Abstract
Many mitochondrial proteins are encoded by nuclear genes and after translation in the cytoplasm are imported via translocases in the outer and inner membranes, the TOM and TIM complexes, respectively. Here, we report the characterization of the mitochondrial protein, Mmp37p (YGR046w) and demonstrate its involvement in the process of protein import into mitochondria. Haploid cells deleted of MMP37 are viable but display a temperature-sensitive growth phenotype and are inviable in the absence of mitochondrial DNA. Mmp37p is located in the mitochondrial matrix where it is peripherally associated with the inner membrane. We show that Mmp37p has a role in the translocation of proteins across the mitochondrial inner membrane via the TIM23-PAM complex and further demonstrate that substrates containing a tightly folded domain in close proximity to their mitochondrial targeting sequences display a particular dependency on Mmp37p for mitochondrial import. Prior unfolding of the preprotein, or extension of the region between the targeting signal and the tightly folded domain, relieves their dependency for Mmp37p. Furthermore, evidence is presented to show that Mmp37 may affect the assembly state of the TIM23 complex. On the basis of these findings, we hypothesize that the presence of Mmp37p enhances the early stages of the TIM23 matrix import pathway to ensure engagement of incoming preproteins with the mtHsp70p/PAM complex, a step that is necessary to drive the unfolding and complete translocation of the preprotein into the matrix.
INTRODUCTION
Mitochondria are essential organelles required for respiration, lipid metabolism, apoptosis, heme metabolism, synthesis of metabolites, free radical processes, metal ion homeostasis, and calcium signaling. Although mitochondria harbor a small genome as well as translational machinery, mitochondrial processes are dependent on hundreds of nuclear-encoded proteins. Consequently, the vast majority of mitochondrial proteins are synthesized on cytosolic polysomes and are posttranslationally imported into mitochondria to one of the four submitochondrial compartments: the outer membrane, the inner membrane, the intermembrane space (IMS), and the matrix. Nuclear encoded proteins destined for the mitochondria contain specific targeting and sorting information to reach the correct mitochondrial subcompartment. In general, proteins targeted to the mitochondrial matrix and many of those directed to the inner membrane include N-terminal cleavable presequences, termed mitochondrial targeting signals (MTS), which have the necessary information to target proteins to the mitochondria (Neupert, 1997; Koehler, 2004; Rehling et al., 2004; Perry and Lithgow, 2005).
All precursor proteins appear to use the general translocase of the outer membrane, the TOM complex, which consists of receptor proteins that recognize and bind the precursor proteins as well as Tom40p, which forms the channel through which mitochondrial proteins are translocated (Neupert, 1997; Pfanner et al., 1997). Passage into and across the inner mitochondrial membrane requires a membrane potential (ΔΨ) across the inner membrane and is facilitated by one of two translocases: the TIM22 complex or the TIM23 complex (Pfanner et al., 1997; Koehler et al., 1999; Truscott et al., 2003; Koehler, 2004). The TIM22 complex mediates the membrane insertion of many inner membrane proteins synthesized without N-terminal cleavable MTSs, such as members of the metabolite carrier family (Koehler et al., 1998; Sirrenberg et al., 1998; Leuenberger et al., 1999; Curran et al., 2002a, 2002b). The TIM23 complex is termed the presequence translocase because it facilitates the transport of precursor proteins bearing N-terminal cleavable presequences into both the inner membrane and the matrix (Neupert, 1997; Pfanner et al., 1997). A precursor protein destined for the mitochondrial matrix can simultaneously span both the outer and inner membranes, as the preprotein passes through the TOM and TIM23 complexes (Schleyer and Neupert, 1985; Rassow et al., 1990; Chacinska et al., 2003). The precursor proteins traverse the import machineries in an extended conformation, such that 50–55 amino acids of the translocation intermediate is required to span both membranes (Rassow et al., 1990; Ungermann et al., 1994).
Unfolding and complete translocation of proteins across the TIM23 complex into the matrix requires the action of the matrix-localized, ATP-dependent, molecular chaperone mtHsp70p/Ssc1p (Kang et al., 1990; Gaume et al., 1998). The presequence assisted motor (the PAM complex), composed of Pam16p/Tim16p, Pam17p, and Pam18p/Tim14p, along with Tim44p and mtHsp70p, act to coordinate the translocation of a preprotein across the TIM23 translocon with the activity of mtHsp70p (D’Silva et al., 2003; Mokranjac et al., 2003b; Truscott et al., 2003; van der Laan et al., 2005). The association of the PAM complex directly at the exit site of the TIM23 channel serves to ensure that the incoming precursor protein engages with and is “trapped” by mtHsp70p, as it emerges from the translocon into the matrix (Gaume et al., 1998; Chacinska et al., 2005). Failure of mtHsp70p to capture the precursor protein may result in the translocation intermediate falling back out of the import channel (Ungermann et al., 1994). After trapping of the preprotein in the import channel in the PAM-assisted manner, subsequent mtHsp70p binding and ATP hydrolysis events are required to drive the unfolding and complete translocation of the precursor protein through the TIM23 channel into the matrix (Gaume et al., 1998; Huang et al., 2000).
In this report we present evidence supporting the identification of a novel protein, Mmp37p (mitochondrial matrix protein of 37 kDa), which is involved in the import of proteins into the matrix via the TIM23 complex. We demonstrate that the Saccharomyces cerevisiae open reading frame (ORF) YGR046w encoding Mmp37p is nonessential for yeast cell viability at 30°C. The presence of Mmp37p is essential, however, for growth at 37°C or in the absence of the mtDNA. Furthermore, shifting mmp37Δ cells to the nonpermissive temperature resulted in the accumulation of uncleaved mitochondrial precursor proteins, suggesting that mmp37Δ cells may be defective in the import of mitochondrial proteins. Using in vitro import assays, we demonstrate that mitochondria isolated from mmp37Δ cells display a reduced mitochondrial import capacity. Specifically, our results indicate that Mmp37p functions in the early stages of import into the matrix through the TIM23-PAM machinery. In particular, we demonstrate here that the import of precursor proteins harboring tightly folded domains close to the N-terminal targeting signals require the presence of Mmp37p to ensure import into the mitochondrial matrix. Furthermore, we report that in the absence of Mmp37p, an alteration in the assembly status of the TIM23 complex is observed. On the basis of our findings, we propose that Mmp37 functions to ensure the integrity of the TIM23 complex such that precursors emerging through the TOM and TIM23 channels can efficiently engage with mtHsp70p to drive their subsequent unfolding and import into the mitochondria.
MATERIALS AND METHODS
Yeast Strains, Media, and Plasmids
Yeast strains used in this study are listed in Table 1. Mating of yeast strains, sporulation and tetrad analysis were performed as previously described (Sherman et al., 1986). The plasmids used in this study are listed in Table 2 and were constructed using standard molecular techniques. As previously described, gene deletions were created in yeast using PCR products generated from plasmid pUG6, whereas the endogenous MMP37 promoter was replaced with the GAL1 promoter using a PCR product generated from plasmid pFA6-kanMX6-GAL1 (Guldener et al., 1996). Yeast cells were either grown in rich (YEP) medium or in defined synthetic medium containing the indicated carbon sources (Rose et al., 1990). Plasmids and PCR products were transformed into yeast cells using lithium acetate (Ito et al., 1983).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| YPH102 | Matα, ura3-52, leu2Δ1, his3Δ-200, ade2-101, lys2-801 | Spencer et al. (1990) |
| Sc252 | Mata, ura3-52, leu2-3,112, ade1, ile | Johnston and Hopper (1982) |
| YRLRS9a | Mata/α, ura3-52/ura3-52, leu2Δ1/leu2-3,112, his3Δ-200/HIS3, ADE1/ade1, ade2-101/ADE2, lys2-801/LYS2, ILE/ile | This work |
| K699b | Mata, ura3, leu2-3,112, his3-11, trp1-1, ade2-1, ho, can1-100 | Jansen et al. (1996) |
| YLM2814b | Mata, ura3, leu2-3,112, his3-11, trp1-1, ade2-1, ho, can1-100, mmp37Δ::KAN | This work |
| YLM2827 | Mata/α, ura3-52/ura3-52, leu2Δ1/leu2-3,112, his3Δ-200/HIS3, ADE1/ade1, ade2-101/ADE2, lys2-801/LYS2, ILE/ile, MMP37/mmp37Δ::KAN | This work |
| YLM3166b | Mata, ura3, leu2-3,112, his3-11, trp1-1, ade2-1, ho, can1-100,GAL1-MMP37::KAN | This work |
a Diploid resulting from YPH102 × Sc252.
b W303-1a genetic background.
Table 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| YCplac22 | A yeast single-copy plasmid containing the TRP1 marker | Gietz and Sugino (1988) |
| YCplac33 | A yeast single-copy plasmid containing the URA3 marker | Gietz and Sugino (1988) |
| YCplac111 | A yeast single-copy plasmid containing the LEU2 marker | Gietz and Sugino (1988) |
| pET15b | A bacterial plasmid for expressing His6 fusion protein | Novagen |
| pFA6-kanMX6-GAL1 | A plasmid for generating PCR cassettes to replace endogenous promoters with the GAL1 promoter | Longtine et al. (1998) |
| pUG6 | A plasmid for generating KAN disruption cassettes by PCR | Guldener et al. (1996) |
| pRL767 | YCplac111 containing MMP37 | This work |
| pRL773 | YCplac111 containing myc6-MMP37 | This work |
| pRL778 | YCplac33 containing MMP37 | This work |
| pRL848 | pET15b containing MMP37 | This work |
Generation of Polyclonal Mmp37p Antiserum
Polyclonal rabbit antiserum was generated against His6-Mmp37p expressed and purified from Escherichia coli. The ORF of MMP37 was cloned into pET15b (Novagen, La Jolla, CA), resulting in a His6 tag at the N-terminus of Mmp37p. This plasmid, pRL848, was subsequently transformed into E. coli strain BL21(DE3)pLysS, and expression of His6-Mmp37p was induced with 1 mM isopropyl-beta-d-thiogalactopyranoside (IPTG). After induction cells were harvested by centrifugation, lysed in presence of NP-40 and His6-Mmp37p purified using Ni-NTA agarose (Qiagen, Valencia, CA) after treatment of inclusion bodies with 6 M guanidine-HCl according to the manufacturer’s instructions. Purified His6-Mmp37p was analyzed by SDS PAGE, and the band corresponding to the His6-Mmp37p protein was excised from the SDS-PAGE gel, combined with Freund’s adjuvant, and injected into a female New Zealand White rabbit.
Isolation and Analysis of Mitochondria
For in vitro protein import analysis and Mmp37p localization analysis, mitochondria were isolated from wild-type and mmp37Δ yeast strains, which had been grown in YEP media containing 2% galactose/0.5% lactate, as described previously (Herrmann et al., 1994). For sublocalization studies, mitochondria were treated with proteinase K, under isoosmotic or hypotonic swelling conditions (mitoplasts) or under Triton X-100 lysis conditions or were treated with 0.1 M sodium carbonate (pH 10.5–12), essentially as previously described (Arnold et al., 1998). For the sonication experiments, wild-type mitochondria (200 μg protein) were preswollen in 60 mM sorbitol, 20 mM HEPES, pH 7.2, reisolated by centrifugation and resuspended in 1 ml of ice-cold 0.6 M sorbitol, 5 mM MgCl2, 20 mM HEPES, pH 7.2. Sonication was performed on ice using a Branson 450 Sonifier, using a microtip with 10 × 10-s pulses (at 100% duty), each with 20-s cooling intervals. After a low speed spin (21,000 × g, 10 min) to remove any nondisrupted mitochondria/mitoplasts, the mitochondrial vesicles were pelleted by centrifugation (100,000 × g, 60 min), and soluble proteins were recovered from the supernatant by TCA precipitation. Steady state levels of mitochondrial proteins were analyzed by Western blotting after separation of mitochondrial proteins by SDS-PAGE.
Mitochondrial membrane complexes were analyzed as follows. Mitochondria (200 μg protein) were solubilized with 1% (wt/vol) digitonin and clarified by centrifugation (30,000 × g, 30 min), and the extract analyzed by blue native gel electrophoresis (BN-PAGE), using 6–16% gels, as described previously (Arnold et al., 1998), followed by a second-dimension resolution by SDS-PAGE, before Western blotting. When indicated, the solubilized protein extracts were supplemented with a further aliquot of digitonin (final concentration 2%), just before BN-PAGE analysis.
Import of Precursor Proteins into Isolated Mitochondria
In vitro mitochondrial protein import was performed essentially as previously described (Stuart et al., 1994). Precursor proteins were synthesized in the presence of [35S]methionine by coupled transcription-translation in a reticulocyte lysate system (Promega, Madison, WI). Before the addition of the lysate containing the radiolabeled precursor protein, mitochondria were preincubated for 10 min at 37°C in import buffer containing 2 mM NADH and an ATP-regenerating system (10 mM ATP, 10 mM MgCl2, 10 mM creatine phosphate, and 100 μg/ml creatine kinase). The preincubation period was included to ensure high ATP levels in the mitochondria; the temperature of 37°C (as opposed to 25°C, did not seem to affect the efficiency of import in the mmp37Δ or wild-type mitochondria (unpublished data). After the preincubation, samples were returned to ice and import was started by addition of the lysate containing the radiolabeled precursor protein together with an additional aliquot of NADH (final concentration 4 mM). Import reactions were performed at 25°C, and aliquots were removed at the indicated time points. Samples were divided and were treated either with proteinase K (50 μg/ml) or were mock-treated for 20 min on ice. After the addition of 2 mM phenylmethylsulfonyl fluoride, mitochondria were reisolated by centrifugation, lysed in SDS-sample buffer, separated by SDS-PAGE, and transferred to nitrocellulose membrane. The levels of radiolabeled protein imported into the mitochondria were quantified using a Storm PhosphoImager analysis system (GE Healthcare Life Sciences, Piscataway, NJ).
RESULTS
YGR046w Is a S. cerevisiae Nonessential ORF
The ORF corresponding to YGR046W was reported to encode an essential mitochondrial protein, and we sought to determine the function for this protein (Entian et al., 1999; Ghaemmaghami et al., 2003). We have given the YGR046w ORF the designation Mmp37p (Mitochondrial matrix protein). As will be described below Mmp37 is located in the mitochondrial matrix and displays an apparent molecular mass of 37 kDa by SDS-PAGE analysis. To gain insight into the function of Mmp37p, we initially attempted to identify temperature sensitive alleles of Mmp37p. While we were constructing the yeast strain necessary to identify such alleles, we encountered difficulties that suggested that MMP37 might be nonessential in a haploid genetic background.
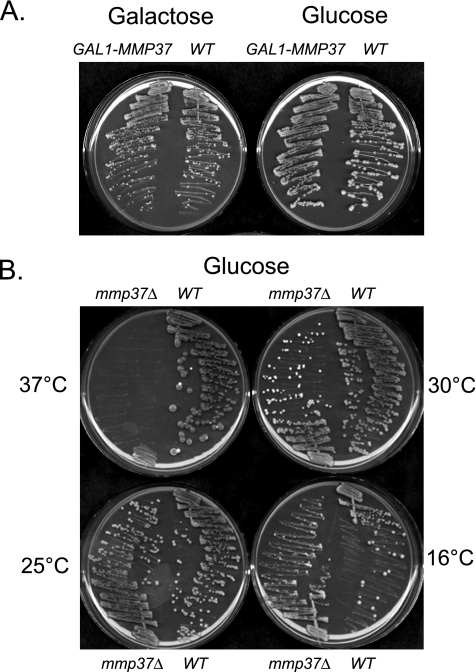
To investigate if expression of Mmp37p is required for viability in yeast, expression of Mmp37p was placed under the control of the GAL1 promoter. If Mmp37p is essential for growth we anticipated that the GAL1-MMP37 strain should grow on galactose containing medium and not grow on glucose containing medium. However, we observed that the GAL1-MMP37 strain was able to grow on glucose containing media (Figure 1A), suggesting that MMP37 is not required for viability.
Figure 1.
Mmp37p is nonessential for viability of haploid S. cerevisiae. (A) Strain K699 (WT) and YLM3166 (GAL1-MMP37) were streaked for single colonies on YEP medium containing either 2% galactose or 2% glucose, and the plates were incubated at 30°C. (B) Single colonies corresponding to yeast strains K699 and YLM2814 (mmp37Δ::kanMX) were picked from solid YEP medium containing 2% glucose propagated at 30°C. Subsequently, these yeast strains were streaked on identical medium, and the plates were incubated at 16, 25, 30, and 37°C.
Because we could not eliminate the possibility that a small amount of Mmp37p could be expressed from the GAL1-MMP37 construct in the presence of glucose, we decided to investigate if MMP37 is essential for growth in the W303-1A haploid genetic background. To delete MMP37 from the genome a mmp37Δ::kanMX deletion cassette was generated that lacked the entire MMP37 ORF. Yeast transformants containing the integrated mmp37Δ::kanMX cassette were selected on YEP 2% glucose medium containing G418, and transformants in which the mmp37Δ::kanMX cassette had replaced the wild-type allele of MMP37 were identified through two independent PCR products. From this standard deletion approach we were able to generate a viable haploid mmp37Δ::kanMX (mmp37Δ) deletion strain (Figure 1B). The ability to generate a mmp37Δ strain was not specifically related to the W303-1A genetic background, because we were also able to construct a mmp37Δ strain in the nonrelated Sc252 genetic background (unpublished data).
Given that it was previously reported that certain mitochondrial proteins (Tim18p, Zim17p, and Yme1p) are essential for growth only at elevated temperatures, we decided to investigate whether Mmp37p was essential for growth at various temperatures (Thorsness and Fox, 1993; Kerscher et al., 2000; Sanjuan Szklarz et al., 2005). The parental wild-type strain and the mmp37Δ strain were streaked on YEP 2% glucose medium, and the plates were incubated at 16, 25, 30, and 37°C (Figure 1B). We observed that the wild-type strain was able to form colonies at all temperatures (Figure 1B). In contrast, the mmp37Δ strain was able to grow only at 16, 25, and 30°C, with a partial growth defect at 16°C (Figure 1B). These results indicate that Mmp37p is essential for growth at 37°C.
We then investigated if the mmp37Δ strain was viable in the absence of mtDNA (Figure 2). Wild-type and mmp37Δ cells were streaked for single colonies on YEP 2% glucose medium in the absence and presence of 25 mg/ml ethidium bromide (EtBr), which leads to the loss of mtDNA. The cells were incubated at 30°C for 3 d, and after the incubation period we observed that the wild-type strain was able to form colonies in the presence and absence of EtBr, whereas the mmp37Δ strain only formed colonies in the absence of EtBr (Figure 2, cf. plate A to plate B). To demonstrate that wild-type cells grown in the presence of EtBr had lost mtDNA, colonies that grew on plates containing EtBr were picked and restreaked on YEP 2% glucose (plate A1) and YEP 3% glycerol (plate A2) media, followed by incubation at 30°C for 7 d. As a control wild-type colonies grown in the absence of EtBr were picked and restreaked on YEP 2% glucose (plate B1) and YEP 3% glycerol (plate B2) media. We observed that wild-type cells exposed to EtBr were viable on fermentable medium (YEP 2% glucose) but were inviable on nonfermentable medium (YEP 3% glycerol), indicating that exposure of the wild-type cells to EtBr leads to the loss of mtDNA. Consequently, these results suggest that Mmp37p is required for viability in the absence of mtDNA.
Figure 2.
The haploid mmp37Δ strain is inviable in the absence of mitochondrial DNA. Strains K699 (WT) and YLM2814 (mmp37Δ:: kanMX) were streaked for single colonies on YEP 2% glucose medium in the presence (A) or absence (B) of 25 μg/ml ethidium bromide (ETBr), and the plates were incubated at 30°C. Colonies that grew on plate A were picked and restreaked on YEP 2% glucose medium (A1) and YEP 3% glycerol medium (A2). Colonies that grew on plate B were picked and restreaked on YEP 2% glucose medium (B1) and YEP 3% glycerol medium (B2).
MMP37 Is Required for Viability after Meiosis
Given that our results concerning whether MMP37 is essential or nonessential were contradictory to previous reports (Giaever et al., 2002), we decided to investigate whether MMP37 is essential for growth after meiosis. We constructed a diploid strain that was heterozygous at the MMP37 locus. One chromosome contained the wild-type MMP37 allele, whereas the homologous chromosome contained a deletion of MMP37. The MMP37 heterozygous diploid strain was sporulated, tetrads were dissected, and meiotic progeny allowed to germinate on YEP 2% glucose medium. After germination of the meiotic progeny, only two of the four spores grew into distinct colonies at 30°C (Figure 3A). On further analysis, the viable spores were determined to be sensitive to G418 (unpublished data), demonstrating that each of these spores harbored the wild-type MMP37 allele. We further examined the mmp37Δ::kanMX-containing spores microscopically. We observed that these spores were able to germinate and form microscopic colonies, but these microscopic colonies never produced macroscopic colonies even after prolonged incubation. From these observations we conclude that MMP37 is essential for viability after meiosis.
Figure 3.
Mmp37p is required for viability of meiotic progeny. (A) Diploid yeast strain YLM2827 heterozygous at the MMP37 locus (MMP37/mmp37Δ::kanMX) was sporulated and meiotic progeny analyzed after tetrad dissection. The spores were allowed to germinate and grow on YEP 2% glucose containing medium at 30°C. Lanes labeled 1–4 show the growth phenotypes for four different asci, and the individual meiotic progeny are labeled A–D. (B) Diploid yeast strain YLM2827 was transformed with plasmid pRL778 (YCplac33/MMP37). A transformant was sporulated and meiotic progeny analyzed after tetrad dissection. The spores were allowed to germinate and grow on YEP 2% glucose medium at 30°C. Lanes labeled 1–4 show the growth phenotypes for four different asci, and the individual meiotic progeny are labeled A–D. All four spores in each tetrad were confirmed to contain pRL778 by complementation of the ura3 phenotype.
We sought to conclusively demonstrate that the inability of mmp37Δ::kanMX spores to form visible colonies was due to the absence of MMP37 and not a defect in a closely linked gene. Consequently, the MMP37/mmp37Δ::kanMX heterozygous diploid strain was transformed with a single-copy plasmid containing URA3 and MMP37. A diploid transformant was sporulated, and tetrads were dissected. After germination of the spores, we observed four visible colonies (Figure 3B). The G418-resistant colonies always contained the URA3 marker, indicating that the mmp37Δ::kanMX deletion was being complemented by the plasmid borne MMP37. The sum of this data indicates that MMP37 is essential for viability after meiosis.
Mmp37p Is Peripherally Attached to the Surface of the Inner Membrane Exposed to the Matrix
Genome-wide localization and proteomic studies have suggested that Mmp37p is a mitochondrial protein (Entian et al., 1999; Ghaemmaghami et al., 2003), and we confirmed these results by immunofluorescence colocalization experiments using both N- and C-terminal myc-tagged derivatives of Mmp37p (unpublished data). To further define the submitochondrial location of Mmp37p, mitochondria and hypotonically swollen mitochondria, termed mitoplasts, were prepared and treated with exogenously added proteinase K (Figure 4A). The proteinase K accessibility profile for Mmp37p was analogous to the profile for the matrix localized control protein, Mge1p, because both proteins were inaccessible to proteinase K in both mitochondria and mitoplast preparations. Efficient hypotonic swelling of mitochondria and rupturing of the outer membrane to generate mitoplasts was confirmed by the loss of the soluble intermembrane space protein, cytochrome c peroxidase (Ccpo), upon reisolation of the mitoplast membranes. The outer membrane protein, Tom70p, and inner membrane proteins, ADP/ATP carrier (AAC) and d-lactate dehydrogenase (d-LD) served as controls and as expected were accessible to the protease after treatment of intact mitochondria and mitoplasts, respectively. When the outer and inner mitochondrial membranes were disrupted by the addition of detergent Triton X-100, Mmp37p and Mge1p were degraded by exogenously added proteinase K (Figure 4A). Note that the soluble intermembrane space-localized control protein Ccpo is a tightly folded protein, so although solubilized by the addition of detergent, it is resistant to proteinase K treatment even when performed in the presence of Triton X-100 (Figure 4A). Taken together these data confirm the mitochondrial localization of Mmp37p and indicate that Mmp37p is localized in the mitochondrial matrix.
Figure 4.
Mmp37p is a matrix-localized protein. (A) Proteinase K (Prot. K) treatment was performed on intact mitochondria (Mitoch.) or mitochondria which had been subjected to hypotonic swelling (Swelling) or Triton X-100 detergent lysis (T X-100) on ice, as indicated. Samples were subjected to SDS-PAGE and Western blotting. (B) Isolated wild-type mitochondria (0.1 mg/ml) were extracted by alkaline treatment using 0.1 M NaCO3 buffer at the indicated pH. Control mitochondria (pH 7.5) were resuspended in 0.6 M sorbitol, 20 mM HEPES-KOH, pH 7.5, buffer. Samples were sedimented by centrifugation and the membrane pellet and supernatant (after TCA precipitation) were subjected to SDS-PAGE and Western blotting. Note, the Ccpo protein displays an inherent protease stability and hence is not degraded by the proteinase K treatment in the presence of Triton X-100. (C) Wild-type mitochondria were subjected to a sonication treatment as described in Materials and Methods. T, total input; P, membrane vesicle pellet; S, soluble matrix proteins.
To ascertain if Mmp37p represents an integral inner membrane protein exposed to the matrix, alkaline extraction of mitochondria was performed across a 7.5–12 pH range (Figure 4B). When the extraction was performed at pH 10.5, Mmp37p along with the control integral inner membrane proteins Tim23p and Su e of the ATP synthase were retained in the membrane fraction, whereas known soluble control proteins, cyclophilin (Cpr3p) and Mge1p (unpublished data), were released into the supernatant. In contrast to the integral inner membrane control proteins, however, a significant fraction of Mmp37p, like Tim44p, a known peripherally bound inner membrane control protein, was released from the membranes to the supernatant when the alkaline extractions were performed at pH values of 11.0–11.5 (Figure 4B). This result indicates that Mmp37p, unlike Tim23p and Su e, is not an integral inner membrane protein. The peripheral association of Mmp37p with the membrane appeared to be somewhat tighter than Tim44p, however, because ∼50% of Tim44p was extracted from the membranes at pH 10.5, whereas efficient extraction of Mmp37p was not observed until pH 11.0 or higher (Figure 4B). Finally, when mitochondrial membranes were disrupted by sonication, Mmp37p cofractionated with the membrane proteins, e.g., Tim23p, as indicated by its recovery with the membrane vesicle pellet fraction, whereas the soluble matrix proteins, such as Mge1p, were recovered in the supernatant fraction (Figure 4C).
The sum of these results indicates that Mmp37p is located in the mitochondrial matrix, where it is associated with the inner membrane. Mmp37p appears not to be an integral inner membrane protein, an observation that is consistent with the lack of predicted transmembrane segments from inspection of the Mmp37p amino acid sequence. Rather, Mmp37p appears to be tightly associated with the inner membrane in a peripheral manner, possibly through hydrophobic- and hydrophilic-based interactions with neighboring inner membrane proteins.
The Precursors for Mdj1p and Hsp60p Accumulate in the Absence of Mmp37p
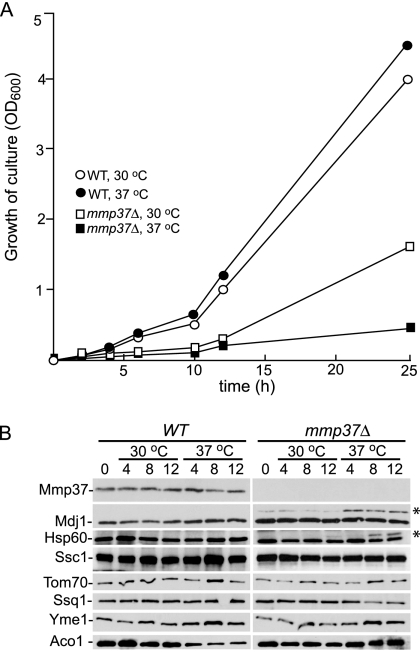
After establishing that Mmp37p is a mitochondrial matrix protein, we sought to determine the mitochondrial function for Mmp37p. To gain insight into the role of Mmp37p, we took advantage of our observation that the mmp37Δ haploid strain grows at 30°C but is inviable at 37°C. Initially we sought to characterize the temporal growth arrest of the mmp37Δ strain after a shift from 30 to 37°C. The parental wild-type and mmp37Δ strains were grown at 30°C until the cells reached midlog phase. Subsequently, the cultures were shifted to 37°C, and the growth of the cells was monitored at the indicated time points (Figure 5A). The mmp37Δ strain exhibited a slow growth phenotype at 30°C, and 12 h after the shift to the nonpermissive temperature, an almost complete cessation in growth was observed for the mmp37Δ strain when compared with the corresponding parental wild-type strain and the mmp37Δ strain grown at 30°C. These results reinforce that the function of Mmp37p is essential after the shift to the restrictive temperature of 37°C.
Figure 5.
The mitochondrial precursors for Mdj1p and Hsp60p accumulate upon shifting mmp37Δ cells to the nonpermissive temperature of 37°C. (A) Strains K699 (WT) and YLM2814 (mmp37Δ) were grown in YEP 2% galactose medium at 30°C until the cells reached midlog phase. Subsequently at time point zero, the cells for each strain were diluted to an OD600 of 0.125 in YEP 2% galactose medium, and each strain was divided into two cultures. One culture was maintained at 30°C, whereas the second culture was shifted to 37°C, and the OD600 for each culture was determined at 2, 4, 6, 8, 10, 12, and 25 h. (B) At time points 0, 4, 8, and 12 h an aliquot of cells was removed from each culture, and mitochondrial lysates were prepared. Ten micrograms of each mitochondrial lysate was separated by SDS-PAGE and analyzed by Western blotting with antisera specific to each of the indicated proteins. The asterisk (*) indicate the position of the Mdj1p and Hsp60p precursors.
After establishing the temporal phenotype for the mmp37Δ strain at the nonpermissive temperature, we were poised to further investigate the function of Mmp37p. We initially investigated if the mitochondrial protein profile was altered upon shift of the mmp37Δ strain to the nonpermissive temperature. The parental wild-type and the mmp37Δ strains were grown at 30°C, and when the cells reached midlog phase, the cultures were divided into two aliquots. One aliquot was maintained at 30°C, whereas the second aliquot was shifted to 37°C. Subsequently, samples were harvested for mitochondrial isolations at the indicated time points, and the protein profile for various mitochondrial proteins was compared by Western blotting (Figure 5B). The steady state levels for the mitochondrial proteins Yme1p, Ssq1p, and aconitase were not greatly perturbed by the absence of Mmp37p or by the shift of the mmp37Δ cells to the restrictive temperature of 37°C. However, we detected the accumulation of the precursor forms for the matrix targeted proteins Mdj1p and Hsp60p in mmp37Δ cells, especially after the shift to the nonpermissive temperature (Figure 5B). The accumulation of Mdj1p and Hsp60p precursors at the nonpermissive temperature implied that Mmp37p may play a role in mitochondrial import, because accumulation of Mdj1p and Hsp60p precursors was previously shown to occur in mutants defective in the mitochondrial protein import pathway (Mokranjac et al., 2003a; Ishikawa et al., 2004; Kozany et al., 2004).
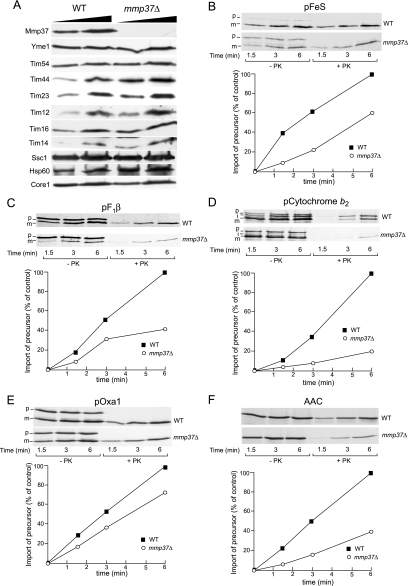
The absence of Mmp37p leads to defects in mitochondrial import. To analyze if Mmp37p has a role in protein import, mitochondria were isolated from wild-type and mmp37Δ strains grown at the permissive temperature of 30°C. The steady state levels for components known to be involved in protein import were compared between wild-type mitochondria and mmp37Δ mitochondria (Figure 6A). The levels of Tim23p, Hsp60p, and Tim44p were slightly elevated in the mmp37Δ mitochondria, whereas the levels of Ssc1p/mtHsp70p, Pam16p/Tim16p, and Pam18p/Tim14p appeared unaffected in mmp37Δ mitochondria. The steady state levels of Tim54p and Tim12p, components involved in the TIM22 import as well as the loading controls, Core1p (a subunit of the cytochrome bc1 complex), and Yme1p (an inner membrane protease) were unaffected in the mmp37Δ mitochondria (Figure 6A). These results indicate that any potential mmp37 defects in mitochondrial import are unlikely to be caused by a decrease in the abundance of the TIM22 and TIM23 transport components.
Figure 6.
mmp37Δ mitochondria are defective for protein import. (A) Mitochondria (25 and 50 μg) isolated from wild-type and mmp37Δ mutant strains grown at 30°C were analyzed by SDS-PAGE and Western blotting. (B–F) Mitochondria isolated from wild-type and mmp37Δ strains were preincubated with an ATP-regenerating system at 37°C for 10 min, after which radiolabeled precursor proteins were added (as indicated). Samples were further incubated at 25°C for the indicated times. After proteinase K (+PK) or mock (−PK) treatment, mitochondrial were reisolated by centrifugation, and samples were subjected to SDS-PAGE and autoradiography. Levels of radioactive precursor proteins imported into mitochondria were quantified, and the amount of preprotein imported into the wild-type mitochondria after 6 min of import was set to 100%.
We next investigated if Mmp37p is required for the in vitro import of proteins into mitochondria using a variety of [35S]methionine labeled mitochondrial precursor proteins with mitochondria isolated from wild-type and mmp37Δ strains grown at the permissive temperature. The requirement for Mmp37p in the import of mitochondrial precursors to the matrix through the TIM23 pathway was analyzed by assaying the import kinetics for pFeS (precursor of Rieske FeS protein of cytochrome bc1 complex), pF1β (precursor of subunit β of the F1-ATPase), precytochrome b2, and pOxa1 (precursor of the inner membrane Oxa1 protein; Figure 6, B–E). Relative to the import into wild-type mitochondria, we observed from mmp37Δ mitochondria that the import of the pFeS (Figure 6B) and pF1β (Figure 6C) precursor proteins was reduced ∼40 and 60%, respectively. Furthermore, the import of precytochrome b2 was reduced 80% in mmp37Δ mitochondria (Figure 6D), whereas the import of pOxa1 was only slightly perturbed in the absence of Mmp37p (Figure 6E). These results suggest that the presence of Mmp37p enhances the import of these proteins into the mitochondria via the TIM23 complex. We also investigated the import of a protein, the ADP/ATP carrier protein (AAC) that depends on the TIM22 translocase. In the absence of Mmp37p, we observed a 60% reduction in the import of AAC (Figure 6F), suggesting that the function of the TIM22 translocase may also be compromised in the absence of Mmp37p.
Given that cytochrome b2 contains a tightly folded heme-binding domain immediately adjacent to the N-terminal presequence (Glick et al., 1993; Stuart et al., 1994), the increased inhibition of cytochrome b2 import into mmp37Δ mitochondria (Figure 6D), relative to the other TIM22 and TIM23 precursors tested, suggested that the import of a precursor protein with a tightly folded domain, may be particularly affected in the absence of Mmp37p. Consistent with this hypothesis, the import of a cytochrome b2 derivative in which the heme-binding domain had been disrupted, pb2(1-167)DHFR, was reduced by only 25% in mmp37Δ mitochondria (unpublished data).
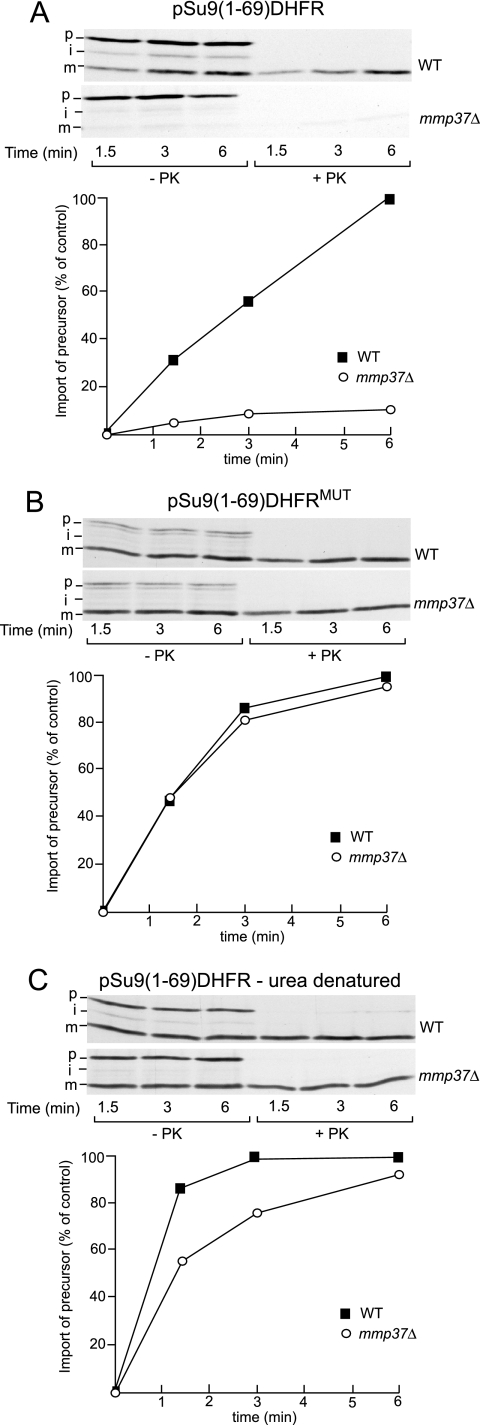
The Requirement for Mmp37p in Mitochondrial Import Is Relieved if the Preprotein Is Unfolded before Import
To directly test the hypothesis that the import of a precursor with tightly folded domain is particularly affected by the absence of Mmp37p, we tested the import of TIM23-dependent precursor proteins that contain a tightly folded mouse dihydrofolate reductase (DHFR) moiety (Figure 7). Import of a DHFR-containing precursor protein into the matrix requires unfolding of the DHFR domain at the mitochondrial surface in a manner dependent on the ability of the N-terminal targeting sequence of the precursor protein to engage with mtHsp70p in the matrix. We initially analyzed the import of the pSu9(1-69)DHFR precursor, which is a well-characterized TIM23/mtHsp70p-dependent import substrate and represents a fusion protein between the mitochondrial targeting signal (residues 1–66) of subunit 9 (Su9) of the Fo-ATP synthase (from Neurospora crassa), plus three amino acids of the mature Su9 protein, followed by the DHFR moiety. Consistent with our hypothesis that tightly folded precursors require Mmp37p for import into the matrix, we observed a nearly complete inhibition of pSu9(1-69)DHFR import into mmp37Δ mitochondria, when compared with wild-type mitochondria (Figure 7A). To confirm that in the absence of Mmp37p the import inhibition of pSu9(1-69)DHFR was due to the presence of a tightly folded DHFR moiety in close proximity to the N-terminal targeting sequence of Su9, we tested the effect of unfolding the DHFR moiety of the pSu9(1-69)DHFR chimera before the import reaction. To do so, we adopted two independent approaches. First, we analyzed the import of a pSu9(1-69)DHFRMUT derivative, which contains a folding incompetent DHFR moiety (Vestweber and Schatz, 1988; Teichmann et al., 1996; Gaume et al., 1998). Import of pSu9(1-69)DHFRMUT into the mmp37Δ mitochondria occurred with very high efficiency (Figure 7B) compared with the wild-type pSu9(1-69)DHFR construct (Figure 7A). When compared with wild-type mitochondria, import of pSu9(1-69)DHFRMUT into mmp37Δ mitochondria was only reduced by 10% (Figure 7B). Second, we analyzed the import of the pSu9(1-69)DHFR preprotein which was chemically denatured in 8 M urea before the mitochondrial import assay. The urea denatured pSu9(1-69)DHFR protein was rapidly and efficiently imported into mmp37Δ mitochondria in a manner nearly identical to wild-type mitochondria (Figure 7C). Therefore, we conclude that the drastic import inhibition observed for the pSu9(1-69)DHFR preprotein into the mmp37Δ mitochondria, can be circumvented if the DHFR moiety of the precursor is unfolded before import.
Figure 7.
Import inhibition observed in mmp37Δ mitochondria can be overcome by unfolding the preprotein before import. (A–C) The import of radiolabeled pSu9(1-69)DHFR preproteins into wild-type and mmp37Δ mitochondria was performed as described in Figure 6. (A) Import of pSu9(1-69)DHFR bearing a wild-type folded DHFR moiety. (B) Import of a pSu9(1-69)DHFRMUT, a derivative bearing an folding incompetent DHFR moiety. (C) Import of pSu9(1-69)DHFR, which had been chemically denatured in 8 M urea before dilution into the import assay. After import, samples were further processed as described above in Figure 6.
Extension of the Preprotein before the Folded DHFR Domain Relieves the Dependency on Mmp37p for Import
Because ∼52 amino acid residues are required to span the outer and inner membrane at translocation contact sites, we reasoned that only 15–17 amino acids of pSu9(1-69)DHFR would be exposed to the matrix, if the DHFR moiety remained folded at the surface of the outer membrane (Rassow et al., 1990; Ungermann et al., 1994). We therefore investigated the possibility that Mmp37p is required for engaging mtHsp70p with precursor proteins that expose only short N-terminal segments to the matrix, due to the proximity of the restrictive folded domain outside. To address this possibility the import of two longer Su9-derived precursor proteins, pSu9(1-79)DHFR and pSu9(1-112)DHFR into isolated mmp37Δ mitochondria was analyzed (Figure 8). In contrast to the pSu9(1-69) derivative, which contains only 3 residues after the N-terminal 66 amino acid targeting signal, the pSu9(1-79) and pSu9(1-112) derivatives contain an additional 13 and 46 residues of the mature Su9 protein sequence, respectively. We observed that import of pSu9(1-79)DHFR (Figure 8A) and pSu9(1-112)DHFR (Figure 8B) into mmp37Δ mitochondria was reduced ∼55 and 30%, respectively. Thus the efficiency of import of both of these slightly longer pSu9-DHFR chimeras in the absence of Mmp37p was substantially greater than that observed for pSu9(1-69)DHFR (Figure 7A), and the degree of import inhibition obtained in the mmp37Δ mitochondria, particularly in the case of the pSu9(1-112)DHFR chimera, was comparable to that observed earlier for the other TIM23-dependent precursor, pOxa1p.
Figure 8.
Import inhibition of the DHFR containing preprotein in mmp37Δ mitochondria can be relieved by extending the N-terminal region of the preprotein. The import of radiolabeled pSu9(1-79)DHFR (A) and pSu9(1-112)DHFR (B) preproteins into wild-type and mmp37Δ mitochondria was performed as described in Figure 6.
On the basis of these data, we conclude that the presence of Mmp37p is required for the import of precursor proteins with tightly folded domains in close proximity to their N-terminal targeting signals. In the absence of Mmp37p, it appears that these precursors fail to efficiently engage with the PAM/mtHsp70p motor, a step that is required to further drive import to the matrix by ensuring unfolding of the import substrate on the outside of the mitochondrial surface by mtHsp70p. The absence of Mmp37p can be negated by allowing the incoming precursor to penetrate further into the matrix and engage with mtHsp70p, either by increasing the length of the N-terminal segment before the folded domain or by prior unfolding of the restricting folded domain.
The Assembly States of the TIM23 Translocase Are Altered in the Absence of Mmp37p
We investigated if the absence of Mmp37p might affect the assembly states of the Tim23p-containing complexes. Tim23p has been reported to exist in complexes of different sizes (Chacinska et al., 2005). The TIM23core complex is ∼90 kDa in size and contains Tim23p and Tim17p. Larger forms of Tim23p-containing complexes, referred to as TIM23* complexes, have recently been reported to exist, and which selectively contain Tim21p as a component of this complex. Tim21p promotes a direct contact between the TIM23* machinery with the TOM machinery in the outer membrane. The TIM23core complex on the other hand, recruits the PAM/mtHsp70p motor complex and does not contain Tim21p (Chacinska et al., 2005).
Mitochondria isolated from wild-type and mmp37Δ strains were solubilized with 1% (wt/vol) digitonin and subjected to blue-native gel electrophoresis (BN-PAGE). Subsequently, the gels were resolved in a second SDS-PAGE dimension, followed by Western blotting (Figure 9A). In wild-type mitochondria the majority of Tim23p was observed in a complex whose size was consistent with that of the TIM23core complex. A smaller fraction of Tim23p was present in larger complexes, consistent with the previously characterized TIM23* complexes (Chacinska et al., 2005). Relative to wild-type mitochondria, the ratio of Tim23p present in the TIM23core and the larger TIM23 complexes appeared altered in mitochondria devoid of Mmp37p (Figure 9A, upper three panels). Specifically, BN-PAGE analysis indicated that a larger proportion of the Tim23p protein was present in TIM23 complexes greater in size than the TIM23core complex, implying the assembly states of the TIM23 complexes are altered in the absence of Mmp37p. In addition we analyzed the assembly of the Pam18p/Tim14p-Pam16p/Tim16p complex, which is reported to be ∼80 kDa in size (van der Laan et al., 2005), and in contrast to that of the TIM23 complexes we did not observe a change in the assembly of the PAM complex in the absence of Mmp37p (Figure 9A, lower two panels). The Mmp37p protein was observed to be assembled into large complexes and did not appear to comigrate with the Tim23p, suggesting that these proteins may not coexist in the same mitochondrial protein complexes (Figure 9A).
Figure 9.
Assembly of TIM23 complexes is altered in absence of Mmp37p. Mitochondria isolated from wild-type and mmp37Δ strains were solubilized with digitonin (1% wt/vol), clarified by centrifugation and either directly analyzed by BN-PAGE (A), or incubated with additional digitonin (final concentration 2% wt/vol), before the BN-PAGE step (B). After separation of solubilized complexes by BN-PAGE, the protein complexes were resolved by a second dimension of SDS-PAGE and were subsequently analyzed by Western blotting with anti-Tim23p, anti-Pam18p/Tim14p, anti-Pam16p/Tim16p, and anti-Mmp37p, as indicated.
In addition we observed that the stability of the Tim23p-containing complexes appeared to be decreased in the absence of the Mmp37p protein. If the detergent-solubilized protein extracts were supplemented with additional digitonin (end concentration 2%), the TIM23 complexes from the mmp37Δ mitochondria became disrupted, as evidence by the presence of Tim23p in complexes of <90 kDa in size (Figure 9B, lower panel). The addition of extra digitonin to the wild-type mitochondrial extract did not have a significant affect on the stability of the TIM23 complexes (Figure 9B, upper panel). No perturbation in the assembly state of the PAM complex by the addition of extra detergent to the mmp37Δ or wild-type mitochondrial extracts was observed (Figure 9B). We conclude from these results that the assembly state and the stability of the TIM23 complexes are altered in the absence of the Mmp37p protein.
DISCUSSION
We present evidence here for the involvement of a new protein, Mmp37p, in the process of mitochondrial protein import. We demonstrate that Mmp37p is localized in the mitochondrial matrix and has an important role in mitochondrial import of precursor proteins across the TIM23 machinery. In particular, the import of precursor proteins containing a tightly folded domain in close proximity to the N-terminal mitochondrial targeting sequence is greatly affected by the absence of Mmp37p. As outlined below, we propose a model in which the function of Mmp37p affects the assembly of the TIM23 complex required to ensure a tight coordination between the activities of the TIM23 channel and the PAM-mtHsp70p motor.
Although it was previously reported that MMP37 (YGR046w) is essential, we discovered in two independent haploid genetic backgrounds that MMP37 is nonessential (Entian et al., 1999; Giaever et al., 2002). The reason for the apparent discrepancy between our observation for haploid mmp37Δ cells and the yeast genome project is not clear. The most likely explanation is that differences in the strain background can influence whether a specific nuclear gene is essential or nonessential. It is known that phenotypes associated with disruptions of other genes, CHC1, HRD2, and TIM18, vary depending on the strain background (Munn et al., 1991; Yokota et al., 1996; Kerscher et al., 2000). Furthermore, MMP37 is not the first gene originally reported by the yeast deletion project to be essential, which upon closer examination is found to be nonessential. TOM13, a component of the TOM complex, was originally deposited as an essential gene in the yeast deletion database, but at a later time was determined to be nonessential (Winzeler et al., 1999; Ishikawa et al., 2004).
Here we report that mmp37Δ cells also exhibit a petite negative phenotype, inviability in the absence of mtDNA, even when grown on a fermentable carbon source such as glucose. A similar phenotype has been observed for yeast cells lacking mitochondrial proteins Tim18p, Tom70p, or Yme1p (Thorsness et al., 1993; Kerscher et al., 2000; Dunn and Jensen, 2003). In addition yeast cells defective in the cardiolipin biosynthetic pathway are unable to grow in the presence of ethidium bromide because of increased osmotic sensitivity, a phenotype that can be suppressed by osmotic stabilization with 1 M sorbitol (Zhong et al., 2005). We observed that mmp37Δ cells were unable to grow on ethidium bromide containing media either in the presence or absence of 1 M sorbitol (unpublished data) and conclude that mmp37Δ cells display a true petite negative phenotype rather than an increased osmotic sensitivity phenotype. The relationship between the various mitochondrial proteins and the petite negative phenotype is currently unclear but may reflect a defect in the import and/or assembly of the ATP/ADP carrier, AAC, or other metabolite carrier proteins. Therefore, it is particularly interesting that we observed a reduction in the efficiency of AAC import in the absence of the Mmp37p.
Even though MMP37 is nonessential for viability, we determined that mmp37Δ haploid cells display a slow growth phenotype at 30°C and a complete failure to grow at the restrictive temperature of 37°C. Using in vitro import assays with mitochondria lacking Mmp37p, we established that Mmp37p influences the efficiency of mitochondrial protein import. Although the import of some TIM23 dependent substrates was only partially affected by the absence of Mmp37p, the import of other substrates, cytochrome b2 and pSu9(1-69)DHFR, was more dramatically affected in mmp37Δ mitochondria. Because cytochrome b2 contains a tightly folded heme-binding domain and pSu9(1-69)DHFR a tightly folded DHFR moiety, in close proximity to their mitochondrial targeting signals, the import of both of these substrates is strongly dependent on the protein unfolding activity of mtHsp70p (Glick et al., 1993; Stuart et al., 1994; Gaume et al., 1998). Due to their limited ability to sufficiently penetrate beyond the TOM and TIM machineries in the absence of their unfolding at the mitochondrial surface, we suggest that these preproteins may not efficiently engage with mtHsp70p in the absence of Mmp37p, to drive their complete translocation into the mitochondrial matrix.
We argue that the import defects observed in mmp37Δ mitochondria are not solely due to a possibly reduced membrane potential in the absence of Mmp37p, because the import of pOxa1, the unfolded pSu9(1-69)DHFR derivatives and pSu9(1-112)DHFR in mmp37Δ mitochondria occurs at levels comparable to wild-type mitochondria. The membrane potential is required to support the initial steps of protein import, i.e., presequence passage across the TIM23 machinery. Although different preproteins may exhibit different levels of membrane potential requirements (possibly due to differences in their N-terminal presequence composition), we note that the pSu9DHFR preproteins (pSu9(1-69)-, pSu9(1-79)- and pSu9(1-112)DHFR) displayed very different levels of dependency on Mmp37p, even though they contain the same N-terminal presequences.
Our data supports the model that the import inhibition observed in the mmp37Δ mitochondria may be due to an altered assembly of the TIM23 import complexes. In the absence of Mmp37p, an increase in the level of TIM23 complexes with sizes consistent with the previously described TIM23* complexes were observed. Tim21p, which is associated with TIM23*, and the PAM complex have been reported to act in an antagonistic manner to influence the switch between a TIM23* complex, which is tethered to the TOM machinery, and the TIM23core complex, which is competent to recruit the PAM machinery (Chacinska et al., 2005). It is therefore possible that Mmp37p influences the ability of TIM23* to form a Tim21p-free TIM23core complex and thus in the absence of Mmp37p, the levels of TIM23core complex competent to recruit the PAM motor may be limiting. Consequently, the import of those precursor proteins with folded domains (such as precytochrome b2 and pSu9(1-69)DHFR), which are dependent on engaging with the PAM/mtHsp70p machinery immediately on their emergence on the trans-side of the TIM23 translocon, to secure them in the import machinery and to promote their subsequent unfolding, will be particularly compromised in mmp37Δ mitochondria. Extension of the linker segment between the targeting signal and the folded domain of the preprotein was observed to alleviate the requirement for Mmp37p in mitochondrial import, suggesting that if the incoming preprotein can penetrate far enough across the TOM and TIM machineries into the matrix to efficiently engage the mtHsp70p motor, its dependency on the function of Mmp37p can be at least partially overcome. This observation may explain the observed nonessential nature of the Mmp37p protein. It is important to note that the TIM23-containing complexes in the mmp37Δ mitochondria were observed to display an inherent instability. The addition of extra detergent to the solubilized TIM23 complexes resulted in their dissociation to smaller complexes in the absence of Mmp37p. We conclude therefore that Mmp37p affects the assembly state of the TIM23 complexes, which in turn may affect their ability to recruit the PAM machinery.
Our proposed model implies that Mmp37p could be a component of the trans-side of the TIM23 complex. Indeed, Mmp37p is localized to the mitochondrial matrix where it appears to interact with the trans-side of the inner membrane in a peripheral manner. However, to date we have been unable to obtain any evidence that Mmp37p is physically associated with the TIM23 or PAM complexes. Using both N- and C-terminal histidine-tagged derivatives of Mmp37p, we purified Mmp37p by Ni-NTA chromatography after mild-detergent lysis with digitonin (a lysis condition known to retain the assembled states of both the TIM23 and PAM complexes) and did not obtain any evidence to support a physical association between Mmp37p and the following components: Tim23p, Tim44p, Pam18p/Tim14p, Pam16p/Tim16p, or mtHsp70p. Consistently, native gel electrophoresis indicated that the mass of Mmp37p is distinct from the TIM23 and PAM complexes. Although we cannot exclude that Mmp37p is a component of, or is transiently associated with the TIM23 or PAM complexes, and despite its demonstrated role in protein import across the inner membrane, we have decided not to refer to Mmp37p as Tim37p at this stage, because of a lack experimental evidence supporting a physical association of Mmp37p with the TIM or PAM machineries.
We also note that the import of a substrate via the TIM22 complex, AAC, is also partially defective in the mmp37Δ mitochondria. It is possible that the observed decrease in import of TIM22 substrates may reflect a translocation defect at the level of the TOM machinery in the mmp37Δ mitochondria. The observed increase in the levels of TIM23* complexes in the absence of Mmp37p may indicate that more TIM23 complexes are physically tethered to the TOM complexes, and this may hinder the accessibility of TIM22 substrates to the TIM22 translocon in the inner membrane. Indeed, the accessibility of presequence-targeted proteins to the TIM23core-PAM machinery may also be hindered for the same reason, which also could contribute to the observed import inhibition of these substrates in the mmp37Δ mitochondria. It is also possible that Mmp37p may play a direct role in the maintenance of the structure of the mitochondrial inner membrane, possibly by affecting the environment of the inner boundary membrane (IBM; that region of the inner membrane that is in close proximity to the outer mitochondrial membrane, i.e., is distinct from the inner cristae membranes) and thus indirectly affects the TIM23 complex stability. The correct positioning of the IBM relative to the outer membrane supports the close relationship between the TOM and TIM import machineries. Thus if the integrity of the IBM is affected by the absence of Mmp37p, the ability of an incoming preprotein to access the TIM23 and TIM22 machineries from the level of the TOM complex may be hindered.
In summary, the data presented here highlight a function for Mmp37p in protein import across the mitochondrial membranes. Specifically, Mmp37p is required to maintain the integrity of the assembled TIM23 complexes such that the coupling of mtHsp70p unfolding and translocation events with transport across the TOM and TIM23 machineries is enhanced. Further characterization of Mmp37 and its relationship to components of the TIM machineries and/or the inner membrane are currently underway in our laboratories.
ACKNOWLEDGMENTS
We are grateful to Lixia Jia for her assistance with the BN-PAGE experiments. We thank the following for their generous gifts of the following antibodies used in this study: Dr. Kai Hell, University of Munich, Tim14p/Pam18p and Tim16p/Pam16p; Dr. Carla Koehler, University of California, Los Angeles Yme1p, Tim54p; and Dr. Roland Lill, University of Marburg, aconitase. This work was supported by research National Institutes of Health Grants R01 GM060392 to R.M.L. and National Science Foundation Grant MCB0347025 to R.A.S.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-04-0366) on July 21, 2006.
REFERENCES
- Arnold I., Pfeiffer K., Neupert W., Stuart R. A., Schägger H. Yeast mitochondrial F1Fo-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 1998;17:7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A., et al. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 2005;120:817–829. doi: 10.1016/j.cell.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Chacinska A., Rehling P., Guiard B., Frazier A. E., Schulze-Specking A., Pfanner N., Voos W., Meisinger C. Mitochondrial translocation contact sites: separation of dynamic and stabilizing elements in formation of a TOM-TIM-preprotein supercomplex. EMBO J. 2003;22:5370–5381. doi: 10.1093/emboj/cdg532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran S. P., Leuenberger D., Oppliger W., Koehler C. M. The Tim9p-Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J. 2002a;21:942–953. doi: 10.1093/emboj/21.5.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran S. P., Leuenberger D., Schmidt E., Koehler C. M. The role of the Tim8p-Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins. J. Cell Biol. 2002b;158:1017–1027. doi: 10.1083/jcb.200205124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Silva P. D., Schilke B., Walter W., Andrew A., Craig E. A. J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc. Natl. Acad. Sci. USA. 2003;100:13839–13844. doi: 10.1073/pnas.1936150100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn C. D., Jensen R. E. Suppression of a defect in mitochondrial protein import identifies cytosolic proteins required for viability of yeast cells lacking mitochondrial DNA. Genetics. 2003;165:35–45. doi: 10.1093/genetics/165.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K. D., et al. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol. Gen. Genet. 1999;262:683–702. doi: 10.1007/pl00013817. [DOI] [PubMed] [Google Scholar]
- Gaume B., Klaus C., Ungermann C., Guiard B., Neupert W., Brunner M. Unfolding of preproteins upon import into mitochondria. EMBO J. 1998;17:6497–6507. doi: 10.1093/emboj/17.22.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O’Shea E. K., Weissman J. S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Giaever G., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Glick B. S., Wachter C., Reid G. A., Schatz G. Import of cytochrome b2 to the mitochondrial intermembrane space: the tightly folded heme-binding domain makes import dependent upon matrix ATP. Protein Sci. 1993;2:1901–1917. doi: 10.1002/pro.5560021112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann J. M., Folsch H., Neupert W., Stuart R. A. Cell Biology: A Laboratory Handbook. San Diego, CA: Academic Press; 1994. [Google Scholar]
- Huang S., Murphy S., Matouschek A. Effect of the protein import machinery at the mitochondrial surface on precursor stability. Proc. Natl. Acad. Sci. USA. 2000;97:12991–12996. doi: 10.1073/pnas.230243097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa D., Yamamoto H., Tamura Y., Moritoh K., Endo T. Two novel proteins in the mitochondrial outer membrane mediate beta-barrel protein assembly. J. Cell Biol. 2004;166:621–627. doi: 10.1083/jcb.200405138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. P., Dowzer C., Michaelis C., Galova M., Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- Johnston S. A., Hopper J. E. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc. Natl. Acad. Sci. USA. 1982;79:6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P. J., Ostermann J., Shilling J., Neupert W., Craig E. A., Pfanner N. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990;348:137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- Kerscher O., Sepuri N. B., Jensen R. E. Tim18p is a new component of the Tim54p-Tim22p translocon in the mitochondrial inner membrane. Mol. Biol. Cell. 2000;11:103–116. doi: 10.1091/mbc.11.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler C. M. New developments in mitochondrial assembly. Annu. Rev. Cell Dev. Biol. 2004;20:309–335. doi: 10.1146/annurev.cellbio.20.010403.105057. [DOI] [PubMed] [Google Scholar]
- Koehler C. M., Jarosch E., Tokatlidis K., Schmid K., Schweyen R. J., Schatz G. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science. 1998;279:369–373. doi: 10.1126/science.279.5349.369. [DOI] [PubMed] [Google Scholar]
- Koehler C. M., Merchant S., Schatz G. How membrane proteins travel across the mitochondrial intermembrane space. Trends Biochem. Sci. 1999;24:428–432. doi: 10.1016/s0968-0004(99)01462-0. [DOI] [PubMed] [Google Scholar]
- Kozany C., Mokranjac D., Sichting M., Neupert W., Hell K. The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat. Struct. Mol. Biol. 2004;11:234–241. doi: 10.1038/nsmb734. [DOI] [PubMed] [Google Scholar]
- Leuenberger D., Bally N. A., Schatz G., Koehler C. M. Different import pathways through the mitochondrial intermembrane space for inner membrane proteins. EMBO J. 1999;18:4816–4822. doi: 10.1093/emboj/18.17.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mokranjac D., Paschen S. A., Kozany C., Prokisch H., Hoppins S. C., Nargang F. E., Neupert W., Hell K. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 2003a;22:816–825. doi: 10.1093/emboj/cdg090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D., Sichting M., Neupert W., Hell K. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 2003b;22:4945–4956. doi: 10.1093/emboj/cdg485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn A. L., Silveira L., Elgort M., Payne G. S. Viability of clathrin heavy-chain-deficient Saccharomyces cerevisiae is compromised by mutations at numerous loci: implications for the suppression hypothesis. Mol. Cell. Biol. 1991;11:3868–3878. doi: 10.1128/mcb.11.8.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu. Rev. Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Perry A. J., Lithgow T. Protein targeting: entropy, energetics and modular machines. Curr. Biol. 2005;15:R423–R425. doi: 10.1016/j.cub.2005.05.031. [DOI] [PubMed] [Google Scholar]
- Pfanner N., Craig E. A., Honlinger A. Mitochondrial preprotein translocase. Annu. Rev. Cell Dev. Biol. 1997;13:25–51. doi: 10.1146/annurev.cellbio.13.1.25. [DOI] [PubMed] [Google Scholar]
- Rassow J., Hartl F. U., Guiard B., Pfanner N., Neupert W. Polypeptides traverse the mitochondrial envelope in an extended state. FEBS Lett. 1990;275:190–194. doi: 10.1016/0014-5793(90)81469-5. [DOI] [PubMed] [Google Scholar]
- Rehling P., Brandner K., Pfanner N. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol. 2004;5:519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P. Methods in Yeast Genetics. A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Sanjuan Szklarz L. K., et al. Inactivation of the mitochondrial heat shock protein zim17 leads to aggregation of matrix hsp70s followed by pleiotropic effects on morphology and protein biogenesis. J. Mol. Biol. 2005;351:206–218. doi: 10.1016/j.jmb.2005.05.068. [DOI] [PubMed] [Google Scholar]
- Schleyer M., Neupert W. Transport of proteins into mitochondria: translocational intermediates spanning contact sites between outer and inner membranes. Cell. 1985;43:339–350. doi: 10.1016/0092-8674(85)90039-x. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Hicks J. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1986. [Google Scholar]
- Sirrenberg C., Endres M., Folsch H., Stuart R. A., Neupert W., Brunner M. Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature. 1998;391:912–915. doi: 10.1038/36136. [DOI] [PubMed] [Google Scholar]
- Spencer F., Gerring S. L., Connelly C., Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart R. A., Gruhler A., van der Klei I., Guiard B., Koll H., Neupert W. The requirement of matrix ATP for the import of precursor proteins into the mitochondrial matrix and intermembrane space. Eur. J. Biochem. 1994;220:9–18. doi: 10.1111/j.1432-1033.1994.tb18593.x. [DOI] [PubMed] [Google Scholar]
- Teichmann U., van Dyck L., Guiard B., Fischer H., Glockshuber R., Neupert W., Langer T. Substitution of PIM1 protease in mitochondria by Escherichia coli Lon protease. J. Biol. Chem. 1996;271:10137–10142. doi: 10.1074/jbc.271.17.10137. [DOI] [PubMed] [Google Scholar]
- Thorsness P. E., Fox T. D. Nuclear mutations in Saccharomyces cerevisiae that affect the escape of DNA from mitochondria to the nucleus. Genetics. 1993;134:21–28. doi: 10.1093/genetics/134.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness P. E., White K. H., Fox T. D. Inactivation of YME1, a member of the ftsH-SEC18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:5418–5426. doi: 10.1128/mcb.13.9.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott K. N., et al. A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J. Cell Biol. 2003;163:707–713. doi: 10.1083/jcb.200308004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C., Neupert W., Cyr D. M. The role of Hsp70 in conferring unidirectionality on protein translocation into mitochondria. Science. 1994;266:1250–1253. doi: 10.1126/science.7973708. [DOI] [PubMed] [Google Scholar]
- van der Laan M., Chacinska A., Lind M., Perschil I., Sickmann A., Meyer H. E., Guiard B., Meisinger C., Pfanner N., Rehling P. Pam17 is required for architecture and translocation activity of the mitochondrial protein import motor. Mol. Cell. Biol. 2005;25:7449–7458. doi: 10.1128/MCB.25.17.7449-7458.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestweber D., Schatz G. Point mutations destabilizing a precursor protein enhance its post-translational import into mitochondria. EMBO J. 1988;7:1147–1151. doi: 10.1002/j.1460-2075.1988.tb02924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Yokota K., et al. cDNA cloning of p112, the largest regulatory subunit of the human 26s proteasome, and functional analysis of its yeast homologue, Sen3p. Mol. Biol. Cell. 1996;7:853–870. doi: 10.1091/mbc.7.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q., Gvozdenovic-Jeremic J., Webster P., Zhou J., Greenberg M. L. Loss of function of KRE5 suppresses temperature sensitivity of mutants lacking mitochondrial anionic lipids. Mol. Biol. Cell. 2005;16:665–675. doi: 10.1091/mbc.E04-09-0808. [DOI] [PMC free article] [PubMed] [Google Scholar]