Abstract
The neurotoxicity of glutamate in the central nervous system is restricted by several (Na+ + K+)-coupled transporters for this neurotransmitter. The astroglial transporter GLT-1 is the only subtype that exhibits high sensitivity to the nontransportable glutamate analogue dihydrokainate. A marked reduction in sensitivity to the blocker is observed when serine residues 440 and 443 are mutated to glycine and glutamine, which, respectively, occupy these positions in the other homologous glutamate transporters. They are located in the ascending limb of the recently identified pore-loop-like structure. Strikingly, mutation of serine-440 to glycine enables not only sodium but also lithium ions to drive net influx of acidic amino acids. Moreover, the efficiency of lithium as a driving ion for glutamate transport depends on the nature of the amino acid residue present at position 443. Mutant transporters containing single cysteines at the position of either serine residue become sensitive to positively as well as negatively charged methanethiosulfonate derivatives. In S440C transporters significant protection against this inhibition is provided both by transportable and nontransportable glutamate analogues, but not by sodium alone. Our observations indicate that the pore-loop-like structure plays a pivotal role in coupling ion and glutamate fluxes and suggest that it is close to the glutamate-binding site.
The role of electrogenic (Na+ + K+)-coupled glutamate transporters, located in the plasma membranes of nerve terminals and glial cells, is to keep the extracellular concentration of the neurotransmitter below neurotoxic levels (1–4). Moreover, at some synapses glutamate transporters appear to be important in limiting the duration of synaptic excitation (5–8). They achieve this by an electrogenic process (9–11) in which the transmitter is cotransported with three sodium ions and one proton (12), followed by countertransport of a potassium ion (12–15). It appears that the three sodium ions are not equivalent. Two of them probably are not entirely specific and can be replaced by lithium, but one is completely specific (16).
The astroglial glutamate transporter GLT-1 has been purified from rat brain to near homogeneity and reconstituted (17, 18). It has been cloned (19) and is related—with around 50% sequence identity—to three different glutamate transporters from the central nervous system (20–22) as well as to one from the retina (23). The physiological importance of GLT-1 for brain function has been illustrated by knock-out experiments (3, 4).
Recently, a number of advances toward our understanding of the structural basis of transporter function have been made. A stretch of 76 aa that contains at least part of the binding site of the nontransportable glutamate analogue dihydrokainate (DHK) has been identified (24). Two adjacent amino acid residues, tyrosine-403 and glutamate-404, appear to be involved in binding of the coupling ions (15, 25), and tyrosine-403 behaves as if it is alternately accessible to either side of the membrane (26). They are located directly in the middle of the stretch of 76 aa that is highly conserved not only between the five glutamate isotransporters, but also in small neutral amino acid transporters (27–30) as well as in bacterial glutamate and dicarboxylic acid transporters (31). The topology of GLT-1 recently has been solved by using a series of functional transporters containing single cysteines. Their topological disposition was determined by using a biotinylated sulfhydryl reagent. The glutamate transporter has eight transmembrane domains long enough to span the membrane as α-helices. Between the seventh and eighth domain a structure reminiscent of a pore loop and an outward-facing hydrophobic linker are positioned (32). The two adjacent residues, tyrosine-403 and glutamate-404, appear to be involved in potassium binding and are close to one of the sodium-binding sites. The essential residues 396–400 are prime candidates for this function (25, 26). All of the above residues are located in the newly identified transmembrane domain 7 (32, 33).
Despite this progress in understanding structure–function relationships, many open questions remain. For example: (i) Where are the binding sites for glutamate located? and (ii) What are the structural determinants for coupling ion and glutamate flux? In this paper we report the identification of a residue of GLT-1, serine-440, which is likely to be close to the binding site for glutamate. It is located in the ascending limb of the pore-loop-like structure (32). Interestingly, mutation of Ser-440 to glycine results in a transporter with broadened ion specificity. This promiscuity is influenced by substitutions at another residue, serine-443, located at the outer edge of the pore-loop-like structure. Our findings suggest that at least part of this structure is crucial for the coupling of sodium and glutamate fluxes.
MATERIALS AND METHODS
Cell Growth and Expression.
HeLa cells were cultured (34), infected with recombinant vaccinia/T7 virus vTF7–3 (35), and transfected with plasmid DNA encoding wild-type or mutant GLT-1, as described (34). Solubilization of transporters expressed in the HeLa cells, their reconstitution in proteoliposomes (15, 19), and transport measurements (15, 19) were done as described. The following radioactive substrates were used: d-[3H]aspartate, 10.5 Ci/mmol, l-[3H]aspartate, 15.5Ci/mmol (both from DuPont/NEN), and l-[3H]glutamate, 60 Ci/mmol (American Radiolabeled Chemicals, St. Louis). Data are presented after subtracting the values obtained from cells transfected with the vector Bluescript SK(−) alone. Inhibition experiments with MTS reagents (Toronto Research Chemicals, Downsview, ON, Canada) were done as described (26). Briefly, cells were plated in 24-well plates and washed with transport medium consisting of 150 mM NaCl/0.5 mM MgSO4/0.3 mM CaCl2/5 mM KPi, pH 7.4. When indicated, 150 mM sodium chloride was substituted with the same concentration of choline chloride or lithium chloride. Each well then was incubated at room temperature with 200 μl of solutions of different ionic composition (see legends to Figs. 4–6) containing the indicated concentration of the specified methanethiosulfonate (MTS) reagent. After 5 min the medium was aspirated and the cells were washed twice with 1 ml of the choline chloride-containing medium, followed by the transport assay, using 200 μl NaCl-containing transport medium supplemented with 0.4 μCi of the radiolabeled amino acid for each well. Each experiment was performed at least three times. Typical experiments are shown. Protein was determined by Lowry’s method (36).
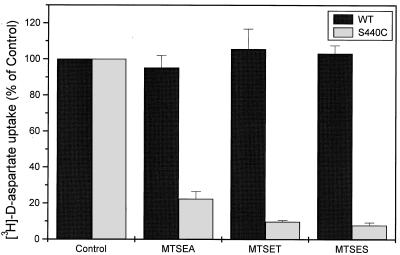
Figure 4.
Effect of MTS reagents on wild-type and S440C mutant transporters. Sodium-dependent d-[3H]aspartate uptake was measured in HeLa cells expressing WT and S440C mutant transporters. Before the transport assay the cells were washed with 150 mM choline chloride-containing medium and subsequently preincubated in the same medium supplemented with or without MTSEA, MTSET, and MTSES at 0.1 mM, 0.2 mM, and 0.25 mM, respectively. After 5 min at room temperature, the cells were washed and assayed for d-[3H]aspartate transport for 10 min. Each bar is mean ± SE of three different experiments. The control value of S440C is 89.5 ± 6.8% of that of wild-type GLT-1.
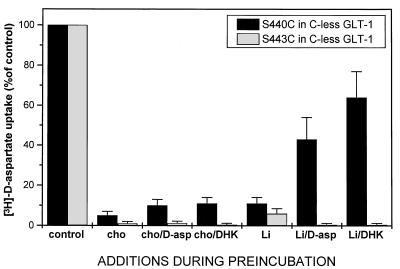
Figure 6.
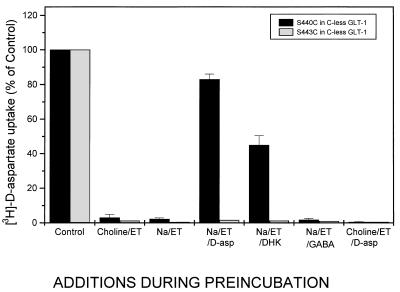
Effect of lithium on protection by d-aspartate and dihdyrokainate against inhibition by MTSET of transport in S440C and S443C. Conditions are the same as those described in the legend to Fig. 5 using, during the preincubation, 150 mM choline chloride (Cho) or 150 mM LiCl (Li) in the presence of 0.15 mM MTSET, with the indicated additions at 1 mM final concentrations. Control, preincubation without MTSET.
Site-Directed Mutagenesis.
Site-directed mutagenesis (37, 38) was done by using uracil-containing single-strand DNA derived from the shortened GLT-1 clone (39) or the cysteine-less GLT-1 construct (26, 32). After verification of the mutants by DNA sequencing, the mutations were subcloned into the original constructs using BsrGI and BstEII. Subcloned DNAs were sequenced in both directions between these two unique restriction sites.
RESULTS
Determinants of Dihydrokainate Sensitivity.
The stretch of residues 364–439 from GLT-1 corresponds to the domain of 76 aa from its human homologue EAAT-2, which has been shown to contain at least part of the determinants for DHK binding (24). Within this stretch of 76 aa GLT-1 differs from the other four glutamate isotransporters in 18 positions. Most of these were individually mutated to the corresponding residues of either GLAST-1 (20) or EAAC-1 (21). All of the mutant transporters exhibited DHK sensitivity similar to that of the wild type (data not shown), suggesting that multiple residues are required for this sensitivity.
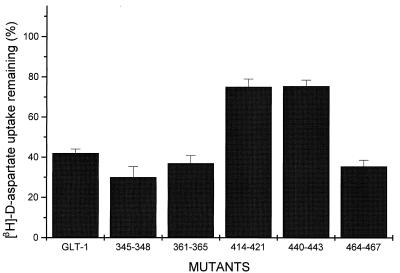
To identify these residues, five clusters in which there are large differences between GLT-1 and GLAST-1 were selected and mutated by introducing the sequence of amino acids from GLAST-1 into GLT-1. These are residues 345–348 (SFFA of GLT-1 are simultaneously mutated to VFIG), 361–365 (ASSAG → SSSSA), 414–421 (MNGVILDG → VNNFDLNF), 440–443 (SIPS → GIPQ), and 464–467 (SLLV → TLII). Two of the clusters, 361–365 and 414–421, correspond to sequences within the 76-aa domain, whereas the three others lie outside this domain. Upon expression in HeLa cells, a significant reduction of DHK sensitivity is observed in two clusters: 414–421 and 440–443 (Fig. 1).
Figure 1.
Effect of dihydrokainate on GLT-1 wild-type and cluster mutant transporters. Sodium-dependent d-[3H]aspartate uptake was measured in HeLa cells expressing wild-type GLT-1 or the indicated cluster mutant transporters in the presence and absence of 120 μM DHK. Values—after 10 min of transport—are expressed as a percentage of those measured in the presence versus absence of DHK. Each bar is the mean ± SE of five different experiments. The uninhibited transport values were (% of wild type): 345–348, 106.4 ± 8.4; 361–365, 66.6 ± 2.8; 414–421, 76.4 ± 3.4; 440–443, 57.4 ± 3.6; and 464–467, 109.3 ± 4.1.
GLT-1 Mutants with Altered Ion Coupling.
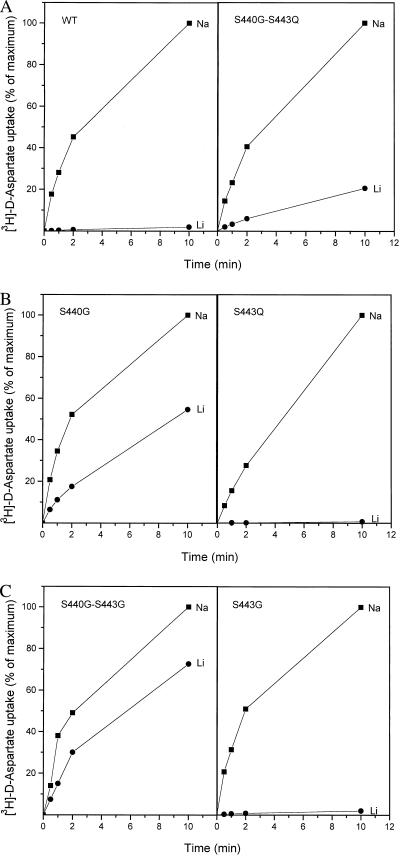
Even though two clusters have been identified that influence the sensitivity of GLT-1 to DHK (Fig. 1), we have continued to study cluster 440–443 in detail. This is because the transporters in which this cluster is mutated to the corresponding GLAST-1 sequence (S440G–S443Q) exhibit increased lithium-supported d-[3H]aspartate uptake in whole cells, relative to the wild type (data not shown). This is not observed when the GLT-1 sequence 414–421 is replaced by that of GLAST-1 (data not shown). Whole-cell uptake may reflect net influx involving countertransport with potassium or exchange with internal glutamate or aspartate (15, 25). The latter could also proceed without external sodium because exchange requires the presence of sodium only on one side of the membrane (13, 14). To determine whether lithium can drive net influx, it is necessary to eliminate “trans” aspartate and glutamate present in intact cells. The S440G–S443Q transporters were solubilized from transfected HeLa cells and reconstituted into liposomes containing internal potassium. A small but very significant net flux of d-[3H]aspartate was observed when the external sodium was replaced with lithium (Fig. 2A). In contrast, influx in the reconstituted wild-type GLT-1 is almost absolutely sodium-dependent: in the presence of lithium uptake is exceedingly low—less than 2% of the levels in the presence of sodium (Fig. 2A)—but it is still significantly higher than background levels observed in the presence of choline. In this medium no uptake at all was detected in the wild type or in the double mutant (data not shown, but see Fig. 3).
Figure 2.
d-[3H]Aspartate transport in proteoliposomes containing wild-type GLT-1 or transporters with mutations at positions 440 and/or 443. HeLa cells were infected with vaccinia/T7 recombinant virus and transfected with wild-type or mutant transporters. Subsequently, the transporters were solubilized and reconstituted as described in Materials and Methods. The internal medium contained 0.12 M potassium phosphate, pH 7.4. Net influx of d-[3H]aspartate was measured at the indicated times using 0.15 M of either NaCl (■), LiCl (●), or choline chloride (not shown, but indistinguishable from the abscissa) and 2.5 μM of valinomycin. (A) Wild type (WT) and S440G–S443Q. (B) S440G and S443Q. (C) S440G–S443G and S443G. For each time point the data are expressed as percentage of the value in sodium-containing medium after 10 min. These values were 2,448, 833, 803, 1,508, 450, and 2,302 pmol/mg protein for wild type, S440G–S443Q, S440G, S443Q, S440G–S443G, and S443G transporters, respectively.
Figure 3.
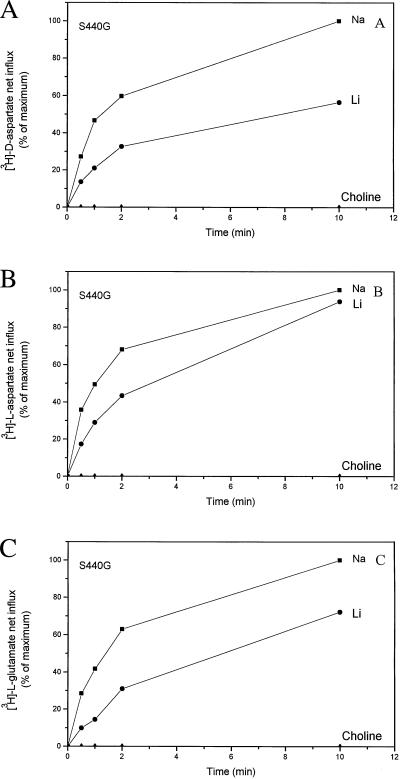
Comparison of d- and l-[3H]aspartate and l-[3H]glutamate transport in proteoliposomes containing S440G transporters. Conditions are the same as those given in the legend to Fig. 2, with 0.15 M of NaCl (■), LiCl (●), and choline chloride (▴) as external media containing the radiolabeled substrates. The 10-min values in sodium-containing media are taken as 100% values. Those were 466, 267, and 26 pmol/mg protein for d- and l-aspartate and l-glutamate, respectively, measured with 0.25, 0.17, and 0.043 μM as external concentrations, respectively. In the case of wild type (not depicted in the figure) the 10-min values were 1,160 (NaCl), 18 (LiCl), and <1 (choline chloride) pmol/mg protein for d-aspartate; 669 (NaCl), 13 (LiCl), and <1 (choline chloride) pmol/mg protein for l-aspartate; and 101 (NaCl), 1.4 (LiCl), and <1 (choline chloride) pmol/mg protein for l-glutamate.
To determine which of the two residues in the S440G–S443Q transporters is a determinant for the broadened ion specificity, the single serine replacement mutants have been analyzed for lithium-supported d-[3H]aspartate net influx in reconstituted proteoliposomes. The data presented in Fig. 2B show that serine-440 is responsible for the change in ion specificity. For the phenomenon to be observed it is essential that serine-440 is replaced by glycine, the amino acid residue present at this position in the other eukaryotic glutamate transporters. Replacement of serine-440 with either cysteine or threonine gives rise to sodium specificity similar to that observed with the wild type. The only other residue at this position with which lithium-supported d-[3H]aspartate influx is observed is alanine, but its extent is much lower than in S440G (data not shown). Replacement of serine-443 with glutamine, present at this position in all other glutamate transporters, yields wild-type-like behavior, and the same is true when it is replaced with glycine (Fig. 2C).
It should be noted that in S440G transporters, which have a serine residue at position 443, the lithium-dependent net flux relative to the sodium (Fig. 2B) is larger than that in the double-mutant S440G–S443Q (Fig. 2A). This suggests that the nature of the residue at the 443 position may modulate the broadening of the ion specificity mediated by the glycine at 440. Indeed, introduction of a glycine at 443 together with the glycine at 440 (S440G–S443G) gives rise to the highest lithium-dependent flux (Fig. 2C), an observation consistently made in all four experiments in which S440G and S440G–S443G were tested together. The lithium-supported net influx has been seen with all substrates tested; with l-[3H]aspartate and l-[3H]glutamate results similar to those with d-[3H]aspartate are obtained (Fig. 3).
Kinetic Analysis of S440G Transporters.
In the wild type, the very low rates of lithium-supported uptake of d-[3H]aspartate and l-[3H]glutamate are a result of effects on both Km and Vmax, the former being the most pronounced (Table 1). S440G transporters have a similar Vmax for both substrates, in lithium as well as in sodium media. These Vmax values are similar to those of the wild type in the presence of lithium. The striking effect of the S440G mutation is that the dramatic increase in the Km for substrate in lithium-containing medium, observed in the wild type, is almost completely eliminated (Table 1). This reduction is more pronounced with d-[3H]aspartate than with l-[3H]glutamate, but also, in the latter case the effect is very significant (Table 1). On the other hand, the affinity of both the wild-type and the S440G transporter for the nontransportable analogue DHK is the same in both media, although the affinity of the S440G transporters is 7-fold lower than that of the wild type. There is no change in the inhibition by DHK in transporters in which serine-443 is replaced by glutamine, which occupies this position in GLAST-1 (data not shown). Because isoleucine-441 and proline-442 are the same in both transporters, serine-440 appears to be the main determinant for DHK sensitivity in the cluster 440–443.
Table 1.
Kinetic constants for d-aspartate and l-glutamate transport and inhibition by dihydrokainate
|
d-Aspartate
|
l-Glutamate
|
DHK
|
|||
|---|---|---|---|---|---|
| Km | Vmax | Km | Vmax | Ki | |
| WT | |||||
| Na | 2.79 ± 0.71 | 987 ± 45 | 5.34 ± 1.3 | 802 ± 71 | 21.6 ± 2.4 |
| Li | 44.4 ± 4.53 | 215 ± 21.2 | 49 ± 1 | 125 ± 4.2 | 36 ± 9.4 |
| S440G | |||||
| Na | 2.83 ± 0.55 | 171 ± 18.9 | 5.63 ± 0.6 | 107 ± 7 | 150 ± 19 |
| Li | 3.98 ± 0.25 | 126 ± 7.1 | 15 ± 1.7 | 118 ± 7 | 216 ± 10.6 |
Influx of d-[3H]aspartate and l-[3H]glutamate was measured for 1 min in proteoliposomes containing wild-type GLT-1 (WT) or S440G transporters in a final volume of 0.38 ml of 0.15 M NaCl- or LiCl-containing medium. In the NaCl containing medium, 4 μCi of d-[3H]aspartate (10.5 Ci/mmol) or l-[3H]glutamate (60 Ci/mmol) was present in each assay supplemented with 1, 2, 4, 6, and 10 μM unlabeled substrate. In the LiCl containing medium 6 μm, 6 μCi labeled substrate together with 2, 4, 6, 10, and 15 μM of the unlabeled amino acid were used or 10 μCi labeled and 20 and 40 μM unlabeled substrate. The Ki for DHK was determined by performing d-[3H]aspartate transport in the absence or presence of DHK. In sodium-containing medium DHK was present at 80 and 400 μM for wild-type GLT-1 and S440G, respectively. In lithium medium the concentrations were 40 and 200 μM, respectively. In the latter medium 5 μCi of d-[3H]aspartate was present together with 2, 4, 5, 10, 15, and 20 μM unlabeled d-aspartate. In the sodium medium 4 μCi labeled together with 1, 2, 4, 6, and 10 μM unlabeled substrate were used. Transport reactions were terminated after 1 min. Km and Ki are expressed in μM and Vmax is expressed as pmol/mg⋅protein/min. Determination ± SEM as done at least three times.
Accessibility to MTS Reagents in Cluster 440–443.
Because DHK is a nontransportable glutamate analogue, it is possible that serine-440, which affects DHK sensitivity, is close to the acidic amino acid-binding site. To explore this possibility, we have substituted serine-440 with cysteine, with the goal of monitoring the accessibility of this cysteine to sulfhydryl reagents, in the presence and absence of glutamate analogues. As can be seen in Fig. 4, S440C transporters are strongly inhibited by small hydrophilic sulfhydryl reagents, such as the positively charged (2-aminoethyl)methanethiosulfonate (MTSEA) and [(2-trimethylammonium)ethyl]methanethiosulfonate (MTSET), as well as by the negatively charged (2-sulfonatoethyl)methanethiosulfonate (MTSES). The wild-type GLT-1 is not inhibited by any of these compounds (Fig. 4). As expected in the case of a close proximity to the acidic amino acid-binding site, the inhibition of S440C transporters by the MTS reagents can be protected by transportable substrates as well as by DHK. In the latter case, the extent of protection with DHK is less than that seen with d-aspartate. The inhibition by the MTS reagents and the protection by the transportable substrates as well as by the nontransportable DHK are the consequence of a direct modification of the cysteine at position 440. This phenomenon is not only observed in a wild-type background, but also with transporters in which this is the only cysteine (S440C in cysteine-less GLT-1, Fig. 5). Sodium, on the other hand, does not confer any protection (Fig. 5). It is of interest to note that the protective effect of d-aspartate as well as of DHK is sodium-dependent; it is not observed in the presence of choline (Figs. 5 and 6). On the other hand, in the presence of lithium they both offer significant protection (Fig. 6). As is the case with d-aspartate, l-glutamate also protects against the inhibition by MTSET (data not shown). γ-Aminobutyric acid (GABA), which is not a substrate, does not reduce inhibition of transport by MTSET (Fig. 5). The results are presented for inhibition by the positively charged impermeant MTSET, but similar data are obtained with the negatively charged MTSES (data not shown). Also, in transporters with a single cysteine introduced at position 443, activity also is inhibited by MTS reagents, here shown for MTSET (Figs. 5 and 6). However, in this mutant transporter no protection is provided by substrate or DHK (Figs. 5 and 6).
Figure 5.
Effect of d-aspartate and dihydrokainate on inhibition of transport of S440C and S443C in a cysteine-less background. HeLa cells expressing single-cysteine S440C or S443C transporters were preincubated with 150 mM choline chloride or 150 mM NaCl for 5 min in the absence or presence of 0.15 mM MTSET. Where indicated, 1 mM d-aspartate, GABA, or DHK was added during the preincubation. It should be noted that only one control (choline chloride without MTSET) is shown, because all the others (without methanethiosulfonate reagent) had similar values. The cells were washed twice and assayed for sodium-dependent d-[3H]aspartate uptake for 10 min. The results are given as the percentage of transport in the presence of MTSET (ET) relative to transport in the absence of the sulfhydryl reagent. Each bar is the mean ± SE of three different experiments. The control values for single-cysteine 440C and 443C transporters were 48.1% ± 7.8 and 77.8% ± 7.8 of that of the cysteine-less GLT-1, respectively. The latter was not inhibited by MTSET under any of the conditions (data not shown).
DISCUSSION
The most remarkable phenotypic change of the S440G mutant is the ability of lithium to drive net flux of acidic amino acids. It should be noted that even in the wild type, the sodium dependence of net influx of transportable substrates is not absolute. Our detection system, reconstituted proteoliposomes, is sensitive enough to detect the very low levels obtained in the wild type in the presence of lithium (Fig. 2A). The replacement of sodium by lithium results both in a 4- to 6-fold reduction of the Vmax, and in a 10- to 16-fold increase in apparent Km, depending on the acidic amino acid tested (Table 1). The striking effect of the mutation is that the apparent Km values for the substrate have become similar in lithium and sodium media, and they resemble the wild-type affinities observed in the presence of sodium.
The Ki for DHK observed with reconstituted wild-type transporter, around 20 μM (Table 1), is in reasonable agreement with the Ki of its human homologue, as seen in oocytes (10 μM, ref. 11). On the other hand, the affinity for the blocker in intact HeLa cells appears to be lower (Fig. 1). This is not fully understood, but is consistent with earlier observations on the two systems (15, 37). Nevertheless, it is obvious that DHK sensitivity in intact cells is a useful diagnostic tool that can be used to screen for mutants with a reduced affinity for the blocker (Fig. 1, Table 1).
It is of interest to note that although the Km of the S440G transporter for transportable substrates is not impaired, there is a 7-fold reduction in its affinity for the nontransportable analogue DHK (Table 1). This indicates that serine-440 is not directly involved in binding of glutamate or its analogues, i.e., it does not ligand the amino or the two carboxyl groups.
Even though position 440 is not at the binding site for glutamate, our accessibility studies indicate that it is not far from this site. When a cysteine is introduced at this position, the transporter becomes sensitive to both positively and negatively charged MTS reagents (Figs. 4 and 5). This inhibition can be protected against by DHK as well as by substrate—as long as sodium is present as well (Fig. 5). Significantly, sodium by itself does not provide any protection (Fig. 5). An alternative explanation for the protection data is that the binding site for substrate and DHK is distant from the 440 position, but binding causes a conformational change that decreases the accessibility of cysteine-440. However, in this case one would have to postulate that the nature of the side chain of the amino acid residue at position-440, hydroxymethyl versus hydrogen, exerts a reciprocal conformational change such that it would modify the distant DHK-binding site (Fig. 1 and Table 1). Although this scenario cannot be ruled out we favor the simple explanation that position 440 is not far from the substrate-binding site. A recent study employing chimeric glutamate transporters identifies one domain as particularly important for substrate binding (40). It is of special interest to note that serine residues 440 and 443 are at the amino-terminal edge of this domain. Introduction of cysteine at the 443 position also renders GLT-1 sensitive to the MTS reagents, but in this case transportable substrates and DHK cannot protect against the inhibition. This suggests that part of the glutamate molecule obstructs the access of the sulfhydryl reagent to the cysteine introduced at the 440 position, but not to that introduced at the 443 position. The extent of protection should be dependent on the shape and dimensions of the glutamate analogue, and this is probably the reason for the different extents of protection by d-aspartate and DHK (Fig. 5). One would predict that serine-443 is located closer to the extracellular space than serine-440. In fact, this is corroborated by the recent determination of the topology of GLT-1 (32): both serines are located in the ascending limb of the pore-loop-like structure, with serine-443 at the extracellular surface (32).
It is important to note two key differences in the accessibility of S440C to MTS reagents, as compared with that of Y403C (26). First, tyrosine-403 is involved in potassium binding (25) and only positively charged MTS reagents can react with and inhibit Y403C, as if it were part of a negatively charged binding pocket for cations. Exposure of S440C transporters to 0.25 mM of the anionic MTSES for 5 min results in an almost complete inhibition (Fig. 4) whereas Y403C transporters are not affected at all by 10 mM of MTSES at similar incubation conditions (26). Second, Y403C transporters can be protected against inhibition by the MTS reagents by transportable substrates, but, in contrast, are sensitized to them by the nontransportable DHK. These and other observations imply alternating access for the 403 position (26). The striking differences in accessibility to MTS reagents between Y403C and S440C (Figs. 4 and 5) indicate that serine-440 is not part of a cationic-binding site.
Changes in ion specificity also have been obtained by conservative mutations at the 403 position (25). In that case, however, potassium binding was compromised and the transporters were locked in the exchange mode. The ability of other ions to replace sodium during exchange was interpreted to mean that sodium- and potassium-binding sites are close. Because it appears that two of the three sodium ions are less specific and replaceable by lithium (16), the ability of a single mutation to render the exchange supportable by lithium probably means that the specific sodium site was modulated. This argument also holds here with one important difference: the mutation is not compromising potassium binding, because the S440G mutants still can catalyze net flux, which is dependent on trans-potassium (12–15). Interestingly, the nature of the substituent at the nearby 443 position can modulate the broadening of the ion specificity (Figs. 2 and 3): net flux of the S440G–S443G mutant is almost as efficient with lithium as with sodium (Fig. 2C).
How can a mutation that is presumably near the glutamate-binding site alter the ion specificity at the specific sodium site? In the absence of a crystal structure we do not know the answer and cannot rule out a long-range conformational change, but we can speculate on two attractive possibilities. The Ki inhibition of transport by the nontransportable analogue (DHK) is not very different in lithium or in sodium media both in the wild type and S440G (Table 1). Furthermore in S440C, d-aspartate and DHK afford protection against inhibition of transport by MTSET in sodium and lithium media but not in those containing choline (Figs. 5 and 6). These observations indicate that sodium and lithium, but not choline, can induce a conformational change in the transporter such that it can bind transportable substrates and DHK. Because transport in the wild type is greatly impaired in lithium, it seems therefore that it is a step after glutamate binding that is responsible for the sodium specificity of transport by GLT-1. At least two, if not all three, of the sodium ions bind before glutamate in the translocation cycle (13–15). Also, lithium can replace sodium in eliciting kainate-sensitive transient currents to EAAT-2, the human homologue of GLT-1 (M. P. Kavanaugh, personal communication). These are thought to reflect conformational changes of the transporter induced by sodium (or lithium) binding (11). This also indicates that the specificity for the sodium somehow is introduced during the translocation step. This idea is also consistent with the decrease in Vmax observed in the S440G mutant relative to the wild type. An attractive possibility is that one of the carboxyl groups of the acidic amino acid may participate in the liganding of the specific sodium and thereby confer the ion specificity. A mutation at the 440 position could cause a small change in the way the substrate is bound, thereby altering the ion selectivity. This idea is consistent with observations on chimeras derived from two related, bacterial, ion-coupled melibiose transporters. In several of these chimeras the ion specificity is dependent on the nature of the sugar substrate used (41), suggesting a tight linkage between the driving ion and the driven substrate. Alternatively, the acidic amino acid substrates may induce a conformational change, so that serine-440 is brought into the proximity of the ion-binding site. It therefore will be important to determine the distance between the residues of the pore-loop-like structure and tyrosine-403 in the presence and absence of substrate. The groundwork for such an approach has been laid in experiments on the proton-coupled lactose permease of Escherichia coli (42, 43). A detailed structure–function study of the pore-loop-like structure and the surrounding transmembrane domains is expected to yield important insights into the coupling mechanism.
Acknowledgments
We thank Mrs. Beryl Levene for expert secretarial assistance and Dr. Lihi Brocke and Estelle R. Bennett for critical reading of the manuscript. Special thanks go to Myriam Grunewald for doing the experiments documented in Fig. 6 in the framework of the revision of this manuscript. This work was supported by the U.S.–Israel Binational Science Foundation, Federal Ministry of Education, Science, Research and Technology, Germany (BMBF), and BMBF’s International Bureau at the Deutsches Zentrum für Luft und Raumfahrt, and the Bernard Katz Minerva Center for Cellular Biophysics.
ABBREVIATIONS
- DHK
dihydrokainate
- MTSEA
(2-aminoethyl)methanethiosulfonate
- MTSET
[(2-trimethylammonium)ethyl]methanethiosulfonate
- MTSES
(2-sulfonatoethyl)methanethiosulfonate
- GABA
γ-aminobutyric acid
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Kanner B I, Schuldiner S. CRC Crit Rev Biochem. 1987;22:1–38. doi: 10.3109/10409238709082546. [DOI] [PubMed] [Google Scholar]
- 2.Nicholls D, Attwell D. Trends Pharmacol Sci. 1990;11:462–468. doi: 10.1016/0165-6147(90)90129-v. [DOI] [PubMed] [Google Scholar]
- 3.Rothstein J D, Dykes Hoberg M, Pardo C A, Bristol L A, Jin L, Kuncl R W, Kanai Y, Hediger M A, Wang Y, Schielke J P, Welty D F. Neuron. 1996;16:675–686. doi: 10.1016/s0896-6273(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka K, Watase K, Manabe T, Yamada K, Watanabe M, Takahashi K, Iwama H, Nishikawa T, Ichihara N, Kikuchi T, et al. Science. 1997;276:1699–1702. doi: 10.1126/science.276.5319.1699. [DOI] [PubMed] [Google Scholar]
- 5.Mennerick S, Zorumski C F. Nature (London) 1994;368:59–62. doi: 10.1038/368059a0. [DOI] [PubMed] [Google Scholar]
- 6.Tong G, Jahr C E. Neuron. 1994;13:1195–1203. doi: 10.1016/0896-6273(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 7.Otis T S, Wu Y C, Trussell L O. J Neurosci. 1996;16:1634–1644. doi: 10.1523/JNEUROSCI.16-05-01634.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamond J S, Jahr C E. J Neurosci. 1997;17:4672–4687. doi: 10.1523/JNEUROSCI.17-12-04672.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanner B I, Sharon I. Biochemistry. 1978;17:3949–3953. doi: 10.1021/bi00612a011. [DOI] [PubMed] [Google Scholar]
- 10.Brew H, Attwell D. Nature (London) 1987;327:707–709. doi: 10.1038/327707a0. [DOI] [PubMed] [Google Scholar]
- 11.Wadiche J I, Arriza J L, Amara S G, Kavanaugh M P. Neuron. 1995;14:1019–1027. doi: 10.1016/0896-6273(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 12.Zerangue N, Kavanaugh M P. Nature (London) 1996;383:634–637. doi: 10.1038/383634a0. [DOI] [PubMed] [Google Scholar]
- 13.Kanner B I, Bendahan A. Biochemistry. 1982;21:6327–6330. doi: 10.1021/bi00267a044. [DOI] [PubMed] [Google Scholar]
- 14.Pines G, Kanner B I. Biochemistry. 1990;29:11209–11214. doi: 10.1021/bi00503a008. [DOI] [PubMed] [Google Scholar]
- 15.Kavanaugh M P, Bendahan A, Zerangue N, Zhang Y, Kanner B I. J Biol Chem. 1997;272:1703–1708. doi: 10.1074/jbc.272.3.1703. [DOI] [PubMed] [Google Scholar]
- 16.Grunewald M, Kanner B. J Biol Chem. 1995;270:17017–17024. doi: 10.1074/jbc.270.28.17017. [DOI] [PubMed] [Google Scholar]
- 17.Danbolt N C, Pines G, Kanner B I. Biochemistry. 1990;29:6734–6740. doi: 10.1021/bi00480a025. [DOI] [PubMed] [Google Scholar]
- 18.Danbolt N C, Storm-Mathisen J, Kanner B I. Neuroscience. 1992;51:295–310. doi: 10.1016/0306-4522(92)90316-t. [DOI] [PubMed] [Google Scholar]
- 19.Pines G, Danbolt N C, Bjoras M, Zhang Y, Bendahan A, Eide L, Koepsell H, Storm-Mathisen J, Seeberg E, Kanner B I. Nature (London) 1992;360:464–467. doi: 10.1038/360464a0. [DOI] [PubMed] [Google Scholar]
- 20.Storck T, Schulte S, Hofmann K, Stoffel W. Proc Natl Acad Sci USA. 1992;89:10955–10959. doi: 10.1073/pnas.89.22.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanai Y, Hediger M A. Nature (London) 1992;360:467–471. doi: 10.1038/360467a0. [DOI] [PubMed] [Google Scholar]
- 22.Fairman W A, Vandenberg R J, Arriza J L, Kavanaugh M P, Amara S G. Nature (London) 1995;375:599–603. doi: 10.1038/375599a0. [DOI] [PubMed] [Google Scholar]
- 23.Arriza J L, Eliasof S, Kavanaugh M P, Amara S G. Proc Natl Acad Sci USA. 1997;94:4155–4160. doi: 10.1073/pnas.94.8.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vandenberg R J, Arriza J L, Amara S G, Kavanaugh M P. J Biol Chem. 1995;270:17668–17671. doi: 10.1074/jbc.270.30.17668. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Bendahan A, Zarbiv R, Kavanaugh M P, Kanner B I. Proc Natl Acad Sci USA. 1998;95:751–755. doi: 10.1073/pnas.95.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zarbiv R, Grunewald M, Kavanaugh M P, Kanner B I. J Biol Chem. 1998;273:14231–14237. doi: 10.1074/jbc.273.23.14231. [DOI] [PubMed] [Google Scholar]
- 27.Arriza J L, Kavanaugh M P, Fairman W A, Wu Y N, Murdoch G H, North R A, Amara S G. J Biol Chem. 1993;268:15329–15332. [PubMed] [Google Scholar]
- 28.Shafqat S, Tamarappoo B K, Kilberg M S, Puranam R S, McNamara J O, Guadano Ferraz A, Fremeau R T., Jr J Biol Chem. 1993;268:15351–15355. [PubMed] [Google Scholar]
- 29.Utsunomiya Tate N, Endou H, Kanai Y. J Biol Chem. 1996;271:14883–14890. doi: 10.1074/jbc.271.25.14883. [DOI] [PubMed] [Google Scholar]
- 30.Kekuda R, Prasad P D, Fei Y J, Torres Zamorano V, Sinha S, Yang Feng T L, Leibach F H, Ganapathy V. J Biol Chem. 1996;271:18657–18661. doi: 10.1074/jbc.271.31.18657. [DOI] [PubMed] [Google Scholar]
- 31.Tolner B, Ubbink Kok T, Poolman B, Konings W N. J Bacteriol. 1995;177:2863–2869. doi: 10.1128/jb.177.10.2863-2869.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grunewald M, Bendahan A, Kanner B I. Neuron. 1998;21:623–632. doi: 10.1016/s0896-6273(00)80572-3. [DOI] [PubMed] [Google Scholar]
- 33.Slotboom D J, Lolkema J S, Konings W N. J Biol Chem. 1996;271:31317–31321. doi: 10.1074/jbc.271.49.31317. [DOI] [PubMed] [Google Scholar]
- 34.Keynan S, Suh Y J, Kanner B I, Rudnick G. Biochemistry. 1992;31:1974–1979. doi: 10.1021/bi00122a011. [DOI] [PubMed] [Google Scholar]
- 35.Fuerst T R, Niles E G, Studier F W, Moss B. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowry O H, Rosebrough N J, Farr A L, Randall R J. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 37.Pines G, Zhang Y, Kanner B I. J Biol Chem. 1995;270:17093–17097. doi: 10.1074/jbc.270.29.17093. [DOI] [PubMed] [Google Scholar]
- 38.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 39.Casado M, Bendahan A, Zafra F, Danbolt N C, Aragon C, Gimenez C, Kanner B I. J Biol Chem. 1993;268:27313–27317. [PubMed] [Google Scholar]
- 40.Mitrovic A D, Amara S G, Johnston G A R, Vandenberg R J. J Biol Chem. 1998;273:14698–14706. doi: 10.1074/jbc.273.24.14698. [DOI] [PubMed] [Google Scholar]
- 41.Hama H, Wilson T H. J Biol Chem. 1993;268:10060–10065. [PubMed] [Google Scholar]
- 42.Wu J, Kaback H R. Proc Natl Acad Sci USA. 1996;93:14498–14502. doi: 10.1073/pnas.93.25.14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun J, Kemp C R, Kaback H R. Biochemistry. 1998;37:8020–8026. doi: 10.1021/bi973192s. [DOI] [PubMed] [Google Scholar]