Abstract
Background
Extracting features from the colonoscopic images is essential for getting the features, which characterizes the properties of the colon. The features are employed in the computer-assisted diagnosis of colonoscopic images to assist the physician in detecting the colon status.
Methods
Endoscopic images contain rich texture and color information. Novel schemes are developed to extract new texture features from the texture spectra in the chromatic and achromatic domains, and color features for a selected region of interest from each color component histogram of the colonoscopic images. These features are reduced in size using Principal Component Analysis (PCA) and are evaluated using Backpropagation Neural Network (BPNN).
Results
Features extracted from endoscopic images were tested to classify the colon status as either normal or abnormal. The classification results obtained show the features' capability for classifying the colon's status. The average classification accuracy, which is using hybrid of the texture and color features with PCA (τ = 1%), is 97.72%. It is higher than the average classification accuracy using only texture (96.96%, τ = 1%) or color (90.52%, τ = 1%) features.
Conclusion
In conclusion, novel methods for extracting new texture- and color-based features from the colonoscopic images to classify the colon status have been proposed. A new approach using PCA in conjunction with BPNN for evaluating the features has also been proposed. The preliminary test results support the feasibility of the proposed method.
Background
In the case of colorectal cancer, abnormal cell growth takes place in the large intestine resulting in the formation of tumors. The detection of any abnormal growth in the colon at an early stage will increase the patient's chance of survival. A few methods, such as sigmoidoscopy, barium x-ray, etc., are available for examination of the colon, but colonoscopy is considered to be the best procedure at present for the detection of abnormalities in the colon [1]. Despite the usefulness of colonoscopic methods, an expert endoscopist is needed to detect colorectal cancer. The endoscopist uses a colonoscope to detect the presence of abnormalities in the colon. The analysis of the endoscopic images is usually performed visually and qualitatively. Consequently, there are constraints such as time-consuming procedures, subjective diagnosis by the expert, interpretational variation, and non-suitability for comparative evaluation. A computer-assisted scheme will help considerably in the quantitative characterization of abnormalities and image analysis, thereby improving overall efficiency in managing the patient.
Computer-assisted diagnosis in colonoscopy consists of colonoscopic image acquisition, image processing, parametric feature extraction, and classification. A number of schemes have been proposed to develop methods for computer-assisted diagnosis for the detection of colonic cancer images. Some researchers use microscopic images [2-4] and others use endoscopic images [5-8].
Esgiar, et al. [2,4] and Todman, et al. [3] have been using microscopic images to analyze and identify features of normal and cancerous colonic mucosa. A number of quantitative techniques for the analysis of images used in the diagnosis of colonic cancer have been investigated. Features based on texture analysis were derived using the co-occurrence matrix, viz., angular second moment, entropy, contrast, inverse difference moment, dissimilarity, and correlation [2]. Orientational coherence metrics have been derived from neurophysiological foundations and applied to the classification of colonic cancer images [3]. Fractal analysis has been also investigated in separating normal and cancerous images [4].
Krishnan, et al. [5-8] have been using endoscopic images to define features of the normal and the abnormal colon. New approaches for the characterization of colon based on a set of quantitative parameters, extracted by the fuzzy processing of colon images, have been used for assisting the colonoscopist in the assessment of the status of patients and were used as inputs to a rule-based decision strategy to find out whether the colon's lumen belongs to either an abnormal or normal category. The quantitative characteristics of the colon are: hue component, mean and standard deviation of RGB, perimeter, enclosed boundary area, form factor, and center of mass [5]. The analysis of the extracted quantitative parameters was performed using three different neural networks selected for classification of the colon. The three networks include a two-layer perceptron trained with the delta rule, a multilayer perceptron with Backpropagation learning and a self-organizing network. A comparative study of the three methods was also performed and it was observed that the self-organizing network is more appropriate for the classification of colon status [6]. A method of detecting the possible presence of abnormalities during the endoscopy of the lower gastro-intestinal system using curvature measures has been developed. In this method, image contours corresponding to haustra creases in the colon are extracted and the curvature of each contour is computed after non-parametric smoothing. Zero-crossings of the curvature along the contour are then detected. The presence of abnormalities is identified when there is a contour segment between two zero-crossings having the opposite curvature signs to those of the two neighboring contour segments. The proposed method can detect the possible presence of abnormalities such as polyps and tumors [7]. Fuzzy rule-based approaches to the labeling of colonoscopic images to render assistance to the clinician have been proposed. The color images are segmented using a scale-space filter. Several features are selected and fuzzified. The knowledge-based fuzzy rule-based system labels the segmented regions as background, lumen, and abnormalities (polyps, bleeding lesions) [8].
Endoscopic images contain rich information of texture and color. Therefore, the additional texture and color information can provide better results for the image analysis than approaches using merely intensity information. In this research work, the definition and extraction of quantitative parameters from endoscopic images based on texture and color information have been proposed. The test results obtained so far by the proposed approach have been encouraging.
Methodology
Endoscopic imaging methods have been used to examine the condition of the colon. The development of an intelligent method for the identification of the colon status is being explored as a computer-aided tool for the early detection of colorectal cancer. It is important to extract the quantitative parameters representing the characteristic properties of the colon from an endoscopic image. The quantitative features are used for detecting the normal or abnormal condition of the colon from the endoscopic images. Endoscopic images contain texture and color information. Features from texture and color are extracted from the endoscopic image to identify a normal colon from an abnormal colon. Krishnan, et al have extracted features from the histogram of the image in the chromatic domain, and the shape of the lumen in the spatial domain and fed them into a feed-forward Neural Network [6]. Four statistical measures, derived from the co-occurrence matrix in four different angles, namely angular second moment, correlation, inverse difference moment, and entropy, have been extracted by Karkanis [9].
Texture-based Feature Extraction
Texture analysis is one of the most important features used in image processing and pattern recognition. It can give information about the arrangement and spatial properties of fundamental image elements. Many methods have been proposed to extract texture features, e.g. the co-occurrence matrix [10], and the texture spectrum in the achromatic component of the image [11]. In this research, a new approach of obtaining quantitative parameters from the texture spectra is proposed both in the chromatic and achromatic domains of the image. These features are evaluated to demonstrate their usefulness and potential for classifying the colon status.
The definition of texture spectrum employs the determination of the texture unit (TU) and texture unit number (NTU) values. Texture units characterize the local texture information for a given pixel and its neighborhood, and the statistics of all the texture units over the whole image reveal the global texture aspects.
Given a neighborhood of δ × δ pixels, which are denoted by a set containing δ × δ elements, P = {P0, P1, ..., P(δ × δ)-1}, where P0 represents the chromatic or achromatic value of the central pixel and Pi {i = 1,2, ..., (δ × δ)-1} is the chromatic or achromatic value of the neighboring pixel i, the TU = {E1, E2, ..., E(δ × δ)-1}, where Ei (i = 1,2, ..., (δ × δ)-1) is determined as follows:
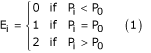
The element Ei occupies the same position as the i-th pixel.
Each element of the TU has one of three possible values; therefore the combination of all the eight elements results in 6561 possible TU's in total. The texture unit number (NTU) is the label of the texture unit and is defined using the following equation:
![]()
The texture spectrum histogram (Hist(i)) is obtained as the frequency distribution of all the texture units, with the abscissa showing the NTU and the ordinate representing its occurrence frequency.
The texture spectra of various image components {I (Intensity), R (Red), G (Green), B (Blue), H (Hue), S (Saturation)} are obtained from their texture unit numbers. Six statistical measures are used to extract new features from each texture spectrum. The statistical measures are energy (α), mean (μ), standard deviation (σ), skew (φ), kurtosis (κ), and entropy (ρ).
Color-based Feature Extraction
Color colonoscopic images exhibited the same color features for the same colon status [1]. Malignant tumors are usually inflated and inflamed. The inflammation is usually reddish and more severe in color than the surrounding tissues. Benign tumors exhibit less intense hues. Redness may specify bleeding and black may be treated as deposits due to laxatives. Green may be the presence of faecal materials, which are not clear during the pre-operative preparation, and yellow relates to pus formation. Based on these properties, some features are extracted from the chromatic and achromatic histograms of the image. In the histograms of each image, certain lower and upper threshold values of the regions of interest are selected for the extraction of the quantitative parameters. The color features are defined as follows:
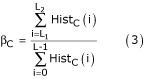
where βC are the color features for various image components C = {I, R, G, B, H, S}, HistC(i) is the histogram amplitude at level I of a particular color component C, L is the number of gray levels, and L1 and L2 are the lower and upper threshold values of the histogram of the ROI.
Features Evaluation
These features are evaluated using BPNN with various training algorithms, viz., resilient propagation (RPROP) [12], scaled conjugate gradient (SCG) algorithm [13], and the Marquardt algorithm [14]. The various training algorithms were originally developed in order to speed up the training phase of the BPNN. Another approach to decrease the training time is to use a principal component analysis (PCA) [15] to substitute the high-dimensional original features set with a low-dimensional set.
Principal component analysis is a feature reduction technique. It is an orthogonal decomposition, which projects data onto the eigenvectors of the covariance matrix of the data. By sorting the eigenvectors by eigenvalue, the projected dimensions can be ranked by variance (which is proportional to eigenvalue). The highest eigenvalues contain the dimensions of highest variance and class separability. By eliminating very small eigenvalues, the dimensionality of the projected space can be reduced without losing much information.
Results and Discussion
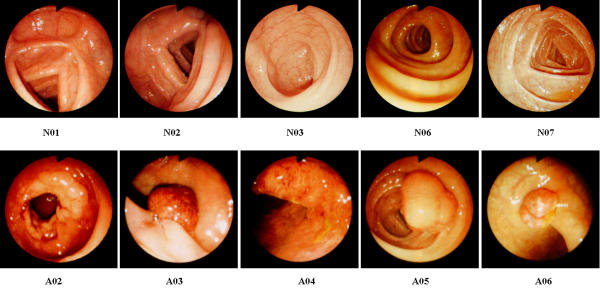
The proposed approaches were evaluated using 66 clinically obtained colonoscopic images (54 abnormal images and 12 normal images). Figure 1 shows the selected colonoscopic images of normal and abnormal cases. Features based on texture and color are defined and extracted from those images for diagnosis of the colon's status.
Figure 1.
The selected colonoscopic images of normal (N01, N02, N03, N06, N07) and abnormal (A02, A03, A04, A05, A06) cases.
Texture spectra were obtained from the raw images in the chromatic and achromatic domains. Novel texture-based features were extracted from the texture spectra. The features were evaluated using Backpropagation Neural Network (BPNN) with various training algorithms, viz., resilient propagation (RPROP), scaled conjugate gradient (SCG), and Marquardt algorithms.
For training the BPNN, half of the sample sets of images were used. The training and testing procedures were carried out with different sets of color components {C} and different numbers of neurons in the hidden layer. The number of input neurons of the BPNN depends on the sets of color components {C}. The BPNN has a Boolean output, which tells whether the colon's status is normal or abnormal. Table 1 shows a number of features having various color component combinations {C}.
Table 1.
The numbers of features from various color component combinations {C}
| {C} | Number of Features |
| {I, R, G, B, H, S} | 36 |
| {I, R, G, B} | 24 |
| {H, S, I} | 18 |
| {H, S} | 12 |
Figure 2 shows the graphs of training time comparison for different training algorithms with various numbers of hidden neurons. The graphs show that the Marquardt algorithm has better performance for the overall number of epochs for training the BPNN than SCG and RPROP algorithms.
Figure 2.
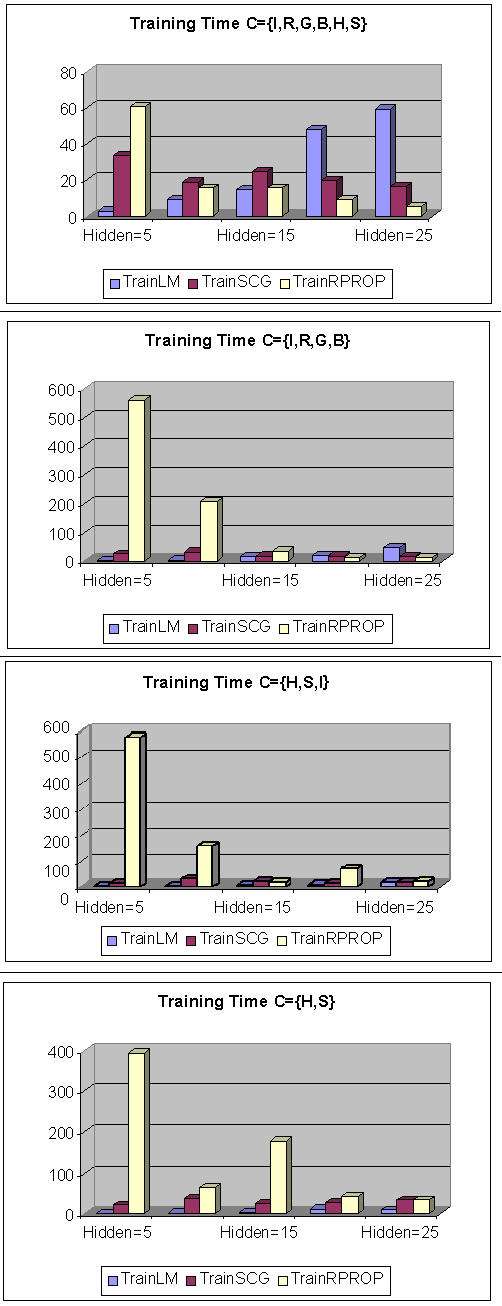
The graphs of training time comparison for various training algorithms with different numbers of hidden neurons.
Table 2 shows the number of epochs needed to train the different algorithms in various color component combinations {C}. The Marquardt algorithm needed fewer epochs than SCG and RPROP algorithms to converge to the minimum mean-squared error of the system.
Table 2.
The number of epochs needed to train the different algorithms in various color component combinations {C}.
| C = {I, R, G, B, H, S} | |||
| Number of Hidden Neurons | Training Epochs | ||
| Marquardt Algorithm | SCG Algorithm | RPROP Algorithm | |
| 5 | 33 | 3294 | 11102 |
| 10 | 28 | 1583 | 2435 |
| 15 | 17 | 1798 | 2090 |
| 20 | 27 | 1256 | 1093 |
| 25 | 19 | 922 | 524 |
| C = {I, R, G, B} | |||
| 5 | 20 | 2281 | 100000 |
| 10 | 23 | 2795 | 36776 |
| 15 | 37 | 1380 | 5294 |
| 20 | 27 | 1184 | 1579 |
| 25 | 37 | 1019 | 1362 |
| C = {H, S, I} | |||
| 5 | 30 | 1293 | 100000 |
| 10 | 23 | 2647 | 28238 |
| 15 | 22 | 1601 | 2337 |
| 20 | 21 | 976 | 9683 |
| 25 | 26 | 961 | 2480 |
| C = {H, S} | |||
| 5 | 17 | 2256 | 75574 |
| 10 | 22 | 3728 | 12624 |
| 15 | 35 | 1948 | 27195 |
| 20 | 58 | 2012 | 6269 |
| 25 | 34 | 2396 | 4403 |
The success of classification of the colon's status is measured by the accuracy. Tables 3, 4, and 5 show the accuracy of the classification for different data sets of color components using various training algorithms, viz., Marquardt, SCG and RPROP training algorithms with different numbers of hidden neurons. The classification performance of the {H, S, I} is better than other data sets {C} with an average of 93.02% accuracy using the Marquardt algorithm.
Table 3.
Accuracy (%) of colon status classification using Marquardt training algorithm
| Number of Hidden Neurons | Classification Accuracy (%) | |||
| C = {I, R, G, B, H, S} | C = {I, R, G, B} | C = {H, S, I} | C = {H, S} | |
| 5 | 89.39 | 84.84 | 93.93 | 87.87 |
| 10 | 92.42 | 84.84 | 92.42 | 84.84 |
| 15 | 90.90 | 95.45 | 95.45 | 90.90 |
| 20 | 95.45 | 92.42 | 92.42 | 89.39 |
| 25 | 95.45 | 92.42 | 90.90 | 96.96 |
Table 4.
Accuracy (%) of colon status classification using SCG training algorithm
| Number of Hidden Neurons | Classification Accuracy (%) | |||
| C = {I, R, G, B, H, S} | C = {I, R, G, B} | C = {H, S, I} | C = {H, S} | |
| 5 | 86.36 | 86.36 | 87.87 | 90.90 |
| 10 | 84.84 | 81.81 | 86.36 | 87.87 |
| 15 | 93.93 | 95.45 | 95.45 | 86.36 |
| 20 | 92.42 | 93.93 | 87.87 | 87.87 |
| 25 | 93.93 | 95.45 | 96.96 | 93.93 |
Table 5.
Accuracy (%) of colon status classification using RPROP training algorithm
| Number of Hidden Neurons | Classification Accuracy (%) | |||
| C = {I, R, G, B, H, S} | C = {I, R, G, B} | C = {H, S, I} | C = {H, S} | |
| 5 | 95.45 | 95.45 | 90.90 | 95.45 |
| 10 | 92.42 | 95.45 | 93.93 | 95.45 |
| 15 | 90.90 | 92.42 | 92.42 | 96.96 |
| 20 | 95.45 | 89.39 | 90.90 | 95.45 |
| 25 | 98.48 | 95.45 | 92.42 | 100 |
Choosing the Marquardt training algorithm results in the lesser number of epochs for the neural network. Another way to help speed up the training time is to choose a significant feature vector. This can be realized by introducing principal component analysis (PCA) in conjunction with BPNN. Principal component analysis reduced the dimension of the features and was applied before using BPNN for classification of the colon status. The BPNN that is used to classify the PCA features has a Boolean output assigned for normal or abnormal colon status. Table 6 shows the number of features after applying PCA with different threshold values, τ, and without applying PCA. For example, τ = 5% means that those principal components whose contribution is less than 5% to the total variance in the feature set were eliminated.
Table 6.
The number of features using principal component analysis (PCA) and without using PCA
| Number of Features | |||||
| {C} | Without PCA | With PCA | |||
| τ = 5% | τ = 2% | τ = 1% | τ = 0.1% | ||
| {I, R, G, B, H, S} | 36 | 3 | 5 | 9 | 15 |
| {I, R, G, B} | 24 | 3 | 6 | 8 | 13 |
| {H, S, I} | 18 | 4 | 7 | 8 | 11 |
| {H, S} | 12 | 4 | 6 | 7 | 9 |
Figure 3 shows the graphs of the Marquardt training time comparison using PCA with different τ (5%, 2%, 1%, 0.1%) and various numbers of hidden neurons. Figure 4 shows the graphs of the Marquardt training (using PCA features, τ = 1%) time comparison for various numbers of hidden neurons. It shows that by using PCA, the overall training time is decreased, thereby increasing the performance of BPNN. The success of the classification of colon status is measured by the accuracy, which is shown in Table 7. The {H, S, I} has better performance for classification than other data sets {C} on average, for both with PCA (96.96%, τ = 1%) and without PCA (93.02%). The classification rate increased by applying PCA, and τ = 1% gave better accuracy rates than other values of τ.
Figure 3.
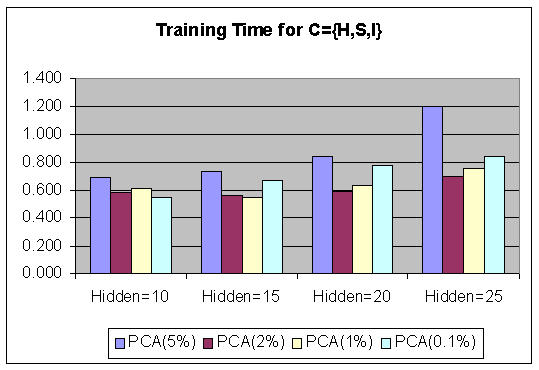
The graphs of Marquardt training time comparison using PCA with different τ (5%, 2%, 1%, 0.1%) and various numbers of hidden neurons
Figure 4.
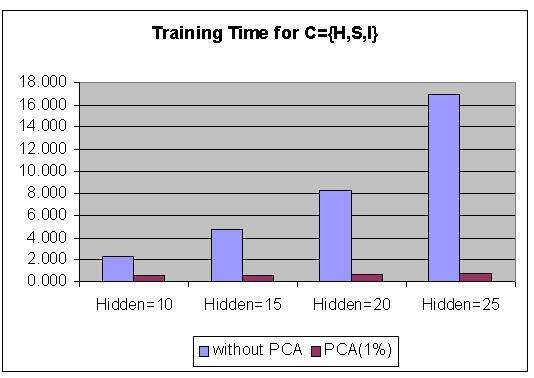
The graphs of Marquardt training time comparison on the usage of PCA with various numbers of hidden neurons (τ = 1%)
Table 7.
Accuracy (%) of the colon status classification using Marquardt training algorithm
| C = {H, S, I} | |||||
| Number of Hidden Neurons | Classification Accuracy (%) | ||||
| Without PCA | With PCA | ||||
| τ = 5% | τ = 2% | τ = 1% | τ = 0.1% | ||
| 10 | 92.42 | 93.94 | 95.45 | 98.48 | 98.48 |
| 15 | 95.45 | 86.36 | 95.45 | 95.45 | 95.45 |
| 20 | 92.42 | 95.45 | 93.94 | 96.96 | 92.42 |
| 25 | 90.90 | 84.85 | 96.96 | 96.96 | 96.96 |
Color features extracted from the histogram of the endoscopic images were also defined. Certain lower and upper threshold values of the region of interest (ROI) were selected for the histograms of each image. Where possible, certain patterns from R, G, B, H, S, and I histograms were identified which could distinguish between normal and abnormal colon conditions. The histograms are plotted in Figures 5,6,7,8,9,10. The selection of the ROI is rather subjective. Based on the histograms of the images, the ROI for each type of histogram is selected as an input for the NN classification. The ROI is chosen such that patterns are seen in the ROI for both normal and abnormal cases.
Figure 5.
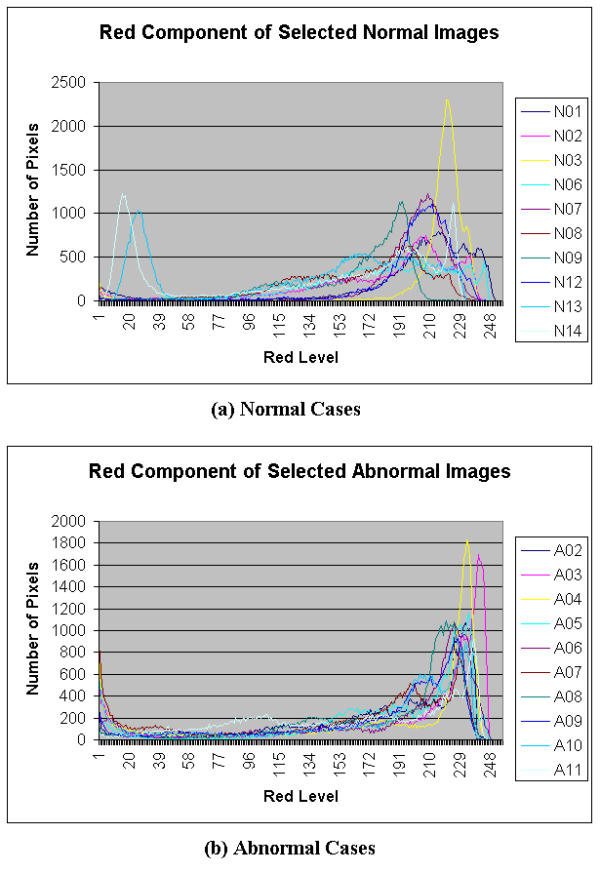
Histograms of red component of Selected Colonoscopic Images
Figure 6.
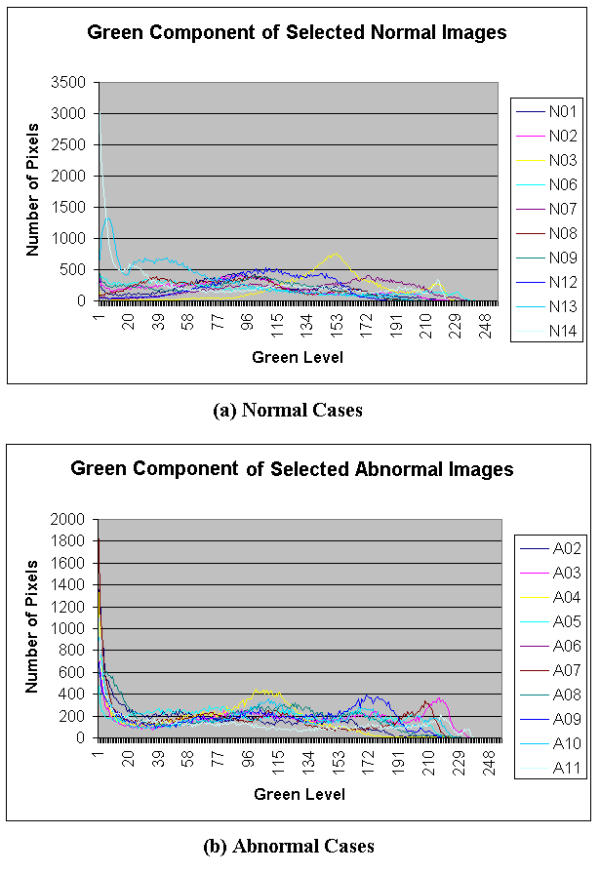
Histograms of green component of Selected Colonoscopic Images
Figure 7.
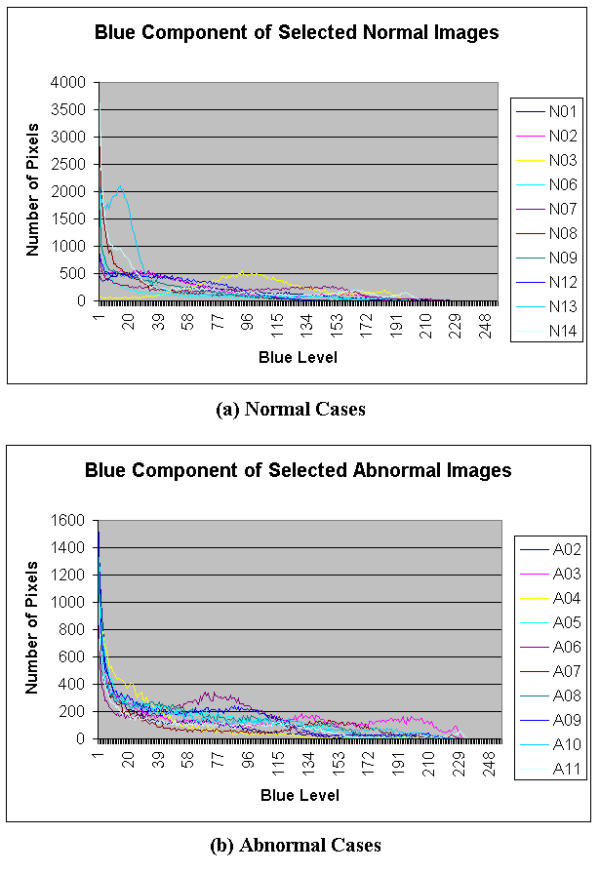
Histograms of blue component of Selected Colonoscopic Images
Figure 8.
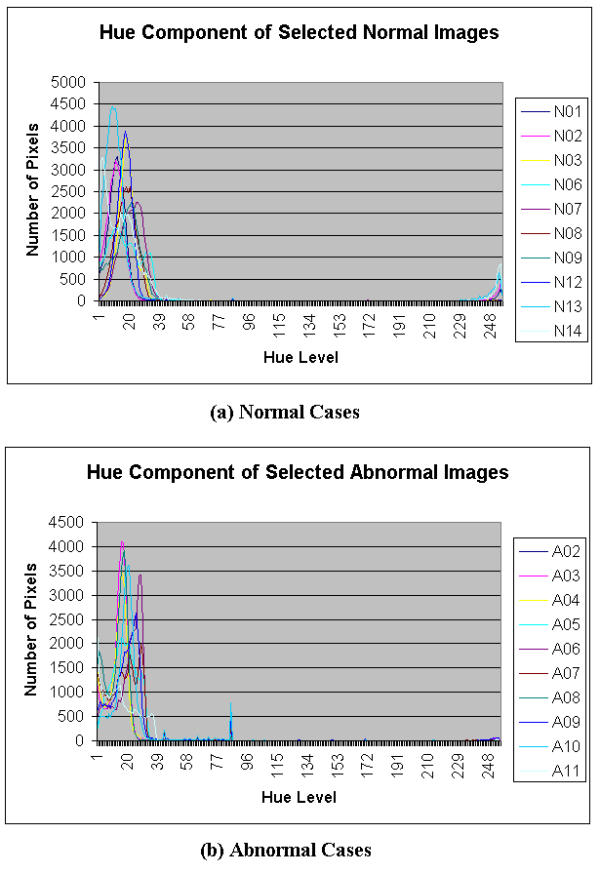
Histograms of hue component of Selected Colonoscopic Images
Figure 9.
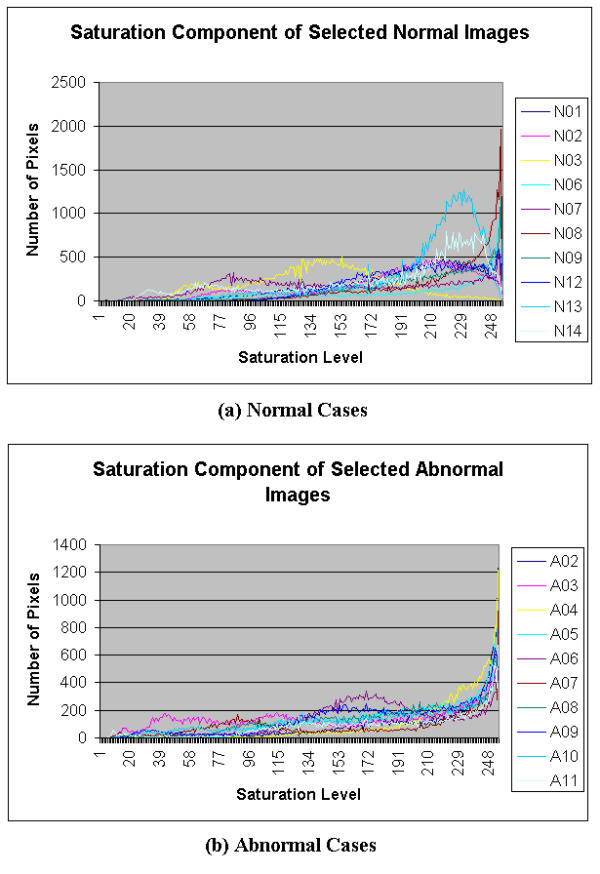
Histograms of saturation component of Selected Colonoscopic Images
Figure 10.
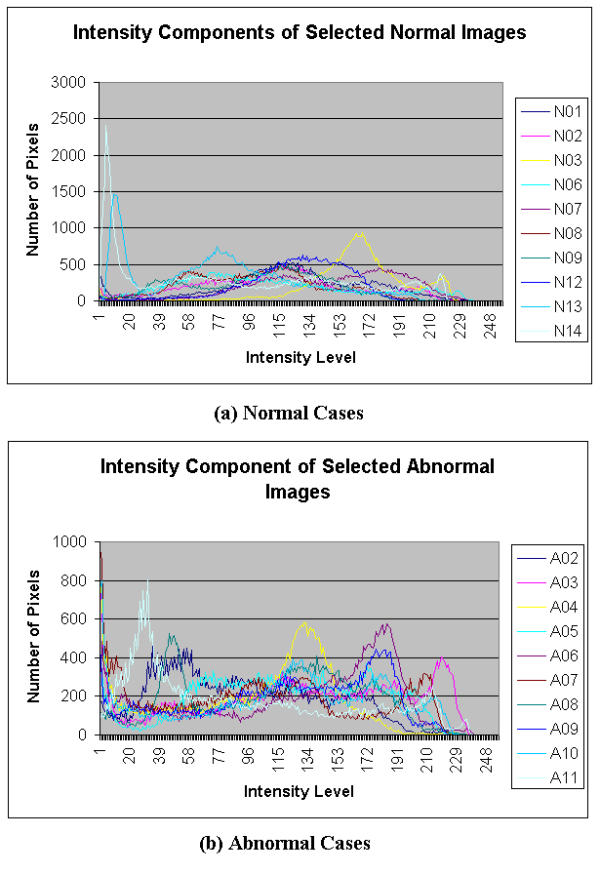
Histograms of intensity component of Selected Colonoscopic Images
Figure 5 shows a histogram for both normal and abnormal endoscopic images. It is observed that for normal and abnormal conditions, the redness is concentrated around the 200 to 255 levels (where 0 is the lowest intensity value and 255 is the greatest intensity value). Figure 6 shows another histogram of both normal and abnormal endoscopic images. It is observed that the histogram has a bell shape pattern for the normal condition and very little pattern is observed for the abnormal condition. The green parameter can be extracted in the ROI from 100 to 255 of the green level. From a similar histogram in Figure 7, the abnormal condition exhibits certain patterns, but the same is not observed in the normal condition. This does not pose a problem to the feature extraction module, as the main objective behind the parameter extraction was to differentiate the normal condition from the abnormal condition. As long as there are some patterns observed in one of the conditions, the parameter extracted from this color space can be considered useful in classification. The selected ROI in the blue level histogram is from 0 to 50.
For the hue component shown in Figure 8, very similar patterns are observed for the normal and abnormal conditions. Its ROI is determined to be in the range of 10 to 50 for both normal and abnormal cases. In the saturation domain shown in Figure 9, the shapes of the histograms for the normal and abnormal cases are also very similar. Generally however, the tail for the normal case has a higher value. The ROI, then, for the extraction of the saturation parameter is set in the range of 100 to 150. In the intensity histogram shown in Figure 10, the pattern observed is very similar to that of the green histograms. In this case, the range is 150 to 255 of the intensity level. Although the highest value of 255 may be included, the lower region begins at 150, which would justify that this extraction intensity parameter will not show the effect of the light source. It may consist of some reflection but the value will be very minimal.
Table 8 shows the classification results for detecting the colon status using color features (βC). A BPNN is used to perform the classification. The procedures for the classification using texture features were applied for the classification using color features. The training was done with the Marquardt algorithm. It was found that the average accuracy using PCA with τ = 0.1% is higher than the classification with PCA, where τ = 5%, 2%, or 1% or without PCA. At τ = 0.1%, PCA does not change the dimension of the features. The number of features will be the same as the number we would have if we were not using PCA at all. The fewer the number of features used, the worse the average classification rate becomes. This shows that all of the extracted color features (βC) are significant.
Table 8.
Accuracy (%) of the colon status classification using color features
| Number of Hidden Neurons | Classification Accuracy (%) | ||||
| Without PCA | With PCA | ||||
| τ = 5% | τ = 2% | τ = 1% | τ = 0.1% | ||
| 10 | 96.96 | 96.96 | 92.42 | 84.84 | 92.42 |
| 15 | 87.87 | 95.45 | 87.87 | 96.96 | 98.48 |
| 20 | 93.93 | 90.9 | 92.42 | 86.36 | 96.96 |
| 25 | 98.48 | 93.93 | 86.36 | 93.93 | 93.93 |
The images are rich with texture and color information. Therefore, it is important to utilize the extracted texture and color features. Experiments have been done that hybrid the texture and color features for colon status classification. It is shown in Table 10 that the average classification accuracy with a hybrid of texture and color features for the classification using PCA with τ = 1% is higher than the classification with PCA, where τ = 5%, 2%, or 0.1% or without PCA.
Table 10.
Average accuracy (%) of the colon status classification
| Features | Average Classification Accuracy (%) | ||||
| Without PCA | With PCA | ||||
| τ = 5% | τ = 2% | τ = 1% | τ = 0.1% | ||
| Texture | 93.02 | 90.15 | 95.45 | 96.96 | 95.83 |
| Color | 94.31 | 94.31 | 89.77 | 90.52 | 95.45 |
| Hybrid | 93.93 | 92.04 | 93.93 | 97.72 | 95.45 |
Table 9.
Accuracy (%) of the colon status classification using hybrid of texture and color features
| Number of Hidden Neurons | Classification Accuracy (%) | ||||
| Without PCA | With PCA | ||||
| τ = 5% | τ = 2% | τ = 1% | τ = 0.1% | ||
| 10 | 95.45 | 93.93 | 90.9 | 98.48 | 92.42 |
| 15 | 90.9 | 93.93 | 95.45 | 96.96 | 98.48 |
| 20 | 93.93 | 89.39 | 92.42 | 98.48 | 93.93 |
| 25 | 95.45 | 90.9 | 96.96 | 96.96 | 96.96 |
Conclusion
In conclusion, new approaches on extracting new texture- and color-based features from colonoscopic images for the analysis of the colon status have been developed. Texture and color are important features, in which some features are able to distinguish the normal and abnormal colon status. However, the classification using only texture or only color features is not as complete a classification as possible since endoscopic image contains both texture and color information. Therefore, the hybrid of texture and color features is proposed to give a better approach to classify colon status.
Principal component analysis is a powerful tool for analyzing these features. It provides a way of identifying patterns in features, and expressing the features in such a way as to highlight their similarities and differences. The performance of backpropagation neural network increases by associating it with principal component analysis. The results obtained verify the effectiveness of the proposed approaches. With refinement, the technique can be tested in clinical setting.
Authors' Contributions
Author 1 MPT carried out the analysis and implementation as well as testing of the software simulations. Author 2 SMK conceived of the study, and participated in its coordination. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
The authors wish to extend their sincere appreciation to Nanyang Technological University in Singapore for their research grant.
Contributor Information
Marta P Tjoa, Email: ph093147@ntu.edu.sg.
Shankar M Krishnan, Email: esmkrish@ntu.edu.sg.
References
- Kato H, Barron JP. Electronic videoendoscopy. Japan: Harwood Academic Publisher; 1993. [Google Scholar]
- Esgiar AN, Naguib RNG, Sharif BS, Bennet MK, Murray A. Microscopic image analysis for quantitative measurement and feature identification of normal and cancerous colonic mucosa. IEEE transactions on information technology in biomedicine. 1998;2:197–203. doi: 10.1109/4233.735785. [DOI] [PubMed] [Google Scholar]
- Todman AG, Naguib RNG, Bennett MK. Orientational coherence metrics: classification of colonic cancer images based on human form perception. Proceedings of the Canadian Conference on Electrical and Computer Engineering. 2001;2:1379–1384. doi: 10.1109/CCECE.2001.933655. [DOI] [Google Scholar]
- Esgiar AN, Naguib RNG, Sharif BS, Bennett MK, Murray A. Fractal analysis in the detection of colonic cancer images. IEEE transactions on information technology in biomedicine. 2002;6:54–58. doi: 10.1109/4233.992163. [DOI] [PubMed] [Google Scholar]
- Krishnan SM, Goh PMY. Quantitative parameterization of colonoscopic images by applying fuzzy technique. Proceedings – 19th International Conference – IEEE/EMBS. 1997.
- Krishnan SM, Yap CJ, Asari KV, Goh PMY. Neural network based approaches for the classification of colonoscopic images. Proceedings of the 20th annual international conference of the IEEE Engineering in Medicine and Biology Society. 1998;20:1678–1680. [Google Scholar]
- Krishnan SM, Yang X, Chan KL, Kumar S, Goh PMY. Intestinal abnormality detection from endoscopic images. International conference of the IEEE on Engineering in Medicine and Biology Society. 1998;2:895–898. doi: 10.1109/IEMBS.1998.745583. [DOI] [Google Scholar]
- Krishnan SM, Yang X, Chan KL, Goh PMY. Region labeling of colonoscopic images using fuzzy logic. Proceedings of the first joint BMES/EMBS conference Serving Humanity, Advancing Technology. 1999.
- Karkanis SA, Iakovidis DK, Maroulis DE, Magoulas GD, Theofanous NG. Tumor Recognition in Endoscopic Video Images using Artificial Neural Network Architectures. In Proceedings of the 26th Euromicro Conference, Workshop on Medical Informatics Netherlands. 2000.
- Gotlieb CC, Kreyszig HE. Texture descriptors based on co-occurrence matrices. Computer Vision, Graphics, and Image Processing. 1990;51:70–86. [Google Scholar]
- He DC, Wang L. Texture features based on texture spectrum. Pattern Recognition. 1991;24:391–399. doi: 10.1016/0031-3203(91)90052-7. [DOI] [Google Scholar]
- Riedmiller M, Braun H. A direct adaptive method for faster Backpropagation learning: the RPROP algorithm. Proceedings of the IEEE International Conference on Neural Networks. 1993. pp. 586–591.
- Moller MF. A scaled conjugate gradient algorithm for fast supervised learning. Neural Networks. 1993;6:525–533. [Google Scholar]
- Hagan MT, Menhaj MB. Training feedforward networks with the marquardt algorithm. IEEE transactions on Neural Networks. 1994;5:989–993. doi: 10.1109/72.329697. [DOI] [PubMed] [Google Scholar]
- Shang C, Brown K. Principal features-based texture classification with neural networks. Pattern Recognition. 1994;27:675–687. doi: 10.1016/0031-3203(94)90046-9. [DOI] [Google Scholar]