Abstract
Coccidioides posadasii is a fungal respiratory pathogen which is responsible for recurrent epidemics of San Joaquin Valley fever (coccidioidomycosis) in desert regions of the southwestern United States. Numerous studies have revealed that the cell wall of the parasitic phase of the fungus is a reservoir of immunoreactive macromolecules and a potential source of a vaccine against this mycosis. A 495-bp fragment of a C. posadasii gene which encodes a putative wall-associated, glycosylphosphatidylinositol (GPI)-anchored β-1,3-glucanosyltransferase was identified by computational analysis of the partially sequenced genome of this pathogen. The translated, full-length gene (GEL1) showed high sequence homology to a reported β-1,3-glucanosyltransferase of Aspergillus fumigatus (70% identity, 90% similarity) and was selected for further study. The GEL1 mRNA of C. posadasii was detected at the highest level during the endosporulation stage of the parasitic cycle, and the mature protein was immunolocalized to the surface of endospores. BALB/c or C57BL/6 mice were immunized subcutaneously with the bacterium-expressed recombinant protein (rGel1p) to evaluate its protective efficacy against a lethal challenge of C. posadasii by either the intraperitoneal or intranasal route. In both cases, rGel1p-immune mice infected with the pathogen showed a significant reduction in fungal burden and increased survival compared to nonimmune mice. The recombinant β-1,3-glucanosyltransferase is a valuable addition to an arsenal of immunoreactive proteins which could be incorporated into a human vaccine against coccidioidomycosis.
Coccidioides is the etiological agent of a human respiratory disease known as coccidioidomycosis or San Joaquin Valley fever. The infectious, air-dispersed spores (arthroconidia) of the filamentous fungus are produced from hyphae that grow primarily in the soil of desert and semiarid regions of North America, Mexico, and disjunct areas of Central and South America (6). Two species of Coccidioides are now recognized (11). C. posadasii, formerly known as the non-California C. immitis strain, is found primarily in Texas, Arizona, and endemic regions outside the U.S., while C. immitis has a smaller biogeographic distribution that is centered in the San Joaquin Valley of California. The parasitic cycles of these two species appear to be morphogenetically identical. Inhaled arthroconidia differentiate into large, multinucleate spherules that give rise to endospores. The latter are responsible for dissemination of the pathogen from original sites of ingress. Infection with C. posadasii is most often asymptomatic or mildly symptomatic, while the disseminated form of the mycosis can be life-threatening and may involve the skin, joints, bone, and meninges in addition to the lungs. The emergence of coccidioidomycosis in nonendemic areas (9) and its occurrence in rising numbers among immunocompromised patients (41) are current issues of concern. An increase in the incidence of coccidioidal infections in pets and wild animals has also been reported (35).
Efforts are under way to develop a human vaccine against this respiratory mycosis (32). Proposed recipients of the vaccine include persons who live in the endemic regions, as well as frequent travelers to the southwestern United States. Vaccination of children in highly endemic regions would provide a major health benefit by reduction of total health care expenditures (2). The feasibility that a coccidioidomycosis vaccine can be developed is based on the observation that natural infection usually confers life-long immunity against the disease (32). The majority of protective reagents that have been characterized to date are products of the parasitic phase of the fungus (22, 32), and the most promising of these has proved to be a cell wall-associated glycoprotein (36). In an attempt to discover additional vaccine candidates, we have used the partial genome database derived from the C. posadasii sequencing project under way at the Institute for Genomic Research for the identification of genes that encode homologs of reported wall-associated proteins (20). Our rationale for this approach was that some of these proteins may be immunogenic and exposed to the host immune system during the parasitic cycle. One mechanism by which cell wall-associated proteins remain bound to the cell surface is via glycosylphosphatidylinositol (GPI) anchors (8). GPI-anchored proteins reported in various microbial pathogens have been shown to be immunogenic and suggested to represent important virulence factors (17, 24). Glucanosyltransferases are representative of such cell wall-bound proteins with GPI linkages, appear to function in the remodeling of newly synthesized polysaccharide polymers of the fungal wall (27), and have been described in both yeast and filamentous fungi (3). In this study we have isolated a gene (GEL1) which encodes a β-1,3-glucanosyltransferase homolog of C. posadasii. We have shown that immunization of BALB/c or C57BL/6 mice with the bacterium-expressed recombinant protein (rGel1p) protects the animals against a lethal challenge of the fungal pathogen.
MATERIALS AND METHODS
Culture conditions and parasitic cell development.
C. posadasii (isolate C735) was used for all experimental procedures reported in this study. The saprobic (mycelial) phase of the fungus was grown on glucose-yeast extract agar (GYE; 1% glucose, 0.5% yeast extract, 2% agar). Arthroconidia (asexual reproductive products of the saprobic phase) were isolated from these plate cultures, suspended in phosphate-buffered saline (PBS), and used to inoculate either liquid GYE medium or a defined glucose-salts medium (21) for growth of the mycelial or parasitic phases, respectively. In vitro growth of parasitic cells (spherules) of C. posadasii demonstrates near-synchronous development during the first generation of the parasitic cycle (17). Spherules obtained from cultures at 72 or 132 h after inoculation with arthroconidia are distinguished by their morphogenetic features; 72-h spherules are in a late stage of isotropic growth and early stage of segmentation, while 132-h spherules are mature and the majority (approximately 80%) have ruptured to release their endospores (6) (see Fig. 3A).
FIG. 3.
(A) Diagrammatic representation of the saprobic and parasitic cycles of C. posadasii. The developmental stages of first- and second-generation parasitic cells are identified by incubation times after inoculation of parasitic cultures, as previously reported (17). (B) RT-PCR amplification of GEL1 and CSA genes expressed in C. posadasii-infected, murine lung tissue. Fluorescence microscopy of Blankofluor-stained whole mounts of infected tissue revealed large numbers of spherules in the endosporulation stage of development (data not shown). The RT-PCR results are representative of three separate preparations of total RNA isolated from three infected mice. (C) QRT-PCR of C. posadasii GEL1 expression at different stages of saprobic and parasitic cell growth in vitro. The relative amounts of GEL1 transcript in the mycelial and spherules (132-h endosporulation stage) were compared to the transcript level in spherules at early segmentation stage (72 h), which was given an arbitrary value of 1. The data indicate that a significant increase in expression of the GEL1 gene occurred during the endosporulation stage of the parasitic cycle.
Genome database analysis and gene discovery.
The C. posadasii genome sequencing project was initiated in 2001 at the Institute for Genomic Research (Rockville, Md.) and is supported by the National Institutes of Health (Bethesda, Md.). The ongoing project involves a whole-genome shotgun strategy for determination of >99% of the 29-Mb genome sequence (30). Only partial coverage of the genome has been completed to date. Genomic libraries of C. posadasii (isolate C735) with inserts of 2 to 10 kb were constructed in the pUC plasmid (Promega, Madison, Wis.) and sequenced from both ends. Each library contained >6 × 105 recombinants, and the combined recombinants of the three libraries have been estimated to be sufficient for sequence analysis of the entire C. posadasii genome (20). Genomic survey sequences have been assembled into unique contigs and incorporated into a public database (available at the website www.tigr.org). Computational analyses of the partial genome database were performed by application of the basic local alignment search tool (BLAST [1]). Sequence alignments were conducted using the translated nucleotide sequences of the contigs and the nonredundant protein database available from the National Center for Biotechnology Information (40; www.ncbi.nlm.nih.gov:80/BLAST/). BLASTX matches (1) were selected with expect (E) values of <10−4 (20). A 495-bp fragment of a single contig was selected on the basis of its high translated sequence homology (E = 10−44) to a reported β-1,3-glucanosyltransferase of Aspergillus fumigatus (Gel1; GenBank accession number AF072700) (14). Sense and antisense primer sequences were selected and synthesized on the basis of regions of the translated 495-bp fragment that aligned with the reported amino acid sequence of the Gel1 protein of A. fumigatus. These nested primers were employed in a PCR with genomic template DNA of C. posadasii isolate C735, and a 391-bp product was amplified. The nucleotide sequences of the sense and antisense primers were 5′-CTCCGGAGGTCTCGTCTAT-3′ and 5′-TAAACCTGCGGCAGCACCCGCGCT-3′, respectively. The amplification conditions included an initial denaturation step at 94°C for 1 min, followed by 30 cycles of denaturation at 94°C for 10 s, annealing at 55°C for 10 s, and extension at 72°C for 1 min. The 391-bp PCR amplicon was ligated into the pCR2.1 TOPO cloning vector (Invitrogen, Carlsbad, Calif.), and the nucleotide sequence of the insert was determined as reported previously (18). This gene fragment of C. posadasii was referred to as GEL1.
More recent BLAST analyses of the partial genome database of C. posadasii have resulted in retrieval of four additional β-1,3-glucanosyltransferase homologs (GEL2 to -5). Preliminary results of these sequence comparisons are also reported here.
GEL1 sequence analysis.
The rapid amplification of cDNA ends (RACE) procedure was used to obtain the full-length cDNA sequence of GEL1, resolve the start of the 5′ untranslated region, and locate the poly(A) addition site of the gene (18). Gene-specific primers were used in combination with a universal primer mix for the SMART RACE procedure, according to the protocol provided by the manufacturer (Clontech, Palo Alto, Calif.). The genome walking method (37) was employed to obtain the full-length genomic sequence of GEL1, using a GenomeWalker kit (Clontech) as described by the manufacturer. Genome walking is a PCR-based method used for sequencing genomic DNA upstream or downstream of a partial gene sequence. Briefly, four restriction libraries of C. posadasii genomic DNA were generated using DraI, EcoRV, PvuI, and StuI restriction enzymes. Adapters were ligated to the restriction fragments, and a two-step, long-distance PCR was performed using the Advantage genomic polymerase mix, as described by the manufacturer (Clontech). The primers used for this first-step PCR amplification were as follows: 5′-AGAATGCGGGGACACAATCTTCTGGAACTG-3′, derived from the sequenced 391-bp amplicon described above, and the adapter primer (AP-1) provided by the manufacturer. The second-step amplification was a nested PCR and was performed using the GEL1-specific primer, 5′-ACTTATTATCTCTTGGCTAAATGCGGATGT-3′, and the nested adapter primer-2 (AP-2) provided by the manufacturer. To obtain the full-length genomic sequence of GEL1, PCR was conducted using the following sense and antisense primers: 5′-CTCCGTTTGAACTCTCCTCTCTTC-3′ and 5′-AAGGAGTTGTCGCCATGCTGC-3′. All PCR products obtained by the RACE and genome walking procedures were separated by agarose gel electrophoresis (1.2%), isolated and ligated into the pCR2.1 TOPO vector, and subjected to nucleotide sequence analysis, as reported elsewhere (18). The NCBI-BLAST and PSI-BLAST programs were used to search for sequences in the GenBank and SWISS-PROT databases with similarities to the translated full-length GEL1 sequence (1). The CLUSTAL X program, version 1.8 (39), was used to align the translated GEL1 of C. posadasii with homologous sequences. The PROSITE algorithm was used to identify conserved motifs in the translated polypeptide with homology to reported proteins (15), and the PSORT and PSORT II algorithms were used for prediction of cellular localization sites of the GEL1 gene product (16, 28).
RT-PCR analysis of GEL1 expression in vivo.
Confirmation that the GEL1 gene was expressed by C. posadasii in vivo was conducted by reverse transcription (RT)-PCR using total RNA as template, which was obtained from abscesses of infected mice. Three C57BL/6 mice (females, 6 weeks old) were challenged by the intraperitoneal (i.p.) route with approximately 100 arthroconidia of C. posadasii as previously described (22). Infected lung tissue obtained from each mouse sacrificed at 12 days postchallenge was used as a source of total RNA for separate RT-PCR assays of GEL1 expression (22). RNA was isolated using the RNeasy protocol, and on-column DNase digestion was conducted as described by the manufacturer (Qiagen, Chatsworth, Calif.). RT was performed using 200 U of Superscript II reverse transcriptase (Invitrogen) in 20 μl of a reaction mixture which contained 50 ng of oligo(dT)17/μl, 0.5 mM deoxynucleoside triphosphates, 10 mM dithiothreitol, 1× first-strand buffer, 17 U of RNAguard RNase inhibitor (Amersham Pharmacia Biotech, Piscataway, N.J.), and 500 ng of template RNA isolated from C. posadasii-infected lung tissue. The RT mixture was incubated at 42°C for 50 min, followed by enzyme inactivation at 70°C for 15 min. The same oligonucleotide primers and PCR conditions for amplification of the 391-bp GEL1 fragment as described above were used for the RT-PCR. To confirm that the murine lung tissue was infected with C. posadasii, simultaneous amplification of the gene that encodes a Coccidioides-specific antigen (CSA; GenBank accession no. AY158466 [31]) was conducted. The sequences of the sense and antisense primers employed for amplification of a 519-bp cDNA fragment of the CSA gene were 5′-AAGTTCTCACTCCTCAGCGCTATCG-3′ and 5′-ACATTAAGGTTCCTCCCCTTCAACC-3′, respectively.
Real-time PCR analysis of GEL1 expression in vitro.
To assess the levels of expression of GEL1 during different stages of the parasitic cycle of C. posadasii, we employed a quantitative real-time PCR (QRT-PCR) assay. Total RNA was separately isolated from near-synchronous, parasitic-phase cultures after 72 and 132 h of incubation in defined glucose salts medium, as previously reported (18) (see Fig. 3A). For purpose of comparison, total RNA was also isolated from the mycelial phase grown in liquid GYE medium for 96 h. Synthesis of cDNA from total RNA obtained from the mycelia and parasitic cells was performed as described above. Oligonucleotide primers used for the real-time PCR assays were designed with software supplied for this purpose by Perkin-Elmer Biosystems (Foster City, Calif.). The sequences of the GEL1-specific sense and antisense primers were 5′-AATCCTTCAGGCGATGGAGG-3′ and 5′-GCGGGAAGAGACTCGCTGT-3′, respectively. This primer pair amplified a 101-bp product using single-stranded template cDNA generated from either the mycelial or parasitic cell-derived RNA preparations. A 191-bp amplicon used for normalization of the assay was derived from the constitutively expressed glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene of C. posadasii (GenBank accession no. AF288134) (18). The sequences of the sense and antisense primers used for amplification of this constitutive gene were 5′-TATGAAGAAGGCCTCTGCCAA-3′ and 5′-ACCTGCGGTAGCATCGAAGAT-3′, respectively. Approximately 500 ng of single-stranded cDNA from each developmental stage was used as template for the QRT-PCR analyses of gene expression. Control PCR and nucleotide sequence analyses confirmed that only single amplicons were generated by each of the above primer pairs, and the products were the appropriate fragments of the GEL1 and GAPDH genes. The QRT-PCR assays were performed using a SYBR Green PCR master mix (PE Biosystems) in the second step of the two-step RT-PCR protocol described by the manufacturer. PCR conditions were as follows: 50°C for 2 min and then 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min. Three assays of GEL1 transcript levels were conducted using separate RNA preparations derived from the mycelial phase and two stages of parasitic cell development. The assays were performed using an ABI PRISM 5700 sequence detection system (PE Biosystems). The data from the QRT-PCR assays were analyzed using the Sequence Detection software package (version 1.7) supplied by the manufacturer of the instrument. The comparative (CT) method (User Bulletin 2, 11 December 1997; PE Biosystems) was used for quantification of gene expression.
Expression of GEL1 by Escherichia coli.
Oligonucleotide primers were designed to amplify a 1.3-kb cDNA fragment of the GEL1 gene which encodes amino acids 19 to 447 (predicted mature protein; see Fig. 2). The nucleotide sequences of the sense and antisense primers were 5′-AGCTCATATGGCCTACGCGGGTG-3′ and 5′-AGGTCGACCTTTAAAGCATTAAAC-3′, which contained engineered NdeI and SalI restriction sites, respectively (nucleotides in boldface type). The amplification parameters were as follows: an initial denaturation step at 94°C for 1 min, followed by 30 cycles of denaturation at 94°C for 10 s, annealing at 50°C for 10 s, and extension at 72°C for 2 min. The 1.3-kb PCR product was digested with NdeI and SalI, separated by agarose gel electrophoresis (1%), excised, and subcloned into the NdeI/SalI site of pET28b (Novagen, Madison, Wis.) to yield the pET28b-GEL1 plasmid construct. Bacterial transformation and purification of the recombinant protein (rGel1p) were performed as previously reported (18). The purified rGel1p was subjected to internal amino acid sequence analysis (18) to confirm its identity. The endotoxin content of the stock solution which contained the recombinant protein (1-mg/ml concentration of PBS [0.1 M, pH 7.4]) was determined using a Limulus amebocyte lysate kit (QCL-1000; BioWhittaker, Walkersille, Md.) as previously reported (22). The stock solution contained 1.0 to 1.5 endotoxin U (5.0 to 7.5 ng of endotoxin) per μg of protein.
FIG. 2.
CLUSTAL X alignment of C. posadasii Gel1p homologs. The sequences of reported β-1,3-glucanosyltransferases of A. fumigatus (AfGel1p), N. crassa (NcGel1p), and S. cerevisiae (ScGas1p) were compared to the translated, full-length ORF of the GEL1 gene of C. posadasii (CpGel1p). Conserved catalytic motifs are boxed (solid line), and the six aligned cysteine residues are indicated by enlarged, bold type. Predicted GPI anchor sites in the C-terminal region of the homologs are boxed (dotted line). The predicted signal peptide cleavage site of the CpGel1p is indicated between S18 and A19. The underlined residues of CpGel1p represent the translated sequence of the 495-bp gene fragment originally identified in the C. posadasii genomic database. GenBank accession numbers are provided in Table 1.
Production of antiserum against rGel1p.
The chromatographically isolated recombinant protein was used to immunize BALB/c mice (6 weeks old) for production of specific antiserum as reported elsewhere (18). The antiserum was used for immunolocalization of the native antigen in parasitic cells of C. posadasii as described below. Preimmune mouse serum was used as a control.
Immunofluorescence.
Freshly isolated, endosporulating spherules obtained from parasitic-phase cultures (132 h) were reacted with either mouse preimmune or anti-rGel1p serum at a 1:200 dilution in PBS. The intact cells were then washed with PBS and prepared for examination by fluorescence microscopy as previously described (17).
Immunization and animal challenge.
Immunoprotection experiments were conducted with BALB/c or C57BL/6 mice (females, 8 weeks old) supplied by the National Cancer Institute (Bethesda, Md.). Mice were challenged with a lethal dose of C. posadasii arthroconidia either by the i.p. or intranasal (i.n.) route. i.p. challenge delivers a consistent number of fungal conidia to the animals and, although not the natural route of infection, has been shown to be a useful method to initially evaluate candidate vaccine reagents (19, 22). Mice were immunized subcutaneously (s.c.) with rGel1p plus adjuvant by using essentially the same protocol as previously described (22). The adjuvant used was unmethylated CpG dinucleotides present in a synthetic oligodeoxynucleotide (ODN) preparation (CpG ODN; Integrated DNA Technologies, Inc., Coralville, Iowa). This same adjuvant has been described in evaluations of other candidate vaccines (22, 29). The CpG ODN sequence used to immunize mice was TCCATGACGTTCCTGACGTT (CpG motifs are underlined). The oligonucleotides were dissolved in PBS (1 mg/ml) and used as stock solution for the vaccination experiments. Three groups of BALB/c mice (20 per group) were examined in the immunization and i.p. challenge experiments. Mice were first immunized s.c. with either adjuvant alone in PBS (10 μg of CpG prepared in 50 μl of PBS plus 50 μl of incomplete Freund's adjuvant [Sigma, St. Louis, Mo.]) (22) or with CpG adjuvant plus two different amounts of rGel1p (1 or 5 μg). The mice were then boosted by s.c. immunization 14 days later with the same amount of immunogen plus adjuvant. The animals were subsequently challenged with 100 viable arthroconidia by the i.p. route 2 weeks after the last immunization and then sacrificed 12 days later to determine the residual CFU in the lungs and spleen, as reported elsewhere (22). The CFU per organ were expressed on a log scale, and the Mann-Whitney U test was used to compare the median numbers as previously described (19). The detection limit of the CFU assay is 100 colonies per organ homogenate (log10 CFU = 2.00). Alternatively, C57BL/6 mice were immunized s.c. with 0.2, 1, or 5 μg of rGel1p plus adjuvant as above and then challenged with 80 viable arthroconidia by the i.n. route. Mice were scored for survival over a 40-day period postchallenge. Survival differences between groups of i.n.-infected mice (12 per group) were analyzed for statistical significance by the Kaplan-Meier method as previously reported (22).
Nucleotide sequence accession numbers.
The C. posadasii GEL1 nucleotide sequences reported in this paper have been submitted to the GenBank database under accession nos. AF288063 (cDNA) and AF395660 (genomic).
RESULTS
Identification of the GEL1 gene in the genome database.
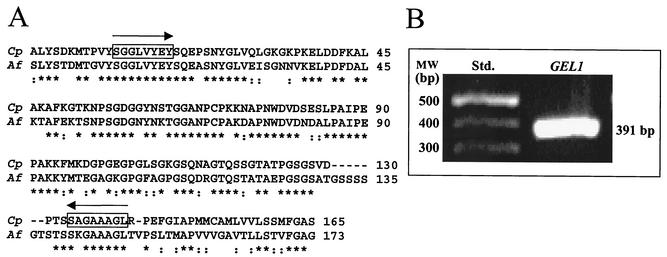
A BLASTX search of the partial C. posadasii genome revealed a 495-bp open reading frame (ORF) which encodes a homolog of an A. fumigatus β-1,3-glucanosyltransferase (14, 26). High sequence homology (64% identity, 78% similarity) was observed between the translated C. posadasii gene fragment and the reported glucan-elongating glucanosyltransferase (GEL1) gene of A. fumigatus (Fig. 1A). Nested PCR primers were synthesized on the basis of nucleotide sequences of the C. posadasii genome fragment which translated N-terminal and C-terminal regions that revealed near-identical alignment with the A. fumigatus Gel1 sequences (Fig. 1A). These primers amplified a 391-bp fragment of the GEL1 homolog in the presence of genomic template DNA of C. posadasii (Fig. 1B). Nucleotide sequence analysis of the 391-bp PCR product confirmed that it was identical to the contig sequence originally identified in the C. posadasii genome database.
FIG. 1.
(A) CLUSTAL X alignment of conserved amino acid sequences of the translated, 495-bp gene fragment of C. posadasii (Cp) and the Gel1 protein of A. fumigatus (Af) reported in GenBank (AF072700). (B) PCR product (391 bp) amplified from C. posadasii genomic DNA using nested primers selected on the basis of the aligned sequences in panel A. The translated primer sequences are boxed in panel A. An asterisk indicates amino acid identity; a colon indicates a conservative substitution (39).
Full-length GEL1 sequence analysis.
The 5′ and 3′ RACE methods yielded cDNA bands of 1.3 and 0.5 kb, respectively. The nucleotide sequences of these bands overlapped with the 495-bp fragment of GEL1 and included the 5′ and 3′ untranslated region sequences of the gene. Application of the 3′ genome walking method resulted in isolation of three genome fragments of 0.4, 1.5, and 3.0 kb. The 1.5-kb band was selected for genomic sequence analysis and was compared to the cDNA sequences obtained from analysis of the RACE products. The complete genomic sequence of the C. posadasii GEL1 gene was obtained as a 3.0-kb PCR product. The ORFs of the GEL1 cDNA and genomic sequences were resolved. The genomic sequence included six introns of 68, 54, 63, 52, 54, and 71 bp. The translated amino acid sequence predicted a protein of 447 residues with a molecular mass of 48.1 kDa and an isoelectric point (pI) of 5.4. Two N-glycosylation sites were predicted by protein sequence analysis using the PROSITE algorithm.
Sequence homology between C. posadasii Gel1p and reported fungal β-1,3-glucanosyltransferases.
Alignment of the predicted C. posadasii protein sequence (CpGel1p) with reported sequences of glucanosyltransferases of filamentous fungi and yeast by CLUSTAL X analysis revealed high levels of homology (Fig. 2; Table 1). The highest sequence similarity and identity were between the Gel1 proteins of C. posadasii, A. fumigatus, and Neurospora crassa (Table 1) and were most evident within the region of the first 300 amino acids (Fig. 2). This fragment of the protein structure includes two conserved motifs that contain the reported active glutamic acid residues (Fig. 2) (25). Sequence analysis of the putative C. posadasii glucanosyltransferase, performed by PSORT, revealed that 18 amino acids at the N terminus have the characteristics of a signal peptide, with a predicted cleavage site between S18 and A19 (Fig. 2). The number and sequence location of the cysteine residues in the C. posadasii protein are highly conserved among all the fungal glucanosyltransferase sequences included in Fig. 2. The relatively high percentage of serine residues and the acidic pI of CpGel1p are common features of reported fungal glucanosyltransferases (33). Examination of the hydropathicity profile of the translated C. posadasii GEL1 gene (data not shown) revealed a hydrophobic C terminus. Sequence analysis of this region predicted a GPI anchor site at G413 (10). This same residue is a predicted GPI anchor site in the aligned Gel1 protein sequences of A. fumigatus and N. crassa, while the predicted site in the reported Saccharomyces cerevisiae glucanosyltransferase (33) is an asparagine residue (Fig. 2).
TABLE 1.
Summary of calculated values for amino acid similarities and identities between C. posadasii Gel1p and glucanosyltransferase sequences of other fungi
| Sequence compared | GenBank accession no. | Similarity (%)a | Identity (%)a |
|---|---|---|---|
| Aspergillus fumigatus Gel1 | AF072700 | 90 | 70 |
| Neurospora crassa Gel1 | CAD21369 | 81 | 58 |
| Schizosaccharomyces pombe Gas1 | CAB46773 | 77 | 46 |
| Aspergillus fumigatus Gel2 | AF208039 | 71 | 36 |
| Candida albicans Phr3 | AF221545 | 66 | 33 |
| Candida glabrata Gas1 | AJ302061 | 62 | 31 |
| Saccharomyces cerevisiae Gas1 | X53424 | 61 | 30 |
| Candida maltosa Epd1 | AB005130 | 61 | 29 |
| Aspergillus fumigatus Gel3 | AF208040 | 58 | 26 |
| Pneumocystis carninii Phr1 | AF191097 | 56 | 26 |
Based on CLUSTAL X alignment of amino acid sequences, taking into account conservative substitutions of residues (39).
Additional putative homologs of glucanosyltransferases identified in the C. posadasii genome database.
Four additional ORFs of C. posadasii have been identified in separate contigs of the genome database, which predict peptides that align with four distinct glucanosyltransferases reported in other fungi (Table 2). Only the highest percent similarity and identity values for each of these translated gene fragments (GEL2 to -5) are presented. Attempts are under way to clone and express these full-length genes, which we suggest encode members of a family of C. posadasii glucanosyltransferases.
TABLE 2.
Summary of calculated values for conserved amino acid similarities and identities between translated sequences of putative glucanosyltransferase genes of C. posadasii and reported glucanosyltransferases of other fungi
| Translated sequences of C. posadasii genes | Homolog (GenBank accession no.) | Similarity/identity (%) |
|---|---|---|
| Gel2 (1.4-kb ORF) | A. fumigatus Gel2 AF208039 | 86/52 |
| Gel3 (0.6-kb ORF) | A. fumigatus Gel3 AF208040 | 77/46 |
| Gel4 (0.7-kb ORF) | C. dubliniensis Phr2 AAG16996 | 80/64 |
| Gel5 (0.6-kb ORF) | C. glabrata Gas3 CAC83346 | 68/27 |
In vivo and in vitro expression of the translated C. posadasii GEL1 gene.
A diagrammatic representation of cell development during the first generation and early second generation of the parasitic cycle of C. posadasii is shown in Fig. 3A. Abscesses obtained from the lungs of C. posadasii-infected C57BL/6 mice at 12 days after i.p. challenge were examined by fluorescence microscopy. Whole mounts were stained with Blankofluor (34), which binds to the glucans and chitin of the fungal cell wall. Spherules were observed in various stages of development. To confirm that the GEL1 homolog was expressed in vivo during the parasitic cycle, RT-PCR was performed using the same primers as above and template cDNA derived from total RNA of the infected lung tissue. A second primer pair, which amplified a 519-bp fragment of a gene that encodes the CSA gene, was included to confirm the presence of C. posadasii in the tissue preparation. The detection of a 391-bp amplicon (Fig. 3B) indicated that the GEL1 gene of the fungal pathogen is expressed in vivo.
QRT-PCR was used to evaluate relative levels of expression of the C. posadasii GEL1 transcript during in vitro growth of both the saprobic and parasitic phases of the fungus (Fig. 3C). QRT-PCR data analysis was conducted by application of the comparative (CT) method. The amounts of GEL1 transcript produced by the saprobic phase and the endosporulation stage of the parasitic cycle (132 h) were compared to those of the segmentation stage of parasitic cell development (72 h), which was given an arbitrary value of 1. On this basis, a 2.3-fold increase in the amount of GEL1 transcript was observed during the endosporulation stage above that of the segmentation growth phase of spherule development (cf. Fig. 3A).
Production of the recombinant protein (rGel1p).
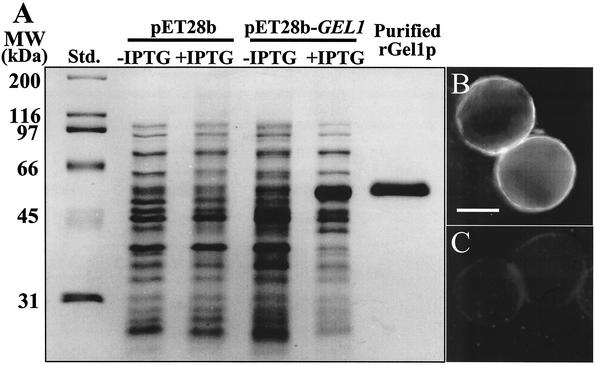
To express the GEL1 gene of C. posadasii, the 1.3-kb PCR-generated cDNA was subcloned into pET28b to yield the pET28b-GEL1 construct. The GEL1 insert lacked the nucleotide sequence which encodes the signal peptide (i.e., residues 1 to 18 were deleted from rGel1p; cf. Fig. 2). The predicted molecular size of the recombinant protein was 48 kDa, which includes the vector-encoded fusion peptide that contained a His tag at its N terminus. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to examine the composition of the cell lysates obtained from E. coli strain BL21(DE3), which had been transformed with either the empty vector or plasmid construct and then induced with IPTG (isopropyl-β-d-thiogalactopyranoside) (Fig. 4A). The molecular size of the recombinant protein (rGel1p) correlated with the predicted size. The 48-kDa rGel1p was isolated by nickel-affinity chromatography, digested with Lys-C, separated by high-pressure liquid chromatography, and subjected to N-terminal sequence analysis (18). The amino acid sequence obtained (DQKVENFTGY) was identical to the predicted sequence of the translated GEL1 gene (residues 246 to 255) (Fig. 2).
FIG. 4.
(A) SDS-PAGE of E. coli-expressed rGel1p. Shown are standards (Std.), lysates of bacteria transformed with either the empty plasmid vector (pET28b), or the vector plus GEL1 gene insert (pET28b-GEL1) in the presence (+) or absence (−) of IPTG, and the nickel-affinity-isolated rGel1p which was subsequently purified by electroelution from an SDS-PAGE gel as previously reported (18). (B and C) Immunofluorescence light micrographs of spherules reacted with murine anti-rGel1p antiserum followed by secondary antibody-fluorescein isothiocyanate (FITC) conjugate (B), or secondary antibody-FITC conjugate alone (C), as previously reported (18). The latter showed no labeling of the cells. The bar in panel B represents 2 μm.
Immunolocalization of the native Gel1p.
The purified rGel1p used to raise specific antiserum in mice recognized the 48-kDa protein in the bacterial lysate (data not shown) and reacted with a surface component of in vitro-grown endospores of C. posadasii (Fig. 4B). The preimmune serum did not bind to the cell surface antigen (Fig. 4C).
Fungal burden in rGel1p-immunized, i.p.-challenged mice.
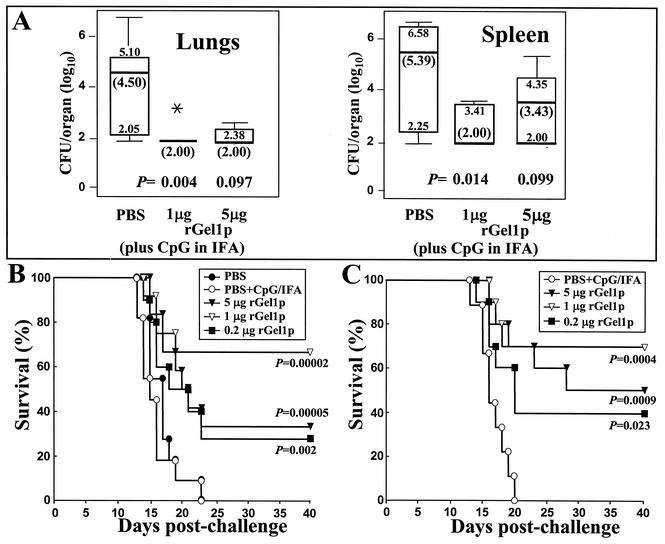
BALB/c mice were immunized with rGel1p, challenged by the i.p. route with a lethal inoculum of C. posadasii (100 viable arthroconidia), and then sacrificed 12 days later to examine residual CFU in their lungs and spleen. Taking into account the detection limit of the CFU assay, the results suggest that few organisms remained in the lungs of Coccidioides-infected mice immunized with two separate doses of 1 μg of rGel1p plus adjuvant. On the other hand, the fungus was less efficiently cleared from the lungs of animals administered 5 μg of the antigen preparation using the same immunization protocol (Fig. 5A). The median number of organisms in the lungs of control mice immunized with adjuvant alone was 4.5 log10 CFU (range, 2.1 to 5.1). The difference between the number of CFU detected in the lungs of control mice and animals immunized with 1 μg of rGel1p was statistically significant (P = 0.004). Mice immunized with 5 μg of rGel1p, on the other hand, did not show a significant reduction in CFU compared to infected control animals. The spleen homogenates of mice immunized with either 1 or 5 μg of rGel1p showed a higher fungal burden and wider range of CFU than the lung homogenates of rGel1p-immune mice. Nevertheless, the group of mice immunized with 1 μg of rGel1p showed a significant reduction of the fungal burden compared to control mice (P = 0.014) and significantly greater clearance of C. posadasii from the spleen than mice immunized with 5 μg of the recombinant protein (P = 0.032).
FIG. 5.
(A) Representative box plot of CFU of C. posadasii detected in dilution plate cultures of lung and spleen homogenates obtained from i.p.-infected BALB/c mice immunized by the s.c. route with rGel1p (1 or 5 μg) plus adjuvant, or immunized with PBS plus adjuvant alone (control). The mice were sacrificed at 12 days postchallenge. The boxes indicate the 25th and 75th percentiles, and the bars show the 5th and 95th percentiles. The horizontal line within the box indicates the median. The asterisk represents a dead mouse. Samples of plated organ homogenates with no CFU were assigned a value of 2.00 log10 based upon the detection limit of the CFU assay. (B and C) Comparisons of the protective efficacy of different amounts of rGel1p (0.2, 1, and 5 μg) plus the same adjuvant as for panel A used to immunize C57BL/6 mice, which were subsequently challenged by the i.n. route with a lethal inoculum (80 arthroconidia) of C. posadassi. Control mice were immunized with PBS alone, or PBS plus adjuvant. Mortality was determined at days 1 through 40 postchallenge for 10 mice in each group. Statistical significance (P values) of the difference between survival plots was determined for mice immunized with each amount of rGel1p plus adjuvant versus mice immunized only with PBS plus adjuvant.
Survival of rGel1p-immunized, i.n.-challenged mice.
Control C57BL/6 mice (12 mice per group) immunized with PBS or PBS plus adjuvant and then challenged via the i.n. route with a lethal inoculum of C. posadasii (80 viable arthroconidia) typically began to die at about 13 days postchallenge (Fig. 5B and C). None of these infected, control mice survived beyond 25 days. Mice immunized with rGel1p (0.2, 1, or 5 μg), on the other hand, all showed significantly greater survival when compared to the control mice (P values are indicated in the figure). However, based on results of the two representative experiments in Fig. 5B and C, there was an apparent trend that mice immunized with 1 μg of rGel1p were better protected than those immunized with either 0.2 or 5 μg of rGel1p, although the difference in percent survival between these groups of mice was not statistically significant. The consistently greater number of survivors among the group of mice immunized with 1 μg of rGel1p suggested that these animals had mounted a more protective immune response against the lethal i.n. challenge of C. posadasii than mice immunized with either the higher or lower dose of the same immunogen. In fact, approximately 40% of the mice immunized with 1 μg of rGel1p that survived to 40 days postchallenge were cleared of coccidioidal infection in their lungs and spleen.
DISCUSSION
The partially sequenced genome of C. posadasii is a valuable resource which can be used to retrieve gene sequences of interest by application of computational tools of bioinformatics to search the genomic database. Similarity between a translated gene sequence and primary structure of a reported protein with known function can provide strong evidence for the presence of a homologous coding region (13). We used the computer program BLASTX (7) to perform conceptual translations of nucleotide sequences in the C. posadasii genomic database, followed by a search of the GenBank protein database (http://www.ncbi.nlm.nih.gov/BLAST/ [40]) to find good alignments between the query and database sequences. Our intent was to identify genes of C. posadasii which encode homologs of wall-associated proteins (8). Evidence from studies of candidate vaccines against coccidioidomycosis has indicated that the most protective antigens are cell wall proteins (19, 36). The rationale for these investigation is based on the fact that genome studies have recently contributed to the discovery of a new array of virulence determinants in pathogenic microbes (7), as well as the identification of novel vaccine candidates (38). The C. posadasii genome consists of approximately 29 Mb arranged in four chromosomes, and it has been suggested to encode about 12,000 proteins (20). Most genes of the pathogen reported to date contain introns (typically <100 bp), which makes identification of ORFs in the genomic DNA database more difficult. Nevertheless, as a result of our BLASTX search of the partial C. posadasii genome we identified a 495-bp ORF which encodes a peptide that aligned with a reported β-1,3-glucanosyltransferase (Gel1) of A. fumigatus (14, 26). The E value (1) obtained from this alignment suggested the presence of a highly homologous coding region in the C. posadasii peptide. The full-length, translated DNA sequence of the C. posadasii gene, obtained by combined genome walking and RACE methods, revealed 90% similarity to the translated sequence of the A. fumigatus Gel1p. The predicted C. posadasii protein contains two conserved glutamate residues identified as the catalytic sites of this family of glycosyl hydrolases (25), six cysteine residues which are positionally conserved in the primary structure of reported fungal glucanosyltransferases, and a hydrophobic C terminus that contains a conserved GPI anchor domain and attachment residue that is homologous to the GPI anchor domains of A. fumigatus and N. crassa. On the basis of these structural similarities, we suggest that the predicted protein of C. posadasii is a homolog of β-1,3-glucanosyltransferases produced by other filamentous fungi.
Glucanosyltransferases are periplasmic and cell wall-associated enzymes which are responsible for the elongation of β-1,3-glucan chains (27). It has been proposed that this enzymatic activity is essential for both mold and yeast morphogenesis and is under the control of a family of functionally related genes (25). In A. fumigatus, a pronounced alteration of the normal phenotype was evident only in the Δgel1/Δgel2 double mutant (3). Deletion of the GEL1 gene of C. posadasii had no effect on development of the parasitic phase in vitro (data not shown). This suggests that Gel1p in this fungus, like the homolog in A. fumigatus, does not play a major role in cell wall morphogenesis, or the loss of GEL1 is compensated by the expression of other, functionally related genes. Recent BLASTX analyses of the C. posadasii genome database have revealed four additional ORFs whose predicted amino acid sequences align with previously reported glucanosyltransferases of A. fumigatus (Gel2p, Gel3p [25]), Candida dubliniensis (Phr2p [12]), and Candida glabrata (Gas3p [33]). We suggest that C. posadasii expresses a family of structurally and functionally related β-1,3-glucanosyltransferases.
GPI-anchored proteins of parasitic microbes which are displayed on the cell surface in vivo have been shown to be highly immunogenic (4, 5). The GEL1 gene of C. posadasii is expressed by the pathogen during infection of host lung tissue, and the highest amount of GEL1 transcript was detected during the endosporulation stage of the parasitic cycle. Endospores are responsible for dissemination of the pathogen from sites of initial colonization in the lungs. These are also the parasitic cells of the pathogen which are small enough to be engulfed by host phagocytes. The recombinant Gel1 protein produced by the expression vector described in this study was recognized in immunoblots of the bacterial lysates by sera from patients with confirmed coccidioidal infection (data not shown). These results suggest that humans infected with C. posadasii are exposed to the native Gel1p during the parasitic cycle. Antibody raised in mice against the purified recombinant protein was used in immunolocalization studies of the native antigen. The Gel1 antigen of C. posadasii was shown to be present at the surface of endospores. On the basis of this preliminary characterization of Gel1p, we concluded that the recombinant antigen was worthy of further examination as a vaccine candidate in our murine model of coccidioidomycosis.
Our initial evaluation of rGel1p as a potential vaccine was conducted by s.c. immunization of BALB/c mice which were subsequently challenged with a lethal inoculation of C. posadasii via the i.p. route. Although i.p. inoculation is not as rigorous as i.n. challenge, it has proved to be a useful method for screening selected antigens. Our immunization protocol, which involved two s.c. injections of either 1 or 5 μg of rGel1p in the presence of CpG adjuvant, resulted in significant clearance of the pathogen from both the lungs and spleen only in mice immunized with the lower dose of the antigen. The degree of clearance was determined at 12 days postinoculation by comparison of CFU in organ homogenates of immunized versus nonimmunized control mice. Apparently, the 1-μg dose of rGel1p elicited a greater protective immune response than the 5-μg dose. Separate groups of C57BL/6 mice immunized with 0.2, 1, or 5 μg of antigen, but challenged by the natural i.n. route, all showed significantly higher numbers of survivors than animals immunized with PBS alone or PBS plus adjuvant. The results of these experiments revealed the same apparent trend in host response to infection as noted above in the immunized, i.p.-challenged mice; groups of animals immunized with 1 μg of rGel1p showed increased numbers of survivors (68 to 70%) compared to those immunized with either 0.2 μg (28 to 40%) or 5 μg (35 to 50%) of the recombinant antigen. Although these differences in survival were not statistically significant, the consistency of results in animals immunized with 1 μg of rGel1p and then challenged by either the i.p. or i.n. route suggests that this vaccine dose provides superior protection in mice against a lethal challenge of the fungal pathogen compared to other immunization protocols tested in this study.
We have examined the nature of the cytokine response in rGel1p-immunized and challenged C57BL/6 mice using methods previously described (22) as well as by comparative analysis of amounts of selected cytokines present in bronchoalveolar lavage fluid of immunized and control mice at 7 and 14 days postchallenge. These data will be presented in a separate publication. In essence, the results support our speculation that immunization with 1 μg of rGel1p stimulated a protective, T helper 1 (Th1) pathway of immune response, which was indicated by an elevated level of gamma interferon (IFN-γ) production and increased ratio of the rGel1p-specific immunoglobulin G2a (IgG2a) titer to the IgG1 antibody titer (23). Immunization with either the lower or higher dose of the recombinant antigen failed to induce the same level of protective response. Failure to protect against coccidioidal infection was associated with reduced levels of IFN-γ production, a corresponding increase in the levels of production of Th2-type cytokines, and high titers of rGel1p-specific IgG1 antibody (22). We conclude that immunization of BALB/c and C57BL/6 mice with the optimal amount of the recombinant β-1,3-glucanosyltransferase homolog provides significant protection against a lethal challenge of C. posadasii, and we suggest that rGel1p is a valuable addition to the arsenal of candidate molecules for a human vaccine against coccidioidomycosis.
Acknowledgments
Support for this study was provided by Public Health Service grants AI19149, AI37232, and U01-AI50910 (Coccidioides genome sequencing project) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, as well as a research grant from the California HealthCare Foundation.
Editor: T. R. Kozel
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnato, A. E., G. D. Sanders, and D. K. Owens. 2001. Cost-effectiveness of a potential vaccine for Coccidioides immitis. Emerg. Infect. Dis. 7:797-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruneau, J.-M., T. Magnin, E. Tagat, R. Legrand, M. Bernard, M. Diaquin, C. Fudali, and J.-P. Latgé. 2001. Proteome analysis of Aspergillus fumigatus identifies glycosylphosphatidylinositol-anchored proteins associated to the cell wall biosynthesis. Electrophoresis 22:2812-2823. [DOI] [PubMed] [Google Scholar]
- 4.Burns, J. M., C. C. Belk, and P. D. Dunn. 2000. A protective glycosylphosphatidylinositol-anchored membrane protein of Plasmodium yoelii trophozoites and merozoites contains two epidermal growth factor-like domains. Infect. Immun. 68:6189-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campos, M. A. S., I. C. Almeida, O. Takeuchi, S. Akira, E. P. Valente, D. O. Procopio, L. R. Travassos, J. A. Smith, D. T. Golenbock, and R. T. Gazzinelli. 2001. Activation of Toll-like receptor-2 by glycosylphosphatidylinositol anchors from a protozoan parasite. J. Immunol. 167:416-423. [DOI] [PubMed] [Google Scholar]
- 6.Cole, G. T., C.-Y. Hung, and N. Delgado. 2002. Parasitic phase-specific gene expression in Coccidioides. ASM News 68:603-611. [Google Scholar]
- 7.Crabb, B. S., and A. F. Coman. 2002. Plasmodium falciparum virulence determinants unveiled. Genome Biol. 3:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Groot, P. W. J., C. Ruiz, C. R. V. de Aldana, E. Duenas, V. J. Cid, F. D. Ray, J. M. Rodriquez-Peña, P. Pérez, A. Andel, J. Caubin, J. Arroyo, J. C. García, C. Gil, M. Molina, L. J. Garcia, C. Nombela, and F. M. Klis. 2001. A genomic approach for the identification and classification of genes involved in cell wall formation and its regulation in Saccharomyces cerevisiae. Comp. Funct. Genom. 2:124-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desai, S. A., O. A. Minai, S. M. Gordon, B. O'Neil, H. P. Wiedemann, and A. C. Arroliga. 2001. Coccidioidomycosis in non-endemic areas: a case series. Respir. Med. 95:305-309. [DOI] [PubMed] [Google Scholar]
- 10.Eisenhaber, B., P. Bork, and F. Eisenhaber. 1999. Prediction of potential GPI-modification sites in proprotein sequences. J. Mol. Biol. 292:741-758. [DOI] [PubMed] [Google Scholar]
- 11.Fisher, M. C., G. L. Koenig, T. J. White, and J. W. Taylor. 2002. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia 94:73-84. [PubMed] [Google Scholar]
- 12.Fonzi, W. A. 1999. PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of β-1,3- and β-1,6-glucans. J. Bacteriol. 181:7070-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gish, W., and D. J. States. 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3:266-272. [DOI] [PubMed] [Google Scholar]
- 14.Hartland, R. P., T. Fontaine, J. P. Debeaupuis, C. Simenel, M. Delepierre, and J.-P. Latgé. 1996. A novel β-(1-3)-glucanosyltransferase from the cell wall of Aspergillus fumigatus. J. Biol. Chem. 271:26843-26849. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann, K., P. Bucher, L. Falquet, and A. Bairoch. 1999. The PROSITE database, its status in 1999. Nucleic Acids Res. 27:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton, P., and K. Nakai. 1997. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Intell. Systems Mol. Biol. 5:147-152. [PubMed] [Google Scholar]
- 17.Hung, C.-Y., J.-J. Yu, K. R. Seshan, U. Reichard, and G. T. Cole. 2002. A parasitic phase-specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory fungal pathogen. Infect. Immun. 70:3443-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung, C.-Y., J.-J. Yu, P. F. Lehmann, and G. T. Cole. 2001. Cloning and expression of the gene which encodes a tube precipitin antigen and wall-associated β-glucosidase of Coccidioides immitis. Infect. Immun. 69:2211-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirkland, T. N., F. Finley, K. Orsborn, and J. N. Galgiani. 1998. Evaluation of the proline-rich antigen of Coccidioides immitis as a vaccine candidate in mice. Infect. Immun. 66:3519-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirkland, T. N., and G. T. Cole. 2002. Gene-finding in Coccidioides immitis: searching for immunogenic proteins, p. 247-254. In K. J. Shaw (ed.), Pathogen genomics: impact on human health. Humana Press, Totowa, N.J.
- 21.Levine, H. B. 1961. Purification of the spherule-endospore phase of Coccidioides immitis. Sabouraudia 1:112-115. [DOI] [PubMed] [Google Scholar]
- 22.Li, K., J. J. Yu, C. Y. Hung, P. F. Lehmann, and G. T. Cole. 2001. Recombinant urease and urease DNA of Coccidioides immitis elicit an immunoprotective response against coccidioidomycosis in mice. Infect. Immun. 69:2878-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magee, D. M., and R. A. Cox. 1995. Roles of gamma interferon and interleukin-4 in genetically determined resistance to Coccidioides immitis. Infect. Immun. 63:3514-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGwire, B. S., W. A. O'Connell, K.-P. Chang, and D. M. Engman. 2002. Extracellular release of the glycosylphosphatidylinositol (GPI)-linked Leishmania surface metalloproteinase, gp63, is independent of GPI phospholipolysis: implications for parasite virulence. J. Biol. Chem. 277:8802-8809. [DOI] [PubMed] [Google Scholar]
- 25.Mouyna, I., M. Monod, T. Fontaine, B. Henrissat, B. Léchenne, and J.-P. Latgé. 2000. Identification of the catalytic residues of the first family of β-(1-3)-glucanosyltransferases identified in fungi. Biochem. J. 347:741-747. [PMC free article] [PubMed] [Google Scholar]
- 26.Mouyna, I., R. Hartland, T. Fontaine, M. Diaquin, C. Simenel, M. Delepieree, B. Henrissat, and J.-P. Latgé. 1998. A 1,3-β-glucanosyltransferase isolated from the cell wall of Aspergillus fumigatus is a homologue of the yeast Bg12p. Microbiology 144:3171-3180.9846753 [Google Scholar]
- 27.Mouyna, I., T. Fontaine, M. Vai, M. Monod, W. Fonzi, M. Diaquin, L. Popolo, R. P. Hartland, and J.-P. Latgé. 2000. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 275:3171-3180. [DOI] [PubMed] [Google Scholar]
- 28.Nakai, K., and M. Kanehisa. 1992. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14:897-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oxenius, N., M. M. A. Martinic, H. Hengartner, and P. Klenerman. 1999. CpG-containing oligonucleotides are efficient adjuvants for induction of protective antiviral immune responses with T-cell peptide vaccines. J. Virol. 73:4120-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan, S., and G. T. Cole. 1992. Electrophoretic karyotypes of clinical isolates of Coccidioides immitis. Infect. Immun. 60:4872-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan, S., and G. T. Cole. 1995. Molecular and biochemical characterization of Coccidioides immitis-specific antigen. Infect. Immun. 63:3994-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pappagianis, D. 2001. Seeking a vaccine against Coccidioides immitis and serologic studies: expectations and realities. Fungal Genet. Biol. 32:1-9. [DOI] [PubMed] [Google Scholar]
- 33.Popolo, L., and M. Vai. 1999. The Gas1 glycoprotein, a putative wall polymer cross-linker. Biochem. Biophys. Acta 1426:385-400. [DOI] [PubMed] [Google Scholar]
- 34.Rüchel, R., M. Schaffrinski, K. R. Seshan, and G. T. Cole. 2000. Vital staining of fungal elements in deep-seated mycotic lesions during experimental murine mycoses using the parenterally applied optical brightener Blankophor. Med. Mycol. 38:231-237. [DOI] [PubMed] [Google Scholar]
- 35.Shubitz, L., M. E. Matz, T. H. Noon, C. C. Reggiardo, and G. A. Bradley. 2001. Constrictive pericarditis secondary to Coccidioides immitis infection in a dog. J. Am. Vet. Med. Assoc. 218:537-540. [DOI] [PubMed] [Google Scholar]
- 36.Shubitz, L., T. Peng, R. Perrill, J. Simmons, K. Orsborn, and J. N. Galgiani. 2002. Protection of mice against Coccidioides immitis intranasal infection by vaccination with recombinant antigen 2/PRA. Infect. Immun. 70:3287-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siebert, P. D., A. Chenchik, E. E. Kellogg, K. A. Lukyanov, and S. A. Lukyanov. 1995. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23:1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suker, J., and J. M. Feavers. 2001. Prospects offered by genome studies for combating meningococcal disease by vaccination. Pharmacogenomics 2:273-283. [DOI] [PubMed] [Google Scholar]
- 39.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wheeler, D. L., C. Chappey, A. E. Lash, D. D. Leipe, T. L. Madden, G. D. Schuler, T. A. Tatusova, and B. A. Rapp. 2000. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 28:10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yorgin, P. D., M. Rewari, A. Y. al-Uzri, A. A. Theodorou, K. M. Scott, and L. L. Barton. 2001. Coccidioidomycosis in adolescents with lupus nephritis. Pediatr. Nephrol. 16:77-81. [DOI] [PMC free article] [PubMed] [Google Scholar]