Abstract
The chromosomal iroBCDEN gene cluster first described for Salmonella enterica is involved in the uptake of catecholate-type siderophore compounds. An orthologous gene cluster has recently been detected in Escherichia coli strains which cause extraintestinal disease. This E. coli iroBCDEN gene cluster has an impact on virulence and has been reported to be located in a pathogenicity island on the chromosome. In this study we characterized an iro gene cluster of a uropathogenic E. coli isolate which is located on a transmissible plasmid related to the R64 plasmid of S. enterica. This cluster is highly homologous to the chromosomal iro cluster of E. coli. When introduced into an E. coli fepA cir fiu aroB mutant, IroN, but not IroBCDE, mediated the utilization of structurally related catecholate siderophores, including 2,3-dihydroxybenzoyl-l-serine, 2,3-dihydroxybenzoyl-d-ornithine, 2,3-dihydroxybenzoic acid, and enterochelin. This study supports the idea of an ongoing horizontal transfer of putative virulence factors and the mobilization of single virulence gene clusters, which lead to a modular assembly of virulence determinants such as pathogenicity islands.
The virulent Escherichia coli strains that cause extraintestinal infections such as urinary tract infection (UTI), bacteremia, and meningitis are distinct from most intestinal commensal E. coli types and diarrheagenic E. coli types (14, 28, 37). These extraintestinal pathogenic E. coli (ExPEC) strains possess specialized virulence factors, including adhesins, siderophores, toxins, polysaccharide coatings (capsules and lipopolysaccharides), protectins, and invasins (18), which allow ExPEC strains to colonize host mucosal surfaces, invade host tissues, foil host defense mechanisms, and incite an injurious host inflammatory response (14, 18, 27, 28). These factors are primarily thought to be inherited vertically within evolutionary lineages but are also thought to be transferred horizontally between lineages, in some instances on plasmids or on pathogenicity-associated islands (PAIs) (6, 21, 26, 35). PAIs are large blocks of established or suspected virulence genes that are inserted into the E. coli genome and which may provide a mechanism for coordinate horizontal transfer of virulence genes between lineages within E. coli and even between species (6, 8, 16, 36). However, the detailed mechanism of horizontal transfer of PAIs and other chromosomal virulence determinants still remains to be elucidated.
As iron is limited in extraintestinal sites of infection, the acquisition of iron is a prerequisite for pathogens encountering the host. The ExPEC strains have multiple iron acquisition mechanisms, including siderophore-mediated iron uptake systems, which contribute to the fully virulent phenotype. The iroBCDENE. coli gene cluster has recently been described for ExPEC strains, with iroNE. coli being orthologous to a catecholate siderophore receptor gene identified in Salmonella spp. (4, 31).
IroN expression was shown to be regulated by the ferric uptake regulator (Fur) and increased by incubating the respective E. coli strains in human urine, ascitic fluid, or blood (31). As with Salmonella, the iroBCDEN gene cluster of E. coli was found to be chromosomally located and in the archetypal uropathogenic E. coli strain 536 part of PAI III (4, 12). The recent work of Russo et al. demonstrated that IroNE. coli enables the uptake of the catecholate siderophore enterobactin in E. coli (32). Using a mouse infection model of ascending UTI, the authors of that study found that the presence of iroNE. coli contributed significantly to the virulence of E. coli (32).
In the present study, we applied a suppressive subtractive hybridization strategy to extraintestinal E. coli strains and detected a gene cluster exhibiting high homology to the E. coli iroBCDEN genes. Unlike the known iroBCDENE. coli cluster, which is located on a chromosomal PAI of uropathogenic E. coli, the iro gene cluster described here is part of a transmissible plasmid related to Salmonella enterica serovar Typhimurium plasmid R64. The characterization of the border revealed the presence of several IS elements. Here we present evidence that iroN mediates the uptake of catecholate siderophores such as 2,3-dihydroxybenzoic acid (DHBA), 2,3-dihydroxybenzoyl-d-ornithine (DHBO), and enterochelin. The data of this study further emphasize the impact of horizontal transfer in the distribution of virulence factors and show that even parts of chromosomal PAIs can be mobilized individually and thus contribute to a modular organization of virulence determinants such as pathogenicity islands.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains, plasmids, and cosmids used in this study are listed in Table 1. Bacteria were grown at 37°C in Luria-Bertani (LB) medium, in Mueller-Hinton II, and in nutrient broth (NB). For iron depletion, NB was supplemented with 50 to 100 μM 2,2-dipyridyl (NBD). For plasmid maintenance, kanamycin (50 μg/ml), ampicillin (100 μg/ml), tetracycline (12 μg/ml), trimethoprim (75 μg/ml), and spectinomycin (50 to 200 μg/ml) were added to the culture medium as required. The E. coli CFT073 strain used in this study is a well-characterized archetypal uropathogenic E. coli (21, 24); the E. coli strain HE300 was isolated from a urine sample of a UTI patient at the diagnostic laboratory of the Max von Pettenkofer-Institut, Munich, Germany. A total of 50 additional E. coli strains from urine samples of patients suffering from UTI and 25 E. coli strains from stool samples of healthy individuals were obtained from the Max von Pettenkofer-Institut diagnostic laboratory and were used as reference strains in PCR and hybridization assays.
TABLE 1.
Bacterial strains, plasmids, cosmids, and oligonucleotides used in this study
| Strain, plasmid, cosmid, or oligonucleotide | Relevant characteristic(s), description, or sequence | Reference and/or source |
|---|---|---|
| Strains | ||
| HE300 | Positive for papA papC fimH fyuA iroN iha bmaE gafD iss | This study; UPEC isolatea |
| DH10B | F−mcrAΔ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 endA1 recA1 deoR (ara leu)7697 araD139 galU galK nupG rpsL λ− | Invitrogen |
| S17-1 | Streptomycin resistance (Strr); trimethoprim resistance (Tmpr); λpir; hsdR pro recA | 23 |
| XL-1 Blue MR′ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac | Stratagene |
| TH2 | supE44 hsdS20 (rB−mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 thi trpR624 | Takara Shuzo Co., Ltd. |
| TH2 spec | Spontaneous spectinomycin mutant | This study |
| H5058 | aroB malT tsx thi cir fiu fepA | 4 |
| Plasmids and cosmids | ||
| p300 | Wild-type plasmid of E. coli HE300; Tcr; Tpr; carrying iroBCDEN genes | This study |
| SuperCos 1 | Cosmid vector; 7.6 kb; Apr Kmr | Stratagene |
| pJS332 | SuperCos 1 with 31.7 kb of p300 carrying iroBCDEN genes | This study |
| pCR4-TOPO | TA cloning vector; 3.9 kb; Apr Kmr | Invitrogen |
| pJS11 | pCR4-TOPO with 196-bp fragment of subtractive cloning (AA107) | This study |
| pJS12 | pCR4-TOPO with 3.8 kb of p300 carrying iroEN genes | This study |
| pJS13 | pCR4-TOPO with 8.6 kb of p300 carrying iroBCDE genes | This study |
| pJS14 | pJS12 with 0.8-kb Tpr cassette in iroN gene | This study |
| pJS15 | pJS12 with 0.8-kb Tpr cassette in iroE gene | This study |
| Oligonucleotides (position [nt]) | ||
| ydfB.for (6808) | 5′-ATATCACGGGAAAGGAAGCAC-3′ | This study |
| ydfB.rev (7215) | 5′-GTCCGGCAAGGTTTATCA-3′ | This study |
| ORF14.for (12886) | 5′-TGGTCAGGTTGCGGAGGCTAT-3′ | This study |
| iroN1.for (13464) | 5′-CCGACGATGATAATGACGAGA-3′ | This study |
| iroN.rev (13806) | 5′-TGTTCCGGTGGCACCCAGTTG-3′ | This study |
| iroN2.for (14668) | 5′-AGATAACATTGAGCCGGTTCC-3′ | This study |
| iroE.for (15734) | 5′-GCATTGACGCCTTTATCTTTC-3′ | This study |
| iroD.rev (16680) | 5′-CTGATTGACGGGATTGGTT-3′ | This study |
| ORF20. rev (23257) | 5′-GCGAAGGCGAGACTATCAGGA-3′ | This study |
| iroB.for (22565) | 5′-CGAGCGGAGGGTAGATGATGA-3′ | This study |
UPEC, uropathogenic E. coli.
Recombinant DNA methods.
Isolation of plasmids, cosmids, and genomic DNA as well as cloning of DNA fragments was performed using standard techniques (3). Using the SuperCos 1 Cosmid vector kit and the Gigapack III gold packaging extract (Stratagene, Heidelberg, Germany), a cosmid library of total DNA from E. coli strain HE300 was constructed according to the manufacturer's recommendations. PCR products representing parts of the iroBCDEN gene cluster were cloned into vector pCR4-TOPO as recommended by the manufacturer (Invitrogen, Karlsruhe, Germany). For DNA hybridization analyses, dot blot or colony blot assays were carried out by the method of Southern by transferring DNA onto a Hybond N+ membrane (Amersham Biosciences Europe, Freiburg, Germany) (3). The DNA probes were generated by PCR from plasmid pJS11 (fragment AA107: forward primer, T7; reverse primer, T3) or from genomic DNA of E. coli HE300 (iroN1.for and iroN.rev) and (ydfB.for and ydfB.rev). The PCR primers used in this study are shown in Table 1. Labeling, hybridization, and detection were carried out by means of the ECL random prime labeling and detection system (Amersham).
Bacterial matings.
E. coli strains HE300, DH10B, and S17-1 were transformed with plasmid p300 and used as donor strains in bacterial mating assays. A spectinomycin-resistant derivative of E. coli TH2 was used as the recipient strain (TaKaRa Bio Inc., Shiga, Japan). Both donor and recipient strains were grown to the late exponential phase, washed, and mixed in a donor/recipient ratio of 1:1. Mating mixture (30 ml) was sedimented by centrifugation, resuspended in 200 μl of LB medium, deposited onto a blood agar plate (BD Biosciences Clontech GmbH, Heidelberg, Germany), and incubated overnight at 37°C. Cells were collected, resuspended by vigorous vortexing, and diluted on liquid medium. E. coli TH2 transconjugants were selected on LB agar plates containing antibiotics (tetracycline [12 μg/ml] and spectinomycin [200 μg/ml]). For calculation of transfer frequencies, donor, recipient, and transconjugant CFU were counted after mating disruption and plating of appropriate dilutions. Donor and recipient spontaneous resistance to selective antibiotics was also determined. E. coli HE300 spectinomycin-resistant strains arose at frequencies of 1:107 or less, whereas resistance to tetracycline in E. coli TH2 alone was undetectable (<1:109).
Selective subtractive hybridization (SSH).
Using E. coli CFT073 as the driver strain and E. coli HE300 as the tester strain, genomic subtractions were carried out by means of the PCR-Select bacterial genome subtraction kit (BD Biosciences Clontech). The genomic DNA from both strains was isolated and digested with RsaI. The tester DNA was ligated separately with two sets of linkers provided by the manufacturer. These two pools were separately hybridized to excess DNA from the driver strain and then mixed together to allow hybridization of the tester sequences. PCR was performed using oligonucleotides complementary to the linker sequences to enrich for sequences that contained different linkers on both ends and thus had not been hybridized to the excess driver DNA. The PCR products were cloned directly into pCR4-TOPO vector (Invitrogen) following the manufacturer's instructions. Using the universal T7 and M13 oligonucleotide primers, sequencing of the cloned products was carried out.
DNA sequencing and phylogenetic analysis.
DNA was sequenced using a BigDye Deoxy termination kit according to the manufacturer's instructions and a model 377 DNA sequencing system (Applied Biosystems, Weiterstadt, Germany). To sequence the pJS332 cosmid, a shotgun library was prepared from mechanically sheared DNA. Fragments with sizes of 1.5 to 2.0 kb were separated by agarose gel electrophoresis, end repaired, and cloned into the pZErO-2.1 vector (Invitrogen). DNAs of random plasmid clones were isolated, purified, and used as templates for shotgun sequencing reactions as described above. Management and analysis of nucleotide sequence data were performed using a Lasergene sequence analysis software system (DNASTAR, Inc., Madison, Wis.). Using the programs BlastN and BlastX (2) (http://www.ncbi.nlm.nih.gov) and FASTA (http://www.ebi.ac.uk/fasta3), homology searches were performed by comparing the sequences with the public DNA and protein databases. The DNA sequences were also compared with the sequences in the unfinished E. coli CFT073 genome DNA database (http://www.genome.wisc.edu; last updated on 2 June 2002).
Sequence analysis.
A MegAlign program from a LASERGENE software package (version 5.01; DNASTAR, Inc.) was used to produce a multiple alignment of amino acid sequences, which included the inferred sequence of IroN determined here together with sequences retrieved from GenBank. Using a MEGA version 2.0 program (22), phylogenetic trees were constructed on the basis of pairwise estimates of the expected number of amino acid replacements per site with the aid of the neighbor-joining algorithm (33).
Construction of iroN and iroE mutants by EZ::TN transposon insertion.
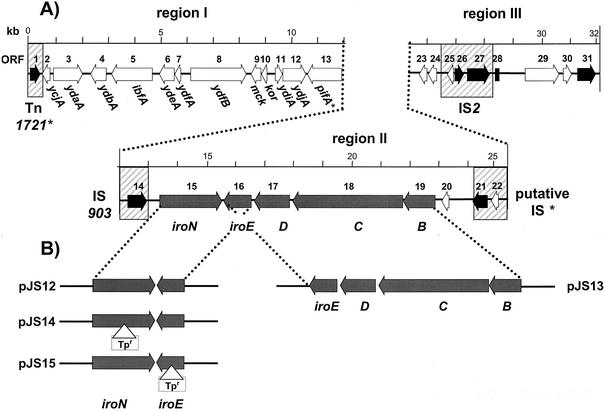
The DNA fragments carrying either iroN and iroE (fragment size, 3,776 bp; primers, ORF14.for and iroD.rev) or iroB, -C, -D, and -E (8,571 bp; iroN2.for and ORF20.rev) were amplified using LA-Taq DNA polymerase (TaKaRa Bio Inc.) with genomic DNA of E. coli HE300 as the template. The resulting PCR products were cloned into pCR4-TOPO vector (Invitrogen), resulting in plasmids pJS12 and pJS13 for iroN and iroBCDE, respectively (Table 1 and Fig. 1). Both plasmids were mutagenized in vitro using a DHFR-1 EZ::TN insertion kit (Epicentre Technologies, Madison, Wis.) and transformed into E. coli TH2, resulting in selection for ampicillin, kanamycin, and trimethoprim resistance. Insertion points were confirmed by restriction analysis and by sequencing with forward and reverse primers provided with the EZ::TN kit (Epicentre). Two plasmids derived from pJS12 with a dihydrofolate reductase 1 EZ::TN insertion in iroN and iroE genes were isolated and designated pJS14 and pJS15, respectively (Table 1 and Fig. 1).
FIG. 1.
Genetic organization of the 31,870-bp inserted DNA of pJS332 with three distinct DNA regions (I to III). (A) The locations and orientations of the ORFs described in this study are indicated by arrows. Genes of the iroBCDEN cluster are indicated by dark gray shading, black arrows indicate transposase genes, and IS elements and transposons are depicted as hatched boxes. Designations below the schemes represent genes with extensive homology to other bacterial alleles as shown in Table 2. (B) Structures of subcloned fragments of the iro gene cluster in pCR4-TOPO giving rise to pJS12 and pJS13 plasmids, as well as the structure of the mutated iroN and iroE genes in plasmids pJS14 and pJS15, respectively. Triangles indicate insertions of trimethoprim transposon to generate mutations of iroN and iroE genes.
RT-PCR.
E. coli H5058 carrying pJS332 was grown to the logarithmic phase at 37°C in nutrient broth containing 50 μM 2,2′-dipyridyl. The RNA was isolated by means of an SV total RNA isolation system (Promega), and reverse transcriptase PCR (RT-PCR) was performed using an Access RT-PCR system (Promega) according to the recommendations of the manufacturer. Briefly, 2 μg of total RNA was treated with 10 U of RQ1 RNase-free DNase (Promega) for 60 min at 37°C followed by heat inactivation of the enzyme at 75°C for 6 min. For cDNA synthesis, the DNase-treated RNA sample was divided among two tubes, the sample and the respective negative control without reverse transcriptase. Avian myeloblastosis virus reverse transcriptase (Promega) was added to the sample together with 0.5 mM deoxynucleoside triphosphates and incubated at 48°C for 45 min. Three pairs of primers were chosen to amplify cDNA and detect transcription of the iro gene cluster: iroN1.for and iroN.rev for iroN (323 bp), iroE.for and iroD.rev for iroED (928 bp), and iroB.for and ORF20.rev for iroB-ORF 20 (674 bp). PCR products were separated by electrophoresis on agarose gel.
Catecholate siderophore uptake assays.
Enterobactin and 2,3-dihydroxybenzoyl-l-serine were purified from the growth supernatant of E. coli AN311 according to a published protocol (39). DHBA was purchased from Sigma Chemicals (Sigma-Aldrich, Munich, Germany), and DHBO was kindly provided by R. Reissbrodt (RKI, Werningerode, Germany). To investigate the functional expression of the identified iroN-iroBCDE gene cluster, the ability to promote the uptake of different catecholate siderophores was tested by a plate assay (30). E. coli strain H5058 (deficient in both catecholate siderophore uptake and enterobactin synthesis) was used as a tester strain for recomplementation of catecholate siderophore-mediated iron uptake (4). For this purpose, plasmids pJS12, pJS13, pJS14, and pJS15 were introduced into the E. coli H5058 strain and recombinant strains were poured into 10 ml of 0.6% NBD agar (NB with 100 μM 2,2′-dipyridyl). Filter paper disks impregnated with enterobactin, DHBS, DHBO, or DHBA (10 μl each of a 1-mg/ml solution) were placed onto the top agar, and after incubation overnight at 37°C, a visible halo around the filter papers appeared, indicative of growth support. Bacteria were stained using a 0.5% solution of 2,3,5-triphenyltetrazoliumchloride (TTC) (9).
Nucleotide sequence accession number.
The nucleotide sequence reported in this study has been deposited in the GenBank database under accession number AY205565.
RESULTS
Detection of a virulence-associated locus by SSH.
To identify new virulence-associated loci in uropathogenic E. coli, we performed SSH with genomic DNA of the uropathogenic archetypal E. coli strain CFT073 and the clinical E. coli isolate HE300 obtained from a patient suffering from severe UTI. A total of 25 subtracted DNA fragments with sizes ranging from 0.1 to 1.8 kb were obtained and subsequently subcloned into pCR4-TOPO vector (data not shown). In total, 13 of the subtractive clones were used as DNA probes in dot blot hybridizations of a reference strain collection to determine the prevalence of subtracted fragments among ExPEC strains. The collection consisted of 50 E. coli strains from blood cultures and urine samples of patients suffering from septicemia and UTIs, respectively. In addition, 25 E. coli strains from stool samples of healthy volunteers were included as control strains. One of the subtracted DNA fragments (AA107) revealed a high level of association with virulence, as it reacted with 23 out of 50 pathogenic E. coli strains but with only 1 out of 25 commensal strains (χ2 = 13.51; P < 0.001). Using the BlastN algorithm of the National Center for Biotechnology Information database at the website http://www.ncbi.nlm.nih.gov (1) as well as the FASTA algorithm of the EMBL database at the website http://www.ebi.ac.uk/fasta33 (22), we determined the nucleotide sequence of fragment AA107 and compared it for sequence homology to known genes. The subtracted fragment AA107 was shown to be a nucleotide fragment 196 bp in length with no significant homology to sequences in the databases.
To further characterize the DNA region adjacent to AA107, we constructed a cosmid library of uropathogenic E. coli strain HE300. Using the fragment AA107 as a DNA probe to screen some 800 cosmid clones by colony blot hybridization, eight cosmid clones were identified. We chose one of the cosmids, pJS332, for further studies and determined the complete sequence by a shotgun approach. To do this, we prepared a random plasmid library and obtained single sequence reads from about 600 clones. Given an average read length of 500 bp, approximately 300 kb of unique reads were generated.
Sequence analysis of cosmid pJS332.
Cosmid pJS332 has 31,870 bp of inserted DNA that appears to be a composite of genes derived from plasmids, a chromosomal pathogenicity island, and additional diverse and unknown sources (Fig. 1 and Table 2). The overall G+C content of the pJS332 DNA region is 49.1%, which is about the average for the E. coli K-12 genome (50.8%). However, the G+C content differs significantly within pJS332 (23 to 72%), displaying peaks and troughs (Fig. 2). These peaks and troughs correspond to different functional units such as transposable elements, indicating a mosaic structure of pJS332 composed of elements of different origin. A total of 31 open reading frames (ORFs) larger than 150 nucleotides (nt), corresponding to a total coding region of 80.1%, were identified (Fig. 1 and Table 2). Seven translated ORFs were found to show no significant similarity or low similarity to protein sequences in the databases, while 24 putative polypeptides were very similar to proteins of S. enterica and E. coli. With regard to the nucleotide homology, the pJS332 DNA region exhibits a modular structure with three distinct regions (Fig. 1 and 2). Region I is located between positions 447 and 11972 and reveals 99.2% homology to 11,528 nt of S. enterica plasmid R64 (accession no. AP005147; nt 29968 to 41495). A 448-bp DNA fragment that represents the end of transposon Tn1721 and that includes the 38-bp repeat unit (IRRII) precedes region I. Immediately downstream of IRRII, region I begins with ORF 2, which shows 100% identity to the ycjA gene of S. enterica serovar Typhimurium plasmid R64. Region I extends from ORF 2 (ycjA) to ORF 13 (pifA*) (Table 2 and Fig. 1).
TABLE 2.
Characteristics of ORFs and deduced amino acid sequences present in the sequenced DNA fragment
| ORF | Gene | Product size (no. of amino acids) | ORF location (nt)a | Protein (description) to which ORF product exhibits homology | Source | Identityb
|
Accession no. | |
|---|---|---|---|---|---|---|---|---|
| % | Range (aa) | |||||||
| 1 | tnpA | 136 | 3-413 | TnpA; transposase of Tn1721, truncated | E. coli | 95 | 135 | P51565 |
| 2 | ycjA | 107 | 791-468 | YcjA; putative bacterial repressor protein | Plasmid R64; S. enterica serovar Typhimurium | 100 | 107 | BAB91595 |
| 3 | ydaA | 377 | 896-2029 | YdaA; putative permease | Plasmid R64; S. enterica serovar Typhimurium | 90 | 377 | BAB91596 |
| 4 | ydbA | 205 | 2939-2322 | YdbA | Plasmid R64; S. enterica serovar Typhimurium | 96 | 205 | BAB91597 |
| 5 | ibfA | 518 | 4686-3130 | lbfA; putative ABC transporter | Plasmid R64; S. enterica serovar Typhimurium | 95 | 518 | BAB91598 |
| 6 | ydeA | 196 | 5539-4949 | YdeA | Plasmid R64; S. enterica serovar Typhimurium | 100 | 196 | BAB91599 |
| 7 | ydfA | 85 | 5796-5539 | YdfA; putative ABC transporter | Plasmid R64; S. enterica serovar Typhimurium | 100 | 85 | BAB91600 |
| 8 | ydfB | 712 | 6150-8288 | YdfB; putative ABC transporter | Plasmid R64; S. enterica serovar Typhimurium | 100 | 712 | BAB91601 |
| 9 | mck | 133 | 8851-8450 | Mck; involved in coordination of plasmid replication with cell division | Plasmid R64; S. enterica serovar Typhimurium | 99 | 133 | BAB91602 |
| 10 | kor | 76 | 9093-8863 | Kor; involved in coordination of plasmid replication with cell division | Plasmid R64; S. enterica serovar Typhimurium | 100 | 76 | BAB91603 |
| 11 | ydiA | 96 | 9389-9679 | YdiA; 93% identical to protein YebA on E. coli plasmid F | Plasmid R64; S. enterica serovar Typhimurium | 100 | 96 | BAB91604 |
| 12 | ydiA | 299 | 9669-10568 | YdjA; 100% identical to protein YebB on E. coli plasmid F | Plasmid R64; S. enterica serovar Typhimurium | 100 | 299 | BAB91605 |
| 13 | pifA* | 443 | 11949-10618 | Truncated ORF; homologous to residues 299 to 714 of 741 residues of protein PifA; phage T7 exclusion protein | Plasmid R64; S. enterica serovar Typhimurium | 100 | 443 | BAB91606 |
| P96329 | ||||||||
| 14 | tnpA | 224 | 12299-12973 | TnpA; transposase of IS903 | Insertion sequence IS903 | 99 | 223 | AAF30382 |
| 15 | iroN | 725 | 13385-15562 | IroN; enterochelin and dihydrobenzoic acid receptor | PAI III of E. coli strain 536 | 99 | 725 | CAC43424 |
| 16 | iroE | 318 | 16563-15607 | IroE | PAI III of E. coli strain 536 | 99 | 318 | CAC43425 |
| 17 | iroD | 409 | 17877-16648 | IroD; putative ferric enterochelin esterase | PAI III of E. coli strain 536 | 99 | 409 | CAC43426 |
| 18 | iroC | 1,261 | 21766-17981 | IroC; putative ABC transporter | PAI III of E. coli strain 536 | 99 | 1,261 | CAC43427.2 |
| 19 | iroB | 371 | 22895-21780 | IroB; putative UDP-glucoronosyl and UDP-glucosyl transferase | PAI III of E. coli strain 536 | 100 | 371 | CAD66179.1 |
| 20 | 89 | 23363-23094 | Hypothetical protein | No significant homology | ||||
| 21 | 173 | 24692-24171 | Hypothetical protein; putative integrase | E. coli strain K5 | 59 | 100 | CAA54707 | |
| 22 | 96 | 25164-24874 | Hypothetical protein | E. coli strain K5 | 59 | 48 | CAA54710 | |
| 23 | 97 | 26026-25733 | Hypothetical protein | E. coli | 80 | 76 | AAB40752 | |
| 24 | iss | 102 | 26347-26039 | Iss (increased serum survival and complement resistance) | E. coli strain 102 | 100 | 102 | AAD41540 |
| 25 | 80 | 26948-26706 | Hypothetical protein | E. coli strain EDL933 | 100 | 26 | AE005326 | |
| 26 | 112 | 26959-27297 | Putative transposase of insertion element IS2 | E. coli | 98 | 109 | P51026 | |
| 27 | 263 | 27404-28195 | Putative integrase of insertion element IS2 | E. coli | 97 | 262 | P51026 | |
| 28 | 54 | 28530-28366 | Hypothetical protein; homologous to gene trp1328 on IS1328 | Yersinia enterocolitica | 63 | 50 | CAB46575 | |
| 29 | 397 | 29418-30611 | Hypothetical protein; putative cobalamin synthesis protein | E. coli strain CFT073 | 65 | 397 | NP_753181 | |
| 30 | 103 | 30750-31061 | Hypothetical protein; putative integrase | Shigella flexneri 2a strain 301 | 59 | 82 | NP_709264 | |
| 31 | 207 | 31248-31869 | Truncated ORF; homologous to gene HCM1.201 on plasmid pHCM1; putative transposase | S. enterica serovar Typhi strain CT18 | 95 | 207 | NP_569402 | |
Nucleotide positions from the start to stop codons in the pJS332 sequence.
Identity is presented as percentage of amino acid identity between the pJS332 ORF and the best hit as determined with BLAST and FASTA. The range is the number of amino acids (aa) over which this identity exists.
FIG. 2.
G+C plot of the DNA sequence of pJS332. Regions I to III are indicated by double-headed arrows; black boxes mark putative transposase genes and IS elements with significantly different G+C content (troughs or peaks). The iroBCDEN gene cluster is shown as a dark gray box. The average G+C content of the E. coli K-12 chromosome is shown with a black line (50.8%).
ORF 13 (pifA*) represents the truncated 3′ part of the pifA gene of R64 and shares 100% identity with nt 893 to 2225. The interruption of pifA is caused by the insertion of a 1,058-bp DNA fragment with 98% identity to insertion sequence IS903. This fragment represents the border with region II and carries ORF 14, which shares 99% identity with the transposase IS903. Region II is located between positions 13078 and 23728 and harbors ORF 15 to ORF 20. It exhibits a high degree of homology to the iroBCDEN gene cluster found in the genome of uropathogenic E. coli. Adjacent to the 3′ end, region II is followed by a small 355-bp DNA mosaic comprised of four fragments of between 30 and 170 nt which is identical to the genomic DNA of E. coli CFT073 neighboring the 3′ end of the iroBCDEN cluster. The border of region II is composed of a truncated putative insertion element carrying ORF 21 and 22, the first of which encodes a conserved amino acid sequence, which is homologous to integrase core domains (Table 2 and Fig. 1).
Region III (located between positions 25287 and 31870) extends from ORF 23 to ORF 31 and reveals sequences related to insertion elements and plasmids or that encode putative products with no significant similarity to sequences in the GenBank database. The presence of a complete copy of insertion element IS2 as well as two further putative transposase genes (ORF 28 and ORF 31) suggests region III to be a hot spot of insertional genetic elements (Fig. 1). The mosaic structure of region III (with DNA fragments of apparently different origins) is also reflected by the G+C content. It displays several troughs, which correspond to different transposable elements (Fig. 2). In particular, the three putative transposase genes (ORF 26, 27, and 31) are visible as deep troughs with a notably lower G+C content than their surroundings (Fig. 1 and 2). Moreover, region III is characterized by a low amount of coding DNA (65.4%), which is probably due to the accumulation of partially truncated IS sequences. The DNA fragment AA107 used as the initial probe was localized between positions 28807 and 28961 as part of the noncoding DNA of region III. In summary, pJS332 is organized in a modular fashion and consists of three regions that appear to be a composite of DNA fragments derived from plasmids, chromosomal loci (PAI III), and diverse IS elements.
The plasmid-carried iroBCDEN cluster is located on a transmissible plasmid related to plasmids of S. enterica.
The homology of the cloned DNA fragment pJS332 to plasmid R64 of Salmonella suggested it for a search for plasmid origins. Plasmid DNA of E. coli HE300 was therefore isolated and subjected to Southern hybridization using fragment AA107 as a DNA probe. The results revealed pJS332 to be part of a plasmid named p300, the size of which is about 78.5 kb as determined by restriction analysis using different enzymes (data not shown). After transformation of the entire plasmid DNA of E. coli HE300 in E. coli S17-1, p300 was mobilized to E. coli TH2 by mating. Thus, the pJS332 region carrying the iroBCDEN cluster is located on a transmissible plasmid that was probably derived from Salmonella plasmid R64, as suggested by the high number of orthologous genes (region I). In addition, both p300 and R64 plasmids confer resistance to trimethoprim, sulfamethoxazole, and tetracycline (15).
RT-PCR experiments detected transcription of the iroBCDE gene cluster.
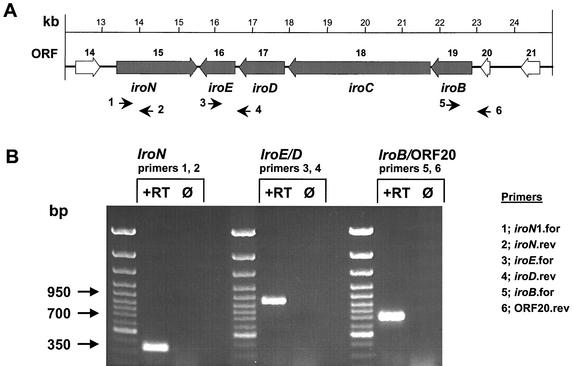
The results of the RT-PCR assays confirmed the transcription of iroN and iroBCDE genes under iron-limited conditions. Interestingly, an amplification product was obtained using primers for the iroB gene and ORF 20 upstream of iroBCDE, suggesting that both form a transcriptional unit transcribed in an operon fashion (Fig. 3). To investigate the functional expression of the identified iroBCDEN gene cluster, the ability to promote the uptake of different catecholate siderophores as iron sources was tested by a plate assay. To do this, the iroBCDEN cluster was subcloned (pJS12 and pJS13; Fig. 1), introduced into the E. coli fepA cir fiu aroB mutant H5058, and examined in a siderophore cross-feeding assay as described previously (4). The results of the feeding assay revealed that as in S. enterica, the iro gene cluster of pJS332 promotes the uptake of enterochelin, DHBS, DHBO, and DHBA. Moreover, IroN alone is sufficient to facilitate catecholate siderophore utilization, since plasmids pJS12 (iroN-iroE) and pJS15 (iroN-iroE::Tpr), but not plasmid pJS13 (iroBCDE) or pJS14 (iroN::Tpr-iroE), promoted siderophore uptake by E. coli H5058 (Fig. 1 and Table 1).
FIG. 3.
RT-PCR analysis of the iroBCDEN cluster. (A) Genetic organization of the iroN, iroBCDE, and upstream genes. Numbered arrows below the map indicate the locations of 5′ and 3′ primers used in RT-PCR. Thick arrows indicate the direction of transcription of each gene. (B) Results of RT-PCR amplification assays. Primers used in each reaction are listed above the corresponding bracket. To verify that DNA was not the amplification template, each RT-PCR was run without the addition of RT (lanes Ø). The size of the expected band for each reaction is indicated.
Phylogenetic analysis of IroN.
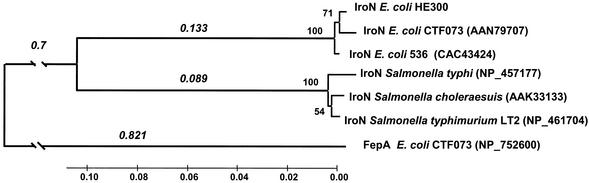
A phylogenetic tree was constructed from the amino acid sequences of IroN for both E. coli and S. enterica by the neighbor-joining algorithm (22) and rooted by the sequence of the E. coli enterobactin receptor FepA (Fig. 4). It revealed two distinct clusters of iroN alleles that grouped by species, with only a little divergence within each cluster. Very recently, Dozois et al. described a plasmid-carried iro gene cluster found in the avian pathogenic E. coli strain χ7122 (13). The amino acid sequence of this IroNχ7122 is 100% identical to that of IroNHE300. In addition, the IroN peptide of the uropathogenic E. coli CP9 strain described by Russo et al. (32) is identical to IroNCFT073 of E. coli CFT073. These results suggest the presence of three phylogenetic lineages of IroN in E. coli (Fig. 4).
FIG. 4.
The phylogenetic tree was constructed using the neighbor-joining algorithm based on the gamma distance with α = 2 and rooted by the sequence of the E. coli enterochelin receptor FepA. The gamma distance assumes that substitutions follow a gamma distribution with the α parameter specifying the amount of variation across amino acid positions. Branch lengths (in number of amino acid substitutions per site) are given in italics above the major branches. Bootstrap confidence levels are given under the nodes, and accession numbers are shown in parentheses.
Distribution of mobilizable iroBCDEN gene clusters among uropathogenic E. coli strains.
To determine the distribution of the plasmid-carried iroBCDEN gene cluster and compare it with the distribution of the chromosomal cluster, dot blot hybridizations were performed using probes derived from either the iroN gene or region I (ydfB gene). In a survey of 50 uropathogenic E. coli isolates and 30 E. coli isolates from the stools of healthy volunteers, we were able to detect iroN in 29 (58%) of the uropathogenic E. coli strains, two of which also proved positive for the ydfB gene (4%). Six of the isolates were positive for ydfB alone. The two isolates positive for both iroN and region I (ydfB) were compared with E. coli strain HE300 with regard to a potential phylogenetic relationship. Interestingly, all three isolates revealed different O antigens, and while the two strains belong to the E. coli phylogenetic group B1, the HE300 strain was assigned to group A by means of a PCR-based typing method (10). Thus, these results favor the idea of a horizontal transmission of this particular virulence factor between only distantly related E. coli strains. These data further demonstrate that the iroN cluster is not linked to plasmid p300 in the vast majority of uropathogenic E. coli strains but rather is chromosomally located or at least linked to a different genetic backbone.
DISCUSSION
In the present report we have described the virulence-associated DNA region pJS332, which was isolated from a uropathogenic E. coli strain. This region, which is 31,870 bp in size, is located on the transmissible plasmid p300 and reveals a composite structure of elements of different origins (chromosomal pathogenicity island, plasmid, and transposable elements) with three distinct DNA regions. Region I resembles a part of plasmid R64 from S. enterica, region II is a copy of the iroBCDEN cluster found on pathogenicity islands of extraintestinal E. coli, and region III comprises different partially truncated insertion sequences. The mosaic nature of p300 is underscored by the observation that the G+C contents of the three DNA regions differ greatly, suggesting that they are of different origins. The iroBCDEN gene cluster encodes a catecholate siderophore uptake system, which is induced in uropathogenic E. coli isolate CP9 upon incubation in urine (31). The recent work of Russo et al. provided evidence that IroN is a virulence factor of E. coli strain CP9 (32). Our study illustrates that the IroN encoded by the plasmid p300 mediates the uptake of different catecholate siderophores such as enterobactin and other linear forms of catecholate siderophores such as DHBA, DHBO, and DHBS. These data broaden the recent results of catecholate siderophore uptake mediated by IroN of E. coli (32) by showing the specific uptake of different purified catecholate siderophore compounds for the first time. In a recent study, Dobrindt et al. analyzed the structures of different PAIs of the uropathogenic E. coli isolate 536 and found the iroBCDEN cluster to be part of PAI III536 (12). Upstream of the iroBCDEN operon, which is homologous to the iro gene cluster of the Salmonella species, an sfa determinant encoding fimbrial adhesins of the S-adhesin family has been detected (11). Interestingly, it has been shown that the iroN gene of strain CP9 is also part of a PAI and is preceded by DNA sequences with homology to the related prs or foc gene cluster (F1C fimbriae) (31). In addition, the recently published genome sequence of the uropathogenic E. coli CFT073 strain reveals a chromosomal iroBCDEN gene cluster on a genomic island neighboring an orthologous sfa/foc gene cluster (38). These data indicate that all iro clusters of E. coli characterized so far are associated with determinants coding for members of the S-adhesin family. However, this is not true for the iro gene cluster of pJS332 described in this study. The homology to the cluster of other E. coli is interrupted upstream of the iroN gene by insertion of IS903, which forms the border with region I. In contrast, the regions downstream of the iroBCDEN cluster bear structural similarity in all E. coli iroBCDEN gene clusters. Different insertion sequences and other partially deleted mobile elements are scattered in the 3′ vicinity of the clusters, suggesting a hot spot for insertion elements. Further elements contributing to the mosaic nature of pJS332 are the transposable element Tn1721 and the insertion sequence IS2, whose origins remain unknown owing to the versatility and widespread distribution of these elements. The presence of insertion sequences adjacent to gene clusters encoding different iron uptake systems has previously been described, and it has been suggested that the insertion sequences mediate the mobilization of the respective cluster. Thus, the genes encoding the aerobactin siderophore system in E. coli are located on the pColV plasmid and flanked by IS1 elements that may facilitate its transposon-like nature (25). The orthologous aerobactin gene cluster of Shigella boydii is part of the genomic island SHI-3 and is framed by IS600 insertion sequences (29). Therefore, the presence of insertion sequences in the direct neighborhood of the iroBCDEN cluster described here is suggestive of a role in its mobilization and transfer. The phylogenetic analysis of different E. coli IroN proteins agrees with the results obtained from sequencing the 3′ end of region II of pJS332, which revealed small nucleotide fragments with a high homology to DNA segments downstream from iroB in E. coli strain CFT073. These data provide circumstantial evidence that the insertion of IS elements followed by site-specific recombination events led to the transfer of the iro gene cluster from the chromosomal location to plasmid p300. During the preparation of the present report, Dozois et al. described an iro gene cluster carried by plasmid pAPEC-1 of the avian pathogenic E. coli strain χ7122 (13). The authors of that study demonstrated that the pAPEC-1 plasmid carries the iutA gene for the ferric aerobactin siderophore receptor. As no iutA gene was detected in E. coli HE300, we suggest a different molecular structure for both pAPEC-1 and p300. Furthermore, the antibiotic resistance (conferred by p300) of E. coli strain HE300 to tetracycline and trimethoprim-sulfamethoxazole supports the structural difference. No comparable resistance pattern is detectable in the avian pathogenic E. coli strain χ7122. In the present report, we show the coexistence of a virulence factor and antibiotic resistance genes on the same transmissible plasmid. Such colocalizations have previously been described for other E. coli pathotypes (34) and suggest that antibiotic-selective pressure also indirectly selects for enhanced virulence.
Pathogenic bacteria have obtained a significant proportion of their genetic diversity through the acquisition of DNA from distantly related organisms (26). The horizontal gene transfers have effectively changed the ecological and pathogenic character of bacterial species, producing extremely dynamic genomes in which substantial amounts of DNA are introduced into and deleted from the chromosome. Different genetic elements such as phages, plasmids, transposons, and PAIs are involved in lateral gene transfer (6, 16).
However, it is not known at present whether pathogenicity islands are acquired intact (e.g., by phage transduction) or by a stepwise insertion of genes into the island. It is also possible that pathogenicity islands use diverse mechanisms of horizontal transfer. The data drawn from this study point to a transfer of single virulence determinants, which may at least modify the structure and function of genomic islands. This also supports the view that virulence determinants are often structured in a modular manner and that parts of PAIs can be distributed individually (19). Furthermore, the detection of a plasmid-carried iro gene cluster corresponds with previous findings that even virulence factors known to be PAI associated in certain strains commonly exhibit divergent patterns of phylogenetic distribution both among lineages and even within a given lineage (7). This is a strong indication that virulence genes are mobile independently of PAIs and that PAIs are subject to continuous, ongoing remodeling (19). Thus, the presence of the E. coli iroBCDEN gene cluster on a transmissible plasmid may explain the particular pattern of phylogenetic distribution described by Johnson et al. (20). The authors of that study found a sporadic appearance of iroNE. coli in many different E. coli serotypes and suggested that either multiple horizontal acquisition events or multiple scattered deletions of iroNE. coli within otherwise iroNE. coli-positive lineages were responsible. These data are consistent with a model in which PAIs, although perhaps occasionally subject to en bloc horizontal transfer (8, 16) or total deletion (5, 6, 17), participate to a much greater extent in the horizontal transfer of single virulence factors by providing genomic regions accessible to the insertion, retention, and release of individual virulence genes.
Our future work is to further investigate the mechanisms of transfer and recombination of the plasmid-carried iroBCDEN gene cluster as a variable virulence factor module. A better understanding of the prevalence and evolutionary origins of the virulence factors of ExPEC strains would accelerate the development of virulence factor-specific preventive measures that are truly needed against these common and morbid infections.
Acknowledgments
We thank Kirsten Weinert for her excellent technical assistance.
This study was supported by a grant from the Bundesministerium für Bildung und Forschung (Kompetenznetzwerk PathogenoMik) to S.S.
Editor: B. B. Finlay
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Bäumler, A. J., T. L. Norris, T. Lasco, W. Voight, R. Reissbrodt, W. Rabsch, and F. Heffron. 1998. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J. Bacteriol. 180:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloch, C. A., and C. K. Rode. 1996. Pathogenicity island evaluation in Escherichia coli K1 by crossing with laboratory strain K-12. Infect. Immun. 64:3218-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blum, G., M. Ott, A. Lischewski, A. Ritter, H. Imrich, H. Tschape, and J. Hacker. 1994. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 62:606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonacorsi, S. P., O. Clermont, C. Tinsley, I. Le Gall, J.-C. Beaudoin, J. Elion, X. Nassif, and E. Bingen. 2000. Identification of regions of the Escherichia coli chromosome specific for neonatal meningitis-associated strains. Infect. Immun. 68:2096-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd, E. F., and D. L. Hartl. 1998. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J. Bacteriol. 180:1159-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brem, D., C. Pelludat, A. Rakin, C. A. Jacobi, and J. Heesemann. 2001. Functional analysis of yersiniabactin transport genes of Yersinia enterocolitica. Microbiology 147:1115-1127. [DOI] [PubMed] [Google Scholar]
- 10.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobrindt, U., G. Blum-Oehler, T. Hartsch, G. Gottschalk, E. Z. Ron, R. Fünfstück, and J. Hacker. 2001. S-fimbria-encoding determinant sfaI is located on pathogenicity island III536 of uropathogenic Escherichia coli strain 536. Infect. Immun. 69:4248-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacker. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dozois, C. M., F. Daigle, and R. Curtiss III. 2003. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 100:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenstein, B. I., and G. W. Jones. 1988. The spectrum of infections and pathogenic mechanisms of Escherichia coli. Adv. Intern. Med. 33:231-252. [PubMed] [Google Scholar]
- 15.Furuichi, T., T. Komano, and T. Nisioka. 1984. Physical and genetic analyses of the Inc-Iα plasmid R64. J. Bacteriol. 158:997-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groisman, E. A., and H. Ochman. 1996. Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87:791-794. [DOI] [PubMed] [Google Scholar]
- 17.Hacker, J., L. Bender, M. Ott, J. Wingender, B. Lund, R. Marre, and W. Goebel. 1990. Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb. Pathog. 8:213-225. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, J. R. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4:80-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, J. R., T. T. O'Bryan, M. Kuskowski, and J. N. Maslow. 2001. Ongoing horizontal and vertical transmission of virulence genes and papA alleles among Escherichia coli blood isolates from patients with diverse-source bacteremia. Infect. Immun. 69:5363-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, J. R., T. A. Russo, P. I. Tarr, U. Carlino, S. S. Bilge, J. C. Vary, Jr., and A. L. Stell. 2000. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroNE. coli, among Escherichia coli isolates from patients with urosepsis. Infect. Immun. 68:3040-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kao, J. S., D. M. Stucker, J. W. Warren, and H. L. Mobley. 1997. Pathogenicity island sequences of pyelonephritogenic Escherichia coli CFT073 are associated with virulent uropathogenic strains. Infect. Immun. 65:2812-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar, S., K. Tamura, and M. Nei. 1994. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput. Appl. Biosci. 10:189-191. [DOI] [PubMed] [Google Scholar]
- 23.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mobley, H. L., D. M. Green, A. L. Trifillis, D. E. Johnson, G. R. Chippendale, C. V. Lockatell, B. D. Jones, and J. W. Warren. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neilands, J. B. 1992. Mechanism and regulation of synthesis of aerobactin in Escherichia coli K12 (pColV-K30). Can. J. Microbiol. 38:728-733. [DOI] [PubMed] [Google Scholar]
- 26.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 27.Oelschlaeger, T. A., U. Dobrindt, and J. Hacker. 2002. Virulence factors of uropathogens. Curr. Opin. Urol. 12:33-38. [DOI] [PubMed] [Google Scholar]
- 28.Orskov, I., and F. Orskov. 1985. Escherichia coli in extra-intestinal infections. J. Hyg. (London) 95:551-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purdy, G. E., and S. M. Payne. 2001. The SHI-3 iron transport island of Shigella boydii 0-1392 carries the genes for aerobactin synthesis and transport. J. Bacteriol. 183:4176-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabsch, W., W. Voigt, R. Reissbrodt, R. M. Tsolis, and A. J. Bäumler. 1999. Salmonella typhimurium IroN and FepA proteins mediate uptake of enterobactin but differ in their specificity for other siderophores. J. Bacteriol. 181:3610-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russo, T. A., U. B. Carlino, A. Mong, and S. T. Jodush. 1999. Identification of genes in an extraintestinal isolate of Escherichia coli with increased expression after exposure to human urine. Infect. Immun. 67:5306-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russo, T. A., C. D. McFadden, U. B. Carlino-MacDonald, J. M. Beanan, T. J. Barnard, and J. R. Johnson. 2002. IroN functions as a siderophore receptor and is a urovirulence factor in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 70:7156-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 34.Scaletsky, I. C., M. S. Gatti, J. F. da Silveira, I. M. DeLuca, E. Freymuller, and L. R. Travassos. 1995. Plasmid coding for drug resistance and invasion of epithelial cells in enteropathogenic Escherichia coli 0111:H. Microb. Pathog. 18:387-399. [DOI] [PubMed] [Google Scholar]
- 35.Schubert, S., B. Picard, S. Gouriou, J. Heesemann, and E. Denamur. 2002. Yersinia high-pathogenicity island contributes to virulence in Escherichia coli causing extraintestinal infections. Infect. Immun. 70:5335-5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schubert, S., A. Rakin, H. Karch, E. Carniel, and J. Heesemann. 1998. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect. Immun. 66:480-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siitonen, A. 1994. What makes Escherichia coli pathogenic? Ann. Med. 26:229-231. [DOI] [PubMed] [Google Scholar]
- 38.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young, I. G., and F. Gibson. 1979. Isolation of enterochelin from Escherichia coli. Methods Enzymol. 56:394-398. [DOI] [PubMed] [Google Scholar]