Abstract
Reactive oxygen species (ROS) generated by the NOX family of NADPH oxidases have been described to act as second messengers regulating cell growth and differentiation. However, such a function has hitherto not been convincingly demonstrated. We investigated the role of NOX-derived ROS in cardiac differentiation using mouse embryonic stem cells. ROS scavengers prevented the appearance of spontaneously beating cardiac cells within embryoid bodies. Down-regulation of NOX4, the major NOX isoform present during early stages of differentiation, suppressed cardiogenesis. This was rescued by a pulse of low concentrations of hydrogen peroxide 4 d before spontaneous beating appears. Mechanisms of ROS-dependent signaling included p38 mitogen-activated protein kinase (MAPK) activation and nuclear translocation of the cardiac transcription factor myocyte enhancer factor 2C (MEF2C). Our results provide first molecular evidence that the NOX family of NADPH oxidases regulate vertebrate developmental processes.
INTRODUCTION
Reactive oxygen species (ROS) are generated either in a nonregulated manner as side products of several enzymatic systems (e.g., cyclooxygenases, nitric oxide [NO] synthases, mitochondrial cytochromes) or in a regulated way as main products of superoxide producing enzymes, the NADPH oxidases. In the mouse, the family of NADPH oxidases includes NOX1, NOX2 (gp91phox), NOX3, and NOX4.
Excessive cellular generation of ROS, such as superoxide anions (O2−) and hydrogen peroxide (H2O2), is potentially destructive and is used by phagocytes to kill invading microorganisms. Under normal conditions, scavenging mechanisms (e.g., superoxide dismutase, catalase, glutathione-glutathione peroxidase system) remove excessive amounts of ROS. Under stress conditions, however, the production of ROS may exceed the reducing capacity of the cell and damage cellular functions. Small amounts of ROS, on the other hand, can function as intracellular second messengers and activate signaling cascades involved in growth and differentiation of many cell types (for review see Rhee, 1999; Laloi et al., 2004). For example, the MAP kinase-signaling pathway is sensitive to ROS (Torres and Forman, 2003). Moreover, distinct signaling pathways have differential sensitivity to oxidative stress, leading to dose-dependent effect on, for example, cardiomyocytes on which ROS can induce hypertrophy or apoptosis (Kwon et al., 2003). Transcription factors such as NF-κB, p53, and AP-1 are redox-sensitive and can be directly modified by ROS, providing a link with the control of gene expression (Morel and Barouki, 1999).
Cardiac differentiation can be studied by differentiating mouse embryonic stem cells (ESC) into embryoid bodies (EB), where the appearance of spontaneously beating cardiomyocytes is observed after 7–8 d of culture. This system thus provides a unique experimental model to study the role of ROS and ROS-generating enzymes in the regulation of cardiomyocyte growth and differentiation in vitro. Previous reports have shown a link between ROS and cardiac differentiation. Indeed, electric field stimulation of EB increases the amount of cardiomyocytes through the augmentation of intracellular ROS and the nuclear translocation of NF-κB (Sauer et al., 1999). Other pathways linking ROS to cardiac development include PI3-kinase as its inhibition impairs ROS production and, as a consequence, cardiac differentiation (Sauer et al., 2000). Signal transduction cascades induced by the cardiogenic cytokine cardiotrophin-1 via Jak/STAT also appear to depend on ROS (Sauer et al., 2004; Ateghang et al., 2006).
In the mouse embryonal carcinoma cell model, activation of p38 MAPK has been shown to be necessary for cardiac differentiation (Davidson and Morange, 2000; Eriksson and Leppa, 2002). However, the precise role and, in particular, the source of ROS in cardiac differentiation remains to be elucidated. Moreover, although in fetal human heart selective expression of NOX2, NOX4, and NOX5 has been reported (Cheng et al., 2001), the expression pattern of NOX isoforms expressed during mouse cardiac differentiation and in mouse cardiomyocytes is not known.
Using the mouse ES cell model, we identified NOX4 as the main NOX expressed in undifferentiated ESC, ES-derived, and neonatal cardiomyocytes. We show that NOX4-dependent ROS are crucial for the induction of cardiac genes and the differentiation of cardiomyocytes from ESC.
MATERIALS AND METHODS
ESC Culture and Differentiation
Mouse ESC CGR8 were cultured in BHK21 medium (GIBCO BRL, Rockville, MD) supplemented with nonessential amino acids, pyruvate, mercaptoethanol, glutamine, penicillin/streptomycin, 10% fetal calf serum (FCS), and LIF-conditioned medium in a humidified 5% CO2 atmosphere at 37°C and maintained at <70% confluency to keep an undifferentiated phenotype (Meyer et al., 2000; Li et al., 2002). The differentiation of CGR8 was performed by the hanging drop method (Maltsev et al., 1994). In brief, EBs were formed within hanging drops (450 cells/20 μl) of differentiation medium (BHK21, as described above), containing 20% FCS and lacking LIF (day 0 of the experiment). After 2 d, EBs were collected and cultured in suspension for 2 d. At day 4 medium was changed and the following day (day 5), EBs were plated to gelatin-coated 24-well plates or glass coverslips (Meyer et al., 2000). At day 8 of differentiation, EBs containing 2 or more beating foci were considered beating EBs (Sauer et al., 1999; Meyer et al., 2000; Li et al., 2002). This scoring technique is based on the fact that an EB containing only a single beating cluster is usually very small and not representative of a typical beating EB in which several beating clusters are usually visible. Three times 24 wells were scored/condition.
Isolation of ESC-derived and Neonatal Mouse Cardiomyocytes
EBs containing cardiomyocytes are detached from culture surfaces by incubating them with 0.05% trypsin-EDTA for 1 min at 37°C. Cells are dissociated with 1 mg/ml collagenase (CLSII, Worthington Biochemical, Lakewood, NJ), and 0.25% mg/ml pancreatin in a buffer containing (in mmol/L) NaCl 117, HEPES 20, NaH2PO4 1.2, KCl 5.4, Mg SO4 1, glucose 5, pH 7.35). EB-derived cardiomyocytes are separated by centrifugation through a discontinuous Percoll gradient and collected at the interface of the two layers. Neonatal cardiomyocytes are isolated from ventricles of 2-d-old mouse neonates as previously described (Jaconi et al., 2000).
Construction and Transfection of Ribozyme-expressing Vector
We designed a ribozyme sequence directed against the mouse NOX4 coding sequence (Irminger-Finger et al., 1998; Castanotto et al., 2002). A single-strand ribozyme sequence and cDNA sequence were synthesized in vitro as oligomers containing two regions of 12–13 nucleotides complementary to the murine NOX4 coding sequence interrupted by the ribozyme loop as follows: 5′-CATCTGCATCTGT-ttcgtcctcacggactcatcag-CTGAACCTCA-3′ (uppercase, murine NOX4 sequences; lowercase, ribozyme loop). This was cloned into the BamHI and SalI sites of pcDNA3.1 vector, resulting in antisense orientation in respect to the cytomegalovirus (CMV) promoter (Invitrogen, Carlsbad, CA). The plasmid containing the anti-NOX4 ribozyme was electroporated into CGR8 cells according to the standard protocol in a Gene Pulser (Bio-Rad, Richmond, CA) at 240 V, 500 μF. Several stable NOX4 ribozyme expressing ES clones, defined as riNOX4 clones, were propagated in the presence of LIF and selected for 10 d using G418 (250 μg/ml). Three stable ES clones were selected for their efficiency in maximally reducing the levels of NOX4 mRNA (measured by RT-PCR and RNase protection assay; see below).
Construction and Transduction of shRNA-expressing Lentivector
Two short hairpin RNAs (shRNAs) targeting the mouse NOX4 mRNA were designed, shNOX4-1: GCTGTCCCTAAACGTTCTACT and shNOX4-2: GGGCCTAGGATTGTGT-TTA. The complementary single-stranded DNA oligonucleotides were annealed and ligated inside the pENTR/U6 vector according to manufacturer’s instructions (Invitrogen). This vector was recombined by the Gateway LR-clonase (Invitrogen) reaction with the pLenti6/Block-it-DEST vector (Invitrogen) to form the shRNA expressing constructs. CGR8 cells stably expressing either shNOX4-1 or -2 were then obtained by transduction of these constructs by lentiviral delivery and subsequent blasticidin selection, as previously described (Suter et al., 2005).
siRNA Transfection
Previously published siRNAs targeting the mouse NOX4 mRNA (Mahadev et al., 2004) as well as a validated negative control siRNA labeled with the Alexa-488 dye were ordered from Qiagen (Chatsworth, CA): siNOX4-a: AACGAAGGGGTTAAACACCTC, siNOX4-b: AAAAGCAAGACTCTACACATC, siRNA-negative control: AATTCTCCGAACGTGTCACGT. Briefly, siRNA-negative control, siNOX4-a or -b (200 pmol), as well as a mix of siNOX4-a and -b (100 pmol each) were complexed with 2 μl of Lipofectamine-2000 in a final volume of 200 μl Opti-MEM medium (Invitrogen). The complexes were added onto freshly passaged ESC in suspension (150,000 cells/well of a 12-well plate). Twenty-four hours after transfection, cells were harvested for EB formation.
ROS Measurement by Amplex Red/Horseradish Peroxidase Method
HEK293 cells constitutively expressing the mouse NOX4 protein were created by transfection with full-length mNOX4 in pcDNA3.1. Neomyicin-resistant clones were selected and verified for functional NOX4-dependent ROS production by Amplex Red. A high-level producing clone was chosen for further experiments. Twenty-four hours before transfection, HEK293-mNOX4 cells were seeded at 4 × 105 cells per well into six-well plates. Plasmids (5 μg/well) expressing the shRNAs as well as a control GFP plasmid were transfected into cells by the calcium phosphate method. The medium was changed the following day. Two days later, transfection efficiency was assessed by evaluating the proportion of GFP-positive cells by fluorescence microscopy. Transfection was considered successful when the efficiency was above 90%. Cells were then detached and seeded into 96-well plates (2 × 104 cells/well). Amplex Red (50 μM) and horseradish peroxidase (HRP; 0.1 U/ml) were added in a final volume of 200 μl/well. Diphenylene iodonium (DPI) was used at a concentration of 2.5 μM. Fluorescence was monitored at 37°C during 1 h with a Fluostar microplate reader (BMG Labtechnologies, Durham, NC) at the excitation wavelength of 544 nm and emission of 590 nm.
RNA Isolation and RT-PCR
Total RNA was isolated from ESC and EBs using RNeasy Mini Kit from Qiagen. RT-PCR was performed in simultaneous one-step by EZrTthRNA PCR Kit (Perkin Elmer-Cetus, Norwalk, CT) according to the manufacturer’s instruction. Briefly, reverse transcription was performed on 1 μg RNA at 60°C for 35 min. After reverse transcription, the cDNAs were used for semiquantitative PCR using sets of specific primers as previously described (Meyer et al., 2000; Li et al., 2002). An initial step of 94°C was used for 5 min followed by 30 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min. RT-PCR was finished with 72°C for 7 min. Fifteen microliters of reaction were run on a 1.5% agarose gel in 1× TAE and photographed on a UV transilluminator using a digital camera.
Real-Time RT-PCR by SYBR Green Detection
The nucleotide sequences of the PCR primers used were as follows: NOX4 forward 5′-TGTTGGGCCTAGGATTGTGTT and reverse 5′-AGGGACCTTCTGTGATCCTCG; MEF2C forward 5′-CCTACATAACATGCCGCCATCT and reverse 5′-GTGGTACGGTCTCCCAACTGA; Nkx2.5 forward 5′-GGATAAAAAAGAGCTGTGCGC and reverse 5′-GGCTTTGTCCAGCTCCACTG; β-tubulin forward 5′-AGACAACTTCGTTTTCGGTCAGT and reverse 5′-CCTTTAGCCCAGTTGTTGCCT. To avoid amplification of genomic DNA, primers were designed to be intron-spanning. Real-time RT-PCR was performed using a TaqMan rapid thermal cycler (Prism 7700, Advanced Biotechnologies [ABI], Columbia, MD). Amplification was carried out as recommended by the manufacturer: 25 μl reaction mixture contained 12.5 μl of SYBR Green PCR Master mix (Applied Biosystems, Foster City, CA), the appropriate primer concentration, and 1 μl of cDNA. The relative cDNA concentrations were established by a standard curve using sequential dilutions of corresponding PCR fragments. The data were normalized to β-tubulin. The amplification program included the initial denaturation step at 95°C for 10 min, 40 cycles of denaturation at 95°C for 10 s, and annealing and extension at 60°C for 1 min. Fluorescence was measured at the end of each extension step. After amplification, melting curves were acquired and used to determine the specificity of PCR products, which were further confirmed using conventional gel electrophoresis.
RNase Protection Assay
RNase protection assays were performed following standard methods (Ma et al., 1996). Briefly, the mouse NOX4 cDNA sequence (nucleotides 1–447 from the start codon) was cloned first into pGEM-T vector and then subcloned into the BamHI and Asp718I sites of pcDNA3.1 vector. The vector was linearized with KpnI. The mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA sequence (nucleotides 393–522 from the start codon) was cloned into pGEM-T vector. The vector was linearized with NotI. The antisense probes against mouse NOX4 and GAPDH were transcribed with T7 polymerase. Total RNA was extracted from wild-type (wt) ESC or from different anti-NOX4 ribozyme clones with the Perfect RNA Mini kit (Eppendorf), and 10 μg of total RNA were hybridized with antisense probes. RNase A, 80 ng/μl, was added in the hybridization mixture to digest free probe and other single-stranded RNA. The protected fragments were purified, resolved on denaturing polyacrylamide gel (8 M urea/5% polyacrylamide), and quantified by autoradiography.
Western Blot Analysis
The ESC and EBs were lysed in a lysis buffer containing 50 mM Tris (pH 7.4), 50 mM NaCl2, 1% Triton X-100, 1 mM EDTA, 10 mM MgCl2 supplemented with a protease inhibitor cocktail (Boehringer Mannheim, Indianapolis, IN) and phosphatase inhibitor, 10 mM sodium orthovanadate (for analysis of p38 MAPK) or a lysis buffer containing 150 mM NaCl, 1 mM EDTA, 20 mM Tris (pH7.4), 1% NP40 (for analysis of MLC2v) and sonicated for 2 s to shear DNA. Cell lysates were centrifuged at 14000 × g (4°C) for 30 min. The insoluble fractions including MLC2v were resuspended in buffer containing 50 mM glycerophosphate (pH 7.4), 1 mM EGTA, 0.3% Triton X-100, and 10% glycerol. Protein concentrations were determined by Bio-Rad Protein Assay. Proteins (60 μg) were separated by SDS-PAGE, transferred to PVDF membrane (Millipore, Bedford, MA), blocked with phosphate-buffered saline (PBS) containing 0.1% Tween 20 and 5% nonfat milk and probed with either rabbit anti-NOX4 (1:500, kindly provided by Dr. E. Ogier-Denis, INSERM U-773, Paris, France), mouse anti-α-tubulin (1:8000, Sigma-Aldrich, St. Louis, MO), rabbit anti-MLC2v (1:500; Meyer et al., 2000), rabbit anti-phospho-p38 MAPK or rabbit anti-p38 MAPK (1:500, Cell Signaling, Beverly, MA) antibodies. The blots were incubated with HRP-conjugated goat anti-rabbit (1:10000) or anti-mouse (1:5000) antibodies (Amersham Biosciences, Piscataway, NJ), followed by detection with Enhanced Chemiluminescence (ECL, Amersham Biosciences).
Indirect Immunofluorescence Staining
The EBs for immunofluorescence staining were plated on gelatin-coated coverslips at day 6 during the differentiation procedure, fixed with 3% paraformaldehyde at day 8, and permeabilized with 0.1% Triton X-100 in PBS. After blocking with 1% BSA in PBS the EBs were incubated for 1 h with monoclonal antibody against α-actinin (Sigma-Aldrich) or rabbit anti-MLC2v antisera (Meyer et al., 2000) and then labeled for 1 h with FITC-conjugated anti-mouse IgG or FITC-conjugated anti-rabbit IgG (Sigma-Aldrich). After being washed three times in PBS, coverslips were mounted on slides with Mowiol, dried overnight, and examined by confocal fluorescence microscopy.
Confocal Imaging
Fixed immunostained EBs were imaged with a Nikon inverted microscope (Melville, NY; TE-2000, objective 60×, 1.4 NA) using a Nipkow-type high-speed confocal microscopy system (Ultraview RS, PerkinElmer, Cambridge, United Kingdom), as described (Tanaami et al., 2002). FITC fluorescence was excited at 488 nm by a Krypton/Argon triple line laser. The emission light (collected above 515 nm through a long-pass barrier filter) was imaged through a relay lens by an intensified high-speed 12-bit cooled CCD camera (Orca ER). Z-sectioning of the EBs was performed (z step = 0.2 μm). Images were recorded on a Dell computer (Round Rock, TX) and 3D reconstruction performed using the software Volocity (San Francisco, CA). The middle section of the 3D stack of images was chosen to represent the fluorescence distribution within the nuclei and throughout the cytoplasm of ES-derived cardiac cells.
In Situ Hybridization
A 526 nucleotides probe for NOX4 was amplified with a specific set of primers (forward: GGATTTCTGGACCTTTGTG and reverse: CAGATAAAGTACAGTCTTCTTA; Vallet et al., 2005) and cloned into pcr4-TOPO vector (Invitrogen). For in situ hybridization experiments the in vitro transcription of digoxigenin-labeled cRNA was performed using the RNA-DIG Labeling Mix (Roche Molecular Biochemicals, Indianapolis, IN) and RNA polymerases T3 and T7 (Imhof et al., 2006). Briefly, paraffin-embedded sections of whole embryos at different embryonal stages were deparaffinized, rehydrated and digested with proteinase K (20 μg/ml 1× PBS buffer) for 30 min at 37°C. After a prehybridization in 20% glycerol for 30 min, sections were then incubated with the digoxigenin-labeled sense and antisense cRNA probes at a dilution of 1:100 in hybridization buffer containing 50% deionized formamide, 10% dextran sulfate, 2× SSC (0.3 M NaCl, 0.03 M trisodium citrate), 1× Denhardt’s solution, 10 μg/ml salmon sperm DNA, and 10 μg/ml yeast tRNA. Hybridization was performed overnight at 55°C in a humid chamber containing 50% formamide in 2× SSC. Posthybridization washes were performed as reported previously (Lewis and Wells, 1992). Tissue samples were incubated overnight at 4°C with an anti-digoxigenin Fab antibody conjugated to alkaline phosphatase (Roche Molecular Biochemicals). Staining was visualized with NBT/5-bromo-4-chloro-3-indolyl phosphate (Dakocytomation, Glostrup, Denmark).
For dual labeling, a set of hybridized sections was additionally subjected to an immunofluorescent labeling of the cardiac tissue by a rabbit polyclonal antibody to α-cardiac actin (Clement et al., 2003; dilution 1:20). The immunoreaction was revealed by rabbit fluorescent anti-IgG conjugated to Alexa 488 (Invitrogen, 1:500; Imhof et al., 2006).
Statistical Analysis
All values are represented as means ± SEM of the indicated number of measurements.
ANOVA test and Student’s t test statistics (two-tailed) were used to determine significance, requiring p < 0.05 for statistical significance.
RESULTS
ROS Scavengers Impair Cardiac Differentiation from ESC whereas Exogenous Hydrogen Peroxide Increases the Percentage of Spontaneously Contracting EBs
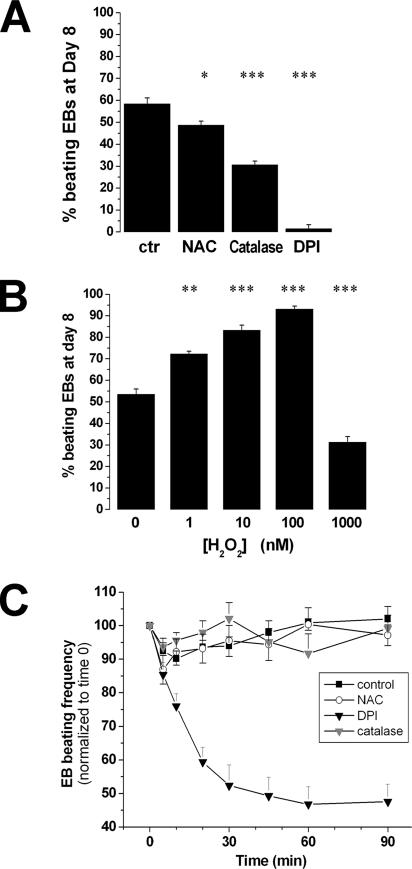
To investigate the role of ROS on cardiomyocyte differentiation, we allowed mouse ESC to differentiate into EBs. We had previously shown that cardiac transcription factors start to be expressed at day 4 of EB differentiation (Meyer et al., 2000). This time point is a critical window for Ca2+-dependent transcriptional processes regulating cardiac differentiation (Meyer et al., 2000; Li et al., 2002). We therefore exposed EBs at day 4 of culture for 2 h to free radical scavengers such as N-acetylcysteine (NAC, 5 mM) and catalase (200 U/ml), as well as to diphenyleneiodonium chloride (DPI, 2.5 μM), an NADPH oxidase inhibitor. We then evaluated the presence of beating activity in EBs at day 8 of culture. As shown in Figure 1A, ROS scavengers significantly decreased the percentage of EBs containing beating cardiomyocytes, suggesting that endogenous ROS generation is involved in cardiac differentiation. On the other hand, exposure of EBs to increasing concentrations of hydrogen peroxide (H2O2, 1–1000 nM) for 2 h at day 4 led to an enhanced beating activity at day 8 (Figure 1B). The most prominent effect was observed with a H2O2 concentration of 100 nM, whereas higher doses (1 μM) depressed cardiomyocyte differentiation compared with control conditions.
Figure 1.
Effect of ROS on cardiac differentiation of mouse ESC. (A) Impairment of EB beating activity at day 8 by a treatment with ROS scavengers such as NAC (5 mM) and catalase (200 U/ml), or with the NOX inhibitor DPI (2.5 μM), applied for 2 h at day 4. (B) Effect of H2O2 treatment (1, 10, 100, 1000 nM applied as a 2-h pulse at day 4), on EB beating activity scored at day 8. (C) Effect of DPI (2.5 μM), NAC (5 mM), and catalase (200 U/ml) on EC-coupling in beating EBs at day 8. Beating frequency (beats/min) was normalized to time 0 and expressed as 100%. Dimethyl sulfoxide was used as control condition. Data are presented as means ± SEM; n = 3 independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001 by two-tailed t test.
To determine whether active NOXs are also required to the maintenance of cardiomyocyte EC coupling, clearly operational at day 8 of culture, we tested the effect of the NADPH-blocker DPI (2.5 μM) or the ROS scavengers NAC (5 mM) and catalase (200 U/ml) on beating EBs at day 8 of culture. DPI decreased the beating frequency of contractile cardiomyocytes to 50% at 30 min, whereas NAC or catalase had no effect (Figure 1C). The DPI effect suggests a possible role of ROS in EC coupling, but we cannot exclude toxicity of DPI on cell contractility, particularly because ROS scavengers had no effect.
These results led us to postulate the involvement of ROS in the regulation of cardiac differentiation.
NOX4 Is the Principal NOX Isoform Expressed in Undifferentiated ESC, Differentiating EBs, and Neonatal Cardiomyocytes
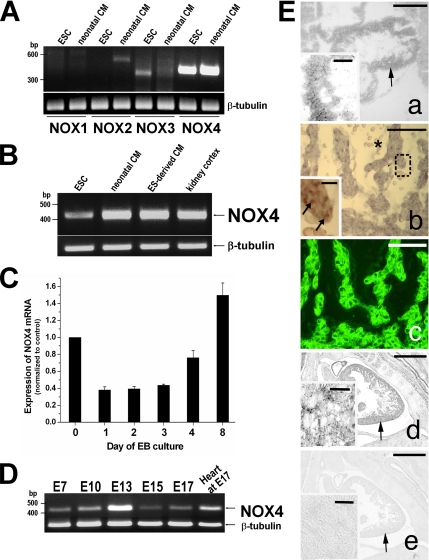
We screened undifferentiated mouse ESC and neonatal mouse cardiomyocytes (CM) for the presence of mRNAs coding for the NOX family members by semiquantitative RT-PCR. As shown in Figure 2A, NOX4 was highly expressed both in ESC and neonatal CM, whereas NOX1 was absent. A weak expression of NOX2 and NOX3 was observed in CM and undifferentiated ESC, respectively. To further confirm the expression of NOX4 in ESC-derived CM, we enzymatically dissociated 10-d-old EBs and isolated CM by percoll gradient separation. In ESC-derived CM NOX4 expression revealed by RT-PCR was comparable to the expression in kidney cortex (Figure 2B), where NOX4 was originally discovered (Shiose et al., 2001).
Figure 2.
In vitro and in vivo expression of NOX4 mRNA. (A) Detection of NOX homologues NOX1, NOX2, NOX3, and NOX4 by RT-PCR in ESC and neonatal mice cardiomyocytes (CM). (B) NOX4 expression by RT-PCR in mouse undifferentiated ESC versus neonatal CM, ESC-derived CM and kidney cortex. (C) Time course analysis by real-time RT-PCR of NOX4 mRNA levels during differentiation of wt ESC (day 0, 4: n = 6; days 1, 2, 3, and 8: n = 3). (D) In vivo NOX4 expression by RT-PCR in total RNA extracted from whole mouse embryos at different stages of development (E7, 10, 13, 15, and 17), and from E17 mouse heart. (E) Localization of NOX4 mRNA in the heart of mouse embryos at E9 (a), E11 (b), and E14 (d) by in situ hybridization (ISH). In panel c, α-cardiac actin immunostaining colocalized with NOX4-positive cells on the section of panel b. Panels d and e, ISH with NOX4 antisense and sense probe, respectively. Arrows in a, d, and e indicate the area enlarged in the inset. Arrow in b indicates diffuse punctiform NOX4 labeling. *Presence of NOX4- negative red blood cells. Scale bars, (a and inset): 300 and 20 μm, respectively; (b and c): 50 μm; (d and e): 700 μm.
Next, using real-time RT-PCR, we analyzed the time course of NOX4 mRNA expression during early stages of EB differentiation. One day after EB formation (day 1), NOX4 levels were diminished by 60% (p < 0.001) compared with undifferentiated cells. Thereafter, NOX4 started to increase at day 4 by 75% (p < 0.01) compared with day 3. It was further augmented at day 8 (Figure 2C).
The in vivo presence of NOX4 in the embryonic heart was also identified by RT-PCR (Figure 2D) and by in situ hybridization at different stages of the mouse embryo development (from embryonic stage (E)9 to E15). Figure 2E shows representative labelings of NOX4 mRNA at E9 (panel a), E11 (panel b), and E14 (panel d). The E11 section was costained with an antibody against α-cardiac actin to confirm the cardiac identity of the cells positive for the NOX4 RNA probe (panel c).
NOX4 Down-Regulation Impaired the Beating Activity of EBs
To assess the involvement of NOX4 in cardiogenesis, we first repressed the levels of NOX4 mRNA using a ribozyme technique. Over several stable ESC clones expressing a cDNA coding for an anti-NOX4 ribozyme (defined as riNOX4 ESC), we selected 3 of 9 clones (clones 1, 6, and 9) for their very low levels of residual NOX4 mRNA, as verified by RNase protection assay (Figure 3A). When quantified by real-time RT-PCR, NOX4 mRNA levels in clone 9 were significantly reduced by more than 65% compared with wt ESC (p < 0.001, n = 6).
Figure 3.
Effect of NOX4 down-regulation on the beating activity of EBs. (A) RNase protection assay for NOX4 on riNOX4 ESC stable clones. (B) EB beating activity at day 8 in wt, mock-transfected, and riNOX4 ESC (clones 1, 6, and 9; n = 3). (C) ROS production quantified by Amplex Red/peroxidase method in HEK293 cells overexpressing mouse NOX4 (HEK-NOX4) and transiently transfected with control shRNA, shNOX4-1 or shNOX4-2 (n = 6). (D) NOX4 mRNA levels in wt and shRNA-transduced ESC quantified by real-time RT-PCR (n = 4). (E) Western blot of NOX4 protein levels assessed in wt and shRNA-expressing EBs at day 4. Alpha-tubulin was used as a loading control. (F) Effect of NOX4 down-regulation by shNOX4 on the EB beating activity at day 8 (n = 3). (G) Transient transfection of ESC with siNOX4-a, -b, -a+b, or siRNA negative control (si-ctr). Top panels, phase-contrast and fluorescent images of ESC 24 h after their transfection with Alexa-488–labeled siRNA-negative control. Bottom panel, beating activity at day 7 in EBs generated after ESC transient transfection (n = 3). Data are presented as means ± SEM. *p < 0.05; **p <0.01; ***p < 0.001, by ANOVA test (C and D) or by t test (B, F, and G).
To functionally correlate altered NOX4 expression with ROS production in ESC, a series of methods (such as luminol, cytochrome C reduction, scopoletin, NBT, and Amplex-Red) did not specifically detect ROS amounts above their respective detection thresholds, suggesting that ESC are low ROS producers (unpublished data).
We next evaluated the effect of ribozyme-dependent NOX4 reduction on the beating activity within EBs at day 8. Importantly, the percentage of spontaneously contracting EBs was reduced by more than 75% in riNOX4 EBs (Figure 3B).
To further confirm results obtained with the ribozyme technique, we used two alternative methods based on RNA interference (RNAi): 1) the stable expression of short-hairpin RNAs (shRNA) targeting NOX4 (defined as shNOX4-1 and -2) into ESC by lentivector-mediated transduction, and 2) the transient expression of anti-NOX4 siRNA oligos. The efficiency of shNOX4-1, shNOX4-2 was first screened by transiently transfecting HEK293 cells constitutively expressing the mouse NOX4 protein (HEK293-mNOX4). NOX4-dependent ROS production (corresponding to the DPI-inhibitable fraction) was quantified by the Amplex Red/peroxidase method. We determined that shNOX4-1, but not shNOX4-2, significantly down-regulated NOX4-dependent ROS production in HEK cells by 40% when compared with the control shRNA (an unrelated sequence; Figure 3C). Because of its lack of effect on ROS production, shNOX4-2 was chosen as a negative control for the ESC experiments. ESC were then transduced by lentivectors expressing shNOX4-1 or shNOX4-2 and subsequently selected by the blasticidin resistance. Analysis by real-time RT-PCR revealed that the amount of NOX4 mRNA was down-regulated by 30% in shNOX4-1–expressing ESC, whereas it remained at the level of control in shNOX4-2 cells (Figure 3D).
We monitored the levels of NOX4 protein in wt and NOX4-down-regulated EBs at day 4 of differentiation, when the cardiac differentiation program is fully activated. Western blot analysis showed a consistent down-regulation of NOX4 protein in shNOX4-1 EBs (Figure 3E, n = 3 independent experiments). At day 8, the percentage of spontaneously contracting shNOX4-1 EBs was reduced by ∼40% when compared with control cells, whereas shNOX4-2 EBs were not different from wt EBs (Figure 3F).
At this point, to further address the involvement of NOX4 in the very early stages of ESC differentiation toward cardiac cell fate, we asked whether even a transient down-regulation of NOX4 could produce a similar effect as the one observed with stable ribozyme or shRNA. To this end, we transiently transfected previously published siRNA oligos targeting NOX4 (siNOX4-a and siNOX4-b), one day before EB formation. By transfecting sister cultures with an irrelevant siRNA-oligo labeled with Alexa-488, we estimated the transfection efficiency to be above 90% (Fig 3G, top panels). At day 7 of EB differentiation, the percentage of beating EBs was reduced by ∼45 and 35% with siNOX4-a and siNOX4-b (each at 200 pmol/ml), respectively, and by 55% when both siNOX4 (each at 100 pmol/ml) were cotransfected (Figure 3G, bottom panels).
In conclusion, these data convincingly depict an early role for NOX4 in the ESC-derived cardiac phenotype.
Rescue of the Beating Activity in riNOX4 EB by Exogenous ROS
To test whether exogenous ROS could rescue the cardiac phenotype in riNOX4 EBs, we chose to apply a 2-h pulse of H2O2 at day 4, a time point at which the cardiac differentiation program is fully activated. This treatment could indeed rescue, in a concentration-dependent manner, the beating activity within riNOX4 EBs at day 8 (Figure 4). On the other hand, doses of H2O2 in the μM range were detrimental (unpublished data) as also observed in wt EBs (Figure 1B). Such deleterious effect was also obtained by overexpressing NOX4 in ESC, which displayed DNA fragmentation observed by DNA laddering (unpublished data). This suggests a dose-response effect of ROS, where low doses are permissive for the appearance of cardiac beating phenotype and high doses lead to apoptosis.
Figure 4.
Rescue of the beating activity in riNOX4 EBs by hydrogen peroxide. H2O2 (1–100 nM) was applied at day 4 for 2 h (n = 3). Data are presented as means ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001, by t test.
Impaired Cardiac Commitment and Myofibrillogenesis in the Absence of NOX4
We investigated whether NOX4 deficiency has an impact on cardiac commitment and, consequently, on the expression levels of transcription factors known to be involved in cardiac differentiation. For this purpose, we quantified Nkx2.5 and myocyte enhancer factor 2 C (MEF2C) transcripts in both wt and riNOX4 differentiating EBs by real-time RT-PCR. When compared with control EBs, riNOX4-differentiating cells showed lower amounts of Nkx2.5 transcripts at day 4 (35% decrease), and this difference was accentuated at day 8 (65% decrease; Figure 5A, left panel). Concerning MEF2C, a 30% reduction was already detected in undifferentiated riNOX4 ESC compared with wt cells (Figure 5A, right panel). MEF2C mRNA transcripts in riNOX4 EBs were also significantly diminished at day 4 (75% decrease), but were comparable with wt EB expression levels at day 8.
Figure 5.
Impact of NOX4 down-regulation on the expression and localization of cardiac transcription factors and sarcomeric proteins. (A) Time-course expression of the cardiac transcription factors Nkx2.5 and MEF2C by real-time RT-PCR (n = 4). (B) Intracellular distribution of MEF2C in differentiating cardiomyocytes within EBs at day 8 by immunofluorescence and confocal microscopy. Bar, 10 μm. (C) Time-course expression of MLC2v mRNA by RT-PCR (a) and protein levels by Western blot (b). Quantification of MLC2v protein levels by densitometry (c, n = 3). (D) Confocal micrographs of α-actinin distribution in wt and riNOX4 EBs at day 8. Bar, 10 μm. Data are presented as means ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001, ANOVA test in A and t test in C.
In addition, we also observed an altered cellular compartmentalization of MEF2C protein. Although wt cardiac-committed cells exhibited a nuclear staining of the protein, MEF2C in riNOX4 cells was restricted to the cytoplasm (Figure 5B), a localization that prevents its role in gene transcription.
We previously found that the expression of ventricular myosin regulatory light chain 2 (MLC2v) was dependent on the expression and the correct localization of MEF2C (Li et al., 2002). Hence, we measured both the mRNA and protein levels of MLC2v during differentiation. In analogy with our previous observations, we detected a decreased MLC2v expression in riNOX4 EBs at day 8 and 12 of differentiation (Figure 5C). To verify if this decrease had an impact also on myofibrillogenesis, we evaluated the organization of the sarcomeric protein, α-actinin in 8-d-old EBs by immunofluorescence. As shown in Figure 5D, wt ESC-derived cardiomyocytes contained α-actinin organized into an interconnected network of myofibrils. In contrast, cardiomyocytes derived from riNOX4 ESC showed an abnormal distribution of α-actinin that was not incorporated into the Z-lines, but remained localized in cytosolic spots (Figure 5D, right panel). Altogether, these data indicate that cardiomyocytes derived from riNOX4 ESC have an altered transcription factor program and do not correctly form sarcomeres and myofibrils.
NOX4-dependent ROS Activate p38 MAPK to Allow Cardiomyocyte Differentiation
To address the role of ROS and in particular which intracellular signaling pathway could lead NOX4-dependent ROS to regulate cardiac differentiation, we investigated the p38 MAPK, a major MAPK activated by H2O2 in other cell types including vascular smooth muscle cells (Ushio-Fukai et al., 1998). We measured the activation of p38 MAPK by Western blot analysis using the anti-phospho-p38 MAPK antiserum. Different concentrations of H2O2 (0.1–10 μM) applied for 1 h could increase the phosphorylation of p38 MAPK in undifferentiated ESC (unpublished data). In contrast, phosphorylated p38 MAPK was completely suppressed in the riNOX4 ESC, whereas the total p38 MAPK levels were similar to that of wt ESC (Figure 6A, Day 0).
Figure 6.
Involvement of NOX4 on p38 MAPK pathway. (A) Levels of p38 MAPK phosphorylation by western blot during differentiation of wt and riNOX4 ESC (clone 6). (B) Levels of phosphorylated p38 MAPK during EB differentiation in the presence of the MAPK inhibitor SB203580 (5 μM). (C) Effect of SB203580 (5 μM) on the percentage of beating EBs scored at day 8 (n = 3). (D) Immunolocalization of MEF2C and α-actinin (day 6 and day 8, respectively) in wt and SB203580-treated EBs by confocal microscopy. Bar, 10 μm. Data are presented as means ± SEM. *p < 0.001, by t test.
We then assessed the time course of p38 MAPK phosphorylation during EB differentiation. In wt EBs, phosphorylated p38 MAPK rapidly increased at day 4 and remained elevated throughout the differentiation procedure. In contrast, riNOX4 EBs (clone 6) exhibited a highly reduced and delayed phosphorylation of p38 MAPK (Figure 6A).
To investigate if cardiac differentiation relies on p38 MAPK activation, possibly via NOX4-dependent ROS production, we used the p38 MAPK inhibitor SB203580, a pyridinyl imidazole that inhibits the activation of the immediate substrate of p38 MAPK, MAPKAP kinase-2 (Cuenda et al., 1995). Either wt ESC or differentiating EBs were cultured in the presence of SB203580 (5 μM). This treatment abolished the phosphorylation of p38 MAPK in both ESC and EBs (Figure 6B), leading to a dramatic reduction of the beating activity in EBs observed at day 8 (Figure 6C). Concomitantly, there was an abnormal cardiac myofibrillogenesis. Alpha-actinin was disorganized inside the cells and showed a spotted distribution instead of a sarcomeric Z-line localization (Figure 6D, bottom panels). Importantly, the inhibition of p38 MAPK by SB203580 prevented the nuclear translocation of MEF2C, as observed at day 6 of differentiation (Figure 6D, top panels). MEF2C was distributed throughout the nuclei of untreated cardiac-committed cells, whereas MEF2C fluorescence was essentially extranuclear in p38-inhibited cells. SB203580 did not have any effect on proliferation of ESC, size, and morphology of EBs during differentiation (unpublished data).
Taken together, these findings point to an involvement of p38 MAPK activation via NOX4-dependent ROS production in the regulation of cardiac differentiation.
DISCUSSION
We used mouse ESC to investigate the source and the role of ROS on cardiomyocyte differentiation. Our results indicate that ROS drive early stages of cardiogenesis, as revealed by an impaired cardiac differentiation in the presence of ROS scavengers. We show that both cardiogenesis and cardiac myofibrillogenesis are impaired when NOX4 is suppressed. Further, our data supports the idea that in ESC the effect of NOX4-mediated ROS (which are below the level of detection with standard methods) is mediated by MEF2C activation via p38 MAPK. Such a sequence of events may be essential for cardiac differentiation.
We showed that nanomolar concentrations of H2O2 administrated to EBs at day 4 enhanced the number of beating EBs. At this time point, cardiac transcription factors start to be fully expressed, but those encoding sarcomeric proteins are not (Li et al., 2002; Hakuno et al., 2005). Interestingly, higher concentrations of H2O2 suppressed the EB beating activity at day 8. This led us to postulate that low doses of ROS are not detrimental but instead elicit dose-dependent biological responses. Indeed, excessive ROS levels repress the expression of several cardiac genes (Morel and Barouki, 1999; Puceat et al., 2003). For instance, Puceat et al. (2003) reported that a daily treatment of EBs with H2O2 prevented the beating activity in differentiating EBs. This is consistent with our observation that NOX4 overexpression in ESC leads to apoptosis (unpublished data). Beside ROS, other signaling molecules have also been implicated in the regulation of cardiac differentiation. A recent article suggests a dose-dependent effect of nitric oxide, with low doses being permissive for differentiation and higher doses leading to apoptosis (Kanno et al., 2004).
Here, we report that in vivo NOX4 mRNA is localized in the early embryonic mouse heart within cardiac cells, as revealed by in situ hybridization coupled with immunofluorescence identification of cardiac markers. Because NOX4 is the main NOX isoform expressed in mouse ESC and its down-regulation by different RNAi methods impairs ESC differentiation into cardiomyocytes, we propose that NOX4-dependent ROS production is critical for cardiogenesis. This is supported by the fact that H2O2 can rescue the beating activity in NOX4-down-regulated EBs when administrated at day 4, a critical time window at which the cardiac differentiation program becomes fully activated (Meyer et al., 2000; Hakuno et al., 2005), but no cardiac cells, identifiable by organized cardiac sarcomeric proteins or by functional automatic contractions, are yet present. Indeed it is at this time point that NOX4 mRNA levels increase. The fact that ROS scavengers do not appear to affect excitation-contraction (EC)-coupling in beating ESC-derived cardiomyocytes at day 8 further points to a developmental role of NOX4-dependent ROS in early cardiogenesis.
These studies do not discriminate whether ROS effect is mainly due to the endogenous production of ROS in cardiac committed-ESC or is rather due to a paracrine effect by neighboring differentiating ESC within the EBs. Only a conditional down-regulation of NOX4 using a very early cardiac promoter could address this question, and we are currently addressing this issue.
Although accumulating evidence indicates that ROS play a critical role as intracellular signaling molecules, the molecular targets of ROS in cardiomyocytes remain unclear (Griendling et al., 2000), in particular during cardiogenesis. Interestingly, a recent publication reports that NADPH-dependent ROS can also play a role in ESC during cardiotrophin-1–induced cardiac proliferation (Sauer et al., 2004; Ateghang et al., 2006).
MEF2C is important in early cardiovascular development (Lin et al., 1997; Lin et al., 1998). This crucial transcription factor is responsible for the activation of several cardiac-specific embryonic genes (Harvey, 1999), including MLC2v (Lin et al., 1997; Liu et al., 2001). Indeed, MEF2C regulates the activity of MLC2v promoter (Chen et al., 1998). We recently demonstrated that the translocation of MEF2C into the nucleus of differentiating ES-derived cardiomyocytes by Ca2+-dependent mechanisms is necessary for the transcription of MLC2v and its insertion into functional myofibrils (Li et al., 2002). Several signaling pathways that activate cardiogenesis converge to MEF2C. Indeed, MEF2C can be modified by phosphatases such as calcineurin, as well as by Ca2+/calmodulin-dependent kinases. Moreover, MEF2C can be directly phosphorylated by p38 MAPK (Han et al., 1997; Zhao et al., 1999; Aikawa et al., 2002; Eriksson and Leppa, 2002; Akazawa and Komuro, 2003) on residues (T293, T300, S387) located within the activation domain of MEF2C (Han et al., 1997; Yang et al., 1999; Zhao et al., 1999).
It has also been shown that ROS can have a direct effect on proteins containing cysteine residues prone to oxidation such as phosphatases (i.e., PTPB; Salmeen et al., 2003; Biswas et al., 2006). Hence, ROS-dependent phosphatase inhibition may result in increased phosphorylations. Conversely, reduced NOX4-dependent ROS may affect phosphorylation-dependent transcription factors upstream to MEF2C. Work is in progress to identify these signaling pathways.
In the absence of NOX4 we observed decreased mRNA levels of MEF2C and NKx2.5 suggesting a reduced cardiac commitment. Moreover, the abolished phosphorylation of p38 MAPK in the absence of NOX4-mediated ROS production leads to a mislocalization of MEF2C and therefore to a reduced MLC2v transcription and disorganized myofibrils.
Collectively our data suggest that NOX-generated ROS leads to p38 phosphorylation and the subsequent MEF2C nuclear translocation necessary for cardiac phenotype determination, transcription of sarcomeric proteins, and myofibrillogenesis. In accordance with our results, other groups have shown that p38 MAPK is activated by ROS, either generated intracellularly or administered exogenously (Raingeaud et al., 1995; Moriguchi et al., 1996; Huot et al., 1997; Seko et al., 1997; Clerk et al., 1998).
Taken together with our previous findings (Li et al., 2002), we propose that at least two lines of signaling act together to induce cardiac differentiation: first, a Ca2+-dependent activation of Ca2+/calmodulin-dependent kinase, and second, a NOX4-dependent activation of p38 MAPK through a moderate increase in ROS. These two signaling pathways converge at the level of MEF2C whose nuclear translocation requires the activation of both pathways (Figure 7).
Figure 7.
Model depicting signaling pathways involved in cardiac differentiation. Calcium and ROS act through CamK and MAPK, resulting in MEF2C transcription and nuclear translocation necessary for cardiac differentiation.
The importance of our observations goes beyond the analysis of cardiac differentiation. Indeed, to our knowledge this is the first molecular demonstration that NOX family NADPH oxidases may play a crucial role in the determination of cell fate. The first NOX family member known, the phagocyte NADPH oxidase NOX2, is, without any doubt, a host defense enzyme. Yet, it is very likely that this is not the only function of NOX family enzymes (e.g., Lambeth, 2002). Aguirre et al. (2005) recently reported in a review that NOX homologues have been identified in microbiological eukaryotes and that they may play important roles in cell differentiation in fungi and other eukaryotic microorganisms.
In support of this notion, our studies demonstrate that NADPH oxidases are not only involved in the host defense, but also in the regulation of developmental processes such as cardiac differentiation through a MAP kinase–dependent pathway.
ACKNOWLEDGMENTS
We are grateful to S. Jaconi for technical assistance with RNase protection assay and chemiluminescence experiments, as well as to Drs. M. Dubois Dauphin, A. Feki, J. Dern-Wieloch, A. Hild, and B. Greggio for technical help with in situ hybridization techniques. We also thank Dr. M. Hauwel for advices on statistical analysis. We also thank Dr. S. Clément for providing α-cardiac actin antibody, Dr. C. Guichard for technical help with Western blot analysis, and in particular Dr. E. Ogier-Denis for providing the newly generated NOX4 antibody. This work was supported by research grants from the Swiss National Science Foundation to M.J. (NRP 4046-058712) and K.-H.K. (31-55805.98).
Abbreviations used:
- CM
cardiomyocytes
- EB
embryoid bodies
- ESC
embryonic stem cells
- EC coupling
excitation-contraction coupling
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-06-0532) on July 14, 2006.
References
- Aguirre J., Rios-Momberg M., Hewitt D., Hansberg W. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 2005;13:111–118. doi: 10.1016/j.tim.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Aikawa R., Nagai T., Kudoh S., Zou Y., Tanaka M., Tamura M., Akazawa H., Takano H., Nagai R., Komuro I. Integrins play a critical role in mechanical stress-induced p38 MAPK activation. Hypertension. 2002;39:233–238. doi: 10.1161/hy0202.102699. [DOI] [PubMed] [Google Scholar]
- Akazawa H., Komuro I. Roles of cardiac transcription factors in cardiac hypertrophy. Circ. Res. 2003;92:1079–1088. doi: 10.1161/01.RES.0000072977.86706.23. [DOI] [PubMed] [Google Scholar]
- Ateghang B., Wartenberg M., Gassmann M., Sauer H. Regulation of cardiotrophin-1 expression in mouse embryonic stem cells by HIF-1α and intracellular reactive oxygen species. J. Cell Sci. 2006;115:1043–1052. doi: 10.1242/jcs.02798. [DOI] [PubMed] [Google Scholar]
- Biswas S., Chida A. S., Rahman I. Redox modifications of protein-thiols: emerging roles in cell signaling. Biochem. Pharmacol. 2006;71:551–564. doi: 10.1016/j.bcp.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Castanotto D., Li J. R., Michienzi A., Langlois M. A., Lee N. S., Puymirat J., Rossi J. J. Intracellular ribozyme applications. Biochem. Soc. Trans. 2002;30:1140–1145. doi: 10.1042/bst0301140. [DOI] [PubMed] [Google Scholar]
- Chen J., Kubalak S. W., Minamisawa S., Price R. L., Becker K. D., Hickey R., Ross J., Jr, Chien K. R. Selective requirement of myosin light chain 2v in embryonic heart function. J. Biol. Chem. 1998;273:1252–1256. doi: 10.1074/jbc.273.2.1252. [DOI] [PubMed] [Google Scholar]
- Cheng G., Cao Z., Xu X., van Meir E. G., Lambeth J. D. Homologs of gp91phox: cloning and tissue expression of Nox3, Nox4, and Nox5. Gene. 2001;269:131–140. doi: 10.1016/s0378-1119(01)00449-8. [DOI] [PubMed] [Google Scholar]
- Clement S., Orlandi A., Bocchi L., Pizzolato G., Foschini M. P., Eusebi V., Gabbiani G. Actin isoform pattern expression: a tool for the diagnosis and biological characterization of human rhabdomyosarcoma. Virchows Arch. 2003;442:31–38. doi: 10.1007/s00428-002-0728-4. [DOI] [PubMed] [Google Scholar]
- Clerk A., Fuller S. J., Michael A., Sugden P. H. Stimulation of “stress-regulated” mitogen-activated protein kinases (stress-activated protein kinases/c-Jun N-terminal kinases and p38-mitogen-activated protein kinases) in perfused rat hearts by oxidative and other stresses. J. Biol. Chem. 1998;273:7228–7234. doi: 10.1074/jbc.273.13.7228. [DOI] [PubMed] [Google Scholar]
- Cuenda A., Rouse J., Doza Y. N., Meier R., Cohen P., Gallagher T. F., Young P. R., Lee J. C. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- Davidson S. M., Morange M. Hsp25 and the p38 MAPK pathway are involved in differentiation of cardiomyocytes. Dev. Biol. 2000;218:146–160. doi: 10.1006/dbio.1999.9596. [DOI] [PubMed] [Google Scholar]
- Eriksson M., Leppa S. Mitogen-activated protein kinases and activator protein 1 are required for proliferation and cardiomyocyte differentiation of P19 embryonal carcinoma cells. J. Biol. Chem. 2002;277:15992–16001. doi: 10.1074/jbc.M107340200. [DOI] [PubMed] [Google Scholar]
- Griendling K. K., Sorescu D., Ushio-Fukai M. NAD(P)H oxidase: role in cardiovascular biology and disease. Circ. Res. 2000;86:494–501. doi: 10.1161/01.res.86.5.494. [DOI] [PubMed] [Google Scholar]
- Hakuno D., Takahashi T., Lammerding J., Lee R. T. Focal adhesion kinase signaling regulates cardiogenesis of embryonic stem cells. J. Biol. Chem. 2005;280:39534–39544. doi: 10.1074/jbc.M505575200. [DOI] [PubMed] [Google Scholar]
- Han J., Jiang Y., Li Z., Kravchenko V. V., Ulevitch R. J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- Harvey R. P. Seeking a regulatory roadmap for heart morphogenesis. Semin. Cell Dev. Biol. 1999;10:99–107. doi: 10.1006/scdb.1998.0277. [DOI] [PubMed] [Google Scholar]
- Huot J., Houle F., Marceau F., Landry J. Oxidative stress-induced actin reorganization mediated by the p38 mitogen-activated protein kinase/heat shock protein 27 pathway in vascular endothelial cells. Circ. Res. 1997;80:383–392. doi: 10.1161/01.res.80.3.383. [DOI] [PubMed] [Google Scholar]
- Imhof A., Charnay Y., Vallet P. G., Aronow B., Kovari E., French L. E., Bouras C., Giannakopoulos P. Sustained astrocytic clusterin expression improves remodeling after brain ischemia. Neurobiol. Dis. 2006;22:274–283. doi: 10.1016/j.nbd.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Irminger-Finger I., Soriano J. V., Vaudan G., Montesano R., Sappino A. P. In vitro repression of Brca1-associated RING domain gene, Bard1, induces phenotypic changes in mammary epithelial cells. J. Cell Biol. 1998;143:1329–1339. doi: 10.1083/jcb.143.5.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaconi M., Bony C., Richards S. M., Terzic A., Arnaudeau S., Vassort G., Puceat M. Inositol 1,4,5-trisphosphate directs Ca(2+) flow between mitochondria and the endoplasmic/sarcoplasmic reticulum: a role in regulating cardiac autonomic Ca(2+) spiking. Mol. Biol. Cell. 2000;11:1845–1858. doi: 10.1091/mbc.11.5.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno S., Kim P. K., Sallam K., Lei J., Billiar T. R., Shears L. L., 2nd. Nitric oxide facilitates cardiomyogenesis in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2004;101:12277–12281. doi: 10.1073/pnas.0401557101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S. H., Pimentel D. R., Remondino A., Sawyer D. B., Colucci W. S. H(2)O(2) regulates cardiac myocyte phenotype via concentration-dependent activation of distinct kinase pathways. J. Mol. Cell. Cardiol. 2003;35:615–621. doi: 10.1016/s0022-2828(03)00084-1. [DOI] [PubMed] [Google Scholar]
- Laloi C., Apel K., Danon A. Reactive oxygen signalling: the latest news. Curr. Opin. Plant Biol. 2004;7:323–328. doi: 10.1016/j.pbi.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Lambeth J. D. Nox/Duox family of nicotinamide adenine dinucleotide (phosphate) oxidases. Curr. Opin. Hematol. 2002;9:11–17. doi: 10.1097/00062752-200201000-00003. [DOI] [PubMed] [Google Scholar]
- Lewis F. A., Wells M. In Situ Hybridization: A Practical Approach. In: Wilkinson D. G., editor. Oxford: IRL Press; 1992. pp. 121–135. [Google Scholar]
- Li J., Puceat M., Perez-Terzic C., Mery A., Nakamura K., Michalak M., Krause K. H., Jaconi M. E. Calreticulin reveals a critical Ca(2+) checkpoint in cardiac myofibrillogenesis. J. Cell Biol. 2002;158:103–113. doi: 10.1083/jcb.200204092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Lu J., Yanagisawa H., Webb R., Lyons G. E., Richardson J. A., Olson E. N. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development. 1998;125:4565–4574. doi: 10.1242/dev.125.22.4565. [DOI] [PubMed] [Google Scholar]
- Lin Q., Schwarz J., Bucana C., Olson E. N. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. P., Nakagawa O., Nakagawa M., Yanagisawa H., Passier R., Richardson J. A., Srivastava D., Olson E. N. CHAMP, a novel cardiac-specific helicase regulated by MEF2C. Dev. Biol. 2001;234:497–509. doi: 10.1006/dbio.2001.0277. [DOI] [PubMed] [Google Scholar]
- Ma Y. J., Dissen G. A., Rage F., Ojeda S. R. RNase protection assay. Methods. 1996;10:273–278. doi: 10.1006/meth.1996.0102. [DOI] [PubMed] [Google Scholar]
- Mahadev K., Motoshima H., Wu X., Ruddy J. M., Arnold R. S., Cheng G., Lambeth J. D., Goldstein B. J. The NAD(P)H oxidase homolog Nox4 modulates insulin-stimulated generation of H2O2 and plays an integral role in insulin signal transduction. Mol. Cell. Biol. 2004;24:1844–1854. doi: 10.1128/MCB.24.5.1844-1854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltsev V. A., Wobus A. M., Rohwedel J., Bader M., Hescheler J. Cardiomyocytes differentiated in vitro from embryonic stem cells developmentally express cardiac-specific genes and ionic currents. Circ. Res. 1994;75:233–244. doi: 10.1161/01.res.75.2.233. [DOI] [PubMed] [Google Scholar]
- Meyer N., Jaconi M., Landopoulou A., Fort P., Puceat M. A fluorescent reporter gene as a marker for ventricular specification in ES-derived cardiac cells. FEBS Lett. 2000;478:151–158. doi: 10.1016/s0014-5793(00)01839-1. [DOI] [PubMed] [Google Scholar]
- Morel Y., Barouki R. Repression of gene expression by oxidative stress. Biochem. J. 1999;342:481–496. [PMC free article] [PubMed] [Google Scholar]
- Moriguchi T., Toyoshima F., Gotoh Y., Iwamatsu A., Irie K., Mori E., Kuroyanagi N., Hagiwara M., Matsumoto K., Nishida E. Purification and identification of a major activator for p38 from osmotically shocked cells. Activation of mitogen-activated protein kinase kinase 6 by osmotic shock, tumor necrosis factor-alpha, and H2O2. J. Biol. Chem. 1996;271:26981–26988. doi: 10.1074/jbc.271.43.26981. [DOI] [PubMed] [Google Scholar]
- Puceat M., Travo P., Quinn M. T., Fort P. A dual role of the GTPase Rac in cardiac differentiation of stem cells. Mol. Biol. Cell. 2003;14:2781–2792. doi: 10.1091/mbc.E02-09-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raingeaud J., Gupta S., Rogers J. S., Dickens M., Han J., Ulevitch R. J., Davis R. J. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J. Biol. Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- Rhee S. G. Redox signaling: hydrogen peroxide as intracellular messenger. Exp. Mol. Med. 1999;31:53–59. doi: 10.1038/emm.1999.9. [DOI] [PubMed] [Google Scholar]
- Salmeen A., Andersen J. N., Myers M. P., Meng T. C., Hinks J. A., Tonks N. K., Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- Sauer H., Neukirchen W., Rahimi G., Grunheck F., Hescheler J., Wartenberg M. Involvement of reactive oxygen species in cardiotrophin-1-induced proliferation of cardiomyocytes differentiated from murine embryonic stem cells. Exp. Cell Res. 2004;294:313–324. doi: 10.1016/j.yexcr.2003.10.032. [DOI] [PubMed] [Google Scholar]
- Sauer H., Rahimi G., Hescheler J., Wartenberg M. Effects of electrical fields on cardiomyocyte differentiation of embryonic stem cells. J. Cell. Biochem. 1999;75:710–723. doi: 10.1002/(sici)1097-4644(19991215)75:4<710::aid-jcb16>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Sauer H., Rahimi G., Hescheler J., Wartenberg M. Role of reactive oxygen species and phosphatidylinositol 3-kinase in cardiomyocyte differentiation of embryonic stem cells. FEBS Lett. 2000;476:218–223. doi: 10.1016/s0014-5793(00)01747-6. [DOI] [PubMed] [Google Scholar]
- Seko Y., Takahashi N., Tobe K., Kadowaki T., Yazaki Y. Hypoxia and hypoxia/reoxygenation activate p65PAK, p38 mitogen-activated protein kinase (MAPK), and stress-activated protein kinase (SAPK) in cultured rat cardiac myocytes. Biochem. Biophys. Res. Commun. 1997;239:840–844. doi: 10.1006/bbrc.1997.7570. [DOI] [PubMed] [Google Scholar]
- Shiose A., Kuroda J., Tsuruya K., Hirai M., Hirakata H., Naito S., Hattori M., Sakaki Y., Sumimoto H. A novel superoxide-producing NAD(P)H oxidase in kidney. J. Biol. Chem. 2001;276:1417–1423. doi: 10.1074/jbc.M007597200. [DOI] [PubMed] [Google Scholar]
- Suter D. M., Cartier L., Bettiol E., Tirefort D., Jaconi M. E., Dubois-Dauphin M., Krause K. H. Rapid generation of stable transgenic embryonic stem cell lines using modular lentivectors. Stem Cells. 2005;24:615–623. doi: 10.1634/stemcells.2005-0226. [DOI] [PubMed] [Google Scholar]
- Tanaami T., Otsuki S., Tomosada N., Kosugi Y., Shimizu M., Ishida H. High-speed 1-frame/ms scanning confocal microscope with a microlens and Nipkow disks. Appl. Opt. 2002;41:4704–4708. doi: 10.1364/ao.41.004704. [DOI] [PubMed] [Google Scholar]
- Torres M., Forman H. J. Redox signaling and the MAP kinase pathways. Biofactors. 2003;17:287–296. doi: 10.1002/biof.5520170128. [DOI] [PubMed] [Google Scholar]
- Ushio-Fukai M., Alexander R. W., Akers M., Griendling K. K. p38 mitogen-activated protein kinase is a critical component of the redox-sensitive signaling pathways activated by angiotensin II. Role in vascular smooth muscle cell hypertrophy. J. Biol. Chem. 1998;273:15022–15029. doi: 10.1074/jbc.273.24.15022. [DOI] [PubMed] [Google Scholar]
- Vallet P., Charnay Y., Steger K., Ogier-Denis E., Kovari E., Herrmann F., Michel J. P., Szanto I. Neuronal expression of the NADPH oxidase NOX4, and its regulation in mouse experimental brain ischemia. Neuroscience. 2005;132:233–238. doi: 10.1016/j.neuroscience.2004.12.038. [DOI] [PubMed] [Google Scholar]
- Yang S. H., Galanis A., Sharrocks A. D. Targeting of p38 mitogen-activated protein kinases to MEF2 transcription factors. Mol. Cell. Biol. 1999;19:4028–4038. doi: 10.1128/mcb.19.6.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., New L., Kravchenko V. V., Kato Y., Gram H., di Padova F., Olson E. N., Ulevitch R. J., Han J. Regulation of the MEF2 family of transcription factors by p38. Mol. Cell. Biol. 1999;19:21–30. doi: 10.1128/mcb.19.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]