Abstract
Many studies have shown that genetic susceptibility plays a key role in determining whether bacterial pathogens successfully infect and cause disease in potential hosts. Surprisingly, whether host genetics influence the pathogenesis of attaching and effacing (A/E) bacteria such as enteropathogenic and enterohemorrhagic Escherichia coli has not been examined. To address this issue, we infected various mouse strains with Citrobacter rodentium, a member of the A/E pathogen family. Of the strains tested, the lipopolysaccharide (LPS) nonresponder C3H/HeJ mouse strain experienced more rapid and extensive bacterial colonization than did other strains. Moreover, the high bacterial load in these mice was associated with accelerated crypt hyperplasia, mucosal ulceration, and bleeding, together with very high mortality rates. Interestingly, the basis for the increased susceptibility was not due to LPS hyporesponsiveness, as the genetically related but LPS-responsive C3H/HeOuJ and C3H/HeN mouse strains were also susceptible to infection. Analysis of the intestinal pathology in these susceptible strains revealed significant crypt epithelial cell apoptosis (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end label staining) as well as bacterial translocation to the mesenteric lymph nodes. Further studies with infection of SCID (T- and B-lymphocyte-deficient) C3H/HeJ mice demonstrated that loss of lymphocytes had no effect on bacterial numbers but did reduce crypt cell apoptosis and delayed mortality. These studies thus identify the adaptive immune system, crypt cell apoptosis, and bacterial translocation but not LPS responsiveness as contributing to the tissue pathology and mortality seen during C. rodentium infection of highly susceptible mouse strains. Determining the basis for these strains' susceptibility to intestinal colonization by an A/E pathogen will be the focus of future studies.
Researchers in bacterial pathogenesis have come to recognize that the course of a microbial infection is not solely directed by the pathogen but rather embodies a dynamic struggle between the infecting microbes and their host (29, 56). The resulting interactions pit the infectious strategies of the pathogen against the host's genetically determined resistance to that infection (7, 20). The outcome of this interplay determines not only whether the infection succeeds but also the resulting clinical manifestation of the disease. Accordingly, elucidating the basis for resistance or susceptibility to human pathogens has been identified as an important step in managing the public health burdens that they impose. The majority of the studies in this area have dealt with host susceptibility to invasive, intracellular pathogens such as Mycobacterium tuberculosis and Salmonella enterica serovar Typhimurium (28, 29, 56). For these and many other microbes, key host susceptibility loci such as the natural resistance-associated macrophage protein 1 (NRAMP1) (28, 29) and Toll-like receptors (TLR) (47, 48) have provided important information regarding bacterial pathogenesis as well as host immunological responses and cell biology.
Curiously, the potential contributions of host genetics to modulating infectious disease have not been examined for entire families of important and relevant bacterial pathogens. Notable among these is the family of attaching and effacing (A/E) bacterial pathogens that includes enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC). Noninvasive gram-negative bacterial pathogens that infect the gastrointestinal tract of their hosts by attaching to the surface of intestinal epithelial cells, EPEC and its hemolytic-uremic syndrome-causing relative EHEC are both major causes of diarrheal illness in children (23, 30, 55). Following their initial adherence, these pathogens form A/E lesions on infected intestinal epithelial cells, characterized by the effacement of microvilli and the formation of pedestal-like structures beneath the adherent bacteria (27, 45). This pathological process is mediated by genes within a genetic pathogenicity island termed the “locus of enterocyte effacement” (LEE) (35, 43). The LEE encodes a type III secretion apparatus as well as several secreted proteins including EspA, EspB, and EspD; the outer membrane adhesin intimin; and the translocated intimin receptor Tir (37, 38). These proteins are all virulence factors, being critical for the intimate attachment of A/E pathogens to host cells, as well as for the ability of these pathogens to cause disease (1, 42).
Despite the recent progress made in understanding at a molecular level how A/E pathogens infect host cells and subvert their function, little is known about how the host responds to these infections. Intestinal biopsy studies have shown that EPEC infection is associated with significant mucosal inflammation and intestinal pathology (17, 18, 33). Although individual hosts likely differ in their immune responses to these infections, this has not been directly examined. Differences have been noted, however, in the ability of hosts to clear EPEC infections. Studies of Brazilian children found that most children infected by EPEC suffered only acute infections with concomitant diarrhea that resolved once the infection was cleared. Interestingly, a smaller population of children suffered from a persistent EPEC infection that resulted in chronic diarrhea (17, 18). These studies suggest that some of the infected individuals were unable to develop sufficient protective immunity against EPEC to clear the pathogen. Similarly, studies conducted with adult volunteers by Tacket et al. found that susceptibility to a standardized EPEC inoculum varied substantially within the volunteer cohort (53). There have been fewer opportunities to study host susceptibility to EHEC infection. However, the severity of the diarrhea and other disease symptoms has been shown to vary among infected individuals during EHEC outbreaks (45), while hemolytic-uremic syndrome, the most severe and dreaded complication, develops in only a fraction of those infected (36, 45). Taken together, these data suggest that host genetics can influence the susceptibility to infection by A/E pathogens.
Unfortunately, identifying host genetic loci that are involved in resistance to A/E pathogens is problematic. As EPEC and EHEC are primarily pediatric pathogens, ethical considerations preclude infecting relevant volunteer populations to identify host resistance genes. Studies examining host susceptibility to A/E pathogens have also been limited by the inability of EPEC and EHEC to readily infect most laboratory animal species, including mice. As a result, we have chosen a surrogate approach, using the related murine pathogen Citrobacter rodentium. A natural pathogen of mice, C. rodentium possesses a LEE pathogenicity island that is very similar to that in EPEC and EHEC and causes A/E lesions on infected epithelial cells in the mouse colon (4, 13, 22, 49, 54). As a result of these similarities, C. rodentium provides a useful model for studying the pathogenesis of both EPEC and EHEC. In this study, we identified the lipopolysaccharide (LPS) nonresponder C3H/HeJ mouse strain as being significantly more susceptible to infection than other mouse strains. Furthermore, infection of two genetically related but LPS-responsive mouse strains determined that the susceptibility occurred irrespective of LPS responsiveness. Additional studies of intestinal pathology identified significant crypt cell apoptosis in the susceptible strains that was dependent on the acquired immune system. We thus describe a related family of mouse strains that demonstrate marked susceptibility to C. rodentium infection. Analysis of these strains provides the potential to identify host factors that influence susceptibility to A/E bacterial pathogens as well as the severity of the associated intestinal mucosal inflammatory response.
MATERIALS AND METHODS
Mice.
Three- to four-week-old C57BL/6, 129S1/SvImJ, C3H/HeJ, and C3H/HeOuJ mice as well as immunodeficient C3H/HeJ SCID mice lacking both T and B lymphocytes (8) were obtained from the Jackson Laboratory (Bar Harbor, Maine). NIH Swiss and BALB/c mice were obtained from Harlan Sprague-Dawley (Indianapolis, Ind.), and C3H/HeN mice were obtained from the National Cancer Institute (Bethesda, Md.). Mice were kept in sterilized cages with filter tops, handled in tissue culture hoods, and fed autoclaved food and water under specific-pathogen-free conditions at our animal facilities. Sentinel animals were routinely tested for common pathogens. The protocols employed were in direct accordance with guidelines drafted by the University of British Columbia's Animal Care Committee and the Canadian Council on the Use of Laboratory Animals.
Bacterial strains and infection of mice.
Mice were orally inoculated with wild-type C. rodentium (formerly Citrobacter freundii biotype 4280) strain DBS100 (49). For inoculations, bacteria were grown overnight with shaking in Luria broth at 37°C. Mice were infected by oral gavage with 0.1 ml of Luria broth containing approximately 2.5 × 108 CFU of C. rodentium. To minimize any differences between infections, mice of different strains were infected with the same bacterial preparation. Mice were sacrificed at various time points postinfection (p.i.), and tissues were prepared for histological analysis, terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end label (TUNEL) staining, or viable bacterial counts as described below.
Survival assessment.
To evaluate the susceptibilities of the different mouse strains to C. rodentium infection, the survival of infected mice was assessed over the course of infection. Mice were monitored every day, and when any mouse showed significant distress or became moribund, it was immediately sacrificed. Survival data are presented as the percentages of the initial 10 mice still surviving at the time point in question.
Tissue collection.
Over the course of the infection, mice were euthanized, and following careful dissection, the first 4 cm of each colon, beginning at the anal verge, was collected. Fecal pellets were removed before the colonic tissue was weighed. Mesenteric lymph nodes (MLNs) were also isolated and removed. Tissues were then placed in 10% neutral buffered formalin (Sigma) for histological analysis, or the colon plus fecal pellets or the lymph nodes were collected in phosphate-buffered saline (pH 7.4) and kept on ice before being processed for viable bacterial counts.
Bacterial counts.
Colonic tissues plus any fecal pellets or lymph nodes were homogenized under low speed with a Kinematica tissue homogenizer (Brinkmann). Homogenates were then serially diluted and plated onto MacConkey agar plates (selective for gram-negative organisms). Bacterial colonies were enumerated the following day. C. rodentium colonies were easily distinguished from colonies derived from commensal flora by their size and appearance. The validity of this approach was verified by PCR analysis for LEE genes. At early time points in infection, a minimum of 104 C. rodentium colonies/colon could be determined via this approach since, at lesser dilutions, the commensal flora from the homogenates of some mice formed a lawn on the agar plates that prevented the identification of individual colonies.
Histology, crypt height measurements, and depth of infection.
Full-thickness colonic tissues were fixed in 10% neutral buffered formalin. Sections (3 μm) were cut and stained with hematoxylin and eosin. Photomicrographs were taken with a Nikon Eclipse E400 microscope. Crypt heights were measured by micrometry by an observer blinded to the experimental condition, with 10 measurements being taken in the distal colon of each mouse. Only well-oriented crypts were measured. Similarly, colonic crypts were examined for the presence of C. rodentium. The number of infected crypts per colonic section was determined, with a crypt considered heavily infected when the bacteria had progressed more than halfway down the crypt.
TUNEL staining.
In brief, tissues were fixed as for histology staining. Apoptosis-related DNA fragmentation was determined by the TUNEL assay with the ApopTag peroxidase in situ apoptosis detection kit (Intergen). Samples were processed according to the manufacturer's instructions. For color development, 3′,3′-diaminobenzidine tetrahydrochloride (Sigma Chemical Co., St. Louis, Mo.) was used. Specimens were lightly counterstained with hematoxylin to identify nuclei. Apoptotic cells were enumerated on TUNEL-stained sections by an observer blinded to the experimental condition. The numbers of TUNEL-positive cells are presented as the average numbers of cells per 10 crypts, with only well-oriented crypts examined. Only TUNEL-positive cells that also showed signs of nuclear shrinkage and other morphological signs of programmed cell death were counted. Based on the low number and sporadic presence of TUNEL-positive cells observed in uninfected tissues and tissues from resistant mice, an arbitrary cutoff of 1 TUNEL-positive cell/10 crypts was determined as the minimum number to report. Photomicrographs were taken with a Nikon camera.
Data presentation and statistical analysis.
All the results are expressed as the means ± 1 standard error of the mean (SEM) of three independent experiments; n refers to the number of mice tested. Statistical significance was calculated by the nonparametric Mann-Whitney test. Multiple comparisons were performed with the Neuman-Keuls multiple comparison test. A P value of <0.05 was considered significant.
RESULTS
C3H/HeJ mice demonstrate increased susceptibility to C. rodentium infection.
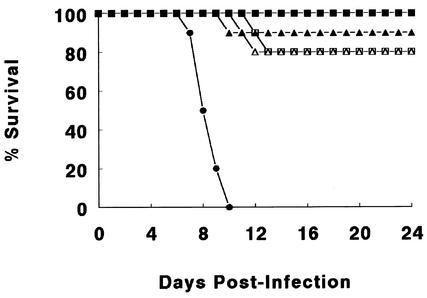
A number of laboratory mouse strains were orally infected with C. rodentium and examined for their susceptibility to this pathogen. All mouse strains showed overt signs of infection, including the defecation of soft stool, a hunched posture, and body weight loss (data not shown). Despite developing overt disease, relatively few mice (<20%) of the C57BL/6, BALB/c, 129S1/SvImJ, or NIH Swiss strains became severely ill from the infection (Fig. 1). The small number of mice that became moribund and required euthanization always did so between days 10 and 14 p.i., in keeping with peak disease symptoms (32, 54). Mice that survived past day 14 p.i. invariably recovered from the infection. In contrast, infection was far more lethal for the C3H/HeJ mouse strain. In our hands, infection resulted in a striking 100% mortality in the C3H/HeJ mice, with the death of the entire cohort occurring between days 6 and 10 p.i. Infection of this strain with C. rodentium was repeated more than a dozen times with the remarkably high incidence of mortality always being observed.
FIG. 1.
Comparison of mortality among different strains of mice orally infected with 2.5 × 108 CFU of C. rodentium. Infection caused only limited mortality in most mouse strains tested, including C57BL/6 (open squares), BALB/c (filled triangles), 129S1/SvImJ (filled squares), and NIH Swiss (open triangles) mice. In contrast, C3H/HeJ mice (filled circles) suffered 100% mortality during the course of infection. Survival data from one representative experiment out of three are shown, with all three experiments showing similar results. Each data point represents the percentage of mice still surviving from an initial population of 10 mice.
C3H/HeJ mice undergo rapid and heavy colonization by C. rodentium.
We began our analysis by examining bacterial colony counts in the colons of infected mice, to determine if the speed or degree of C. rodentium colonization was abnormal in C3H/HeJ mice. Previous studies have shown that extensive colonization of the colon by C. rodentium can take up to 6 days in most mouse strains (32, 54). Since the majority of C. rodentium-infected C3H/HeJ mice died between days 7 and 9 p.i., colonic tissues from all mouse strains were analyzed prior to this, at days 4 and 6 p.i. As expected, most mouse strains were poorly colonized at day 4 p.i. It was not until day 6 p.i. that the C57BL/6, BALB/c, 129S1/SvImJ, and NIH Swiss mouse strains (referred to hereafter as the resistant strains) were extensively colonized. At this point, the mean number of colonic C. rodentium bacteria ranged from 1.2 × 108 to 5.1 × 108 CFU/mouse (Table 1). In contrast, C3H/HeJ mice were colonized more rapidly with significant numbers of C. rodentium bacteria (1.1 × 109) found on day 4 p.i. and even higher numbers (8.7 × 109) found at day 6 p.i. At both time points, the bacterial numbers recovered from the C3H/HeJ mice were significantly greater than the numbers found in all other mouse strains tested.
TABLE 1.
C. rodentium colonic colony counts of the mice in this studya
| Strain |
C. rodentium CFU at day:
|
|
|---|---|---|
| 4 | 6 | |
| NIH Swiss | (1.5 ± 0.3) × 104 | (5.1 ± 1.9) × 108 |
| C57BL/6 | (2.4 ± 0.5) × 104 | (4.3 ± 1.1) × 108 |
| BALB/c | <1.0 × 104 | (1.2 ± 0.4) × 108 |
| 129S1/SvImJ | <1.0 × 104 | (3.9 ± 0.6) × 108 |
| C3H/HeJ | (1.1 ± 0.2) × 109b | (8.7 ± 1.4) × 109b |
All mouse strains studied developed a time-dependent increase in C. rodentium colony counts in the colon following oral inoculation of this pathogen. Colonization occured more rapidly and to a higher level in the C3H/HeJ strain. Values are the mean colonic C. rodentium CFU ± 1 SEM from three independent experiments, each with groups of four to five mice.
Significantly higher bacterial load than those of all other mouse strains at the same time point (P < 0.05).
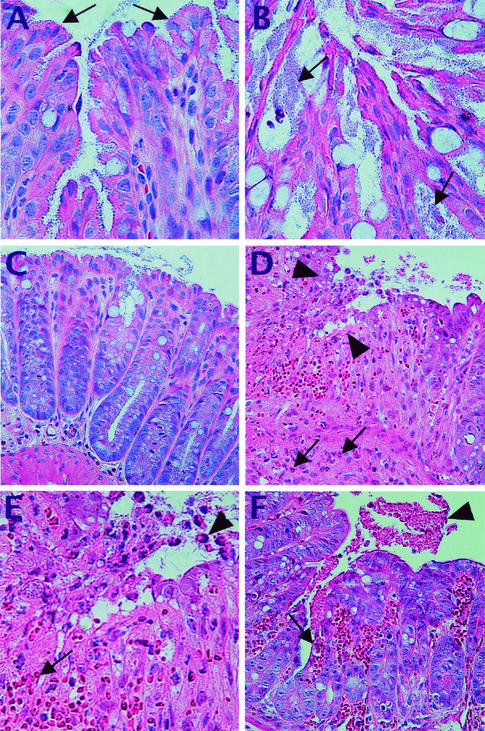
Histological analysis revealed that the basis for the 15- to 70-fold-greater bacterial loads carried by the C3H/HeJ mice was due to differences in the sites of C. rodentium colonization. Previous studies have shown that, early in the infection, C. rodentium bacteria colonize only the most superficial colonic epithelial cells, with few crypts infected to more than half their depth (54). Interestingly, far more crypts were heavily infected at day 6 p.i. in C3H/HeJ mouse tissues (19.9 ± 1.9 crypts) than in other strains (5.6 ± 2.4 in NIH Swiss, 4.3 ± 1.8 in C57BL/6, 2.7 ± 1.7 in BALB/c, and 6.3 ± 4.9 in 129S1/SvImJ mice). Furthermore, the infected crypts appeared to contain more bacteria that penetrated deeper into each crypt (Fig. 2). In addition, while most strains showed significant bacterial colonization only in the distal colon (39, 54), the entire length of the colon of C3H/HeJ mice was heavily infected (data not shown). Additional studies found that colonization differences were not simply due to a delay in the resistant strains. Even at the peak of colonization and disease symptoms (days 10 to 14) in the resistant strains (data not shown), the numbers of C. rodentium were only moderately increased over day 6 numbers (ranging from 3.4 × 108 to 8.9 × 108 CFU). Taking this into account, the C3H/HeJ mice still carried bacterial loads some 10- to 20-fold greater than the heaviest bacterial loads carried by any of the resistant mouse strains at any point during their infection.
FIG. 2.
C. rodentium infection is heavier and leads to mucosal ulceration and friability in susceptible mice. C. rodentium infection at day 6 p.i. was predominantly superficial in resistant strains, as seen in colonic tissue taken from an infected 129S1/SvImJ mouse (A). Note the superficial location of C. rodentium in the colon (arrows). In contrast, the bacterial load was much heavier in the C3H/HeJ mice, with larger numbers of bacteria (arrows) that penetrated more deeply into colonic crypts (B). The inflammatory response was fairly mild in resistant strains at day 6 p.i., shown here with colonic tissue from an infected C57BL/6 mouse that shows well-preserved tissue morphology (C). In contrast, the inflammatory response was much stronger in the susceptible C3H/HeJ mice (D), resulting in heavy inflammatory cell infiltration and ulceration (arrowheads), as well as the loss of crypt morphology. Various inflammatory cells including neutrophils (arrows) can be seen. Shown at higher magnification, the histology from panel D shows the loss of epithelial cells (arrowhead) and the denuding of the mucosal surface. The mucosal ulceration is accompanied by hyperemia and hemorrhage with numerous red blood cells (arrow) observed in the ulcerated region (E). Aside from overt ulceration, the tissue damage seen in C3H/HeJ mice often led to extensive bleeding into the colonic lumen (F) (see arrow), with the blood in the lumen being indicated by the arrowhead.
C3H/HeJ mice undergo more rapid tissue pathology than do other mouse strains.
We next examined the macroscopic and microscopic changes in the colons of C3H/HeJ mice at day 6 p.i. to determine if the susceptible mice suffered aggravated pathology. Interestingly, the pathological changes in the various mouse strains reflected the speed and degree of bacterial colonization. Resistant mouse strains showed little inflammation or intestinal pathology at day 6 p.i. Despite being colonized by C. rodentium, their colons were filled with well-formed stool pellets and showed no significant increases in colon weights or crypt heights (Table 2). In contrast, by day 6 p.i., the colons of most C3H/HeJ mice were thickened and hyperemic, containing only a few large and soft stool pellets. There was also evidence of extensive fluid and mucus accumulation in the colonic lumen (data not shown). Along with the increase in colon weight, there was a marked increase in crypt heights (Table 2). All these changes were associated with a strong mucosal inflammatory response, with overt infiltration of the colonic mucosa by polymorphonuclear cells and various mononuclear cells (Fig. 2D). There was also significant tissue damage with frequent submucosal hyperemia and focal areas of mucosal ulceration (Fig. 2E). The mucosal ulceration was often associated with mucosal hemorrhage and large amounts of blood in the colonic lumen (Fig. 2F).
TABLE 2.
Colon weights and crypt heights during C. rodentium infection of the mice in this studya
| Strain | Mean colon wt (mg) at day:
|
Mean crypt ht (μm) at day:
|
||||
|---|---|---|---|---|---|---|
| 0 | 4 | 6 | 0 | 4 | 6 | |
| NIH Swiss | 102 ± 10 | 113 ± 8 | 140 ± 12 | 127 ± 13 | 153 ± 19 | 170 ± 23 |
| C57BL/6 | 99 ± 4 | 109 ± 7 | 123 ± 6 | 123 ± 10 | 147 ± 15 | 145 ± 12 |
| BALB/c | 96 ± 7 | 115 ± 7 | 121 ± 10 | 138 ± 15 | 140 ± 12 | 137 ± 16 |
| 129S1/SvImJ | 100 ± 5 | 115 ± 12 | 136 ± 9 | 125 ± 8 | 145 ± 12 | 157 ± 19 |
| C3H/HeJ | 105 ± 7 | 143 ± 9b | 223 ± 17b | 134 ± 12 | 210 ± 27b | 265 ± 22b |
C. rodentium infection resulted in a time-dependent increase in both colon weights and crypt heights in all three mouse strains studied, but the increases were more rapid in the C3H/HeJ strain. Values are the mean colon weight or crypt height ± 1 SEM from three independent experiments, each with groups of four to five mice.
Significantly higher value than those for all other mouse strains at the same time point (P < 0.05).
Loss of LPS responsiveness or NRAMP1 function is not responsible for the susceptibility.
C3H/HeJ mice are poorly responsive to LPS, reflecting an inactivating mutation in their TLR4 gene (47). As a result, this mouse strain is highly susceptible to a number of gram-negative bacterial pathogens such as Salmonella serovar Typhimurium (6). The TLR4 mutation in C3H/HeJ mice impairs their innate immune response to these pathogens, permitting unrestrained systemic bacterial replication, often leading to the death of the host (15). Considering that C. rodentium as well as EPEC and EHEC is a gram-negative bacterium, it seemed plausible that defective TLR4 signaling in C3H/HeJ mice was the basis for their susceptibility. To test this hypothesis, we obtained the two strains of mice most closely related to C3H/HeJ mice, namely, the C3H/HeN and C3H/HeOuJ strains. Both strains were originally derived from the same stock as the C3H/HeJ strain but exhibit normal responsiveness to LPS, and both have been used repeatedly as genetic controls for the C3H/HeJ mice (34).
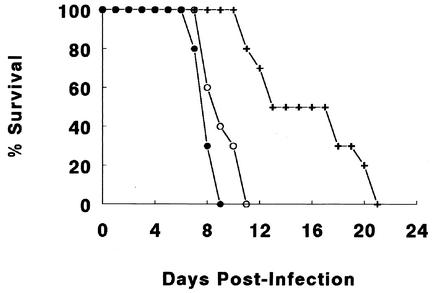
We infected all three mouse strains with C. rodentium and studied their survival as an indicator of their susceptibility to infection. Interestingly, despite their normal innate responses to LPS, the C3H/HeOuJ mice rapidly became ill from the infection, showing similar signs of disease as those seen in the C3H/HeJ mice. The C3H/HeOuJ mice survived only slightly longer (average, 8.9 ± 1.2 days) than the C3H/HeJ mice (average, 7.8 ± 1.0 days), with the differences not reaching significance (Fig. 3). In contrast, the normally LPS-responsive C3H/HeN mice initially appeared more resistant to the infection; however, this relative resistance was not maintained. While the C3H/HeN mice took significantly longer to develop disease symptoms and thus survived longer than did the C3H/HeJ and C3H/HeOuJ mice (16.4 ± 3.1 days; P < 0.05), they all eventually become lethally ill from the infection. This resulted in all (100%) of the C3H/HeN mice requiring euthanization between days 11 and 21 p.i., thus identifying the C3H/HeN mice as also being highly susceptible to C. rodentium infection.
FIG. 3.
C. rodentium infection caused rapid mortality in C3H/HeJ (filled circles) and C3H/HeOuJ (open circles) mice, with these two strains suffering 100% mortality by day 12 of infection. In comparison, severe illness was slower to develop in the C3H/HeN mice (crosses), but between days 10 and 21 p.i., all mice of this strain succumbed to the infection. Survival data from one representative experiment out of three are shown, with all three experiments showing similar results. Each data point represents the percentage of mice still surviving from an initial population of 10 mice.
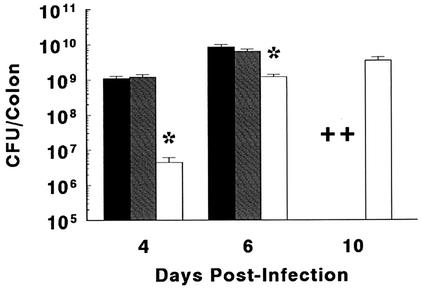
We next looked at bacterial colonization levels as well as colonic tissue damage in these three related strains. The speed and levels of bacterial colonization were similar between the C3H/HeJ and C3H/HeOuJ mice (Fig. 4), while bacterial colonization occurred more slowly in the C3H/HeN strain. Similarly colon weights and crypt heights increased more rapidly in the two most susceptible mouse strains (Table 3). While the tissue pathology was delayed in the C3H/HeN mice, it progressed more rapidly than in the resistant mouse strains, with mucosal ulceration seen by day 10 p.i. Taken together, these results identify differences among these three mouse strains in their relative susceptibilities to C. rodentium infection. Even so, all three strains proved to be highly susceptible to infection by this A/E pathogen. While we are reluctant to argue that TLR4 does not play any role during C. rodentium infection, these studies suggest that TLR4 signaling is not the basis for the increased susceptibility of the C3H/HeJ strain to C. rodentium colonization.
FIG. 4.
Following oral infection, all three C3H strains were heavily colonized by C. rodentium. The mean numbers of bacteria or CFU recovered from the colons of infected C3H/HeJ (black bars), C3H/HeOuJ (shaded bars), and C3H/HeN (open bars) mice are shown in this figure. While C3H/HeJ and C3H/HeOuJ mice showed similar colonization levels at days 4 and 6 p.i., C3H/HeN mice carried significantly smaller bacterial loads (asterisks; P < 0.05) at these time points. By day 10 p.i., colony counts were performed only on C3H/HeN mice, as the majority of the C3H/HeJ and C3H/HeOuJ mice required euthanization (crosses) by this time. Values are the mean colonic C. rodentium CFU ± 1 SEM from three independent experiments, each with groups of four to five mice.
TABLE 3.
Colon weights and crypt heights during C. rodentium infection of the susceptible mice in this studya
| Strain | Mean colon wt (mg) at day:
|
Mean crypt ht (μm) at day:
|
||||
|---|---|---|---|---|---|---|
| 0 | 4 | 6 | 0 | 4 | 6 | |
| C3H/HeJ | 105 ± 7 | 143 ± 9b | 223 ± 17b | 134 ± 12 | 210 ± 27b | 265 ± 22b |
| C3H/HeOuJ | 110 ± 4 | 145 ± 9b | 208 ± 21b | 145 ± 9 | 190 ± 18b | 249 ± 19b |
| C3H/HeN | 104 ± 10 | 128 ± 11 | 165 ± 14b,c | 129 ± 14 | 139 ± 9c | 195 ± 22b,c |
C. rodentium infection resulted in a time-dependent increase in both colon weights and crypt heights in all three related C3H mouse strains studied. Values are the mean colon weight or crypt height ± 1 SEM from three independent experiments, each with groups of four to five mice.
Significantly elevated over values from uninfected mice of the same strain (P < 0.05).
Significantly lower value than those for the other susceptible mouse strains at the same time point (P < 0.05).
Another important host genetic locus identified as mediating significant effects on host susceptibility to many bacterial pathogens is NRAMP1. While its function is still controversial, it appears to regulate the environment of the phagosome, starving phagocytosed bacteria of ions necessary for growth (21, 29, 56). Previous studies with Salmonella serovar Typhimurium and Mycobacterium have shown that mouse strains suffering an inactivating mutation in the NRAMP1 gene (D169) are highly susceptible to these intracellular pathogens (29, 56). Since C. rodentium rarely encounters macrophages or neutrophils, the two cell types known to express NRAMP1, it seemed un likely that NRAMP1 function would underlie host susceptibility to C. rodentium. Even so, the mouse strains tested were chosen in part to address this issue. Interestingly, the BALB/c and C57BL/6 mouse strains, which lack functional NRAMP1, carried a similar bacterial load and followed a similar course of infection as the NRAMP1-expressing 129S1/SvImJ strain. Furthermore, the highly susceptible C3H strains are known to express functional NRAMP1 (15). Taken together, the pattern of strain susceptibility to C. rodentium infection showed no apparent role for NRAMP1 involvement.
Susceptible mouse strains exhibit significant crypt cell apoptosis.
As mentioned above, the susceptible mice exhibited frequent areas of mucosal ulceration as well as friability with bleeding into the colonic lumen during infection. As this tissue damage probably contributed to the severe morbidity and mortality seen in these mouse strains, we were interested in determining the underlying pathological mechanisms. Histological examination under high magnification revealed several interesting observations. The first was that, along with considerable colonic inflammation and tissue damage in the susceptible mice, there were also substantial numbers of cells displaying signs of shrinkage and nuclear fragmentation suggestive of programmed cell death (apoptosis). We utilized the TUNEL staining approach to assay for apoptotic cells in the different strains of mice before and during infection. Under uninfected conditions, very few TUNEL-positive cells were observed in the colonic mucosa. A small number of epithelial cells being sloughed into the colonic lumen or those that had already been sloughed were TUNEL positive, and this apoptotic cell population was seen in the colons of both susceptible and resistant mouse strains.
We next examined tissues from infected mice. As mentioned in an earlier study, C. rodentium infection is accompanied by the increased shedding of mature colonocytes into the colonic lumen, often with bacteria still attached (54). This large population of sloughed cells was frequently TUNEL positive in both resistant and susceptible mouse strains. Interestingly, despite the heavy colonization of the colonic epithelium with C. rodentium, very few superficial epithelial cells showed signs of apoptosis (Fig. 5). Although few TUNEL-positive cells were observed at the mucosal surface, more TUNEL-positive cells were observed at the base of the crypts. Although they were not numerous in the resistant strains, the number of TUNEL-positive crypt cells was dramatically greater in the susceptible mouse strains (Fig. 5), showing a time-dependent increase that matched the tissue pathology (Table 4). While the identity of a portion of these apoptotic cells was difficult to determine, the majority appeared to be crypt epithelial cells based on their location and size. While it is not clear if the apoptosis was induced through a bacterial or host-dependent trigger, apoptotic cells were observed at the base of both uninfected and infected crypts. This suggests that the induction of programmed cell death did not require direct bacterial attachment.
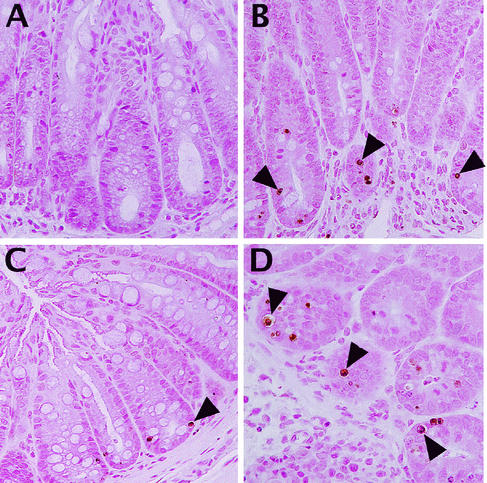
FIG. 5.
TUNEL staining identified significant numbers of apoptotic cells at the base of the colonic crypts in susceptible, but not resistant, mice. As shown in panel A, very few colonic cells were TUNEL positive in tissue sections from C. rodentium-infected resistant mouse strains (shown here with an infected C57BL/6 mouse at day 6 p.i.). In contrast, TUNEL staining (brown) identified numerous cells as TUNEL positive at the base of the crypts in C3H/HeJ mice at day 6 p.i. (B). While not as numerous, TUNEL-positive cells were also seen in the same location in the C3H/HeN mice, also shown at day 6 p.i. (C). At higher magnification, with colonic tissue from a C3H/HeOuJ mouse at day 6 p.i., the location and size of the TUNEL-positive cells identify them as being crypt epithelial cells (D).
TABLE 4.
Numbers of TUNEL-positive cells per 10 colonic crypts in different mouse strainsa
| Strain | No. of TUNEL-positive cells/ 10 colonic crypts at day:
|
||
|---|---|---|---|
| 0 | 4 | 6 | |
| NIH Swiss | <1 | <1 | <1 |
| C57BL/6 | <1 | <1 | <1 |
| BALB/c | <1 | <1 | <1 |
| 129S1/SvImJ | <1 | <1 | <1 |
| C3H/HeJ | <1 | 4.6 ± 1.1b | 16.4 ± 3.3b |
| C3H/HeOuJ | <1 | 3.7 ± 1.3b | 13.1 ± 2.1b |
| C3H/HeN | <1 | 1.4 ± 0.3b | 4.5 ± 0.6bc |
| SCID (C3H/HeJ) | <1 | <1 | 2.4 ± 0.9b,c |
C. rodentium infection resulted in a time-dependent increase in the number of TUNEL-positively staining cells at the base of colonic crypts in susceptible mice only. Values represent the mean number of TUNEL-positive cells per 10 colonic crypts ± 1 SEM from three independent experiments, each with groups of four to five mice.
Significantly elevated over values from uninfected mice of the same strain (P < 0.05).
Significantly lower value than that for C3H/HeJ mice at the same time point (P < 0.05).
Susceptible mouse strains suffer greater bacterial translocation to the MLNs.
The second observation made from infected susceptible mice was that, along with the massive numbers of bacteria found in the gut lumen, small numbers of C. rodentium bacteria could be identified within the mucosal and submucosal regions of the colon. These bacteria were often close to areas of mucosal ulceration or near heavily infected crypts. Previous studies have shown that commensal bacterial overgrowth in the colon as well as inflammation and other forms of colonic tissue damage can lead to the translocation of luminal bacteria from the colon to the MLNs and other internal organs (2, 12). In some cases this translocation was found to cause sepsis and the eventual death of the host. Interestingly, a recent study found that although C. rodentium is primarily a luminal pathogen, small numbers of bacteria translocated to the MLNs during the infection (25), although the bacterial numbers were extremely low relative to the luminal burden.
Based on the greatly increased bacterial load and tissue damage seen in infected susceptible mice, we theorized that C. rodentium translocation might contribute to their greater morbidity and mortality during infection. We therefore removed MLNs from infected mice of the different strains tested in this study and assessed the C. rodentium burdens carried in these tissues. The MLN counts were all negative for uninfected mice, whereas at day 6 p.i. significant differences in bacterial numbers were seen. The resistant mice, including the NIH Swiss ([4.3 ± 1.3] × 105 CFU), C57BL/6 ([2.9 ± 0.8] × 105 CFU), BALB/c ([9.3 ± 2.7] × 104 CFU), and 129S1/SvImJ ([7.7 ± 2.3] × 105 CFU) mice, had bacterial MLN levels ranging from approximately 105 to 106 CFU/mouse. While these numbers were not insignificant, they represented a very small percentage (roughly 0.1%) of the C. rodentium bacteria found in the colon at the same time. Interestingly, there were substantially more bacteria found in the MLNs of the susceptible mice, with (6.0 ± 3.3) × 107 CFU found in the C3H/HeJ mice, while the C3H/HeOuJ mice carried a slightly smaller bacterial load ([3.4 ± 0.8] × 107 CFU). The C3H/HeN mice carried even fewer bacteria than did their related strains ([1.4 ± 0.4] × 106 CFU). These numbers indicate that roughly 10- to 500-fold-more bacteria were in the MLNs of the susceptible mice than in those of the resistant mice, also corresponding to an increased percentage of bacteria (between 0.5 and 1%) in the MLNs relative to the luminal load carried by these mice. The importance of these findings is probably best understood when put into the context of the well-characterized Salmonella serovar Typhimurium murine typhoid model. The load of C. rodentium in the MLNs of the most susceptible mice (C3H/HeJ and C3H/HeOuJ) was as high as the number of Salmonella serovar Typhimurium bacteria recovered from the internal organs of Salmonella-infected mice just prior to the onset of sepsis and death (56). Based on this comparison, the translocation of C. rodentium from the gut into the MLNs must be considered a potential factor in the heightened morbidity and mortality seen in the susceptible mice during infection.
T and B lymphocytes influence tissue pathology but not bacterial colonization.
As earlier outlined, the loss of TLR4 and NRAMP1-dependent innate immune defenses did not appear to be the underlying causes for increased susceptibility to C. rodentium infection. We therefore decided to examine the role of the host's acquired immune response in mediating susceptibility. This was accomplished by infecting C3H/HeJ mice suffering from the SCID mutation and thus lacking mature T and B lymphocytes (8). In an earlier study, we found that RAG1-deficient mice (also lacking T and B lymphocytes) on a C57BL/6 genetic background suffered less tissue pathology, but died from the infection with a much higher frequency, than wild-type mice (54). Surprisingly, in the present study we found that SCID mice on a C3H/HeJ background survived 2 to 3 days longer (10.9 ± 1.3 days) than did wild-type C3H/HeJ mice (7.8 ± 1.0 days; P < 0.05). When we examined bacterial colonization levels in the colon at day 6 p.i., the SCID mice had bacterial numbers similar to those seen for wild-type mice (7.1 × 109 versus 8.7 × 109 CFU). In contrast, the number of C. rodentium bacteria that had translocated to the MLNs of the SCID mice was (8.9 ± 1.0) × 106, significantly less than the (6.0 ± 3.3) × 107 CFU in the C3H/HeJ mice, but still highly elevated compared to resistant mouse strains.
Increases in crypt heights and colon weights were moderately attenuated in the SCID mice (210 ± 24 μm and 168 ± 12 mg, respectively) compared to immunocompetent C3H/HeJ mice (265 ± 20 μm and 223 ± 27 mg, respectively). When we performed TUNEL staining, we also observed a significant attenuation in the number of apoptotic cells in the colons from SCID mice (Table 4) and little mucosal ulceration (data not shown). These observations support the hypothesis that the apoptosis seen in this model is mediated through the host immune response rather than being directly triggered by C. rodentium or by a bacterial product. Furthermore, these results suggest that host lymphocytes may contribute to the bacterial translocation seen in susceptible mice, perhaps through the induction of epithelial cell apoptosis and disruption of the epithelial barrier. Even so, in the absence of T and B cells, bacterial translocation still occurred, and mortality was only moderately delayed, indicating that other factors probably play a more important or direct role in these features of the infection.
DISCUSSION
Despite widespread interest in the mechanisms of mammalian host resistance to bacterial pathogens, studies in this area have almost exclusively focused on intracellular bacterial pathogens such as Salmonella serovar Typhimurium, Legionella pneumophila, and Mycobacterium (14, 24, 29, 46, 56). As a result, the host susceptibility loci already identified have been those expressed primarily by macrophages and other phagocytes. While these studies have proven invaluable, phagocytes represent only one aspect of host antimicrobial defenses, and while important in combating intracellular and systemic pathogens, they play less of a protective role against mucosal pathogens. This is particularly true with regard to A/E bacteria, which rarely penetrate the intestinal mucosa but instead reside within the intestinal lumen (51, 55). This pathogenic strategy has proven highly successful, ensuring that A/E bacteria are relatively protected from the host's phagocytes. However, the gastrointestinal tract possesses a number of other innate and acquired defenses (44, 51), together with physical barriers to infection, that undoubtedly influence the ability of A/E bacteria to infect their hosts. Determining which factors are critical in host protection and their contribution to disease pathology offers great potential for improving present therapies as well as developing vaccines directed against these important human and veterinary pathogens.
To date, the study of EPEC and EHEC has been limited because of the severity of the disease and because the normally susceptible populations are children. It has also been particularly difficult to advance these studies in tissue culture, since susceptibility to A/E pathogens may manifest only within the intact host. We therefore chose to use the surrogate pathogen C. rodentium in a murine model. This approach permitted the relatively easy screening of a number of genetically defined mouse strains for their susceptibility to infection. While preliminary studies by Barthold et al. in the 1970s originally identified C3H/HeJ mice as suffering high mortality during C. rodentium infection (5), the basis had not been examined until now. This study is the first to examine the basis for host susceptibility to an A/E pathogen. Moreover, it is the first to link this susceptibility to a more rapid and extensive bacterial colonization of the colon, severe colonic tissue damage, bacterial translocation to the MLNs, and host cell apoptosis.
While C. rodentium infection of most inbred and outbred strains of mice caused only limited mortality, almost 100% of the C3H/HeJ, C3H/HeOuJ, and C3H/HeN strain mice needed to be euthanized following infection. This study identifies the primary cause for the high death rate as increased susceptibility to C. rodentium colonization and the associated consequences of this colonization. Compared to resistant mouse strains, the susceptible mice underwent more rapid and extensive colonization resulting in bacterial colony counts elevated up to 50-fold over those in less susceptible strains. Bacterial colonization not only occurred more rapidly and involved a larger bacterial load but also spread more deeply into colonic crypts throughout the length of the colon. A second cause for the high lethality was the more rapid and severe colonic pathology in susceptible mice than in resistant mice. In concert with an accelerated onset of inflammation and crypt hyperplasia, the colonic pathology in the susceptible mice involved extensive crypt cell apoptosis, mucosal ulceration, and friability with bleeding into the colonic lumen. While C. rodentium infection of less susceptible mouse strains has been described elsewhere as modeling some aspects of inflammatory bowel diseases (31, 32), the pathology seen in susceptible mice may prove more relevant to the severe tissue damage seen in these inflammatory diseases.
Based on the key role of the host immune system in controlling many other bacterial pathogens, we initially examined host immunity in the susceptibility to C. rodentium infection. Recently, several groups have tested the immune system in protecting the host against C. rodentium, finding that mice lacking interleukin 12, gamma interferon (52), and the receptor for tumor necrosis factor (26), as well as mice suffering severe immunodeficiencies (i.e., lacking both T and B lymphocytes) (54), were more susceptible to infection than immunocompetent mice were. Interestingly, the C3H/HeJ and C3H/HeOuJ mice in the present study suffered heavier bacterial loads and more rapid and severe tissue pathology than any of the immunodeficient mouse strains described previously. Thus, the underlying basis for these strains' susceptibility to C. rodentium infection is unlikely to be one of the aforementioned immunodeficiencies. Based on the speed of colonization in susceptible mice, we postulated that the susceptibility might reflect a selective defect in innate immunity, impairing their ability to recognize and deal with C. rodentium.
Based on this hypothesis, and because C3H/HeJ mice are defective in TLR4 signaling, we tested the role of this innate pathway in the susceptibility to C. rodentium infection. We infected two strains of mice that are closely related to C3H/HeJ but respond normally to LPS. Historically, the C3H strain originated from a cross between a Bragg albino female and a DBA/J male in 1920. Progeny of these mice were transferred to the National Institutes of Health, where the name C3H/HeN arose, and then to the Jackson Laboratory in 1948, where the strain became identified as C3H/HeJ. The C3H/HeOuJ strain resulted from a transfer of C3H/HeJ mice to Outzen from the Jackson Laboratory in 1952, and these mice were transferred back to the Jackson Laboratory in 1981 (34). Between 1952 and 1962 the C3H/HeJ strain suffered a mutation in the TLR4 gene, becoming LPS hyporesponsive. Because they were derived from the same original stock, the C3H/HeN and C3H/HeOuJ strains have been repeatedly used as controls for C3H/HeJ mice in a variety of studies. These studies have shown that C3H/HeJ mice are highly susceptible to infection by invasive gram-negative bacteria such as Salmonella serovar Typhimurium, in part because of a delayed neutrophil recruitment to sites of bacterial replication (19, 50). This impaired inflammatory response permits the rapid systemic spread of bacterial pathogens, ultimately leading to the death of the host (11, 40).
In contrast to systemic infections, very little is known about the host's innate immune response to enteric luminal pathogens such as C. rodentium. Our results showed that the LPS-responsive C3H/HeOuJ mouse strain was as susceptible to infection as the C3H/HeJ strain was. The C3H/HeN strain mice, while less susceptible, still underwent significant bacterial colonization and colon pathology and ultimately succumbed to the infection between days 10 and 21 p.i. These results may reflect the more recent separation of the C3H/HeJ and C3H/HeOuJ strains, since their genetics should be more similar to each other than to those of the C3H/HeN strain. Based on the intermediate susceptibility of the C3H/HeN strain, more than one genetic locus may be involved in modulating the susceptibility phenotype. While insufficient to fully determine if TLR4 plays any role during C. rodentium infection, these studies suggest that susceptibility to C. rodentium infection is independent of TLR4. Similarly, since C3H/HeJ and related strains express functional NRAMP1, this host resistance protein also appears to be uninvolved. We speculate that an as-yet-unidentified host defense pathway may be the basis for the susceptibility to C. rodentium infection. Since this pathogen primarily interacts with intestinal epithelial cells, impaired production of antimicrobial effectors by these cells or perhaps goblet cells is a plausible basis for the susceptibility to this pathogen. The demonstration that C3H/HeJ SCID mice carried similar bacterial loads as wild-type C3H/HeJ mice confirms that the acquired immune system plays little role in controlling C. rodentium colonization of the host intestine.
Aside from rapid C. rodentium colonization and high bacterial loads, a second aspect of the susceptibility was the exaggerated tissue pathology and intestinal inflammation seen in the C3H mouse strains. Colon weights and crypt heights were already significantly elevated by day 4 p.i., at least 4 to 6 days more rapidly than in resistant mouse strains. Previous studies have shown that the epithelial hyperplasia is dependent on the host immune response (31, 54). In agreement with those findings, the SCID defect attenuated the epithelial hyperplasia, inflammation, and other aspects of tissue pathology without affecting colonic bacterial numbers. This reduction in tissue damage presumably contributed to the SCID mice surviving longer than wild-type mice. This is in contrast to our previous study where RAG1-knockout mice on the resistant C57BL/6 background died because they were unable to clear the infection, despite suffering less colon damage than wild-type C57BL/6 mice that ultimately survived (54). In the present study, the reduced tissue damage seen in SCID C3H/HeJ mice may have been responsible for the reduced bacterial translocation seen in these mice. Indeed, the reduction in bacterial translocation is a plausible basis for the delayed onset of morbidity and mortality in infected SCID mice. Taken together, these results highlight the role of the immune system in causing the tissue pathology and disease symptoms seen during this infection.
Another significant aspect of the tissue pathology was the mucosal ulceration and tissue friability observed by day 6 p.i. in both the C3H/HeJ and C3H/HeOuJ strains and by day 10 p.i. in the C3H/HeN strain. Lesions occurred in both the distal colon and the cecum of susceptible mice but rarely in the more resistant mouse strains. While epithelial hyperplasia occurs in all mouse strains tested so far, frequent and severe mucosal ulceration has been described only for the three susceptible mouse strains in this report. There could be many reasons underlying the ulceration, including the extensive apoptosis of host cells observed in the colons of susceptible mice. While a number of bacterial pathogens cause host cells to undergo programmed cell death, the role of A/E pathogens in this pathology is unclear. Studies with tissue culture suggest that EPEC and EHEC can kill host cells, although the mode of death remains controversial (3, 10). Despite this in vitro finding, we observed that very few C. rodentium-infected colon cells were TUNEL positive. We did see a substantial increase in TUNEL-positive cells at the base of many colonic crypts in infected susceptible mice. The location and appearance of the apoptotic cells were consistent with crypt epithelial cells. The loss of these basal crypt cells could trigger mucosal ulceration by impairing the ability to replace epithelial cells that are shed during the infection. Since C. rodentium bacteria were rarely found at the base of crypts, it seems unlikely that the apoptosis was caused directly by the bacteria. Despite this, we cannot rule out a role for a soluble bacterial factor. The observation of apoptotic cells in uninfected crypts as well as reduced crypt cell apoptosis in SCID mice also suggests that the acquired immune system is involved and thus provides a solid basis for future studies characterizing the mechanisms underlying the crypt cell apoptosis in this model.
We also speculate that C. rodentium translocation out of the gut may be responsible in part for the morbidity and mortality seen in this model. The large numbers of bacteria found in the MLNs of susceptible mice provide a plausible explanation for the morbidity and mortality in these strains. While not as numerous, the several hundred thousand bacteria in the MLNs of the resistant mouse strains could in part underlie the morbidity that they suffered as well. While the exact mechanisms by which bacteria translocate from the gut are unclear, translocation is usually associated with intestinal tissue damage, inflammation, and/or bacterial overgrowth. All these factors were present in the C. rodentium infection model. Since translocation to the MLNs can occur even with commensal organisms, we doubt that this process reflects a specific pathogenic strategy, but rather a more passive event. Similarly, while the C3H mice might be predisposed to excessive bacterial translocation, the high bacterial numbers in their MLNs are probably a simple reflection of the huge bacterial loads carried in their colons. It remains to be determined if the increased tissue damage and apoptosis seen in the C3H mice permitted the egress of more bacteria out of the colon, or alternatively, whether the increased bacterial translocation triggered the excessive tissue damage seen in these mice.
Taken together, the results of this study demonstrate that susceptible mouse strains develop a more rapid and severe colonic pathology during C. rodentium infection than do resistant mice. It remains to be determined whether the pathology observed is simply commensurate with the heavier bacterial loads or whether these susceptible strains develop an overexuberant and ultimately maladaptive response to infection. While there are no published reports identifying the three C3H strains as being highly susceptible to other bacterial pathogens, models of inflammatory bowel disease have shown that C3H/HeJ mice develop inflammation more rapidly and with a greater penetrance than do other strains (9, 41). In most of these models, the development of intestinal inflammation is dependent on the presence of normal bacterial flora. Furthermore a substrain of C3H/HeJ, the HeJBir mouse, develops a spontaneous colitis requiring the presence of bacteria (16). This suggests that the C3H genetic background may be predisposed to respond maladaptively to enteric bacteria, through excessive colonic inflammation and tissue damage. The C. rodentium infection model may therefore also prove useful in elucidating the mechanisms underlying immunological dysregulation in the intestine.
In summary, our results identify a family of mice that display heightened susceptibility to infection by the A/E pathogen C. rodentium. The susceptibility manifests as a dramatic increase in the bacterial load as well as severe tissue pathology including host cell apoptosis and bacterial translocation to the MLNs, ultimately leading to the death of the host. While the genetic basis for the susceptibility remains to be determined, we have shown that it is not based on previously characterized host resistance loci such as TLR4 and NRAMP1. Based on those studies so far published, the LPS-responsive C3H/HeOuJ and C3H/HeN mouse strains have not demonstrated an increased susceptibility to systemic bacterial infections. Therefore, we hypothesize that the underlying defect in host defense may be specific to the gastrointestinal tract and possibly involves the intestinal epithelium, since intestinal epithelial cells are the host cells that primarily interact with this pathogen. The fact that C. rodentium is a natural mouse pathogen and that it dwells almost exclusively within the colonic lumen provides us with a unique opportunity to study host-bacterial pathogen interactions and hopefully identify novel host resistance mechanisms at this critical interface with the environment.
Acknowledgments
We thank Valerie Templeman, Jutta Kaiser, and Mehran Ghoreishi for technical assistance and Carrie Rosenberger for critical reading of the manuscript.
This work was supported by operating grants from the CIHR, the HHMI, and NSERC (B.B.F.) as well as the CCFC (K.J.). B.A.V. is supported by a CDDF/CIHR postdoctoral fellowship and is an honorary Isaak Walton Killam Fellow. W.D. was supported by an MRC postdoctoral fellowship. B.B.F. is a Howard Hughes International Research Scholar, a CIHR Distinguished Investigator, and the UBC Peter Wall Distinguished Professor.
Editor: A. D. O'Brien
REFERENCES
- 1.Abe, A., U. Heczko, R. G. Hegele, and B. B. Finlay. 1998. Two enteropathogenic Escherichia coli type III secreted proteins, EspA and EspB, are virulence factors. J. Exp. Med. 188:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asfaha, S., W. K. MacNaughton, C. B. Appleyard, K. Chadee, and J. L. Wallace. 2001. Persistent epithelial dysfunction and bacterial translocation after resolution of intestinal inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 281:G635-G644. [DOI] [PubMed] [Google Scholar]
- 3.Barnett Foster, D., M. Abul-Milh, M. Huesca, and C. A. Lingwood. 2000. Enterohemorrhagic Escherichia coli induces apoptosis which augments bacterial binding and phosphatidylethanolamine exposure on the plasma membrane outer leaflet. Infect. Immun. 68:3108-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barthold, S. W., G. L. Coleman, P. N. Bhatt, G. W. Osbaldiston, and A. M. Jonas. 1976. The etiology of transmissible murine colonic hyperplasia. Lab. Anim. Sci. 26:889-894. [PubMed] [Google Scholar]
- 5.Barthold, S. W., G. W. Osbaldiston, and A. M. Jonas. 1977. Dietary, bacterial, and host genetic interactions in the pathogenesis of transmissible murine colonic hyperplasia. Lab. Anim. Sci. 27:938-945. [PubMed] [Google Scholar]
- 6.Bernheiden, M., J. M. Heinrich, G. Minigo, C. Schutt, F. Stelter, M. Freeman, D. Golenbock, and R. S. Jack. 2001. LBP, CD14, TLR4 and the murine innate immune response to a peritoneal Salmonella infection. J. Endotoxin Res. 7:447-450. [PubMed] [Google Scholar]
- 7.Bloom, B. R. 2000. On the particularity of pathogens. Nature 406:760-761. [DOI] [PubMed] [Google Scholar]
- 8.Bosma, M. J., and A. M. Carroll. 1991. The SCID mouse mutant: definition, characterization, and potential uses. Annu. Rev. Immunol. 9:323-350. [DOI] [PubMed] [Google Scholar]
- 9.Bristol, I. J., M. A. Farmer, Y. Cong, X. X. Zheng, T. B. Strom, C. O. Elson, J. P. Sundberg, and E. H. Leiter. 2000. Heritable susceptibility for colitis in mice induced by IL-10 deficiency. Inflamm. Bowel Dis. 6:290-302. [DOI] [PubMed] [Google Scholar]
- 10.Crane, J. K., B. P. McNamara, and M. S. Donnenberg. 2001. Role of EspF in host cell death induced by enteropathogenic Escherichia coli. Cell. Microbiol. 3:197-211. [DOI] [PubMed] [Google Scholar]
- 11.Cross, A., L. Asher, M. Seguin, L. Yuan, N. Kelly, C. Hammack, J. Sadoff, and P. Gemski, Jr. 1995. The importance of a lipopolysaccharide-initiated, cytokine-mediated host defense mechanism in mice against extraintestinally invasive Escherichia coli. J. Clin. Investig. 96:676-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deitch, E. A., R. Berg, and R. Specian. 1987. Endotoxin promotes the translocation of bacteria from the gut. Arch. Surg. 122:185-190. [DOI] [PubMed] [Google Scholar]
- 13.Deng, W., Y. Li, B. A. Vallance, and B. B. Finlay. 2001. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect. Immun. 69:6323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diez, E., S. H. Lee, S. Gauthier, Z. Yaraghi, M. Tremblay, S. Vidal, and P. Gros. 2003. Birc1e is the gene within the Lgn1 locus associated with resistance to Legionella pneumophila. Nat. Genet. 33:55-60. [DOI] [PubMed] [Google Scholar]
- 15.Eden, C. S., R. Shahin, and D. Briles. 1988. Host resistance to mucosal gram-negative infection. Susceptibility of lipopolysaccharide nonresponder mice. J. Immunol. 140:3180-3185. [PubMed] [Google Scholar]
- 16.Elson, C. O., Y. Cong, and J. Sundberg. 2000. The C3H/HeJBir mouse model: a high susceptibility phenotype for colitis. Int. Rev. Immunol. 19:63-75. [DOI] [PubMed] [Google Scholar]
- 17.Fagundes Neto, U., V. C. Ferreira, F. R. Patricio, V. L. Mostaco, and L. R. Trabulsi. 1989. Protracted diarrhea: the importance of the enteropathogenic E. coli (EPEC) strains and Salmonella in its genesis. J. Pediatr. Gastroenterol. Nutr. 8:207-211. [PubMed] [Google Scholar]
- 18.Fagundes-Neto, U., M. R. Kallas, and F. R. Patricio. 1997. Morphometric study of the small bowel mucosa in infants with diarrhea due to enteropathogenic Escherichia coli strains. Hepatogastroenterology 44:1051-1056. [PubMed] [Google Scholar]
- 19.Fierer, J., M. A. Swancutt, D. Heumann, and D. Golenbock. 2002. The role of lipopolysaccharide binding protein in resistance to Salmonella infections in mice. J. Immunol. 168:6396-6403. [DOI] [PubMed] [Google Scholar]
- 20.Finlay, B. B., and P. Cossart. 1997. Exploitation of mammalian host cell functions by bacterial pathogens. Science 276:718-725. [DOI] [PubMed] [Google Scholar]
- 21.Forbes, J. R., and P. Gros. 2001. Divalent-metal transport by NRAMP proteins at the interface of host-pathogen interactions. Trends Microbiol. 9:397-403. [DOI] [PubMed] [Google Scholar]
- 22.Frankel, G., A. D. Phillips, M. Novakova, H. Field, D. C. Candy, D. B. Schauer, G. Douce, and G. Dougan. 1996. Intimin from enteropathogenic Escherichia coli restores murine virulence to a Citrobacter rodentium eaeA mutant: induction of an immunoglobulin A response to intimin and EspB. Infect. Immun. 64:5315-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 24.Frehel, C., F. Canonne-Hergaux, P. Gros, and C. De Chastellier. 2002. Effect of Nramp1 on bacterial replication and on maturation of Mycobacterium avium-containing phagosomes in bone marrow-derived mouse macrophages. Cell. Microbiol. 4:541-556. [DOI] [PubMed] [Google Scholar]
- 25.Ghaem-Maghami, M., C. P. Simmons, S. Daniell, M. Pizza, D. Lewis, G. Frankel, and G. Dougan. 2001. Intimin-specific immune responses prevent bacterial colonization by the attaching-effacing pathogen Citrobacter rodentium. Infect. Immun. 69:5597-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goncalves, N. S., M. Ghaem-Maghami, G. Monteleone, G. Frankel, G. Dougan, D. J. Lewis, C. P. Simmons, and T. T. MacDonald. 2001. Critical role for tumor necrosis factor alpha in controlling the number of lumenal pathogenic bacteria and immunopathology in infectious colitis. Infect. Immun. 69:6651-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goosney, D. L., S. Gruenheid, and B. B. Finlay. 2000. Gut feelings: enteropathogenic E. coli (EPEC) interactions with the host. Annu. Rev. Cell Dev. Biol. 16:173-189. [DOI] [PubMed] [Google Scholar]
- 28.Govoni, G., and P. Gros. 1998. Macrophage NRAMP1 and its role in resistance to microbial infections. Inflamm. Res. 47:277-284. [DOI] [PubMed] [Google Scholar]
- 29.Gruenheid, S., and P. Gros. 2000. Genetic susceptibility to intracellular infections: Nramp1, macrophage function and divalent cations transport. Curr. Opin. Microbiol. 3:43-48. [DOI] [PubMed] [Google Scholar]
- 30.Hecht, G. 2001. Microbes and microbial toxins: paradigms for microbial-mucosal interactions. VII. Enteropathogenic Escherichia coli: physiological alterations from an extracellular position. Am. J. Physiol. Gastrointest. Liver Physiol. 281:G1-G7. [DOI] [PubMed] [Google Scholar]
- 31.Higgins, L. M., G. Frankel, I. Connerton, N. S. Goncalves, G. Dougan, and T. T. MacDonald. 1999. Role of bacterial intimin in colonic hyperplasia and inflammation. Science 285:588-591. [DOI] [PubMed] [Google Scholar]
- 32.Higgins, L. M., G. Frankel, G. Douce, G. Dougan, and T. T. MacDonald. 1999. Citrobacter rodentium infection in mice elicits a mucosal Th1 cytokine response and lesions similar to those in murine inflammatory bowel disease. Infect. Immun. 67:3031-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill, S. M., A. D. Phillips, and J. A. Walker-Smith. 1991. Enteropathogenic Escherichia coli and life threatening chronic diarrhoea. Gut 32:154-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hopkins, W., A. Gendron-Fitzpatrick, D. O. McCarthy, J. E. Haine, and D. T. Uehling. 1996. Lipopolysaccharide-responder and nonresponder C3H mouse strains are equally susceptible to an induced Escherichia coli urinary tract infection. Infect. Immun. 64:1369-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaper, J. B., T. K. McDaniel, K. G. Jarvis, and O. Gomez-Duarte. 1997. Genetics of virulence of enteropathogenic E. coli. Adv. Exp. Med. Biol. 412:279-287. [DOI] [PubMed] [Google Scholar]
- 36.Karch, H. 2001. The role of virulence factors in enterohemorrhagic Escherichia coli (EHEC)-associated hemolytic-uremic syndrome. Semin. Thromb. Hemost. 27:207-213. [DOI] [PubMed] [Google Scholar]
- 37.Kenny, B. 2002. Mechanism of action of EPEC type III effector molecules. Int. J. Med. Microbiol. 291:469-477. [DOI] [PubMed] [Google Scholar]
- 38.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 39.Luperchio, S. A., and D. B. Schauer. 2001. Molecular pathogenesis of Citrobacter rodentium and transmissible murine colonic hyperplasia. Microbes Infect. 3:333-340. [DOI] [PubMed] [Google Scholar]
- 40.MacVittie, T. J., A. D. O'Brien, R. I. Walker, and S. R. Weinberg. 1982. Inflammatory response of LPS-hyporesponsive and LPS-responsive mice to challenge with Gram-negative bacteria Salmonella typhimurium and Klebsiella pneumoniae. Adv. Exp. Med. Biol. 155:325-334. [DOI] [PubMed] [Google Scholar]
- 41.Mahler, M., I. J. Bristol, E. H. Leiter, A. E. Workman, E. H. Birkenmeier, C. O. Elson, and J. P. Sundberg. 1998. Differential susceptibility of inbred mouse strains to dextran sulfate sodium-induced colitis. Am. J. Physiol. 274:G544-G551. [DOI] [PubMed] [Google Scholar]
- 42.Marches, O., J. P. Nougayrede, S. Boullier, J. Mainil, G. Charlier, I. Raymond, P. Pohl, M. Boury, J. De Rycke, A. Milon, and E. Oswald. 2000. Role of tir and intimin in the virulence of rabbit enteropathogenic Escherichia coli serotype O103:H2. Infect. Immun. 68:2171-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagler-Anderson, C. 2000. Tolerance and immunity in the intestinal immune system. Crit. Rev. Immunol. 20:103-120. [PubMed] [Google Scholar]
- 45.Nataro, J., and J. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.North, R. J., R. LaCourse, L. Ryan, and P. Gros. 1999. Consequence of Nramp1 deletion to Mycobacterium tuberculosis infection in mice. Infect. Immun. 67:5811-5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. V. Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 48.Qureshi, S. T., L. Lariviere, G. Leveque, S. Clermont, K. J. Moore, P. Gros, and D. Malo. 1999. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4). J. Exp. Med. 189:615-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schauer, D. B., and S. Falkow. 1993. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect. Immun. 61:2486-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shahin, R. D., I. Engberg, L. Hagberg, and C. Svanborg Eden. 1987. Neutrophil recruitment and bacterial clearance correlated with LPS responsiveness in local gram-negative infection. J. Immunol. 138:3475-3480. [PubMed] [Google Scholar]
- 51.Simmons, C. P., S. Clare, and G. Dougan. 2001. Understanding mucosal responsiveness: lessons from enteric bacterial pathogens. Semin. Immunol. 13:201-209. [DOI] [PubMed] [Google Scholar]
- 52.Simmons, C. P., N. S. Goncalves, M. Ghaem-Maghami, M. Bajaj-Elliott, S. Clare, B. Neves, G. Frankel, G. Dougan, and T. T. MacDonald. 2002. Impaired resistance and enhanced pathology during infection with a noninvasive, attaching-effacing enteric bacterial pathogen, Citrobacter rodentium, in mice lacking IL-12 or IFN-gamma. J. Immunol. 168:1804-1812. [DOI] [PubMed] [Google Scholar]
- 53.Tacket, C. O., M. B. Sztein, G. Losonsky, A. Abe, B. B. Finlay, B. P. McNamara, G. T. Fantry, S. P. James, J. P. Nataro, M. M. Levine, and M. S. Donnenberg. 2000. Role of EspB in experimental human enteropathogenic Escherichia coli infection. Infect. Immun. 68:3689-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vallance, B. A., W. Deng, L. A. Knodler, and B. B. Finlay. 2002. Mice lacking T and B lymphocytes develop transient colitis and crypt hyperplasia yet suffer impaired bacterial clearance during Citrobacter rodentium infection. Infect. Immun. 70:2070-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vallance, B. A., and B. B. Finlay. 2000. Exploitation of host cells by enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 97:8799-8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zaharik, M. L., B. A. Vallance, J. L. Puente, P. Gros, and B. B. Finlay. 2002. Host-pathogen interactions: host resistance factor Nramp1 up-regulates the expression of Salmonella pathogenicity island-2 virulence genes. Proc. Natl. Acad. Sci. USA 99:15705-15710. [DOI] [PMC free article] [PubMed] [Google Scholar]