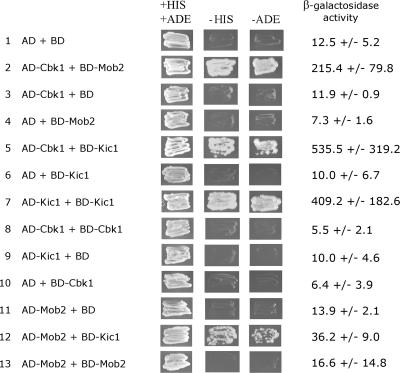
The novel phenotypes conferred by mutation of the RAM genes in C. neoformans may indicate different protein–protein interactions between the components of this pathway compared with interactions previously studied in S. cerevisiae. To test for pairwise physical interactions we performed yeast two-hybrid assays using C. neoformans Cbk1, Kic1, and Mob2 proteins. Robust interactions were observed between the two kinases Cbk1 and Kic1, between Cbk1 and its binding partner Mob2, Kic1 and itself, and a weaker interaction between Kic1 and Mob2 (Figure 8). These results suggest that the protein–protein complex architecture is similar in the two divergent yeasts, despite differing output from the signaling pathway.
Figure 8.
Cbk1, Kic1, and Mob2 physically interact. The coding regions of the CBK1, KIC1, and MOB2 genes were fused adjacent to the activating domain (AD) or binding domain (BD) of S. cerevisiae Gal4 and expressed in the S. cerevisiae strain PJ69–4a, which has the ADE2, HIS3, and lacZ genes under the control of Gal4. Protein–protein interactions were detected by growth of transformed strains on media lacking histidine (−HIS) or adenine (−ADE), and by increased β-galactosidase activity. Average β-galactosidase activities of three independent experiments with the SE of the mean (in Miller units) are reported.
We also tested the ability of C. neoformans Cbk1, Kic1, and Mob2 to complement the corresponding S. cerevisiae RAM mutants. Although these constructs functioned in two-hybrid assays, they failed to complement the cell separation defects of S. cerevisiae mutants (unpublished data). This is not completely unexpected, as homologues from S. pombe also failed to complement S. cerevisiae mutants (Racki et al., 2000). These findings provide further confirmation of functional divergence between the C. neoformans and S. cerevisiae RAM pathways and may suggest that differences in protein structure between species might mediate interactions with different upstream or downstream components of the pathway that lead to distinct phenotypes.
DISCUSSION
The regulation of cellular morphology is critical to all organisms. Fundamental processes from cell division to the development of specialized tissues and organs in higher eukaryotes require the coordinated generation of cell polarity. One would assume that because the generation of polarity is so fundamental for life that there would be many conserved proteins and functions. In yeast, the polarity of dividing cells is precisely regulated by a number of pathways, including the recently elucidated RAM signaling pathway, which comprises the proteins Cbk1, Mob2, Kic1, Hym1, Sog2, and Tao3 (Nelson et al., 2003; Irazoqui and Lew, 2004). S. cerevisiae strains harboring mutations in any of these genes display cell separation defects and a loss of polarity or are lethal, depending on the genetic background.
In this study, we identified five components of the homologous RAM-signaling pathway in C. neoformans, a human pathogenic fungus that grows as a yeast, yet which lies within the phylum Basidiomycota and is highly divergent from ascomycete yeasts like S. cerevisiae. RAM in C. neoformans also controls cell separation and polarity. However, in this fungus disruption of this pathway leads to hyperpolarization instead of depolarization, indicating that the pathway functions to negatively regulate polarization in this organism rather than to positively regulate it as it does in S. cerevisiae and S. pombe. Furthermore, the RAM genes are not essential in the C. neoformans standard serotype A strain background (H99/KN99a/α), whereas they are essential in S. cerevisiae (unless the SSD1 gene has also been mutated [Kurischko et al., 2005]) and in S. pombe. A third difference is that the RAM pathway acts in parallel with the Ca2+-calmodulin–activated protein phosphatase calcineurin in C. neoformans but not in S. cerevisiae. Simultaneous inhibition of both pathways is lethal, as observed when C. neoformans RAM mutants fail to grow in the presence of the drugs FK506 or cyclosporin A, which each function to inhibit calcineurin, and by the inability to obtain cbk1 cna1 double mutants through genetic crosses.
The RAM pathway is an abbreviation referring to both morphology and localization of Ace2, a C2H2 zinc finger transcription factor that specifically localizes to daughter cells in S. cerevisiae. Some of the differences between S. cerevisiae and C. neoformans may be accounted for by unique Ace2 functions. One major question about the C. neoformans RAM pathway is the function it has in regulating the equivalent of Ace2 in this fungus. We searched the completed genome for matches to S. cerevisiae Ace2. The closest match (e-11), CNA01450, is predicted to contain two C2H2 Zn finger domains at its C-terminal end (like Ace2). However, a reciprocal BLAST search of S. cerevisiae revealed that this protein is more similar to Crz1 or Swi5 than to Ace2. Furthermore, CNA01450 is also the closest match when S. cerevisiae Crz1 is used to search the C. neoformans genome. Thus at present, no clear Ace2 homolog has been found in the C. neoformans genome and an equivalent functioning gene has not been identified, although it is the object of ongoing research.
Despite these differences in RAM pathway function in the basidiomycete C. neoformans compared with model ascomycete fungi, some similarities exist to their homologues in more distant relatives. Homologues of proteins in the RAM pathway have been identified in diverse species, including fungi, the fruit fly D. melanogaster, the nematode C. elegans, and humans (Table 4). The other fungi generally have phenotypes equivalent to the S. cerevisiae and S. pombe mutants. In Neurospora crassa, mutation of the Cbk1 homolog (Cot-1) results in defective hyphal elongation and increased branching (Yarden et al., 1992). In U. maydis, mutation of the Cbk1 homolog (ukc1) causes a distorted, compact, highly branched morphology and defects in pathogenesis (Durrenberger and Kronstad, 1999). In Aspergillus nidulans, mutation of the HYM1 homolog hymA blocks conidiophore development (Karos and Fischer, 1996). Also, homologues of CBK1 have recently been identified in the human fungal pathogens Pneumocystis carinii, where its transcript level is regulated by pH, and C. albicans, where deletion of CBK1 leads to an inability to produce hyphae and a consequent aggregation of round cells (McNemar and Fonzi, 2002; Kottom and Limper, 2004).
Table 4.
Characterized RAM pathway homologs
| Cryptococcus neoformans | Cbk1 | Kic1 | Mob2 | Tao3 | Sog2 | |
|---|---|---|---|---|---|---|
| Saccharomyces cerevisiae | Cbk1 | Kic1 | Mob2 | Tao3 | Hym1 | Sog2 |
| Schizosaccharomyces pombe | Orb6 | Orb3/Nak1 | Mob2 | Mor2 | Pmo25 | |
| Aspergillus nidulans | HymA | |||||
| Neurospora crassa | Cot-1 | |||||
| Ustilago maydis | Ukc1 | |||||
| Caenorhabditis elegans | Sax-1 | |||||
| Drosophila melanogaster | Trc | Dmob | Fry | Mo25a | ||
| Homo sapiens | Ndr1 | Mst3 | Mob |
Cbk1 is a serine/threonine kinase in the AGC (protein kinases A, G, and C) family of kinases, which includes the mammalian Ndr and WARTS/LATS kinases (Bidlingmaier et al., 2001). Kic1 is also a serine/threonine protein kinase that is related to the PAK/Ste20 family of proteins (Sullivan et al., 1998). Mob2, Sog2, Hym1, and Tao3 interact with at least one of these kinases to mediate their functions in S. cerevisiae (Nelson et al., 2003).
a Mo25 is the closest D. melanogaster homolog of Hym1, but it does not appear to function in a pathway with other RAM homologs in Drosophila (He et al., 2005).
Homologues of the RAM pathway have been recently characterized in higher eukaryotes. The D. melanogaster homologues of Cbk1 and Tao3, tricornered (Trc) and furry (Fry), respectively, control wing hair development and the positioning and dendritic branching of neurons (Emoto et al., 2004; He et al., 2005). Similarly, the C. elegans Cbk1 homolog (Sax-1) regulates neuronal cell shape and appears to function as an inhibitor of neurite initiation and spreading (Zallen et al., 2000). Interestingly, the neuronal cells of sax-1 mutants are irregularly expanded and extended instead of displaying a normal compact, spherical morphology, a phenotype qualitatively more similar to that observed in RAM mutants of C. neoformans than in S. cerevisiae. Furthermore, neurite initiation in C. elegans is regulated in parallel by both the Sax-1 RAM pathway and a calcium/calmodulin-dependent protein kinase, Unc-43 (Zallen et al., 2000), similar to the parallel regulation of viability in C. neoformans by RAM and calcineurin.
In mammals, the homolog of Cbk1 appears to be the NDR1 protein kinase, which is regulated by a Kic1 homolog and Ca2+ phosphatase activity. The NDR family of protein serine/threonine kinases is a subclass of the AGC kinases, which includes the mammalian, fly, and worm large tumor suppressor (LATS) kinases and S. cerevisiae Dbf2 and Dbf20 in addition to the Cbk1 homologues (Tamaskovic et al., 2003a). LATS/WARTS, a close relative of NDR1, is required as a tumor suppressor in flies and mammals. NDR1 physically interacts with S100B, a Ca2+-binding protein, which affects its phosphorylation state and kinase activity (Millward et al., 1998; Tamaskovic et al., 2003b). S100B is overexpressed in more than 80% of metastatic melanomas, and up-regulated NDR1 transcription indicates a higher risk of invasive breast cancer (Hauschild et al., 1999; Adeyinka et al., 2002). A mammalian Kic1 homolog, Mst3, has been recently identified and it acts to regulate NDR through phosphorylation (Stegert et al., 2005). Both Mst3 and Ndr activity appear to be regulated by the mammalian protein phosphatase PP2A (Stegert et al., 2005). It will be important to elucidate how phosphatases like PP2A in mammals and calcineurin in C. neoformans fit into the RAM signaling pathway.
The NDR1 pathway in mammals and the homologous RAM pathway in other eukaryotes from fungi to flies are both key regulators of cellular morphogenesis and proliferation. Cell morphology and viability in C. neoformans appears to be regulated in parallel by the RAM and calcineurin pathways, indicating that cross-talk between the RAM signaling pathway and intracellular calcium signaling may be conserved with multicellular eukaryotes. Thus, a better understanding of the function of this pathway may result from studies in C. neoformans in addition to the ascomycete model fungi such as S. cerevisiae or S. pombe. Elucidation of the molecular mechanism of this regulation, its cross-talk with other signaling pathways, and its upstream and downstream components represent important future research avenues in elucidating the function of the RAM signaling pathway in the eukaryotic kingdom.
ACKNOWLEDGMENTS
We thank Francis Luca and Cornelia Kurischko for the generous gift of S. cerevisiae RAM mutants. We thank Connie Nichols for the DsRED plasmid and assistance with actin staining. We also thank Andrew Alspaugh, Daniel Lew, Connie Nichols, and Julian Rutherford for critical reading of the manuscript. Gene identification was greatly facilitated by the following C. neoformans genome sequencing projects: Stanford Genome Technology Center (http://www-sequence.stanford.edu), The Institute for Genomic Research (http://www.tigr.org/tdb/e2k1/cna1), Duke University (http://cneo.genetics.duke.edu), and the Broad Institute (http://broad.mit.edu/annotation/fungi/cryptococcus_neoformans/). Felicia Walton was supported by a research fellowship from the American Society for Microbiology. This work was supported by the National Institute of Allergy and Infectious Diseases R01 Grants AI042159, AI063443, and AI50438 to J.H.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-02-0125) on June 14, 2006.
REFERENCES
- Adeyinka A., Emberley E., Niu Y., Snell L., Murphy L. C., Sowter H., Wykoff C. C., Harris A. L., Watson P. H. Analysis of gene expression in ductal carcinoma in situ of the breast. Clin. Cancer Res. 2002;8:3788–3795. [PubMed] [Google Scholar]
- Bidlingmaier S., Weiss E. L., Seidel C., Drubin D. G., Snyder M. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:2449–2462. doi: 10.1128/MCB.21.7.2449-2462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölker M. Ustilago maydis—a valuable model system for the study of fungal dimorphism and virulence. Microbiology. 2001;147:1395–1401. doi: 10.1099/00221287-147-6-1395. [DOI] [PubMed] [Google Scholar]
- Cardenas M. E., Hemenway C. S., Muir R. S., Ye R., Fiorentino D., Heitman J. Immunophilins interact with calcineurin in the absence of exogenous immunosuppressive ligands. EMBO J. 1994;13:5944–5957. doi: 10.1002/j.1460-2075.1994.tb06940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Perfect J. Cryptococcus neoformans. Washington, DC: American Society for Microbiology Press; 1998. [Google Scholar]
- Chang Y. C., Kwon-Chung K. J. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect. Immun. 1998;66:2230–2236. doi: 10.1128/iai.66.5.2230-2236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin D. G., Nelson W. J. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Durrenberger F., Kronstad J. The ukc1 gene encodes a protein kinase involved in morphogenesis, pathogenicity and pigment formation in Ustilago maydis. Mol. Gen. Genet. 1999;261:281–289. doi: 10.1007/s004380050968. [DOI] [PubMed] [Google Scholar]
- Emoto K., He Y., Ye B., Grueber W. B., Adler P. N., Jan L. Y., Jan Y. N. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 2004;119:245–256. doi: 10.1016/j.cell.2004.09.036. [DOI] [PubMed] [Google Scholar]
- Feig L. A. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–425. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- Fraser J. A., Subaran R. L., Nichols C. B., Heitman J. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell. 2003;2:1036–1045. doi: 10.1128/EC.2.5.1036-1045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries B. C., Goldman D. L., Cherniak R., Ju R., Casadevall A. Phenotypic switching in Cryptococcus neoformans results in changes in cellular morphology and glucuronoxylomannan structure. Infect. Immun. 1999;67:6076–6083. doi: 10.1128/iai.67.11.6076-6083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., et al. Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc. Natl. Acad. Sci. USA. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesbrecht P., Kersten T., Maidhof H., Wecke J. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol. Mol. Biol. Rev. 1998;62:1371–1414. doi: 10.1128/mmbr.62.4.1371-1414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschild A., Engel G., Brenner W., Glaser R., Monig H., Henze E., Christophers E. S100B protein detection in serum is a significant prognostic factor in metastatic melanoma. Oncology. 1999;56:338–344. doi: 10.1159/000011989. [DOI] [PubMed] [Google Scholar]
- He Y., Emoto K., Fang X., Ren N., Tian X., Jan Y. N., Adler P. N. Drosophila Mob family proteins interact with the related tricornered (Trc) and warts (Wts) kinases. Mol. Biol. Cell. 2005;16:4139–4152. doi: 10.1091/mbc.E05-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang L. M., Maguire J. A., Doyle P., Fyfe M., Roscoe D. L. Cryptococcus neoformans infections at Vancouver Hospital and Health Sciences Centre (1997–2002): epidemiology, microbiology and histopathology. J. Med. Microbiol. 2004;53:935–940. doi: 10.1099/jmm.0.05427-0. [DOI] [PubMed] [Google Scholar]
- Idnurm A., Reedy J. L., Nussbaum J. C., Heitman J. Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot. Cell. 2004;3:420–429. doi: 10.1128/EC.3.2.420-429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui J. E., Lew D. J. Polarity establishment in yeast. J. Cell Sci. 2004;117:2169–2171. doi: 10.1242/jcs.00953. [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karos M., Fischer R. hymA (hypha-like metulae), a new developmental mutant of Aspergillus nidulans. Microbiology. 1996;142:3211–3218. doi: 10.1099/13500872-142-11-3211. [DOI] [PubMed] [Google Scholar]
- Kojima K., Bahn Y.-S., Heitman J. Calcineurin, Mpk1, and Hog1 MAPK pathways independently control fludioxonil antifungal sensitivity in Cryptococcus neoformans. Microbiology. 2006;152:591–604. doi: 10.1099/mic.0.28571-0. [DOI] [PubMed] [Google Scholar]
- Kottom T. J., Limper A. H. Pneumocystis carinii cell wall biosynthesis kinase gene CBK1 is an environmentally responsive gene that complements cell wall defects of cbk-deficient yeast. Infect. Immun. 2004;72:4628–4636. doi: 10.1128/IAI.72.8.4628-4636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurischko C., Weiss G., Ottey M., Luca F. C. A role for the Saccharomyces cerevisiae regulation of Ace2 and polarized morphogenesis signaling network in cell integrity. Genetics. 2005;171:443–455. doi: 10.1534/genetics.105.042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Co-regulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen. Int. J. Med. Microbiol. 2002;292:299–311. doi: 10.1078/1438-4221-00215. [DOI] [PubMed] [Google Scholar]
- Lo H. J., Kohler J. R., DiDomenico B., Loebenberg D., Cacciapuoti A., Fink G. R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Lodge J. K., Jackson-Machelski E., Toffaletti D. L., Perfect J. R., Gordon J. I. Targeted gene replacement demonstrates that myristoyl-CoA: protein N-myristoyltransferase is essential for viability of Cryptococcus neoformans. Proc. Natl. Acad. Sci. USA. 1994;91:12008–12012. doi: 10.1073/pnas.91.25.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus B. J., et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland C. M., Chang Y. C., Kwon-Chung K. J. High frequency transformation of Cryptococcus neoformans and Cryptococcus gattii by Agrobacterium tumefaciens. Fungal Genet. Biol. 2005;42:904–913. doi: 10.1016/j.fgb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- McNemar M. D., Fonzi W. A. Conserved serine/threonine kinase encoded by CBK1 regulates expression of several hypha-associated transcripts and genes encoding cell wall proteins in Candida albicans. J. Bacteriol. 2002;184:2058–2061. doi: 10.1128/JB.184.7.2058-2061.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse C. B., Hooykaas P.J.J., van den Hondel C.A.M.J.J., Ram A.F.J. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr. Genet. 2005;48:1–17. doi: 10.1007/s00294-005-0578-0. [DOI] [PubMed] [Google Scholar]
- Millward T. A., Heizmann C. W., Schafer B. W., Hemmings B. A. Calcium regulation of Ndr protein kinase mediated by S100 calcium-binding proteins. EMBO J. 1998;17:5913–5922. doi: 10.1093/emboj/17.20.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell T. G., Perfect J. R. Cryptococcosis in the era of AIDS-100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitnaul L. J., Castrucci M. R., Murti K. G., Kawaoka Y. The cytoplasmic tail of influenza A virus neuraminidase (NA) affects NA incorporation into virions, virion morphology, and virulence in mice but is not essential for virus replication. J. Virol. 1996;70:873–879. doi: 10.1128/jvi.70.2.873-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson J. B., Ivey M. H., Bulmer G. S. Cryptococcus neoformans: pseudohyphal forms surviving culture with Acanthamoeba polyphaga. Infect. Immun. 1978;20:262–266. doi: 10.1128/iai.20.1.262-266.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B., et al. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell. 2003;14:3782–3803. doi: 10.1091/mbc.E03-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols C. B., Fraser J. A., Heitman J. PAK kinases Ste20 and Pak1 govern cell polarity at different stages of mating in Cryptococcus neoformans. Mol. Biol. Cell. 2004;15:4476–4489. doi: 10.1091/mbc.E04-05-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K., Cox G. M., Wang P., Toffaletti D. L., Perfect J. R., Heitman J. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 2003;71:4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr M. C., Huffnagle G. B. Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect. Immun. 2004;72:6206–6210. doi: 10.1128/IAI.72.11.6206-6210.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L., Glick B. S., Rothman J. E. A new type of coated vesicular carrier that appears not to contain clathrin: its possible role in protein transport within the Golgi stack. Cell. 1986;46:171–184. doi: 10.1016/0092-8674(86)90734-8. [DOI] [PubMed] [Google Scholar]
- Racki W. J., Becam A. M., Nasr F., Herbert C. J. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J. 2000;19:4524–4532. doi: 10.1093/emboj/19.17.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusnak F., Mertz P. Calcineurin: form and function. Physiol. Rev. 2000;80:1483–1521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- Saville S. P., Lazzell A. L., Monteagudo C., Lopez-Ribot J. L. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell. 2003;2:1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini T., Orci L., Amherdt M., Brunner M., Kahn R. A., Rothman J. E. ADP-ribosylation factor is a subunit of the coat of Golgi-derived COP-coated vesicles: a novel role for a GTP-binding protein. Cell. 1991;67:239–253. doi: 10.1016/0092-8674(91)90176-y. [DOI] [PubMed] [Google Scholar]
- Stegert M. R., Hergovich A., Tamaskovic R., Bichsel S. J., Hemmings B. A. Regulation of NDR protein kinase by hydrophobic motif phosphorylation mediated by the mammalian Ste20-like kinase MST3. Mol. Cell. Biol. 2005;25:11019–11029. doi: 10.1128/MCB.25.24.11019-11029.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan D. S., Biggins S., Rose M. D. The yeast centrin, cdc31p, and the interacting protein kinase, Kic1p, are required for cell integrity. J. Cell Biol. 1998;143:751–765. doi: 10.1083/jcb.143.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaskovic R., Bichsel S. J., Hemmings B. A. NDR family of AGC kinases—essential regulators of the cell cycle and morphogenesis. FEBS Lett. 2003a;546:73–80. doi: 10.1016/s0014-5793(03)00474-5. [DOI] [PubMed] [Google Scholar]
- Tamaskovic R., Bichsel S. J., Rogniaux H., Stegert M. R., Hemmings B. A. Mechanism of Ca2+-mediated regulation of NDR protein kinase through autophosphorylation and phosphorylation by an upstream kinase. J. Biol. Chem. 2003b;278:6710–6718. doi: 10.1074/jbc.M210590200. [DOI] [PubMed] [Google Scholar]
- Toffaletti D. L., Rude T. H., Johnston S. A., Durack D. T., Perfect J. R. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F., Wiley D. J., Nurse P. Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc. Natl. Acad. Sci. USA. 1998;95:7526–7531. doi: 10.1073/pnas.95.13.7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton F. J., Idnurm A., Heitman J. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol. Microbiol. 2005;57:1381–1396. doi: 10.1111/j.1365-2958.2005.04779.x. [DOI] [PubMed] [Google Scholar]
- Weinzierl G., Leveleki L., Hassel A., Kost G., Wanner G., Bölker M. Regulation of cell separation in the dimorphic fungus Ustilago maydis. Mol. Microbiol. 2002;45:219–231. doi: 10.1046/j.1365-2958.2002.03010.x. [DOI] [PubMed] [Google Scholar]
- Weiss E. L., Kurischko C., Zhang C., Shokat K., Drubin D. G., Luca F. C. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 2002;158:885–900. doi: 10.1083/jcb.200203094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggin G. R., Fawcett J. P., Pawson T. Polarity proteins in axon specification and synaptogenesis. Dev. Cell. 2005;8:803–816. doi: 10.1016/j.devcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Yarden O., Plamann M., Ebbole D. J., Yanofsky C. cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J. 1992;11:2159–2166. doi: 10.1002/j.1460-2075.1992.tb05275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen J. A., Peckol E. L., Tobin D. M., Bargmann C. I. Neuronal cell shape and neurite initiation are regulated by the Ndr kinase SAX-1, a member of the Orb6/COT-1/warts serine/threonine kinase family. Mol. Biol. Cell. 2000;11:3177–3190. doi: 10.1091/mbc.11.9.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]