Abstract
The Sec61/SecY translocon mediates translocation of proteins across the membrane and integration of membrane proteins into the lipid bilayer. The structure of the translocon revealed a plug domain blocking the pore on the lumenal side. It was proposed to be important for gating the protein conducting channel and for maintaining the permeability barrier in its unoccupied state. Here, we analyzed in yeast the effect of introducing destabilizing point mutations in the plug domain or of its partial or complete deletion. Unexpectedly, even when the entire plug domain was deleted, cells were viable without growth phenotype. They showed an effect on signal sequence orientation of diagnostic signal-anchor proteins, a minor defect in cotranslational and a significant deficiency in posttranslational translocation. Steady-state levels of the mutant protein were reduced, and when coexpressed with wild-type Sec61p, the mutant lacking the plug competed poorly for complex partners. The results suggest that the plug is unlikely to be important for sealing the translocation pore in yeast but that it plays a role in stabilizing Sec61p during translocon formation.
INTRODUCTION
Translocation of proteins across and integration into the membrane of the endoplasmic reticulum (ER) are both mediated by the Sec61 translocon complex (Johnson and van Waes, 1999). Proteins are targeted to this complex by hydrophobic signal sequences (von Heijne, 1990). Uncleaved signals and subsequent hydrophobic segments of the polypeptide are integrated into the bilayer as transmembrane helices. Neither the determinants of protein topology in the membrane nor the mechanism of integration are fully understood. The simplest case of protein topogenesis in the membrane is the orientation of the signal sequence upon entering the translocon. Signal sequences may translocate either the C-terminal end (cleavable signals and signal-anchors) or the N-terminal end (reverse signal-anchors). The best established determinant of signal orientation is the distribution of charged residues on either side of the hydrophobic core of the signal. According to the “positive-inside rule” (von Heijne, 1986; Hartmann et al., 1989), the more positive end is generally cytosolic. Additional determinants are the folding of sequences N-terminal to an internal signal that may sterically hinder N-translocation (Denzer et al., 1995), and the hydrophobicity of the apolar signal core (Sakaguchi et al., 1992; Wahlberg and Spiess, 1997; Eusebio et al., 1998; Harley et al., 1998; Rösch et al., 2000; Goder and Spiess, 2003).
The Sec61 translocon consists of Sec61α (Sec61p in yeast) with 10 transmembrane domains, and the single spanning proteins Sec61β (Sbh1p) and Sec61γ (Sss1p), corresponding in bacteria to the SecY complex (SecY/SecG/SecE). The recent crystal structure of the archaebacterial SecY complex of Methanococcus jannaschii (van den Berg et al., 2004) revealed a surprisingly compact helix bundle with a potential hydrophilic channel pore and a lateral exit site made of transmembrane segments TM2 and TM7, consistent with previous cross-linking data (Plath et al., 1998). It suggests that the translocation channel is formed by a single heterotrimer. The structure clearly represents the closed state, because the central passage is blocked by a lumenal plug domain and a central constriction ring. To open, the plug needs to move out and the ring must expand to widen the central passage. By cysteine mutagenesis and disulfide cross-linking, it was demonstrated that translocating polypeptides indeed localize to the center of SecY in Escherichia coli (Cannon et al., 2005). The plug domain is likely to be involved in gating the translocon triggered by the signal sequence to allow entry and transfer of the polypeptide to be translocated and to seal the channel when idle (Tam et al., 2005).
In a systematic analysis of conserved charged residues in Sec61p that were candidates to be responsible for the positive-inside rule, three residues, R67, R74, and E382, were identified to fulfill the criteria of increasing N-translocation of a model signal-anchor protein with more positive N-terminal flanking charges when mutated and to decrease N-translocation of a model protein with an opposite charge difference (Goder et al., 2004). Based on the structure of the archaebacterial SecY complex, E382 is located at the cytoplasmic end of transmembrane segment TM8, and both R67 and R74 are in the plug domain, positions seemingly appropriate to influence the orientation of a signal entering the closed channel. To test the role of the plug domain in signal orientation and its importance for translocon functionality and cell viability, specific mutations were introduced to disturb its structure, including a full deletion. Surprisingly, mutation or deletion of the plug domain did not affect viability or growth, despite reduced translocation efficiency and translocon formation.
MATERIALS AND METHODS
Yeast Strains and Model Protein Constructs
The yeast strain RSY1293 (matα, ura3-1, leu2-3,-112, his3-11,15, trp1-1, ade2-1, can1-100, sec61::HIS3, [pDQ1]) was a gift from R. Schekman (University of California, Berkeley, Berkeley, CA) (Pilon et al., 1997). pDQ1 is YCplac111 (LEU2 CEN) containing SEC61 with its own promoter, except that codons 2–6 were replaced by a sequence encoding H6RS. This tagged Sec61p functionally corresponds to wild-type Sec61p (Pilon et al., 1997). In strain VGY61, pDQ1 was replaced by YCPlac33 (URA3 CEN) with the same SEC61 gene, to be used to introduce mutant sec61 generated by PCR using appropriate mutagenic primers and Vent polymerase (New England Biolabs, Beverly, MA) in YCplac111 (LEU2 CEN) by plasmid shuffling using 5-fluoroorotic acid (5-FOA) (Goder et al., 2004). VGY61 with a disruption of SSH1 was described previously (Goder et al., 2004).
Model proteins 40[Leu16](+5), 60[H1](+1), and [Leu16](−3) were described previously (Goder et al., 2004). The model proteins were expressed in pRS426 (URA3 2μ) with a glyceraldehyde-3-phosphate dehydrogenase (GPD) promotor. To analyze membrane insertion of dipeptidyl aminopeptidase B (DPAPB), the coding sequences of DAP2 was amplified by PCR, fused to a C-terminal triple-hemagglutinin (HA) epitope tag, and cloned into pRS426 with a GPD promotor. After pulse labeling of expressing cells, DPAPB was immunoprecipitated with a mouse monoclonal anti-HA antibody. Translocation of endogenous carboxypeptidase Y (CPY) was analyzed using a rabbit anti-CPY antiserum.
Labeling and Immunoprecipitation
For in vivo pulse labeling, an overnight yeast culture was diluted and grown to OD600 ∼1. Cells equivalent to 1.5 OD were resuspended in 200 μl of medium, incubated for 15 min at 30°C, and labeled for 5 min with 100 μCi/ml [35S]methionine (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). At 4°C cells were supplemented with 10 mM azide, pelleted, resuspended in 50 mM Tris, pH 7.5, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and lysed with glass beads for 7 min in a bead-beater, supplemented with 1% SDS, and heated at 95°C for 5 min. Cell remnants were removed by centrifugation for 10 min in a microfuge, and the supernatant was used for immunoprecipitation using anti-HA antibodies. Immune complexes were isolated with protein A-Sepharose (GE Healthcare) and analyzed by SDS-gel electrophoresis and autoradiography. Signals were quantified with a PhosphorImager (GE Healthcare). For deglycosylation, immune complexes were boiled in 50 mM Na citrate, pH 6, 1% SDS, and incubated with 1 mU endo-β-D-N-acetyl glucosaminidase H for 1 h at 37°C before gel electrophoresis.
Analysis of Sec61p Stability
To determine steady-state levels of Sec61p by immunoblot analysis, 10 OD600 equivalents of yeast cells were lysed in SDS-sample buffer with glass beads and boiled for 10 min. Aliquots of equal total protein were separated by SDS-gel electrophoresis, blotted onto nitrocellulose, and decorated with a rabbit antiserum against the C terminus of Sec61p, a gift from C. Stirling (University of Manchester, Manchester, United Kingdom; Stirling et al., 1992). Antibody was detected using horseradish peroxidase-conjugated anti-mouse secondary antibody and the enhanced chemoluminescence kit (GE Healthcare). Equal protein loading was based on Coomassie blue staining of a separate gel.
To compare protein levels in cells expressing wild-type and mutant Sec61p simultaneously, YTX57 (matα, ura3-1, leu2-3,-112, his3-11,15, trp1-1, ade2-1, can1-100), the corresponding wild type of strain RSY1293, was transformed with YCplac111 (LEU2 CEN) containing the Δplug (codons 52–74 replaced by that of a glycine) or the ΔTM2 sec61 mutant (deletion of codons 77–107) and subjected to immunoblot analysis as described above. To test the effect of overexpressed β and/or γ subunit of the Sec61 complex, the resulting strain was transformed in addition with YEplac195 (URA3 2μ) containing SBH1, YEplac112 (TRP1 2μ) containing SSS1, or both, or with pRS426 (URA3 2μ) or YEplac112 (TRP1 2μ) containing SBH1 and SSS1, respectively, driven by a GPD promoter.
To determine the half-lives of Sec61p, cells expressing wild-type or mutant Sec61p were grown overnight, diluted to OD600 = 0.5, and incubated at 30°C with 100 μg/ml cycloheximide for up to 8 h. A blocking concentration of cycloheximide was maintained by addition of further 50 μg/ml every 2 h. Aliquots were subjected to immunoblot analysis as described above.
Growth Analysis
For serial dilution experiments, yeast strains were grown in YPDA medium at 30°C to mid-log phase and diluted to 0.1 OD600. Aliquots of fivefold serial dilutions were transferred onto YPDA plates with or without 0.3 μg/ml tunicamycin or 4 mM dithiothreitol and incubated at 15, 30, or 39°C. Alternatively, liquid cultures were inoculated and incubated at 30°C, and OD600 was measured as a function of time.
Sequence Alignment and Protein Structure Modeling
Seventy-six eukaryotic Sec61p homologues were identified in the UniProt database (release 6.1; Bairoch et al., 2005) by using BLAST (Altschul et al., 1997), and a multiple sequence alignment was generated using T_COFFEE (Notredame et al., 2000). Residue conservation was quantified using ScoreCons applying the Valdar01 scoring scheme with the default PET91 substitution matrix (Valdar, 2002).
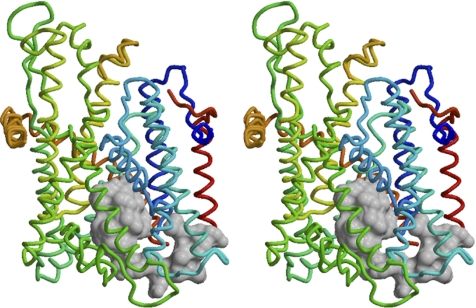
A comparative protein structure model for the yeast Sec61 heterotrimeric complex was built on the experimental structure of the M. jannaschii SecYEβ translocon (van den Berg et al., 2004; PDB 1RHZ) solved by x-ray crystallography at 3.5-Å resolution. Alignment between the yeast Sec61 target and M. jannaschii template sequences for all three chains (Sec61p, Sbh1p, and Sss1p) was based on a multiple sequence alignment generated by T_COFFEE (Notredame et al., 2000) by using 10 intermediate homologous sequences. The sequence identity between the yeast target and the archaeal template was 37% for Sec61p, 23% for Sbh1p, and 31% for Sss1p. The positioning of alignment insertions and deletions in the structural context was visually inspected using DeepView (Guex and Peitsch, 1997). Alternative alignments were derived for segments Sec61p 54-57, 140-149, 201-239, 318-333, 403-412, and 469-480 and used to generate three-dimensional models by using SWISS-MODEL (Schwede et al., 2003) and Modeler (Sali and Blundell, 1993). Segments P200-E212 and D227-N240 of Sec61p could not be modeled reliably and were therefore excluded from the final model. Graphical representations were generated with Molscript (Kraulis, 1991), molecular surfaces were calculated using MSMS (Sanner et al., 1996), and combined graphical objects were rendered with Raster3D (Merritt and Bacon, 1997).
RESULTS
Disruption or Deletion of the Sec61p Plug Domain Does Not Affect Viability and Growth
Based on the experimental structure of the M. jannaschii SecYEβ translocon, a comparative structure model for the yeast Sec61 complex was built (Figure 1). In the plug domain, mutations R67E and R74E have previously been shown to affect topology in a manner consistent with a charge effect according to the positive-inside rule. The two arginines are not conserved in the M. jannaschii SecY, but there is an arginine (R66 in SecY) close to the position corresponding to R74 of Sec61p. Interestingly, R67 and R74 in the yeast model and R66 of SecY seem to be facing the solvent below the plug domain, raising the possibility that their positive charge might not directly act on signal sequences and that their mutation might take effect by disturbing the plug's structure. To test this hypothesis, specific mutations expected to disrupt the plug were introduced. L63, L66, and L70, which form the hydrophobic core of the plug domain, were replaced individually or in combination by asparagine, which is small and hydrophilic, but uncharged (L63N, L66N, L70N, and LLLNNN). Furthermore, L70 was deleted (ΔL70), or the six residues at the very tip of the plug (residues 67–72) or the entire plug domain (residues 52–74; highlighted in Figure 1) were replaced by a glycine residue (Δtip and Δplug, respectively).
Figure 1.
Plug domain of the yeast Sec61 complex. A stereo representation of the structure of the yeast Sec61 complex as modeled on the experimental structure of the M. jannaschii SecYEβ translocon (van den Berg et al., 2004) is shown with the cytosolic side facing up. The model is shown as a backbone in stereo with the plug domain (residues 52–74), which was replaced by a glycine in the Δplug mutant, displayed as a space-filling contour. Sbh1p is shown in red, Sss1p in orange, and Sec61p is colored from N to C terminus in blue to yellow. Segments P200-E212 and D227-N240 of Sec61p are not present in the model.
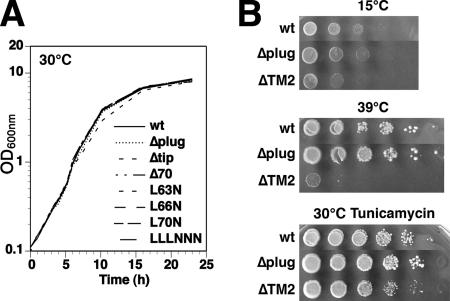
Wild-type SEC61 was replaced by these plug mutants in yeast cells lacking the second nonessential translocon complex consisting of Ssh1p, Sbh2p, and Sss1p due to a disruption of SSH1, Surprisingly, the resulting mutant cells were all viable and showed no growth defect at any temperature (Figure 2). There was also no growth difference in the presence of tunicamycin, which causes protein misfolding in the ER and increased retrotranslocation. The plug domain is thus not essential for general Sec61p function.
Figure 2.
Mutations in the plug domain have no growth defect. (A) Growth curves were measured for liquid cultures of Δssh1 cells expressing wild-type Sec61p or the indicated plug mutations. (B) Δssh1 cells expressing wild-type Sec61p, or the mutants lacking the entire plug domain (Δplug) or TM2 (ΔTM2), were plated at serial dilutions onto YPDA plates and incubated for 3 d at 15 or 39°C, or onto YPDA plates containing 0.3 μg/ml tunicamycin and incubated for 3 d at 30°C.
Plug Deletion Affects Signal Orientation and Protein Translocation
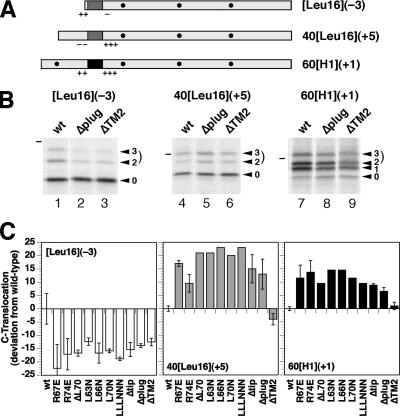
To analyze the plug mutants with respect to signal orientation, their effect on three model proteins was determined that insert with mixed orientations and are thus sensitive reporters of changes in topogenesis (Goder et al., 2004). Construct [Leu16](−3) (Figure 3A) has an N-terminal signal-anchor of 16 consecutive leucines with positive N-terminal and negative C-terminal flanking charges [Δ(C–N) = −3, according to the rules by Hartmann et al., 1989]. Constructs 40[Leu16](+5) and 60[H1](+1) have internal signal-anchors and an opposite charge difference of +5 and +1, respectively. Resulting topologies are reflected in the glycosylation patterns. Addition of two or three glycans represents polypeptides with a translocated C terminus. All mutations in the plug domain caused a decrease of glycosylated [Leu16](−3) by ∼15% (Figure 3, B and C), and a similar increase of glycosylated 40[Leu16](+5). The effects are essentially the same as those previously observed for mutations R67E and R74E (Goder et al., 2004). These results indicate that disturbing the plug structure is equivalent to deleting the entire domain.
Figure 3.
All plug mutations have the same effect on the topology of model proteins. (A) Diagnostic protein substrates to analyze topology changes are shown schematically with the signal-anchor sequence in dark gray (Leu16) or black (of the asialoglycoprotein receptor H1), flanking charges as + and −, and glycosylation sites as black dots. Their names indicate the length of the N-terminal hydrophilic sequence preceding the signal, the hydrophobic signal core (H1 or Leu16) in brackets, and the charge difference Δ(C–N) according to Hartmann et al. (1989) in parentheses. (B) The three substrates were expressed in Δssh1 cells with wild-type, Δplug, or ΔTM2 Sec61p, pulse labeled for 5 min with [35S]methionine, and analyzed by immunoprecipitation, SDS-gel electrophoresis, and autoradiography. The forms corresponding to the polypeptides with zero, one, two, or three glycans are indicated with arrowheads. The dash designates the position of the 37-kDa molecular weight standard. (C) Effect of indicated Sec61p mutants on the orientation of the model proteins was determined by PhosphorImager quantitation of labeling experiments like those shown in B. Two- and threefold glycosylated species together represent polypeptides with a translocated C terminus. Their fraction of the total (in case of 60[H1](+1) excluding the unglycosylated, not integrated products) are represented as the deviation from the value for the wild-type in percentage points). The results of single determinations (no error bars) or of two to six measurements (average with SD) are shown. The absolute fraction of C-terminally translocated products with wild-type Sec61p was 29.1% for [Leu16](−3), 46.3% for 40[Leu16](+5), and 60.2% for 60[H1](+1).
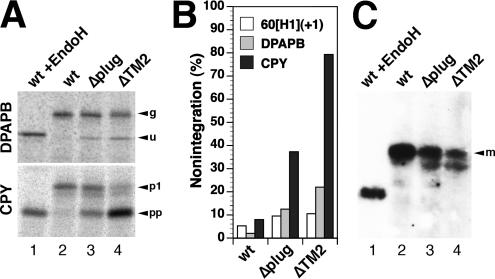
Because the N-terminal extension of 60[H1](+1) contains a glycosylation site, this construct also distinguishes between products with a translocated N terminus (once glycosylated) and those that were not integrated into the membrane at all (unglycosylated proteins). In cells with wild-type Sec61p, 5.3 ± 1.4% (n = 8) of the products were unglycosylated. The plug mutants produced slightly increased levels of nonintegration of ∼9% (Δplug; 9.5 ± 2.4%; n = 4), suggesting a minor insertion defect. Because of the hydrophobicity of the signal-anchors of our substrate proteins, they are very likely inserted cotranslationally. Analysis of DPAPB, an established cotranslational substrate of the translocon, similarly revealed the production of some unglycosylated polypeptides in cells expressing Δplug (Figure 4, A and B). On introduction of a glycosylation site into the N-terminal domain, they remained unglycosylated (our unpublished data), confirming an insertion defect rather than inverted integration. Posttranslational translocation of CPY, however, was much more severely compromised by the plug deletion, because almost one-half the products remained as unglycosylated pre-pro-CPY (Figure 4, A and B). The deficiency in CPY translocation was also reflected in reduced steady-state levels of mature CPY in these cells (Figure 4C).
Figure 4.
Co- and posttranslational integration defects of Sec61p mutations. Integration of DPAPB as a cotranslational substrate and of CPY as a posttranslational substrate of the Sec61 translocon was analyzed in a Δssh1 background by pulse labeling for 5 min with [35S]methionine, immunoprecipitation, gel electrophoresis, and autoradiography (A). The different products correspond to glycosylated (g) and unglycosylated (u) forms of DPAPB and to the glycosylated first proform (p1) and the unglycosylated pre-pro-form (pp) of CPY. As a control, labeled protein from cells expressing wild-type Sec61p (wt) were analyzed after deglycosylation with endoglycosidase H (EndoH). The unglycosylated forms correspond to nonintegrated polypeptides and were quantified as a fraction of the total in B. In C, the steady-state levels of mature CPY (m) was determined by immunoblot analysis. Equal amounts of cell lysates were analyzed in lanes 2–4.
The plug domain connects transmembrane segments TM1 and TM2. TM2 is part of the exit site through which signal and stop transfer sequences laterally leave the translocon into the lipid membrane. Signal sequences have been cross-linked to TM2 and TM7, which led to the model that these TMs form the signal binding site in the translocon (Plath et al., 1998). Interestingly, it has previously been reported that Sec61p function is retained upon deletion of either TM2 (residues 73–107) or TM3 (residues 108–143), but not of both (Wilkinson et al., 2000). We confirmed this finding for a deletion of TM2. Cells with Sec61p lacking residues 77–107 (ΔTM2) were viable and grew almost as well as wild type, except at 39°C (Figure 2B). This deletion only affected the topology of [Leu16](−3) (Figure 3), produced some unglycosylated DPAPB, and was strongly deficient in CPY translocation (Figure 4). The combined deletion of the plug domain and TM2 was lethal as judged by the inability of transformed cells to lose the wild-type SEC61 plasmid on 5-FOA plates and by tetrad analysis.
Plug Deletion Impairs Translocon Stability
By immunoblot analysis, the steady-state levels of Sec61p were found to be reduced for the Δplug deletion in comparison with the wild-type (Figure 5A), but similarly to ΔTM2 (for which this was previously observed by Wilkinson et al., 2000). This could be the result of reduced folding efficiency and/or reduced half-life of the folded protein. To estimate the half-life, the Sec61p levels were determined after inhibition of new synthesis with cycloheximide (Figure 5B). Whereas the wild-type protein was degraded with a half-life of ∼2 h, ΔTM2 disappeared more than twice as rapidly. In contrast, no change in half-life was detected upon deletion of the plug domain, indicating that after assembly into the translocon complex Δplug is as stable as wild-type Sec61p.
Figure 5.
Plug deletion impairs translocon levels. (A) Steady-state amounts of wild-type and mutant Sec61p were determined by immunoblot analysis of equal amounts of protein. (B) The half-lives of wild-type Sec61p and the Δplug and ΔTM2 mutants were estimated by immunoblot analysis of cells incubated for 0, 1, 2, 4, or 8 h in the presence of the translation inhibitor cycloheximide. Similar starting signals of Δplug and ΔTM2 versus wild type are shown. (C) Sec61p levels were determined by immunoblot analysis for cells simultaneously expressing wild-type Sec61p from a chromosomal SEC61 gene, and the Δplug or ΔTM2 mutants from a CEN plasmid (wt/Δplug and wt/ΔTM2, respectively). The cells contained no additional plasmid (−), or a 2μ plasmid with SBH1 (+β), SSS1 (+γ), or both (+β+γ) with their natural promoters, or a 2μ plasmid with SBH1 or SSS1 with a GPD promoter (+β* and +γ*, respectively). Alternatively, the cells were incubated for 3 h with 0.3 μg/ml tunicamycin (+Tun) to induce an unfolded protein response. As controls, cells expressing only wild-type Sec61p (lanes 1, 9, 11, 13, and 15) or the Δplug or ΔTM2 mutants (lanes 2 and 16, respectively) were analyzed. Lanes 9–14 are from the identical immunoblot.
It has previously been observed that the ΔTM2 mutant in the presence of a wild-type copy of SEC61, i.e., in a heterozygous situation, could hardly be detected (Wilkinson et al., 2000). We also found this to be the case for the plug deletion (Figure 5C, lanes 1–8), indicating that the mutants compete with wild-type Sec61p for a limiting component(s). Obvious candidates are the β and γ subunits of the translocon complex, Sbh1p and Sss1p, respectively. Indeed, expression of both SBH1 and SSS1 from a 2μ plasmid, or of either coding sequence with a strong GPD promoter increased steady-state levels of Δplug and ΔTM2 to detectable levels. This is not an indirect effect of producing an excess of unassembled subunits limiting the capacity of protein degradation, because treatment with tunicamycin, which increases the load of ER-associated degradation even more, did not rescue Δplug Sec61p (lanes 9–14). However, that increasing the copy number of SBH1 and SSS1 did not restore Δplug levels to those of wild-type Sec61p suggests that they are not the only limiting components. The results indicate that the absence of the plug domain reduces the ability of Sec61p to form stable translocons by interacting with partner proteins, among them the β and γ subunits.
DISCUSSION
It was unexpected that the complete deletion of the plug domain did not affect viability or growth at any temperature. Even though the primary sequence of the plug domain and the hinge segments are not well conserved, the structure as such is well conserved. The plug seems to close the pore in its idle state to small molecules. In E. coli, the synthetic lethality of prlA3 (F67C in the plug domain of SecY) with prlG3 (S120C at the periplasmic end of SecE) supported this notion (Flower et al., 1995; Harris and Silhavy, 1999), because formation of a disulfide bridge between the two cysteines locks the plug outside in a permanently open state. In yeast, the consequence of ion leakage from the ER is unclear, because the major Ca2+ stores are the vacuole and the Golgi, and leakage may be compensated by increased activity of Ca2+ ATPases. In addition, the constriction ring alone may block ion passage, and there is evidence in the mammalian system for an alternative sealing of the translocon by the lumenal chaperone BiP (Hamman et al., 1998; Haigh and Johnson, 2002; Alder et al., 2005).
The plug domain has also been proposed to play a role in gating the translocation channel for the translocation of polypeptides. This function does not seem to be essential, but its absence may be responsible for the effects on the topology of our model proteins. Mutations in the plug or at the interface to the plug are good candidates to destabilize the closed state of the translocon by disturbing the plug's fit into the lumenal cavity. Consistent with such a general mechanism of influencing topogenesis, all plug mutations analyzed, including the complete deletion, have similar effects on signal orientation. Previously, R67 and R74 in the plug were identified to contribute to orienting signals according to the positive-inside rule (Goder et al., 2004), and their general position in the structure seemed to be compatible with a charge effect on signal sequences. Because any mutation disturbing the plug also affects the correct positioning of these arginines, it is not formally possible to distinguish whether the topology effects are due to an altered electrical potential and/or to destabilization of the closed state of the translocon.
However, as a result of an increased flexibility of the structure in mutant translocons, the hydrophilic portions of substrate polypeptides may more easily insert into the pore, open it, and pass through, and hydrophobic segments may more readily exit into the lipid bilayer. As a consequence, the initial orientation in which a polypeptide enters the translocon might be favored. For the N-terminal signal-anchor of construct [Leu16](−3), there is evidence in mammalian cells that it initially engages with the translocon in an Nexo/Ccyt direction (exoplasmic N terminus, cytoplasmic C terminus), before it inverts its orientation (Goder and Spiess, 2003). Plug mutations indeed cause an increase in Nexo/Ccyt products of [Leu16](−3) (Figure 3) consistent with this expectation. Because there is no experimental data on how internal signals initially enter the translocon, the effect of increased translocon flexibility on their integration cannot be predicted.
Our results on the stability of plug mutants of Sec61p also point to a different role for the plug. In the closed state as represented in the crystal structure, the translocon is a compact helix bundle, supported and stiffened from the inside by the plug domain. In the active state, in contrast, it must have considerable flexibility as the transmembrane helices move to expand the central constriction and to open laterally to allow access to the lipid. The transition to the open state is accompanied or even triggered by the plug moving out. Deletion of the plug results in diminished steady-state levels of Sec61p, but the half-life of the assembled protein is not reduced. The deletion thus does not render the final translocon more sensitive to degradation. Instead, the efficiency of folding and heterooligomerization seems to be reduced. The Δplug mutant is essentially undetectable in the presence of wild-type Sec61p, except when the oligomerization partners Sbh1p and/or Sss1p are overexpressed. Sec61p without the plug domain seems to compete poorly with the wild-type protein for limiting components, among them the β and γ subunits, and is rapidly degraded. In the same vein, the increased flexibility of assembled plugless translocons may compromise association with the Sec62/63/71/72 complex for posttranslational translocation and explain the defect in CPY translocation.
Not surprisingly, the structurally more drastic deletion of TM2 has similar and more severe effects on translocon assembly and posttranslational insertion. ΔTM2 and ΔTM3 have been reported to have reduced binding to the Sec62 complex and to produce synthetic lethality with temperature-sensitive sec62–1 (Wilkinson et al., 2000). In addition, the assembled ΔTM2 translocon also has a reduced half-life.
In summary, Sec61p is surprisingly resistant to mutagenesis, because it can tolerate the deletion of entire transmembrane segments (TM2 or TM3; Wilkinson et al., 2000) or the whole plug domain, or mutation of many well conserved residues (Goder et al., 2004; Cheng et al., 2005) without dramatic growth defects. These mutations sufficiently preserve the essential functions of the translocon to provide a passage through the membrane and into the lipid bilayer and to connect to the ribosome and the Sec62/63/71/72 complex for the targeting of co- and posttranslational substrates.
ACKNOWLEDGMENTS
We thank Drs. E. Hartmann, H. Riezman, K. Römisch, R. Schekman, and C. Stirling for strains and reagents; Dr. Lorenza Bordoli for sequence alignment; and Thierry Schmidlin for critically reading the manuscript. This work was supported by Grant 3100A0-109424/1 from the Swiss National Science Foundation.
Abbreviations used:
- 5-FOA
5-fluoroorotic acid
- CPY
carboxypeptidase Y
- DPAPB
dipeptidyl aminopeptidase B
- ER
endoplasmic reticulum
- GPD
glyceraldehyde-3-phosphate dehydrogenase
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-03-0200) on July 5, 2006.
REFERENCES
- Alder N. N., Shen Y., Brodsky J. L., Hendershot L. M., Johnson A. E. The molecular mechanisms underlying BiP-mediated gating of the Sec61 translocon of the endoplasmic reticulum. J. Cell Biol. 2005;168:389–399. doi: 10.1083/jcb.200409174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon K. S., Or E., Clemons W. M., Jr., Shibata Y., Rapoport T. A. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J. Cell Biol. 2005;169:219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Jiang Y., Mandon E. C., Gilmore R. Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J. Cell Biol. 2005;168:67–77. doi: 10.1083/jcb.200408188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzer A. J., Nabholz C. E., Spiess M. Transmembrane orientation of signal-anchor proteins is affected by the folding state but not the size of the aminoterminal domain. EMBO J. 1995;14:6311–6317. doi: 10.1002/j.1460-2075.1995.tb00321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eusebio A., Friedberg T., Spiess M. The role of the hydrophobic domain in orienting natural signal sequences within the ER membrane. Exp. Cell Res. 1998;241:181–185. doi: 10.1006/excr.1998.4042. [DOI] [PubMed] [Google Scholar]
- Flower A. M., Osborne R. S., Silhavy T. J. The allele-specific synthetic lethality of prlA-prlG double mutants predicts interactive domains of SecY and SecE. EMBO J. 1995;14:884–893. doi: 10.1002/j.1460-2075.1995.tb07070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goder V., Junne T., Spiess M. Sec61p contributes to signal sequence orientation according to the positive-inside rule. Mol. Biol. Cell. 2004;15:1470–1478. doi: 10.1091/mbc.E03-08-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goder V., Spiess M. Molecular mechanism of signal sequence orientation in the endoplasmic reticulum. EMBO J. 2003;22:3645–3653. doi: 10.1093/emboj/cdg361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N., Peitsch M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Haigh N. G., Johnson A. E. A new role for BiP: closing the aqueous translocon pore during protein integration into the ER membrane. J. Cell Biol. 2002;156:261–270. doi: 10.1083/jcb.200110074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamman B. D., Hendershot L. M., Johnson A. E. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell. 1998;92:747–758. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]
- Harley C. A., Holt J. A., Turner R., Tipper D. J. Transmembrane protein insertion orientation in yeast depends on the charge difference across transmembrane segments, their total hydrophobicity, and its distribution. J. Biol. Chem. 1998;273:24963–24971. doi: 10.1074/jbc.273.38.24963. [DOI] [PubMed] [Google Scholar]
- Harris C. R., Silhavy T. J. Mapping an interface of SecY (PrlA) and SecE (PrlG) by using synthetic phenotypes and in vivo cross-linking. J. Bacteriol. 1999;181:3438–3444. doi: 10.1128/jb.181.11.3438-3444.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E., Rapoport T. A., Lodish H. F. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc. Natl. Acad. Sci. USA. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. E., van Waes M. A. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- Kraulis P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:936–950. [Google Scholar]
- Merritt E. A., Bacon D. J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- Notredame C., Higgins D. G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Pilon M., Schekman R., Römisch K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 1997;16:4540–4548. doi: 10.1093/emboj/16.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K., Mothes W., Wilkinson B. M., Stirling C. J., Rapoport T. A. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- Rösch K., Naeher D., Laird V., Goder V., Spiess M. The topogenic contribution of uncharged amino acids on signal sequence orientation in the endoplasmic reticulum. J. Biol. Chem. 2000;275:14916–14922. doi: 10.1074/jbc.M000456200. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M., Tomiyoshi R., Kuroiwa T., Mihara K., Omura T. Functions of signal and signal-anchor sequences are determined by the balance between the hydrophobic segment and the N-terminal charge. Proc. Natl. Acad. Sci. USA. 1992;89:16–19. doi: 10.1073/pnas.89.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A., Blundell T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sanner M. F., Olson A. J., Spehner J. C. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Schwede T., Kopp J., Guex N., Peitsch M. C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling C. J., Rothblatt J., Hosobuchi M., Deshaies R., Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol. Biol. Cell. 1992;3:129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P. C., Maillard A. P., Chan K. K., Duong F. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 2005 doi: 10.1038/sj.emboj.7600804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W. S. Scoring residue conservation. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- van den Berg B., Clemons W. M., Jr., Collinson I., Modis Y., Hartmann E., Harrison S. C., Rapoport T. A. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- von Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. The signal peptide. J. Membr. Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- Wahlberg J. M., Spiess M. Multiple determinants direct the orientation of signal-anchor proteins: the topogenic role of the hydrophobic signal domain. J. Cell Biol. 1997;137:555–562. doi: 10.1083/jcb.137.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson B. M., Tyson J. R., Reid P. J., Stirling C. J. Distinct domains within yeast Sec61p involved in post-translational translocation and protein dislocation. J. Biol. Chem. 2000;275:521–529. doi: 10.1074/jbc.275.1.521. [DOI] [PubMed] [Google Scholar]