Abstract
Differentiation of mammary epithelium in vivo requires signaling through prolactin- and ErbB4/HER4-dependent mechanisms; how these pathways intersect is unknown. We show herein that HC11 mouse mammary cells undergo ErbB4-dependent lactational differentiation. Prolactin and the ErbB4 ligand HB-EGF each induced STAT5A activation, expression of lactogenic differentiation markers, and lumen formation in three-dimensional Matrigel cultures in HC11 cells. ErbB4 undergoes ligand-dependent transmembrane domain cleavage at Val-675, releasing a soluble 80-kDa intracellular domain (s80HER4) that localizes to nuclei; the physiological relevance of s80HER4 is unknown. A HER4V675A mutant abolishing transmembrane cleavage impaired STAT5A activity, lactogenic gene expression, and lumen formation. Kinase-dead HER4KD was neither cleaved nor able to induce differentiation of HC11 cells. Without treating HC11 cells with prolactin or HB-EGF, s80HER4 (expressed from a cDNA construct) localized to the nucleus, activated STAT5A, and induced three-dimensional lumen formation. Nuclear localization of exogenous s80HER4 required intact kinase activity of s80HER4, as did activation of STAT5A. In contrast, nuclear localization of s80HER4 and STAT5A activation did not require the 16-amino acid region of the ErbB4 intracellular domain specific to the Cyt-1 isoform of ErbB4, and absent in the Cyt-2 isoform. These results suggest that s80HER4 formation contributes to ErbB4-dependent differentiation of mammary epithelial cells.
INTRODUCTION
Growth factors and hormones control the signaling networks that regulate the physiological cycles of proliferation, differentiation, and involution in the mammary epithelium. Alterations in these signaling networks could therefore affect the regulation of these processes, resulting in pathological consequences including cancer (Chodosh, 2002). Each member of the HER/ErbB family of receptor tyrosine kinases (RTKs) regulates different aspects of mammary development (Stern, 2003). The four family members, ErbB1/EGFR/HER1 (epidermal growth factor receptor), ErbB2/Neu/HER2, ErbB3/HER3, and ErbB4/HER4, each contain ligand-binding extracellular, transmembrane domain and intracellular tyrosine kinase domains. The ErbB receptors exhibit ligand-inducible dimerization, transphosphorylation, and tyrosine kinase activity, with the exception of ErbB3, for which tyrosine kinase activity has not been observed. Either homo- or heterodimers between family members can occur, with ErbB2 being the preferred heterodimeric partner. ErbB2 does not bind any conventional ligand, relying on ligand binding to ErbB1, ErbB3, or ErbB4 for tyrosine kinase induction. The multitude of potential ligand-driven ErbB dimers generates a variety of cellular responses.
ErbB receptors are activated by two ligand families: the epidermal growth factor (EGF) family (EGF, transforming growth factor-α [TGFα], amphiregulin [AR], heparin binding-EGF [HB-EGF], epiregulin, and betacellulin) and the neuregulins (heregulin [neuregulin-1], neuregulin-2, -3, and -4). In the breast epithelium, ErbB1 is activated by EGF, AR, and TGFα, and together they contribute primarily to ductal proliferation. HB-EGF binds to both ErbB1 and ErbB4, and contributes to differentiation via ErbB4. The neuregulins bind to both ErbB3 and ErbB4. Heregulin (HRG) regulates both proliferation and differentiation of breast epithelial cells (Marte et al., 1995; Yang et al., 1995; Jones et al., 1996).
ErbB4 signaling impairs growth of many human breast cancer cells (Sartor et al., 2001) and promotes differentiation of murine mammary glands (Jones et al., 1999; Long et al., 2003; Tidcombe et al., 2003). In vivo, ErbB4 expression and activity are lowest during phases of epithelial cell proliferation (puberty and early pregnancy) and highest during phases of differentiation (late pregnancy and early lactation; Schroeder and Lee, 1998). Mammary glands from mice that lack ErbB4 activity have lactational defects due to an impaired program of differentiation, measured by decreased expression of milk proteins and decreased activity of STAT5A, a transcription factor required for lactation (Jones et al., 1999; Long et al., 2003; Tidcombe et al., 2003). STAT5A deficiency results in failed lactogenesis, a phenotype reminiscent of ErbB4-deficient mammary glands (Liu et al., 1997; Shillingford et al., 2001; Cui et al., 2004).
Many ErbB4-positive breast cancers, as well as normal human and mouse mammary tissue, exhibit nuclear localization of ErbB4 (Srinivasan et al., 2000; Abd El-Rehim et al., 2004; Junttila et al., 2005). Recent work by several groups has described the mechanism by which specific ErbB4 splice variants translocate to the nucleus. Treatment of cells with ligand results in stepwise proteolysis of the JMa splice variant of ErbB4 (Elenius et al., 1997; Rio et al., 2000; Cheng et al., 2003), beginning with cleavage by TACE (tumor necrosis factor-α converting enzyme) and shedding of the ectodomain (Carpenter, 2003). This leaves an 80-kDa ErbB4 fragment tethered to the membrane (m80HER4). A second proteolytic event in the transmembrane domain of ErbB4, carried out by a γ-secretase (Ni et al., 2001, 2003; Lee et al., 2002), liberates the soluble cytoplasmic domain (s80HER4) with an active tyrosine kinase into the cytosol (Linggi et al., 2005). The JMb splice variant lacks the TACE cleavage sequence (Elenius et al., 1997; Rio et al., 2000). In a recently published study, mutation of a transmembrane valine residue (a putative γ-secretase cleavage site) impaired ErbB4 cleavage (Vidal et al., 2005). Because TACE-mediated cleavage of ErbB4 is prerequisite for γ-secretase cleavage, it is predicted that the JMb isoform would not yield s80HER4.
Localization of s80HER4 to both the cytosol and nucleus has been previously described. There are three nuclear localization sequences (NLS) within s80HER4, and three putative nuclear export sequences (Carpenter, 2003). Mutation of one of these NLS sequences prevented nuclear translocation of s80HER4 (Williams et al., 2004). The function of nuclear s80HER4 is not known, although recent reports suggest that s80HER4 may associate with the transcription factors STAT5A in the nucleus and transactivate gene expression (Williams et al., 2004; Vidal et al., 2005). Additionally, nuclear association of ErbB4 with the transcription factor YAP suggests that s80HER4 may participate in transcriptional regulation (Komuro et al., 2003).
In this report we describe experiments using the mouse mammary epithelial cell line HC11 as a model of mammary differentiation to investigate ErbB4 and its cleavage product, s80HER4, in transmitting differentiation signals. Multiple aspects of differentiation required ErbB4 kinase activity and cleavage of ErbB4 to produce s80HER4. Expression of s80HER4 was itself sufficient to initiate differentiation, whereas kinase-dead s80KD was unable to associate with or phosphorylate STAT5A, to differentiate HC11 cells, and accumulate in nuclei.
MATERIALS AND METHODS
Cell Culture
HC11 cells and all derivatives were cultured at 5% CO2 in growth media (DMEM/F12 [Invitrogen Life Sciences, Carlsbad, CA] supplemented with 10% fetal bovine serum [FBS], EGF [10 ng/ml; Invitrogen], insulin [5 μg/ml; Sigma-Aldrich, St. Louis, MO]), and hydrocortisone (1 μg/ml; Sigma-Aldrich) unless otherwise indicated. For some experiments, cells were cultured overnight in priming media [serum-free/EGF-free DMEM-F12 media supplemented with hydrocortisone (1 μg/ml) and insulin (5 μg/ml)] for 2 d and then treated for the indicated time points (30 min-2 d) with PRL (5 μg/ml; Sigma-Aldrich), EGF (10 ng/ml; R&D Systems, Minneapolis, MN), HRGβ1 (10 ng/ml; a gift from Genentech, San Francisco, CA), HB-EGF (10 ng/ml; Sigma-Aldrich). For three-dimensional (3D) cultures, 5 × 104 cells in 100 μl priming media supplemented with the indicated factors were resuspended in 100 μl cold growth factor-reduced Matrigel (BD Biosciences, San Jose, CA) and plated in one well of a 96-well plate precoated with growth-factor–reduced Matrigel. Cultures were allowed to polymerize at 37°C for 1 h, then overlaid with 50 μl priming media to prevent drying. Cells were cultured 10 d, then fixed in 1% paraformaldehyde (PFA) in phosphate-buffered saline (PBS), pH 7.4, stained with DAPI (Vector Laboratories, Burlingame, CA), and photographed with a Zeiss LCM210 using Scion Image 2.0 software (Thornwood, NY).
Western Analysis and Immunoprecipitation (IP)
Western analysis and immunoprecipitations (IPs) from whole cell lysates were performed as previously (Sartor et al., 2001) using the following antibodies: rabbit polyclonal anti-ErbB1, ErbB3, and ErbB4 (C-terminus) were generated in this laboratory and have been previously described (Sartor et al., 2001); ErbB2 (Neomarkers, Fremont, CA); STAT5A and phospho-Tyr 694 STAT5 A/B (Zymed Laboratories, South San Francisco, CA), ErbB4 (HFR; Neomarkers), GFP, β-casein, α-tubulin, c-jun, and phosphotyrosine (PY20; Santa Cruz Biotechnologies, Santa Cruz, CA), p44/42 (Cell Signaling Technology, Beverly, MA). NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce Biotechnology, Rockford, IL) were used to separate nuclear and cytoplasmic compartments of cells, according to the manufacturer’s protocol. For membrane versus cytosolic separation, cells (108) were lysed in ice-cold buffer A (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, and 250 mM sucrose, 1 mM sodium vanadate, and 1× protease inhibitor cocktail; Roche, Indianapolis, IN) using a 19-gauge needle. The samples were centrifuged at 1500 × g, 4°C for 10 min to pellet nuclei and cell debris. Membranes were pelleted from supernatants by centrifugation for 1 h at 10,000 × g in a Beckman ultracentrifuge. The resultant supernatant represented cytosolic fractions. The pellets, representing membrane fractions, were resuspended in 10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 250 mM sucrose, 1 mM sodium vanadate, and 1× protease inhibitor cocktail for 45 min at 4°C and then cleared by centrifugation.
Immunohistochemistry
Cells grown on glass chamber slides were fixed in 1% PFA, permeabilized in methanol:dimethylsulfoxide (1:1), washed twice with PBS, blocked for 1 h in 3% gelatin/0.2% Tween-20 in PBS (PBST), and then incubated overnight at 4°C in STAT5A antibody or ErbB4 antibody (1:70 in PBST). Slides were washed in PBST, incubated for 1 h at room temperature in rhodamine or Alexa488-conjugated anti-mouse IgG, washed, and mounted with DAPI+ Vectashield (Vector Laboratories). Cells were photographed using the Zeiss LCM 210 microscope and Scion Image 2.0 software (Scion, Frederick, MD).
Plasmids and Mutagenesis
pLXSN-HER4 and pLXSN-HER4KD cDNA have been described previously (Sartor et al., 2001). The V675A mutation of the P1′ valine (residue 675) was performed by PCR-assisted site-directed mutagenesis using the primer 5′-TTATGCTAGAAGGAAGAGCATCAAA-3′ (mutated residue underlined) and the Quick-change Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The intracellular domain of HER4, s80HER4 (residues 676-1308) fused at the NH2-terminus with GFP was subcloned into pcDNA4 (Invitrogen). Mutagenesis of residue 751 in the ATP-binding pocket of s80 was performed in pcDNA4-s80 as described above using the primer 5′ AGAATCCGAATAGCCACAGGAATCTTCAC-3′, resulting in a K-to-R mutation (mutated nucleotides underlined). PcDNA4-s80Cyt2 was generated from a PmeI fragment of pcDNA3–4ICD-Cyt2, provided by Dr. Klaus Elenius (Junttila et al., 2005), encoding s80Cyt2.
Transfections and Luciferase Assays
Cells, 5 × 105, were seeded into a 10-cm dish 24 h before transfection. Cells were transfected in 5 ml serum-free DMEM with 12 μl Fugene6 Transfection Reagent (Roche) and 5 μg plasmid DNA. For generation of stable cell lines, cells were selected with either G418 (500 μg/ml) or zeocin (1 μg/ml). For transient transfection assays, cells were treated with factors in serum-free media 24 h after transfection. Cells were harvested 48 h after transfection. Luciferase assays were performed using 200 μg protein (Luciferase Assay Kit; Promega, Madison, WI) according to the manufacturer’s instructions.
Reverse Transcription-Polymerase Chain Reaction
Total RNA was extracted using TRIzol Reagent (Invitrogen). RT-PCR was performed using total RNA (1 μg per 50 μl reaction) in the One-Step RT-PCR kit (Qiagen, Valencia, CA) according to manufacturer’s suggestions using gene-specific primers: mouse β-casein (forward 5′ACT ACA TTT ACT GTC TCC TCT GAG and reverse 5′ GTG CTA CTT GCT GCA GAA AGT ACA G, producing a 534-base pair amplicon); mouse keratin 18 (forward 5′ CGC ATC GTC TTG CAG ATC GAC A, and reverse 5′ GCT GAG ACC AGT ACT TGT CCA G, producing a 375-base pair amplicon). RT-PCR products were resolved on 2% agarose gels.
RESULTS
ErbB4 Ligands Induce Differentiation of HC11 Cells
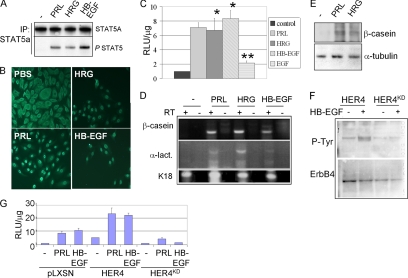
Evidence from transgenic mouse models demonstrated the requirement for ErbB4 in lactogenic differentiation of the mammary gland. We used the HC11 cell line, derived from the mammary gland of a female Balb/C mouse at midpregnancy, as an ex vivo model of mammary epithelial cell differentiation to determine the role of s80HER4 in this process. HC11 cells have been widely used as a model of prolactin (PRL)-induced mammary epithelial cell differentiation. To determine if ErbB4-activating ligands were able to induce a differentiated phenotype in HC11 cells, we initially treated cells with HRG, HB-EGF, and PRL to observe the effects of these ligands on STAT5A activation, using phosphorylation at Tyr-694 and nuclear translocation as surrogate markers of STAT5A activity. STAT5A was phosphorylated at Tyr-694 after a 30 min treatment with PRL and with the ErbB4 ligands HRG and HB-EGF (Figure 1A). Although STAT5A was primarily cytoplasmic in serum-starved HC11 cells, treatment with PRL for 30 min resulted in nuclear translocation of STAT5A (Figure 1B). HRG and HB-EGF also induced nuclear translocation of STAT5A (Figure 1B). Ligand-induced STAT5A-mediated transcriptional activation was examined in transient transfection assays using a reporter construct comprised of residues (−294 to +1) of the human β-casein gene driving the Photinus luciferase cDNA (pβcasein-lux). This region of the β-casein gene harbors STAT5A DNA-binding sites (Winklehner-Jennewein et al., 1998). PRL treatment for 24 h increased luciferase activity 7.1-fold compared with untreated cells, as did HRG (6.7-fold) or HB-EGF (8.3-fold; Figure 1C). Cells treated with EGF, an ErbB1-specific ligand, induced luciferase activity to a lesser extent (2.1-fold), suggesting that ErbB1 homodimers are not responsible for maximal induction of STAT5-mediated transcription.
Figure 1.
ErbB4 ligands activate STAT5A in HC11 cells. (A) STAT5A immunoprecipitates (IP) from HC11 cells cultured in serum-free (SF) media with PRL, HRG, or HB-EGF for 30 min were analyzed by immunoblot (IB) with indicated antibodies at the right of each panel. (B) Immunohistochemistry detecting STAT5A localization in serum-starved HC11 cells treated with indicated factors for 30 min. Scale bar, 100 μm. (C) HC11 cells transfected with the STAT5A-responsive promoter, pβcasein-lux, and cultured 48 h in SF media. Agonists were added as indicated for the final 24 h of the experiment. Luciferase activity is shown as the average relative light units (RLU) per μg of total protein ± SD. Experiments were repeated three times, with each sample being analyzed in triplicate. *p < 0.003 versus control. **p < 0.01 versus control. Student’s unpaired t test. (D and E) Confluent HC11 cells cultured in priming media for 2 d and then in priming media ± PRL, HRG, or HB-EGF (where indicated) for an additional 2 d. (D) RT-PCR analysis used primer sets to detect expression of β-casein, α-lactalbumin (α-lact.), or keratin 18 (K18). For each sample analyzed, the reverse transcriptase (RT) was either included (+) or left out (−) of the reaction. (E) Whole cell extracts were analyzed by Western analysis using antibodies against β-casein and α-tubulin. (F) HC11 cells were stably transfected with a pLXSN-derived construct encoding human ErbB4/HER4 or a variant harboring a point mutation at lysine 751 [rendering the product kinase-dead (HER4KD)]. Pooled clones of HC11-HER4 and HC11-HER4KD cells were analyzed by Western blot to detect tyrosine phosphorylation of ErbB4 immunoprecipitates. Serum-starved cells were treated with HB-EGF for 10 min. Blots were stripped and reprobed with a mAb against ErbB4. (G) Pooled clones of HC11-pLXSN, HC11-HER4, or HC11-HER4KD cells were transfected with pβcasein-lux and then treated with PRL or HB-EGF for 24 h, measuring luciferase activity as described above.
β-casein and α-lactalbumin are milk proteins whose expression is induced upon lactogenic differentiation of HC11 cells. Thus, we examined RNA expression of the endogenous β-casein and α-lactalbumin genes as a physiological marker of lactogenic differentiation. RT-PCR analysis demonstrated that PRL treatment (48 h) induced β-casein and α-lactalbumin mRNAs. HRG and HB-EGF also induced β-casein and α-lactalbumin mRNAs (Figure 1D). Expression of keratin 18 (K18) transcripts was detected in all samples. Expression of the β-casein protein was induced in HC11 cells treated 48 h with PRL or HRG (Figure 1E). These results suggest that ErbB4-activating ligands (HRG and HB-EGF) result in activation of STAT5A similar to PRL. Because neither ErbB2 nor ErbB3 bind to HB-EGF and EGF did not result in high levels of STAT5A-mediated transcription or β-casein expression, this indirectly suggests that ErbB4 signaling is responsible for the nuclear translocation and transcriptional activity of STAT5A in response to these ligands. The results, however, do not discriminate between the possibilities that ErbB4 is functioning as a homodimer or in heterodimeric concert with other ErbB receptors.
To determine the effects of impaired ErbB4 activity on HRG and HB-EGF–induced signaling in HC11 cells, we generated pools of stable cell lines expressing overexpressing wild-type HER4 or kinase-dead HER4KD. Ligand-induced ErbB4/HER4 tyrosine phosphorylation was impaired in cells overexpressing HER4KD (HC11-HER4KD; Figure 1F). HC11-HER4KD cells were transfected with the pβcasein-lux (Figure 1G). As a control for ectopic overexpression of HER4KD, HC11-HER4 cells were examined in parallel. Treatment of HC11-HER4 cells with PRL resulted in a 12.4-fold increase in luciferase activity, somewhat greater than the levels of PRL-induced luciferase activity seen in HC11-pLXSN cells (Figure 1G). This suggests that wild-type HER4 overexpression may enhance PRL-mediated STAT5A activity. In contrast, treatment of HC11-HER4KD cells with PRL induced luciferase activity 4.8-fold, a decrease when compared with luciferase acitivity from PRL-treated HC11-pLXSN cells, suggesting that HER4KD expression partially impaired PRL-dependent activation of this promoter. HB-EGF-induced STAT5A activation was similarly increased in HC11-HER4 cells, and expression of HER4KD abolished promoter transactivation in response to HB-EGF (Figure 1G).
Morphological Differentiation of HC11 Cells in Response to ErbB4
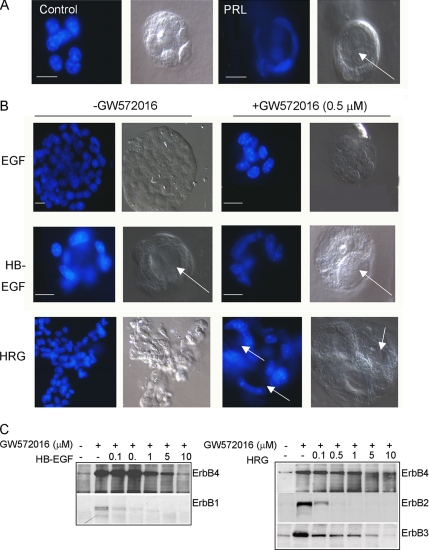
HC11 cells were embedded in serum-free, growth factor-reduced Matrigel (an extracellular matrix) for 10 d to examine morphological differentiation in three dimensions. Cells grown in serum-free media containing insulin and hydrocortisone grew as disorganized colonies, consisting of 14.8 ± 4.5 cells per colony on average (Figure 2A). Addition of PRL reduced the number of cells per colony (10.7 ± 4.2) and resulted in the organization of cells into a single cell layer surrounding a single lumen.
Figure 2.
ErbB4 activity is required for 3D lumen formation. (A) 3D culture of HC11 cells in growth factor-reduced Matrigel ± PRL. Cells were cultured 10 d, stained with DAPI, and photographed using a Zeiss LCM-210; DAPI and DIC images are shown. Arrows indicate lumen formation. Scale bars, 25 μm. (B) 3D culture of HC11 cells in growth factor-reduced Matrigel ± EGF, HB-EGF, or HRG, and ±GW572016 (0.5 μM). Cells were cultured 10 d, stained with DAPI, and photographed using a Zeiss LCM-210; DAPI and DIC images are shown. Arrows indicate lumen formation. Scale bars, 25 μm. (C) HC11 cells were serum/EGF-starved in the presence or absence of increasing concentrations of GW572016 (0–10 μM) for 16 h, and treated for 30 min with HB-EGF or HRG. ErbB1, ErbB2, ErbB3, and ErbB4 immunoprecipitates (IPs) were analyzed by immunoblot for phosphotyrosine.
The addition of EGF to HC11 cells in 3D culture for 10 d resulted in increased cell number per colony (38.6 ± 9.1) compared with untreated controls (Figure 2B). The EGF-treated colonies remained cellular masses without lumens. The EGF-induced increase in cell number per colony was inhibited by the addition of the ErbB1/ErbB2 inhibitor GW572016 at 0.5 μM (12.8 ± 3.8 cells per colony; Figure 2B). Similar to PRL, addition of HB-EGF decreased the number of cells per colony (9.4 ± 3.2) compared with untreated controls and resulted in the organization of cells into a single epithelial layer surrounding a lumen. Lumen formation in response to HB-EGF was not inhibited by 0.5 μM GW572016. HRG increased the number of cells per colony (36.9 ± 11.6) compared with untreated controls and caused colonies to grow as branched, solid cords. The increased growth of HRG-treated cells was inhibited by 0.5 μM GW572016 (16.8 ± 7.2; Figure 2B). Interestingly, HRG-treated cultures grown with 0.5 μM GW572016 formed multilobulated structures comprised of a single-cell layer surrounding multiple lumens. This suggests that reduction in HER2 signaling (see below) allows the HRG-dependent HER4 morphological differentiation signal to predominate.
Published data indicate that GW572016 inhibits human ErbB1 and ErbB2 enzymatic activity more potently than it does ErbB4 (5–10-fold higher Ki; Xia et al., 2002), but this has not been tested with regard to mouse ErbB receptors. We incubated cells with increasing concentrations of GW572016 for 16 h, treated cells with HRG or HB-EGF for 30 min, and examined receptor phosphorylation. Treatment of HC11 cells with GW572016 at 0.1 μM significantly inhibited ErbB1 and ErbB2 tyrosine phosphorylation and at 0.5 μM reduced tyrosine phosphorylation of ErbB3, but not that of ErbB4 (Figure 2C). Note that ErbB3 is itself devoid of tyrosine kinase activity, but supports the activity of other ErbB receptors. ErbB4 tyrosine phosphorylation was observed with concentrations of GW572016 as high as 5 μM. Although EGF-dependent activation of p44/42 MAP kinase was inhibited in cells treated with 0.5 μM GW572016, PRL-mediated MAP kinase activation was not impaired by GW572016 (unpublished data), demonstrating the specificity of the inhibitor. GW572016 (0.5 μM) did not impair lumen formation by HB-EGF and actually enhanced it in HRG-treated cells; this suggests that ErbB4, but not ErbB1 or ErbB2, plays a prominent role in structural differentiation of mammary epithelial cells in response to these ligands.
Differentiation of HC11 Cells Is Induced by Expression of s80HER4
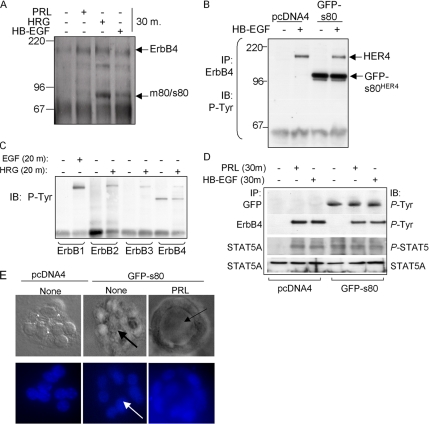
Previous reports demonstrate that ErbB4 undergoes HRG-inducible proteolytic processing, releasing the soluble ErbB4 80-kDa intracellular domain (s80HER4) into the cytoplasm. We determined if HB-EGF or HRG could result in cleavage of endogenous ErbB4 to generate s80HER4 by treating serum-starved HC11 cells with ligand (30 min) and then analyzing ErbB4 immunoprecipitates by Western blot with an antibody against the ErbB4 intracellular domain. A single immunoreactive species was detected at 180 kDa in untreated cells, consistent with the size of endogenous ErbB4 (Figure 3A). Within 30 min of HRG treatment, 180- and 80-kDa immunoreactive products were identified. To a lesser extent, HB-EGF induced the formation of the 80-kDa product; we were unable to detect the 80-kDa isoform in response to PRL after 30 min.
Figure 3.
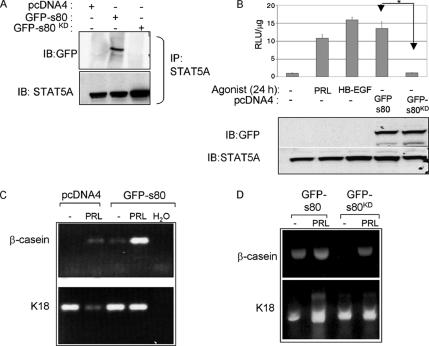
Expression of s80HER4 induces ligand-independent STAT5A phosphorylation. (A) HC11 cells serum-starved overnight, treated with PRL, HRG, or HB-EGF for 30 min. ErbB4 was immunoprecipitated with a polyclonal ErbB4 antibody; immunocomplexes were analyzed by Western blot using a monoclonal ErbB4 antibody. Molecular weights (kDa) are indicated at left. The predicted sizes of the membrane bound full-length ErbB4/HER4 and an 80-kDa cleavage product that is either membrane-bound or soluble (m80HER4/s80HER4) are indicated at right. (B) HC11 cells were stably transfected with pcDNA4 or pcDNA4-s80, encoding GFP-tagged s80HER4. Pooled clones of stably transfected cells were grown in serum-free media overnight and then treated ± HB-EGF for 30 min. ErbB4 was immunoprecipitated using a polyclonal antibody. Immunoprecipitates were analyzed by Western with an antibody against phosphotyrosine. (C) Western analysis to detect tyrosine phosphorylation of ErbB1, ErbB2, or ErbB3 immunoprecipitates from HC11-GFP-s80 cells treated ± EGF or HRG where indicated. (D) Pools of HC11-pcDNA4 or HC11-GFP-s80 cells were serum-starved and then treated with PRL or HB-EGF for 30 min. Western analysis of ErbB4 or GFP immunoprecipitates to detect phosphotyrosine resides; Western analysis of STAT5A immunoprecipitates to detect total STAT5A and phosphotyrosine 694 STAT5A/B. (E) 3D culture of HC11 cells in growth factor-reduced Matrigel ± PRL. Cells were cultured 10 d, stained with DAPI, and photographed; DAPI and DIC images are shown. Arrows indicate lumen formation.
HC11 cells were stably transfected with a cDNA encoding the entire intracellular domain of human HER4 in frame with green fluorescent protein (GFP; Figure 3B). The GFP tag was at the NH2-terminus of s80HER4, so as not to disrupt the COOH-terminal PDZ-binding domain, which may be important in signaling (Ni et al., 2003). In these cells, GFP-s80HER4 was constitutively tyrosine phosphorylated (Figure 3B), consistent with recent reports that s80HER4 is a constitutively active tyrosine kinase capable of autophosphorylation (Linggi et al., 2005); however, endogenous full-length ErbB4 was not basally phosphorylated by GFP-s80HER4, nor did GFP-s80HER4 expression interfere with or enhance full-length ErbB4 phosphorylation in response to HB-EGF. Phosphorylation of ErbB1 in response to EGF or of ErbB2 and ErbB3 in response to HRG was not affected by expression of GFP-s80HER4 in HC11 cells (Figure 3C), nor was GFP-s80HER4 found to coprecipitate with ErbB1, -2, or -3 (Figure 3C and unpublished data). Although PRL induced the Tyr-694 phosphorylation of STAT5 in HC11-pcDNA4 cells, STAT5A phosphorylation was not detected in untreated HC11-pcDNA4 cells (Figure 3D), similar to parental HC11 cells (see Figure 1A). In contrast, constitutive STAT5A phosphorylation was seen in HC11-GFP-s80 cells in the absence of exogenous ligand (Figure 3D). This suggests that expression of s80HER4 is sufficient to induce STAT5A phosphorylation under basal conditions. Interestingly, these studies found that ErbB4 became tyrosine phosphorylated in response to PRL, although the mechanism for this is currently unknown (Figure 3D).
In 3D Matrigel cultures, HC11-pcDNA4 cells formed disorganized colonies in the absence of additional factors (Figure 3E), similar to untreated parental HC11 cells (as shown in Figure 2A). HC11-GFP-s80 cells grown in 3D colonies formed rudimentary lumens in the absence of exogenous ligand (Figure 3E) and contained fewer cells than untreated HC11-pcDNA4 cells (9.1 ± 2.6 vs. 14.8 ± 4.5, respectively; p = 0.02, Student’s unpaired t test). Lumen formation was enhanced by the addition of exogenous PRL, without statistically altering the number of cells per colony versus untreated GFP-s80HER4 cells (8.3 ± 3.4; p = 0.26, Student’s t test). These results suggest that expression of s80HER4 leads to growth inhibition and enhances acinar formation of HC11 cells in 3D cultures.
Kinase Activity of s80HER4 Is Required for Nuclear Translocation and STAT5A Activation
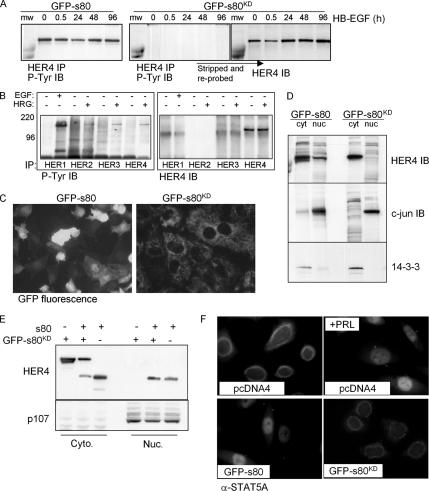
We constructed pcDNA4-s80KD, encoding GFP-tagged kinase-dead s80HER4 (s80KD), harboring a single Lysine 751 to Arginine point mutation in the ATP-binding pocket to disrupt kinase activity. Stable transfection of HC11 cells with this construct generated HC11-GFP-s80KD cells. Although GFP-s80HER4 was tyrosine phosphorylated in the presence or absence of HB-EGF, GFP-s80KD was not basally tyrosine phosphorylated, nor was it tyrosine phosphorylated in cells treated with HB-EGF for up to 96 h (Figure 4A). This is consistent with a recent report demonstrating that Lys 751 to Ala mutation within s80HER4 impairs tyrosine phosphorylation in vivo and kinase activity in vitro (Linggi et al., 2005). Although GFP-s80KD was not tyrosine phosphorylated, expression of GFP-s80KD did not interfere with ligand-induced tyrosine phosphorylation of full-length ErbB4 or ErbB1, ErbB2, or ErbB3 (Figure 4B). Consistent with previous reports describing nuclear localization of s80HER4, GFP fluorescence of live cells revealed cytoplasmic and nuclear localization of GFP-s80HER4 (Figure 4C). In contrast, GFP-s80KD was strictly localized to the cytoplasmic compartment of cells. Biochemical separation of HC11-GFP-s80KD cells into nuclear and cytoplasmic compartments also revealed that GFP-s80KD was strictly limited to the cytoplasm, but that GFP-s80HER4 was present in both nuclei and cytoplasm (Figure 4D). This is the first demonstration that kinase activity of s80HER4 is required for its nuclear translocation. To determine if kinase-dead GFP-s80KD could translocate to the nucleus in the presence of active s80HER4, we coexpressed an untagged s80 together with GFP-tagged s80KD (Figure 4E). Although untagged s80HER4 was found in both nuclear and cytoplasmic compartments, GFP-s80KD was never detected in the nucleus, regardless of coexpression with s80HER4, confirming that kinase activity of s80 is required for its nuclear localization. Immunofluorescent localization of phopsho-STAT5 was examined in HC11-GFP-s80 and HC11-GFP-s80KD cells. Although 16.7 ± 3.4% of HC11-GFP-s80 cells displayed basal nuclear localization of phospho-STAT5, nuclear localization of phospho-STAT5 was not detected in HC11-GFP-s80KD cells (Figure 4F).
Figure 4.
Kinase activity of s80HER4 is required for its nuclear localization and tyrosine phosphorylation. (A) HC11-GFP-s80 and HC11-GFP-s80KD cells were serum-starved overnight and then treated with HB-EGF for 0–96 h. IP was performed using an anti-HER4 polyclonal antibody and then analyzed by IB for phosphotyrosine. Blots were stripped and reprobed with a monoclonal anti-HER4 antibody. n.s., nonspecific band. (B) HC11-GFP-s80KD cells were serum-starved overnight and then treated ± ligand for 30 min before the immunoprecipitation of ErbB1, -2, -3, and -4 from low-salt lysates. Immunoprecipitates were analyzed by Western analysis with a phosphotyrosine antibody. (C) HC11-GFP-s80 and HC11-GFP-s80KD cells were analyzed for expression of GFP by live cell fluorescence microscopy. (D) HC11-GFP-s80 and HC11-GFP-s80KD cells were separated into cytoplasmic and nuclear fractions, which were analyzed by Western analysis using HER4, c-jun, and 14–3-3 antibodies. (E) HC11 cells were transiently transfected with pcDNA4-s80 (not GFP-tagged), pcDNA4-GFP-s80KD, or with both together. Cells were separated in cytoplasmic and nuclear fractions then analyzed by Western blot with antibodies against ErbB4 and p107. (F) Immunofluorescence detection of STAT5A in HC11-GFP-s80 and HC11-GFP-s80KD cells.
STAT5A immunoprecipitates were examined by Western analysis to detect a potential STAT5A-s80HER4 association. Although STAT5A coprecipitated with GFP-s80HER4 from HC11-GFP-s80 cells, GFP-s80KD was not detected in STAT5A immunocomplexes (Figure 5A), despite equal levels of STAT5A immunoprecipitation. This is consistent with the idea that SH2-domains within STAT5A mediate interactions with phosphotyrosine residues within ErbB4. We examined STAT5A-mediated transcription using the pβcasein-lux reporter in parental HC11 cells (Figure 5B). Expression of GFP-s80HER4 resulted in a 12.6-fold increase in luciferase activity in the absence of ligand. In contrast, expression of the kinase-dead GFP-s80KD was unable to increase luciferase activity, even though GFP-s80HER4 and GFP-s80KD were expressed at similar levels. These data suggest that kinase activity is critical for ligand-free, s80HER4-mediated activation of this STAT5-dependent promoter.
Figure 5.
Expression of s80HER4 is sufficient to induce differentiation of HC11 cells. (A) Low-salt lysates (125 mM NaCl) of cells grown in serum-free media were immunoprecipitated with a monoclonal STAT5A antibody. Immunoprecipitates were analyzed by Western using a rabbit GFP antibody or a mouse STAT5A antibody. (B) Cells were transiently transfected with pβcasein-lux and then treated ± PRL or HB-EGF (HC11-pcDNA4 cells only) or were maintained in serum-free media for the final 24 h of culture. Top, luciferase activity was determined as described above. *p < 0.001. Student’s t test. n = 3. Top, Western analysis of cell extracts using a GFP antibody detected GFP-tagged s80HER4 or s80KD (top); Western analysis using a STAT5A antibody demonstrated equal amounts of STAT5A in each lane (bottom). (C and D) RT-PCR analysis to detect expression of β-casein or K18. (C) HC11-pcDNA4 and HC11-GFP-s80 were cells grown to confluence, serum-starved, and then treated for 2 d in the presence or absence of PRL. Where indicated, water was added to the RT-PCR reaction as a negative control. (D) RNA harvested from HC11-GFP-s80 and HC11-GFP-s80KD cultured ± PRL were analyzed for expression of K18 or β-casein as described above.
Endogenous β-casein gene expression was examined in HC11-GFP-s80HER4 cells (Figure 5C). β-casein mRNA was detected in HC11-pcDNA4 cells treated with PRL, but not in untreated cells. However, β-casein expression was detected in untreated HC11-GFP-s80HER4 cells and was further elevated with PRL. In contrast, HC11-GFP-s80KD cells did not exhibit basal expression of β-casein, whereas PRL-induced expression of β-casein was observed in HC11-GFP-s80KD cells (Figure 5D). These results suggest that kinase activity of s80HER4 is required for STAT5A-mediated differentiation. Because tyrosine kinase activity is required for both s80HER4 nuclear localization and for interaction with STAT5A, it is possible that inhibition of s80HER4 nuclear localization per se would be sufficient to impair STAT5A nuclear localization and transcriptional activation.
Differentiation of HC11 Cells Requires Transmembrane Cleavage of ErbB4
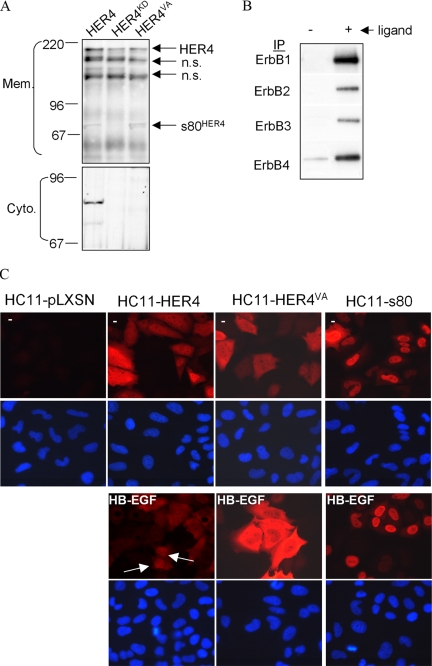
The human and mouse ErbB4 transmembrane domains harbor a canonical γ-secretase cleavage sequence also found in Notch and amyloid precursor protein (APP), among others (Carpenter, 2003). In proteins that are processed by γ-secretase, the P1′ Val residue (nearest the cytoplasmic domain) is critical for γ-secretase–mediated processing, such that its mutation abolishes transmembrane cleavage by γ-secretase. We generated a Val 675 (the P1′ Val) to Ala point mutation within the transmembrane domain of human ErbB4/HER4 (JMa/Cyt-1 isoform) in pLXSN, thus eliminating the putative ErbB4 γ-secretase cleavage site. Wild-type human HER4, kinase-dead HER4KD (Sartor et al., 2001), and HER4VA were overexpressed in pooled clones of HC11 cells by stable transfection (Figure 6A). We treated serum-starved HC11-HER4, HC11-HER4KD, and HC11-HER4VA cells with HB-EGF (30 min) and then separated cells into membrane and cytosolic fractions (Figure 6A). The 180- and 80-kDa ErbB4 products were detected in membrane preparations of HC11-HER4. We observed an accumulation of the 80-kDa ErbB4 product in membrane preparations of HC11-HER4VA cells. In contrast, only the 180-kDa isoform of ErbB4 was detected in cells expressing HER4KD. This is consistent with previous reports that preliminary cleavage events may require kinase activity of ErbB4. Although membrane-bound 80-kDa ErbB4 was detected in HC11-HER4 and to a lesser extent in HC11-HER4VA cells, cytoplasmic 80-kDa ErbB4 was only detected in HC11-HER4 cells, suggesting that the γ-secretase cleavage site was altered by mutation of Val 675. This is similar to results of other groups, in which a Val to Ile point mutation was introduced into the human HER4 sequence at Val 673 (Vidal et al., 2005). Other groups have also shown the accumulation of a membrane bound 80-kDa HER4 product when cells were treated with a chemical γ-secretase inhibitor, which inhibited the formation of soluble s80HER4 (Maatta et al., 2006).
Figure 6.
A cleavage of ErbB4 is required for differentiation of HC11 cells. (A) Pools of HC11 cells stably transfected with pLXSN-HER4, -HER4KD, or -HER4VA were cultured overnight in serum-free media and treated for 30 min with HB-EGF. Membrane and cytoplasmic extracts were fractionated and analyzed by immunoblot (IB) using an ErbB4 mAb (Neomarkers). Molecular weights are shown at left. n.s., nonspecific band. (B) Western analysis to detect tyrosine phosphorylation of ErbB1, -2, -3, and -4 IPs from serum-starved HC11-HER4VA cells treated with EGF (ErbB1) or HRG (ErbB2–4) for 10 min. (C) Serum-starved cells cultured 30 min ± HB-EGF. Arrows indicate nuclear localization. DAPI staining of nuclei is shown in bottom panels. Indirect immunofluorescence to detect ErbB4 is shown in the top panels.
ErbB1-4 immunoprecipitates from HC11-HER4VA cells were examined for tyrosine phosphorylation by Western analysis. EGF increased tyrosine phosphorylation of ErbB1 (Figure 6B), whereas HRG increased tyrosine phosphorylation of ErbB2 and ErbB3. Basal levels ErbB4 tyrosine phosphorylation were observed and may reflect the high level of ErbB4/HER4 expression in these cells.
Immunofluorescence detecting ErbB4/HER4 was used to examine its subcellular localization. ErbB4 was below the level of detection in HC11-pLXSN cells (Figure 6C). In the absence of stimulation, ErbB4/HER4 was diffusely cytoplasmic in HC11-HER4 and HC11-HER4VA cells, but was cytoplasmic and nuclear in HC11-s80 cells (Figure 6C, top panels). Although treatment of HC11-HER4 cells with HB-EGF resulted in nuclear accumulation of ErbB4 in 28.9 ± 6.4% of all cells, ErbB4 was never detected in the nucleus of HC11-HER4VA cells in response to HB-EGF (Figure 6C, bottom panels), nor was nuclear ErbB4 detected in TPA-treated HC11-HER4VA cells (unpublished data).
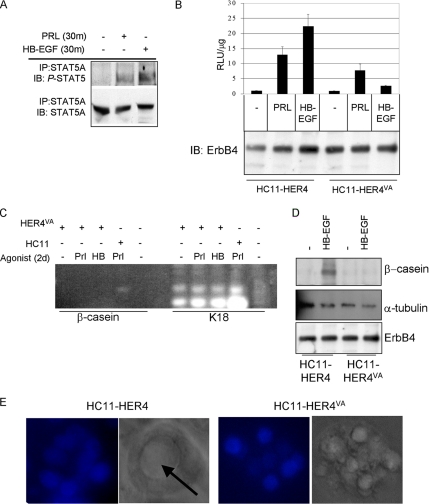
HC11-HER4VA cells displayed STAT5A phosphorylation in response to PRL and HB-EGF (Figure 7A). However, STAT5-dependent transcription in HC11-HER4VA cells was impaired in response to HB-EGF compared with HC11-HER4 cells, despite equal levels of HER4/ErbB4 expression in each cell line (Figure 7B). Additionally, PRL-induced transcription was moderately decreased in HC11-HER4VA cells compared with HC11-HER4 cells. Although keratin 18 mRNA was constitutively expressed in HC11-HER4VA cells, the induced expression of β-casein mRNA was not observed in response to HB-EGF or PRL (Figure 7C). Although HB-EGF induced the expression of β-casein protein in HC11-HER4 cells, β-casein was not detected in protein extracts from HB-EGF–treated HC11-HER4VA cells (Figure 7D). Finally, HC11-HER4 cells were able to organize into lumen-containing structures in Matrigel supplemented with HB-EGF, whereas HC11-HER4VA cells continued to grow as disorganized colonies in the presence of HB-EGF (Figure 7E). Taken together, all the results suggest that s80HER4 production from ErbB4 is necessary to elicit full differentiation of mammary epithelial cells in response to ErbB4 ligands. These data are consistent with reports from others suggesting s80HER4 may act as a nuclear chaperone to STAT5A, because HER4VA does not allow for nuclear translocation of the ErbB4 intracellular domain (Williams et al., 2004).
Figure 7.
Cleavage of ErbB4 at V675 is required for ErbB4-mediated differentiation. (A) Pooled clones of HC11-HER4VA cells were serum-starved overnight and then treated for 30 min with PRL or HB-EGF. STAT5A immunoprecipitates (IP) were analyzed by immunoblot (IB) to detect phospho-STAT5A/B or STAT5A. (B) HC11-HER4 and HC11-HER4VA cells were transiently transfected with pβcasein-lux. Cells remained in serum-free media or were treated with the indicated factors for the final 24 h of the experiment. Luciferase activity was determined in extracts as described above (top panel). Extracts were further examined for total levels of HER4/ErbB4 expression by immunoblot using a monoclonal ErbB4 antibody (bottom panel). (C) RT-PCR analysis to detect expression of β-casein or K18 in HC11 or HC11-HER4VA cells treated with PRL or HB-EGF (HB). (D) Western analysis to detect expression of total HER4 levels and β-casein or α-tubulin in extracts from HC11-HER4 and HC11-HER4VA cells cultured in priming media for 48 h and then in priming media ± HB-EGF for an additional 48 h. (E) 3D culture of HC11-HER4 or HC11-HER4VA cells grown in growth factor-reduced Matrigel in the presence of HB-EGF for 10 d. Arrow indicates lumen formation. DAPI and DIC images are shown.
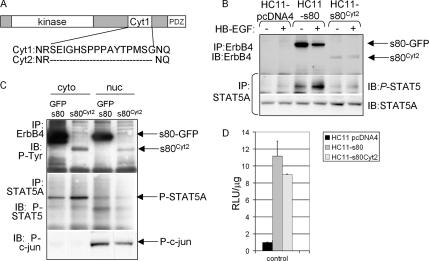
STAT5A Activation via s80HER4 Does Not Require the 16-Amino Acid Region Specific for the s80Cyt1 Variant, but Lacking in the s80Cyt2 Splice Variant
Two intracellular ErbB4 splice variants have been described, Cyt1 (encoding the entire intracellular domain) and Cyt2, which lacks a 16-amino acid sequence (Figure 8A; Kainulainen et al., 2000). All data presented thus far are from experiments using the Cyt1 isoform, referred to as s80HER4. To determine if the 16 amino acids present in s80HER4 but absent in s80Cyt2 are required for s80HER4-mediated STAT5A activation, we expressed s80Cyt2 in HC11 cells by stable transfection with pcDNA4-s80Cyt2 (Figure 8B). As with GFP-s80HER4, constitutive tyrosine phosphorylation of s80Cyt2 was observed (note that s80Cyt2 was not tagged with GFP). Also like GFP-s80HER4, expression of s80Cyt2 resulted in basal, ligand-free Tyr-694 phosphorylation of STAT5A, whereas HC11-pcDNA4 cells displayed only ligand-dependent STAT5A phosphorylation (Figure 8B). We also observed nuclear localization of s80Cyt2 (Figure 8C). Nuclear STAT5A was detected in untreated HC11-s80Cyt2 cells, albeit at lower levels than what was seen in HC11-GFP-s80 cells. Although this observation may be due to intrinsic differences between s80HER4 and s80Cyt2, it is possible that this may have been due to the observed lower expression levels of s80Cyt2 compared with GFP-s80HER4. We examined STAT5A-mediated transcription using the pβcasein-lux reporter in parental HC11 cells (Figure 8D). Expression of GFP-s80HER4 resulted in a 11.1-fold increase in luciferase activity in the absence of ligand, whereas expression of s80Cyt2 increased luciferase activity 8.8-fold higher than untreated HC11-pcDNA4 cells. These data suggest that the 16-amino acid sequence absent in s80Cyt2 is not necessary for s80HER4-mediated activation of this STAT5-dependent promoter.
Figure 8.
Expression of s80Cyt2 induces STAT5A activity in HC11 cells. (A) Schematic diagram of s80HER4 (i.e., the intracellular domain of HER4) indicating the two naturally occurring cytoplasmic splice variants of human ErbB4/HER4. (B) Western analyses of ErbB4 or STAT5A immunoprecipitates from cells cultured overnight in serum-free media and then treated ± HB-EGF for 30 min. ErbB4 immunoprecipitates (made with a polyclonal ErbB4 antibody) were analyzed by Western with a monoclonal ErbB4 antibody. Stat5A immunoprecipitates were analyzed by Western with antibodies against phospho-STAT5 or STAT5A. (C) HC11-s80 and HC11-s80Cyt2 cells were separated into cytoplasmic and nuclear lysates, which were used for immunoprecipitation (described below) or directly for Western with an antibody against phospho-c-jun. ErbB4 immunoprecipitates were analyzed by Western analysis for phospho-tyrosine; STAT5A immunoprecipitates were analyzed by Western for phospho-STAT5. (D) Cells (indicated in legend) were transiently transfected with pβCasein-lux. Cells were maintained in serum-free media for the final 24 h of culture. Luciferase activity was measured as described above. n = 3.
DISCUSSION
Genetically engineered mouse models demonstrate that ErbB4 is required for mammary differentiation and STAT5A activation (Jones et al., 1999; Long et al., 2003; Tidcombe et al., 2003). However, the mechanism by which the ErbB4 pathway signals differentiation has not been well-defined. Using an ex vivo model of differentiation, we demonstrate that ErbB4 signaling induces mammary cell differentiation and that ligand-induced cleavage to a soluble, intracellular/nuclear fragment is both necessary and sufficient to trigger differentiation in response to ErbB4 ligands.
Although the biochemical details of ErbB4 proteolytic processing resulting in ErbB4 ectodomain shedding and nuclear accumulation of s80HER4 have been known and nuclear ErbB4 has been observed in many cell types, the identification of a physiological role for s80HER4 has remained elusive. Our results demonstrate that one function of s80HER4 is to direct STAT5A-mediated lactogenic differentiation in response to ErbB4 ligands. Previous reports suggest that nuclear accumulation of the ErbB4 intracellular domain (ICD) may be required for STAT5A-mediated transcription in transient reporter systems. It is known that the carboxy-terminal acidic regions of ErbB4, as well as ErbB1 and ErbB2, contain an intrinsic transactivation function and are able to transactivate a GAL4-VP16 luciferase-based reporter system (Lin et al., 2001; Ni et al., 2001; Wang et al., 2004). Our results using the HC11 model of mammary differentiation extend these findings to include multiple physiological aspects of lactogenic differentiation, such as endogenous milk protein expression and structural acinar formation in three dimensions, underscoring the physiological relevance of s80HER4.
Expression of s80HER4 is detected in the nucleus and cytoplasm of HC11 cells and in human breast cancer samples, consistent with the presence of protein sequences within s80HER4 that govern nuclear import and export and suggesting a complex pattern of intracellular shuttling that we are only beginning to understand. At least one factor that regulates nuclear accumulation of s80HER4 is its tyrosine kinase activity, because kinase-dead GFP-s80KD was strictly restricted to the cytoplasm of cells, as shown using live cell fluorescence (Figure 4C) and biochemically (Figure 4D). The mechanism by which the kinase domain of s80HER4 dictates nuclear localization is unknown, but one possibility is that tyrosine phosphorylation of s80HER4 facilitates its association with a nuclear importin or somehow facilitates the retention of s80HER4 within the nucleus. Kinase activity of s80HER4 was also required for ligand-free phosphorylation and nuclear translocation of STAT5A. The idea that s80HER4 functions as a nuclear chaperone for STAT5A has been previously reported in studies in which mutation of a nuclear localization sequence within s80HER4 completely impaired STAT5A nuclear translocation (Williams et al., 2004). The results presented here are not inconsistent with this, in that cytoplasmic retention of GFP-s80KD also correlated with cytoplasmic retention of STAT5A. However, it is not yet clear if it is loss of nuclear localization, loss of kinase activity per se, or loss of both together that prevents the ligand-free nuclear accumulation of STAT5A in HC11-GFP-s80KD cells.
Lactogenic gene expression and morphological changes in three dimensions were induced either by PRL, HRG, or HB-EGF (Figure 2). Although HB-EGF stimulates both ErbB1 and ErbB4, inhibition of ErbB1 (using 0.5 mM GW572016) did not impair differentiation; in contrast, inhibition of ErbB4 activity (by overexpression of kinase-dead HER4KD) blocked differentiation of HC11 cells in response to HB-EGF. Similarly, HRG-induced activation of ErbB2 was unnecessary and may even inhibit structural differentiation of HC11 cells, because GW572016-mediated inhibition of ErbB2 allowed HC11 lumen formation in Matrigel. Interestingly, when allowed to proceed in the absence of the ErbB inhibitor, HRG-mediated stimulation of ErbB2, ErbB3, and ErbB4 tyrosine kinase activity increased the cellular content of HC11 colonies and induced the formation of multilobulated solid cords of cells. The summary effect of HRG on morphological structure appears, then, to depend on the complex ratio of ErbB2 to ErbB4 signaling. For example, inhibition of ErbB2 in the presence of HRG-dependent ErbB4 signal allowed a level of HRG/ErbB4-dependent lumen formation and differentiation. ErbB2-mediated morphological changes in three dimensions have been demonstrated by others using MCF10A cells (Muthuswamy et al., 2001).
Each of the ErbB receptors has been observed within nuclei. ErbB1 (Lin et al., 2001), ErbB2 (Wang et al., 2004), and ErbB3 (Offterdinger et al., 2002) have been reported to translocate to the nucleus upon ligand stimulation as full-length proteins. In contrast, a proteolytic fragment of ErbB4 with an active tyrosine kinase is known to localize to the nucleus, but not the entire receptor. ErbB4 is first processed by TACE and next by γ-secretase. We demonstrate that the resulting proteolytic fragment, s80HER4, accumulated in the nuclei of mammary epithelial cells, induced STAT5A phosphorylation and expression of β-casein and stimulated acinar lumen formation. The requirement for ligand-inducible proteolysis of ErbB4 was demonstrated by the V675A mutation in the γ-secretase cleavage site of ErbB4, which impaired cytoplasmic release and nuclear accumulation of ErbB4, and at the same time impaired the biological consequences of ErbB4 signaling in HC11 cells. ErbB4 signaling from the V675A mutant may mimic the signaling effects of the JMb splice variant of ErbB4, which lacks the short juxtamembrane TACE cleavage site, and therefore cannot produce s80HER4. Given the role of ErbB4 in mammary gland development and the effects of ErbB4 proteolysis on downstream signaling, the balance of JMa versus JMb variants could have important physiological implications in development, differentiation, and cancer of the mammary gland.
Our results presented here suggest that ErbB4 may differentiate mammary cells through nuclear accumulation of s80HER4, thus identifying a physiological role for nuclear ErbB4 and supporting a mechanism by which ErbB4 differentiates the mammary epithelium.
ACKNOWLEDGMENTS
This work was supported in part by a grant from the Breast Cancer Research Foundation and National Institutes of Health Grant CA112553.
Abbreviations used:
- EGF
epidermal growth factor
- HB-EGF
heparin-binding EGF-like growth factor
- PRL
prolactin
- TACE
tumor necrosis factor-α converting enzyme
- PBS
phosphate-buffered saline
- RLU
relative light units
- RT-PCR
reverse transcription polymerase chain reaction
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05.02-0101) on July 12, 2006.
REFERENCES
- Abd El-Rehim D. M., Pinder S. E., Paish C. E., Bell J. A., Rampaul R. S., Blamey R. W., Robertson J. F., Nicholson R. I., Ellis I. O. Expression and co-expression of the members of the epidermal growth factor receptor (EGFR) family in invasive breast carcinoma. Br. J. Cancer. 2004;91:1532–1542. doi: 10.1038/sj.bjc.6602184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter G. ErbB-4, mechanism of action and biology. Exp. Cell Res. 2003;284:66–77. doi: 10.1016/s0014-4827(02)00100-3. [DOI] [PubMed] [Google Scholar]
- Cheng Q. C., Tikhomirov O., Zhou W., Carpenter G. Ectodomain cleavage of ErbB-4, characterization of the cleavage site and m80 fragment. J. Biol. Chem. 2003;278:38421–38427. doi: 10.1074/jbc.M302111200. [DOI] [PubMed] [Google Scholar]
- Chodosh L. A. The reciprocal dance between cancer and development. N. Engl. J. Med. 2002;347:134–136. doi: 10.1056/NEJMcibr020712. [DOI] [PubMed] [Google Scholar]
- Cui Y., Riedlinger G., Miyoshi K., Tang W., Li C., Deng C. X., Robinson G. W., Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell. Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenius K., Corfas G., Paul S., Choi C. J., Rio C., Plowman G. D., Klagsbrun M. A novel juxtamembrane domain isoform of HER4/ErbB4, isoform-specific tissue distribution and differential processing in response to phorbol ester. J. Biol. Chem. 1997;272:26761–26768. doi: 10.1074/jbc.272.42.26761. [DOI] [PubMed] [Google Scholar]
- Jones F. E., Welte T., Fu X.-Y., Stern D. F. ErbB4 signaling in the mammary gland is required for lobuloalveolar development and Stat5 activation during lactation. J. Cell Biol. 1999;147:77–87. doi: 10.1083/jcb.147.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones F. E., Jerry D. J., Guarino B. C., Andrews G. C., Stern D. F. Heregulin induces in vivo proliferation and differentiation of mammary epithelium into secretory lobuloalveoli. Cell Growth Differ. 1996;7:1031–1038. [PubMed] [Google Scholar]
- Junttila T. T., Sundvall M., Lundin M., Lundin J., Tanner M., Harkonen P., Joensuu H., Isola J., Elenius K. Cleavable ErbB4 isoform in estrogen receptor-regulated growth of breast cancer cells. Cancer Res. 2005;65:1384–1393. doi: 10.1158/0008-5472.CAN-04-3150. [DOI] [PubMed] [Google Scholar]
- Kainulainen V., Sundvall M., Maatta J. A., Santiestevan E., Klagsbrun M., Elenius K. A natural ErbB4 isoform that does not activate phosphoinositide 3-kinase mediates proliferation but not survival or chemotaxis. J. Biol. Chem. 2000;275:8641–8649. doi: 10.1074/jbc.275.12.8641. [DOI] [PubMed] [Google Scholar]
- Komuro A., Nagai M., Navin N. E., Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J. Biol. Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- Lee H.-J., Jung K.-M., Huang Y. Z., Bennett L. B., Lee J. S., Mei L., Kim T.-W. Presenilin-dependent γ-secretase-like intramembrane cleavage of ErbB4. J. Biol. Chem. 2002;277:6318–6323. doi: 10.1074/jbc.M110371200. [DOI] [PubMed] [Google Scholar]
- Lin S. Y., Makino K., Xia W., Matin A., Wen Y., Kwong K. Y., Bourguignon L., Hung M. C. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- Linggi B., Cheng Q. C., Rao A. R., Carpenter G. The ErbB-4 s80 intracellular domain is a constitutively active tyrosine kinase. Oncogene. 2005;25:160–163. doi: 10.1038/sj.onc.1209003. [DOI] [PubMed] [Google Scholar]
- Liu X., Robinson G. W., Wagner K. U., Garrett L., Wynshaw-Boris A., Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- Long W., Wagner K.-U, Lloyd K.C.K., Binart N., Shillingford J. M., Hennighausen L., Jones F. E. Impaired differentiation and lactational failure in ErbB4-deficient mammary glands identify ERBB4 as an obligate mediator of Stat5. Development. 2003;130:5257–5268. doi: 10.1242/dev.00715. [DOI] [PubMed] [Google Scholar]
- Maatta J. A., Sundvall M., Junttila T. T., Peri L., Laine V. J., Isola J., Egeblad M., Elenius K. Proteolytic cleavage and phosphorylation of a tumor-associated ErbB4 isoform promote ligand-independent survival and cancer cell growth. Mol. Biol. Cell. 2006;17:67–79. doi: 10.1091/mbc.E05-05-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marte B. M., Jeschke M., Graus-Porta D., Taverna D., Hofer P., Groner B., Yarden Y., Hynes N. E. Neu differentiation factor/heregulin modulates growth and differentiation of HC11 mammary epithelial cells. Mol. Endocrinol. 1995;9:14–23. doi: 10.1210/mend.9.1.7760847. [DOI] [PubMed] [Google Scholar]
- Muthuswamy S. K., Li D., Lelievre S., Bissell M. J., Brugge J. S. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat. Cell Biol. 2001;3:3785–3792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni C.-Y., Murphy M. P., Golde T. E., Carpenter G. γ-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- Ni C. Y., Yuan H., Carpenter G. Role of the ErbB-4 carboxyl terminus in γ-secretase cleavage. J. Biol. Chem. 2003;278:4561–4565. doi: 10.1074/jbc.M210504200. [DOI] [PubMed] [Google Scholar]
- Offterdinger M., Schofer C., Weipoltshammer K., Grunt T. W. c-erbB-3, a nuclear protein in mammary epithelial cells. J. Cell Biol. 2002;157:929–939. doi: 10.1083/jcb.200109033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio C., Buxbaum J. D., Peschon J. J., Corfas G. Tumor necrosis factor-alpha-converting enzyme is required for cleavage of erbB4/HER4. J. Biol. Chem. 2000;275:10379–10387. doi: 10.1074/jbc.275.14.10379. [DOI] [PubMed] [Google Scholar]
- Sartor C. I., et al. HER4 mediates ligand-dependent antiproliferative and differentiation responses in human breast cancer cells. Mol. Cell. Biol. 2001;21:4265–4275. doi: 10.1128/MCB.21.13.4265-4275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder J. A., Lee D. C. Dynamic expression and activation of ERBB receptors in the developing mouse mammary gland. Cell Growth Differ. 1998;9:451–464. [PubMed] [Google Scholar]
- Shillingford J. M., Miyoshi K., Flagella M., Shull G. E., Hennighausen L. Signal transducer and activator of transcription (Stat) 5 controls the proliferation and differentiation of mammary alveolar epithelium. J. Cell Biol. 2001;155:531–542. doi: 10.1083/jcb.200107065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R., Gillett C. E., Barnes D. M., Gullick W. J. Nuclear expression of the c-erbB-4/HER4 growth factor receptor in invasive breast cancers. Cancer Res. 2000;60:1483–1487. [PubMed] [Google Scholar]
- Stern D. F. ErbBs in mammary development. Exp. Cell Res. 2003;284:89–98. doi: 10.1016/s0014-4827(02)00103-9. [DOI] [PubMed] [Google Scholar]
- Tidcombe H., Jackson-Fisher A., Mathers K., Stern D. F., Gassmann M., Golding J. P. Neural and mammary gland defects in ErbB4 knockout mice genetically rescued from embryonic lethality. Proc. Natl. Acad. Sci. USA. 2003;100:8281–8286. doi: 10.1073/pnas.1436402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal G. A., Narres A., Marrero L., Jones F. E. Presenilin-dependent gamma-secretase processing regulates multiple ERBB4/HER4 activities. J. Biol. Chem. 2005;280:19777–19783. doi: 10.1074/jbc.M412457200. [DOI] [PubMed] [Google Scholar]
- Wang S. C., et al. Binding at and transactivation of the COX-2 promoter by nuclear tyrosine kinase receptor ErbB-2. Cancer Cell. 2004;6:251–261. doi: 10.1016/j.ccr.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Williams C. C., Allison J. G., Vidal G. A., Burow M. E., Beckman B. S., Marrero L., Jones F. E. The ERBB4/HER4 receptor tyrosine kinase regulates gene expression by functioning as a STAT5A nuclear chaperone. J. Cell Biol. 2004;167:469–478. doi: 10.1083/jcb.200403155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winklehner-Jennewein P., Geymayer S., Lechner J., Welte T., Hansson L., Geley S., Doppler W. A distal enhancer region in the human beta-casein gene mediates the response to prolactin and glucocorticoid hormones. Gene. 1998;217:127–139. doi: 10.1016/s0378-1119(98)00380-1. [DOI] [PubMed] [Google Scholar]
- Xia W., Mullin R. J., Keith B. R., Liu L. H., Ma H., Rusnak D. W., Owens G., Alligood K. J., Spector N. L. Anti-tumor activity of GW572016, a dual tyrosine kinase inhibitor blocks EGF activation of EGFR/erbB2 and downstream Erk1/2 and AKT pathways. Oncogene. 2002;21:6255–6263. doi: 10.1038/sj.onc.1205794. [DOI] [PubMed] [Google Scholar]
- Yang Y., Spitzer E., Meyer D., Sachs M., Niemann C., Hartmann G., Weidner K. M., Birchmeier C., Birchmeier W. Sequential requirement of hepatocyte growth factor and neuregulin in the morphogenesis and differentiation of the mammary gland. J. Cell Biol. 1995;131:215–226. doi: 10.1083/jcb.131.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]