Abstract
The gram-negative bacterium nontypeable Haemophilus influenzae (NTHI) is the predominant pathogen in chronic otitis media with effusion and, with Streptococcus pneumoniae and Moraxella catarrhalis, is a causative agent of acute otitis media. To identify potential virulence determinants, bacterial gene expression was monitored by differential fluorescence induction during early disease progression in one specific anatomical niche of a chinchilla model of NTHI-induced otitis media. Genomic DNA fragments from NTHI strain 86-028NP were cloned upstream of the promoterless gfpmut3 gene. NTHI strain 86-028NP served as the host for the promoter trap library. Pools of 2,000 transformants were inoculated into the left and right middle ear cavities of chinchillas. Middle ear effusions were recovered by epitympanic tap at 24 and 48 h, and clones containing promoter elements that were induced in vivo and producing green fluorescent protein were isolated by two-color fluorescence-activated cell sorting. Insert DNA was sequenced and compared to the complete genome sequence of H. influenzae strain Rd. In a screen of 16,000 clones, we have isolated 44 clones that contain unique gene fragments encoding biosynthetic enzymes, metabolic and regulatory proteins, and hypothetical proteins of unknown function. An additional eight clones contain gene fragments unique to our NTHI isolate. Using quantitative reverse transcription-PCR, we have confirmed that 26 clones demonstrated increased gene expression in vivo relative to expression in vitro. These data provide insight into the response of NTHI bacteria as they sense and respond to the middle ear microenvironment during early events of otitis media.
Otitis media (OM) is a highly prevalent pediatric disease worldwide and is the primary cause for emergency room visits by children (24). While it is rarely associated with mortality any longer, the morbidity associated with OM is significant. Hearing loss is a common problem associated with this disease, often affecting a child's behavior, education, and development of language skills (7, 23, 43). The socioeconomic impact of OM is also great, with direct and indirect costs of diagnosing and managing OM exceeding $5 billion annually in the United States alone (25).
Whereas antibiotic therapy is common and the surgical placement of tympanostomy tubes has been successful in terms of draining effusions, clearing infection, and relieving pain associated with the accumulation of fluids in the middle ear, the emergence of multiple antibiotic-resistant bacteria and the invasiveness of tube placement have revealed the need for more effective and accepted approaches to the management and, preferably, the prevention of OM. Progress in vaccine development is most advanced for Streptococcus pneumoniae, the primary causative agent of acute OM (AOM), as evidenced by the recent approval and release of a heptavalent pneumococcal conjugate vaccine (19). While this vaccine has been highly efficacious for invasive pneumococcal disease and pneumococcal AOM, reports indicate an increased number of AOM cases due to serotypes not included in the vaccine (10, 20, 36, 40). Less progress has been made for nontypeable Haemophilus influenzae (NTHI), the gram-negative pathogen that predominates in chronic OM with effusion (27, 41). Contributing to this delay is our lack of understanding of the dynamic interplay between microbe-expressed virulence factors and the host's immune response as the disease progresses from a condition of host immunological tolerance of a benign nasopharyngeal commensal to the presence of an opportunistic invader in the normally sterile middle ear space.
We hypothesize that NTHI expresses a number of as yet unidentified genes important to the ability of this organism to cause OM and further that NTHI utilizes a set of genes that are known but whose role in pathogenesis is unclear or unsuspected. A number of molecular biology techniques have recently been developed that facilitate the identification and analysis of previously uncharacterized virulence-associated factors, both those that are constitutively expressed and those that are differentially expressed during disease progression. Thus, in order to identify genes that are differentially expressed in a chinchilla model of NTHI infection of the middle ear, we constructed a green fluorescent protein (GFP) promoter trap system. Differential fluorescence induction (DFI) is an enrichment strategy that takes advantage of the expression of GFP from the jellyfish Aequorea victoria to identify genes expressed under various environmental conditions. When DFI is combined with fluorescence-activated cell sorting (FACS), fluorescent bacterial clones are isolated from a library of clones that contain the gfp reporter gene downstream of random fragments of H. influenzae DNA, thus identifying a subset of Haemophilus inserts containing promoters that are regulated by environmental signals.
DFI has been successfully utilized for many microorganisms. Dhandayuthapani and coworkers employed a GFP reporter system and flow cytometry to study mycobacterial gene expression upon interaction with macrophages (16). Falkow and coworkers constructed a promoter trap system to identify genes whose transcription was increased when salmonellae were subjected to environments simulating in vivo growth and when Salmonella enterica serovar Typhimurium was internalized by cultured macrophage-like cells (45-47). Recently, DFI has been used to identify promoters expressed in S. pneumoniae and Staphylococcus aureus when these organisms were grown under varied in vitro conditions simulating infection (32, 39). In addition, DFI has been utilized to study gene regulation in Bacillus cereus in response to environmental stimuli (17), in S. pneumoniae in response to a competence stimulatory peptide (9), and in Bartonella henselae (28), Listeria monocytogenes (53), Brucella abortus (21), Escherichia coli (2), and Shigella flexneri (37) upon interaction with and invasion of host cells.
In this work, bacteria from middle ear fluids recovered by epitympanic tap were fluorescently “tagged” with a fluorochrome-labeled antibody specific for bacterial outer membrane proteins to aid in our ability to isolate these clones from effusions. Bacteria were isolated by sorting cells positive for GFP fluorescence. Using this DFI methodology, we have identified a set of clones with increased promoter activity in vivo and confirmed gene expression by quantitative reverse transcription-PCR (RT-PCR).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
NTHI strain 86-028NP is a minimally passaged clinical isolate obtained from a pediatric patient who underwent tympanostomy and tube insertion for chronic OM with effusion at Columbus Children's Hospital. Strain 86-028NP has been extensively characterized in chinchilla models of OM (5, 26, 42). The strain has been maintained frozen in skim milk containing 20% (vol/vol) glycerol. Plasmid pGZRS-39A, obtained from S. West and coworkers, is an E. coli-Actinobacillus pleuropneumoniae shuttle vector that contains the origin of replication from an A. pleuropneumoniae plasmid, the lacZα region containing the multiple cloning site from pUC19, and the kanamycin resistance gene from Tn903 (52). The promoter trap vector was constructed by cloning the gfpmut3 gene (14) as a BamHI-to-EcoRI fragment into pGZRS-39A to form pRSM2169. This plasmid was transformed by electroporation into NTHI strain 86-028NP, generating the parent-plasmid strain 86-028NP/pRSM2169. Strains 86-028NP/pRSM2211 and 86-028NP/pKMM4B5 are modifications of the promoter trap vector containing either the strong promoter for outer membrane protein P2 or a 152-bp gene fragment from strain 86-028NP cloned into a unique BamHI site upstream of the gfpmut3 gene, respectively. NTHI was grown on chocolate agar containing 20 μg of kanamycin/ml (Gibco BRL Life Technologies) or in brain heart infusion (BHI) broth (Difco Laboratories) supplemented with 2 μg of hemin chloride/ml (dissolved in 20 mM NaOH; Sigma) and 2 μg of NAD (Sigma)/ml (sBHI) and was incubated at 37°C under 5% CO2.
Construction of an NTHI gfp library.
Random genomic DNA fragments were prepared for ligation into the promoter probe vector. Genomic DNA was isolated from strain 86-028NP by using the Puregene DNA isolation kit (Gentra Systems) according to the manufacturer's protocol. DNA was partially digested with Sau3AI (0.25 U/μg of DNA; New England Biolabs) for 1 h at 37°C and then separated by gel electrophoresis, and DNA fragments of 0.5 to 1.5 kb were recovered using the Qiagen gel extraction kit. For vector preparation, pRSM2169 was isolated from an overnight culture by using the Wizard Plus Maxiprep DNA purification system (Promega) according to the manufacturer's protocol. Plasmid DNA was linearized by BamHI digestion, and 5′ phosphate groups were removed by treatment with calf intestinal alkaline phosphatase (Gibco BRL Life Technologies). Genomic DNA fragments were ligated with the linearized, phosphatase-treated vector (38) and electroporated into competent NTHI strain 86-028NP prepared for electroporation according to a modified protocol (34). Briefly, cells were grown to an optical density at 600 nm (OD600) of 0.3 in sBHI broth at 37°C and 220 rpm. Cells were chilled on ice for 30 min and subsequently washed with an equal volume of 0.5× SG (1× SG is 15% glycerol plus 272 mM sucrose) at 4°C. Washes were repeated a total of three times. Following washes, cells were suspended in 1× SG to a 100-fold concentrated volume. Cells were electroporated using the Bio-Rad Gene Pulser II set at 200 Ω, 2.5 kV, and 25 μF. Cells were diluted in 1 ml of prewarmed sBHI, incubated for 2 h at 37°C under 5% CO2, and plated on chocolate agar for overnight growth of transformants. Transformants were selected and frozen in pools of 1,000 clones in skim milk containing 20% (vol/vol) glycerol. A 68,000-member gfp promoter probe library was generated for use in this study. According to the probability calculation of Clarke and Carbon (13), in order to achieve a 99% probability of having a given DNA sequence represented in a library of 300-bp fragments of strain 86-028NP DNA (1.8 × 106 bp/genome), we would need a library of 27,629 clones. Our library represents 2.5-fold coverage of the 86-028NP genome.
Animal infection model.
Healthy adult chinchillas (Chinchilla lanigera) with no evidence of middle ear infection by either otoscopy or tympanometry were used to screen the library for promoter activity in vivo. This chinchilla model of NTHI-induced OM has been well characterized (3, 4, 42) and has been used to determine the protective efficacy of several NTHI outer membrane proteins, combinations of outer membrane proteins, chimeric synthetic peptide vaccine components, and adjuvant formulations as vaccinogens against OM (5, 6, 26). Two pools of the NTHI/pRSM2169 library (1,000 clones each) were grown overnight on chocolate agar containing kanamycin. The library was combined and subjected to one round of FACS analysis, collecting the 50% of the population that was least fluorescent. Sorted clones were plated on chocolate agar overnight and subsequently diluted in cold 10 mM phosphate-buffered saline (PBS) to 3.3 × 106 CFU/ml, and 300 μl (1.0 × 106 CFU; 500 CFU/clone) each was used to inoculate the left and right transbullar cavities (2,000 clones/ear) of two chinchillas. Twenty-four and 48 h later, middle ear fluids were retrieved by epitympanic tap and prepared for FACS.
Flow cytometry and FACS.
Chinchilla middle ear fluids were diluted, if necessary, to 250 μl with sterile saline. An equal volume of N-acetyl-l-cysteine (0.5%, wt/vol) in Dulbecco’s buffered saline (pH 7.4) was added for 5 min at room temperature as a mucolytic agent (35). Fluids were centrifuged (at 300 × g for 5 min) to remove cellular debris, red blood cells, and inflammatory cells, and supernatants containing bacteria were transferred to a fresh tube. Bacteria were incubated with a chinchilla antiserum (1:50 dilution) directed against a whole-outer-membrane-protein preparation derived from NTHI strain 86-028NP for 45 min at 4°C, pelleted by centrifugation (at 2000 × g for 5 min), and washed twice with cold DPBS containing 0.05% bovine serum albumin. Bacteria were subsequently labeled with a cross-reactive, Phycoprobe R-phycoerythrin (R-PE)-conjugated anti-human IgG (heavy plus light chains) antibody (10 μg/ml in 100 μl of PBS; Biomeda Corp) for 30 min at 4°C. After three successive washes to remove unbound antibody, cells were resuspended in 300 μl of DPBS for FACS analysis. As controls for sorting, strains 86-028NP/pRSM2169 (negative control), 86-028NP/pKMM4B5 (intermediate fluorescent control), and 86-028NP/pRSM2211 (highly fluorescent control) were resuspended from growth on chocolate agar and labeled as described above. These control preparations were used to set the appropriate size and fluorescence gates by using a Coulter Epics Elite flow cytometer equipped with an argon laser emitting at 488 nm. Bacteria were gated for size based on log forward angle and side scatter detection and for sorting by fluorescein isothiocyanate and PE labeling of bacteria. Sorted cells were collected into cold sBHI and plated on chocolate agar. After overnight growth, cells were collected for a secondary round of infection or were individually selected and grown overnight, screened by individual clone for fluorescence when grown in vitro, and frozen in skim milk containing 20% (vol/vol) glycerol prior to plasmid isolation and sequencing of insert DNA. The efficiency of sorting of control strains and screening of isolated clones were confirmed by using a Coulter EPICS flow cytometer.
Plasmid isolation and sequencing of insert DNA.
Those clones isolated by FACS analysis (positive for GFP fluorescence in vivo) and screened for the absence of fluorescence in vitro were grown overnight on chocolate agar plates containing kanamycin and prepared for plasmid isolation by using the Qiaprep Miniprep Kit (Qiagen) according to the manufacturer's protocol. Plasmid insert DNA was sequenced using primer 5′-TGCCCATTAACATCACCATCTA-3′, which is complementary to the gfpmut3 gene and downstream of the insert DNA. Sequencing reactions were performed using the ABI Prism BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems) according to manufacturer's protocol with a GeneAmp PCR System 9700 (Applied Biosystems). The reaction products were then purified by passage through Sephadex G-50 in a 96-well multiscreen HV plate (Millipore) and subsequently analyzed on an ABI Prism 3100 DNA analyzer (Applied Biosystems). Insert sequences were analyzed against the complete annotated sequence of H. influenzae strain Rd. Those inserts with no nucleotide homology to strain Rd were subsequently analyzed using the BlastN and BlastX algorithms via the National Center for Biotechnology Information (NCBI) web server. Further sequence analysis was performed with DNAstar (Madison, Wis.) software.
RNA isolation.
RNAs were isolated from strain 86-028NP that had been grown to mid-log phase in sBHI and from bacteria and cells contained in chinchilla middle ear fluids recovered 48 h postinoculation by using the TRIzol LS reagent (Gibco Life Technologies) according to the manufacturer's protocol. DNA was removed from the RNA preparation by using a DNA-free kit (Ambion) according to the manufacturer's protocol. DNase I-treated RNA samples were purified by passage through a Qiagen RNeasy column. RNA purity and integrity were assessed by 260- and 280-nm spectrophotometer readings and on the Agilent 2100 Bioanalyzer (Agilent Technologies), respectively. RNA was aliquoted and stored at −80°C.
Quantitative RT-PCR.
The relative gene expression of RNA samples obtained in vitro versus that of RNA samples obtained in vivo was assessed by quantitative RT-PCR using the one-step QuantiTect SYBR Green RT-PCR kit (Qiagen) according to the manufacturer's protocol. By using primers designed to amplify a portion of the open reading frame (ORF) downstream of the putative in vivo-induced promoters identified by FACS analysis, gene-specific mRNA was reverse transcribed and amplified by RT-PCR on the ABI Prism 7700 sequence detection system (Applied Biosystems). The amount of product was calculated using a standard curve generated by amplifying a fragment of the gyrase gene (gyr) from known amounts of bacterial genomic DNA (102 to 107 genomic copies of DNA) by amplifying a fragment of the gyrase (gyr) gene. Controls were analyzed in parallel to verify the absence of DNA in the RNA preparation as well as the absence of primer dimers in control samples lacking template RNA. In addition, RT-PCR products were analyzed by gel electrophoresis, and in all cases, a single product was observed at the appropriate base pair size. Amounts of bacterial RNA in different samples were normalized relative to expression of gyr, which was shown to be constitutively expressed under various growth conditions that we tested in vitro (data not shown).
RESULTS
Construction of an NTHI promoter trap library.
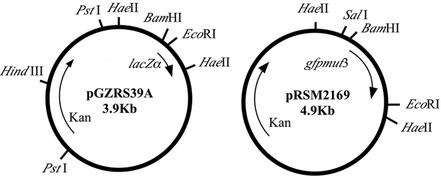
Plasmid pGZRS39A, a derivative of pGZRS-1 isolated from A. pleuropneumoniae, is an A. pleuropneumoniae-E. coli shuttle vector (52). This plasmid contains the origin of replication from A. pleuropneumoniae, the lacZα gene from pUC19, and the kanamycin resistance gene from Tn903. Falkow and coworkers constructed a promoter probe construct to screen a Salmonella library in vitro for promoters up-regulated under conditions simulating the phagosomal environment as well as for those induced in vivo in a macrophage-like cell line (45, 47). These studies used a GFP mutant (gfpmut3) containing two amino acid changes, S65G and S72A, that displayed enhanced fluorescence emission when excited at 488 nm, had high solubility, and showed fast kinetics of chromophore formation (14). We cloned the gfpmut3 gene into pGZRS39A to produce pRSM2167 (Fig. 1).
FIG. 1.
Plasmid pGZRS39A, a derivative of pGZRS-1 isolated from A. pleuropneumoniae, is an A. pleuropneumoniae-E. coli shuttle vector (52). This plasmid contains the lacZα gene from pUC19 and the kanamycin resistance gene from Tn903. The gfpmut3 gene (14) was cloned as a BamHI-to-EcoRI fragment into pGZRS39A to produce pRSM2169.
For library generation, a Sau3AI partial digest of NTHI strain 86-028NP genomic DNA was prepared and ligated into BamHI-digested and alkaline phosphatase-treated pRSM2169. Due to restriction barriers, we found it necessary to isolate plasmid DNA from strain 86-028NP and use this for library generation. When plasmid DNA was electroporated back into NTHI strain 86-028NP, the transformation efficiency was improved 1,000-fold. Thus, plasmids containing random genomic DNA inserts 5′ to the gfpmut3 gene were electroporated into strain 86-028NP, generating a library of 68,000 clones. In order to assess the quality of the library, 50 clones were selected at random and grown overnight on chocolate agar, plasmids were isolated, and insert DNA was sequenced. A majority (64%) of the selected clones had inserts between 200 and 500 bp, while those of 32% exceeded 500 bp. The majority of inserts showed homology to H. influenzae strain Rd ORFs, and 15 clones had sequences unique to strain 86-028NP DNA. Of those clones with homology to strain Rd, 60% were in the correct orientation, 36% of which contained sequence upstream of an ORF (data not shown). Although a majority of clones had inserts smaller than 500 bp, no correlation was found between small insert size and increased gfp expression. In fact, four clones exhibited slight to moderate fluorescence in vitro; of these, three had inserts between 200 and 500 bp and one had an insert larger than 700 bp.
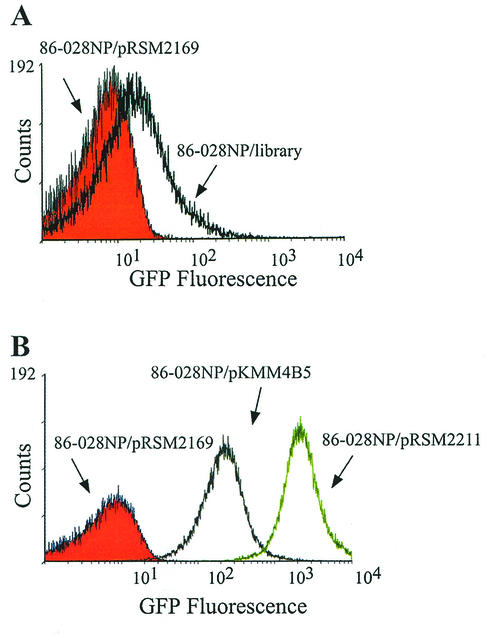
A fraction of the library (approximately 1,000 clones) was grown on chocolate agar, harvested in PBS, and analyzed by flow cytometry for GFP fluorescence (Fig. 2A). Compared to strain 86-028NP/pRSM2169, which contains the promoter trap vector without insert DNA, the pool of library clones displays increased fluorescence intensity, resulting in a shift of the histogram. Thus, the library contains clones with promoters at varying levels of activity.
FIG. 2.
FACS analysis of GFP-expressing derivatives. (A) A mixture of 1,000 clones from the promoter trap library (86-028NP/library) (black overlay) was compared to strain 86-028NP/pRSM2169 (containing the promoter trap vector without insert DNA) (red histogram) for GFP fluorescence. The library contains random gene fragments containing promoters at various levels of activity. (B) H. influenzae strains were grown for 16 h on chocolate agar supplemented with kanamycin. Cultures were diluted in cold PBS (pH 7.4), adjusted to similar OD490 values, and analyzed with a Coulter Epics flow cytometer. The following strains were analyzed for GFP expression: strain 86-028NP/pRSM2211 (high fluorescence) (green overlay), strain 86-028NP/pKMM4B5 (intermediate fluorescence) (gray overlay), and strain 86-028NP/pRSM2169 (no fluorescence) (red histogram).
FACS analysis of 86-028NP derivatives expressing GFP at low, intermediate, and high levels.
In order to establish the FACS parameters necessary to identify and sort GFP-expressing bacteria, we utilized a panel of isolates demonstrating varying levels of GFP expression. Background fluorescence was assessed using strain 86-028NP/pRSM2169; therefore, any observed fluorescence would be due to the lacZα promoter driving gfp. However, this strain does not produce detectable levels of GFP (see Fig. 2B) and in fact does not demonstrate increased fluorescence relative to the parent strain 86-028NP (data not shown). We generated a high-level GFP-expressing isolate by cloning a 500-bp fragment containing the strong promoter for outer membrane protein P2 expression into SalI-BamHI-digested pRSM2169. This plasmid was transformed into 86-028NP by electroporation, generating the high-level GFP-expressing strain 86-028NP/pRSM2211. This strain demonstrates a ∼100-fold increase in GFP fluorescence relative to strain 86-028NP/pRSM2169 (Fig. 2B). An intermediately fluorescent derivative clone, 86-028NP/pKMM4B5, was isolated by FACS analysis and used both in preliminary experiments and as a control for cell sorting. Clone 86-028NP/pKMM4B5 exhibits a ∼10-fold increase in fluorescence relative to strain 86-028NP/pRSM2169 (Fig. 2B).
Many plasmids are segregated rapidly in vitro in the absence of antibiotic selection. Thus, in order to assess whether the promoter trap vector used here is prone to this event, a single colony of strain 86-028NP/pRSM2211 was isolated on chocolate agar and passaged 20 times in the absence of antibiotic selection. No significant decrease in fluorescence intensity was observed relative to that in bacteria grown in the presence of an antibiotic (data not shown). In addition, the plasmid was maintained in the absence of antibiotic selection in vivo. Similar bacterial counts were observed when bacteria-containing middle ear fluids collected from a chinchilla were plated on chocolate agar with or without kanamycin (data not shown). These data demonstrate that the promoter trap vector was stably maintained in the absence of antibiotic selection. In addition to problems with plasmid stability, early studies on the use of GFP as a reporter to study host-pathogen interactions demonstrated that GFP could be continuously synthesized as a cytoplasmic protein with low toxicity, having minimal effects on bacterial cell surface dynamics (12). The construction of a high-level GFP-expressing derivative allowed us to assess GFP toxicity on NTHI. Growth curves of the wild-type strain (86-028NP) and the high-level GFP-producing strain 86-028NP/pRSM2211 grown under similar conditions were compared. The growth rates were similar, indicating that GFP expression was not toxic to the cells (data not shown).
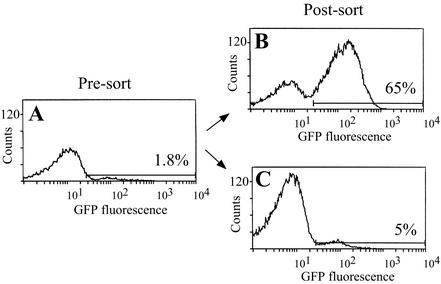
The 86-028NP GFP-expressing derivatives were used to define the parameters for efficient cell sorting. Strain 86-028NP/pRSM2169 was mixed with the intermediate-level GFP-expressing derivative, strain 86-028NP/pKMM4B5, at a 100:1 ratio, simulating the in vivo environment, which is expected to contain a small percentage of GFP-expressing clones relative to the total bacterial population. This mixture was subjected to FACS analysis, collecting the 1.8% of the population that was most fluorescent and the 52% of the population that was least fluorescent (Fig. 3). Flow cytometric analysis of the sorted populations revealed an enrichment of strain 86-028NP/pKMM4B5 to 65% of the bacterial population, a phenomenon that was not observed in sorting on the negative population. Subsequent rounds of sorting would be expected to further enrich for this intermediate fluorescent population. The inability to decrease the amount of fluorescent bacteria in the negative sort was attributed to the size of the gate set for negative sorting. We can enrich for cells with low-level or no GFP expression by gating on the 10% of the population that is least fluorescent (data not shown).
FIG. 3.
FACS separation of a mixed bacterial population enriches for intermediately fluorescent bacteria. Strain 86-028NP/pRSM2169 was mixed with strain 86-028NP/pKMM4B5 at a 100:1 ratio (presort) and subjected to one round of FACS (A). Bacterial cells were sorted by gating on the intermediately fluorescent (B) or low-fluorescence (C) population and were analyzed for fluorescence by flow cytometry (postsort). The percentage of the population within the marker gate is given for each histogram.
Direct labeling of bacteria from middle ear fluids increases sorting efficiency.
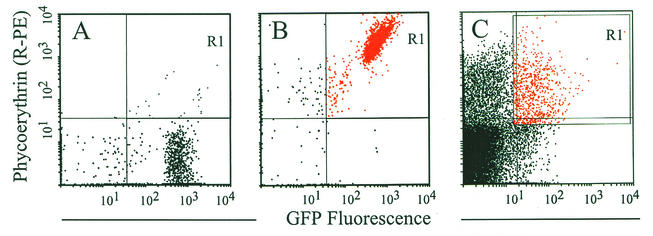
Having established the ability to sort fluorescent from nonfluorescent clones in vitro, we used a similar strategy to sort fluorescent clones from effusions obtained from the chinchilla middle ear during AOM. Our ability to use DFI in vivo was dependent on our ability to sort GFP-expressing bacteria from nonfluorescent bacteria, fluorescent and nonfluorescent cellular debris, and eukaryotic cells. Bacteria constitute a low percentage (20%) of the total events detected in an effusion sample as analyzed by FACS (see Fig. 4C). This makes it difficult to sort based on GFP fluorescence even if one also gates on bacteria by forward and side scatter. For this reason we found it advantageous to also label the bacteria with another fluorochrome, R-PE, and collect only those particles that were double labeled, specifically the bacteria that were producing GFP and were labeled with R-PE. Bacteria were thus incubated with a chinchilla-derived NTHI-specific outer membrane protein antiserum and were subsequently labeled with an R-PE-conjugated anti-human antibody that was found to cross-react with the chinchilla antiserum (Fig. 4B). Labeling the NTHI proved useful in collecting these GFP-expressing bacteria. An effusion spiked with strain 86-028NP/pKMM4B5 (0.4%) was subjected to FACS analysis. A sorting gate was set on this subpopulation (0.4%), and it was collected for subsequent analysis. A 100-fold increase in this subpopulation was observed after only one round of sorting (data not shown). We applied this bacterial labeling technique to the DFI isolation strategy described below to identify promoters induced in early events of OM.
FIG. 4.
Labeling of bacteria in effusions improves FACS analysis. (A and B) GFP-expressing bacteria (strain 86-028NP/pKMM4B5) were incubated with a 1:50 dilution of a high-titer chinchilla antiserum directed against 86-028NP outer membrane proteins. Following incubation and successive washes, the bacterial pellet was resuspended in buffer without (A) or with (B) R-PE-labeled goat anti-human IgG (heavy plus light chains) at a concentration we had previously demonstrated to facilitate optimal cross-reactivity with the chinchilla antiserum (data not shown). GFP-expressing bacteria tagged with the PE-labeled antibody were detected in quadrant R1 (compare panels A and B). (C) Labeling of bacteria from effusions. This labeling methodology was used to identify GFP-expressing bacteria present in middle ear effusions, separating their signal from that of background, debris, and eukaryotic cells. Bacteria were separated from the effusions by setting the sort gate (R1) as shown.
DFI strategy to identify promoters induced in vivo in AOM.
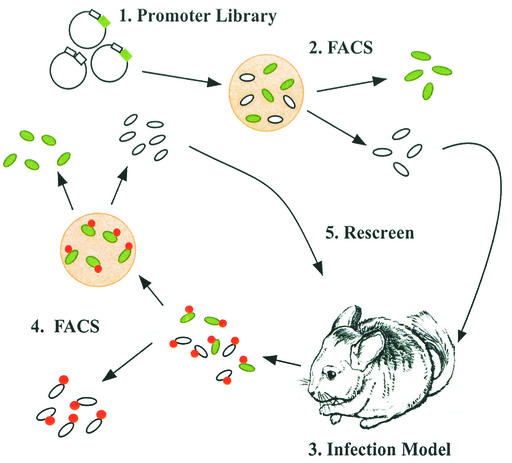
The promoter trap library was grown overnight on chocolate agar. To select against those clones containing promoters that induced GFP expression in vitro, the library was subjected to one round of FACS analysis, collecting only those clones expressing low levels of GFP (shown schematically in Fig. 5). These clones were pooled and used to inoculate the chinchilla middle ear transbullarly. Following 24 and 48 h of infection, bacterium-containing effusions were removed by epitympanic tap. Bacteria were indirectly labeled with an R-PE-labeled antibody and subjected to FACS analysis by gating on R-PE-tagged bacteria but sorting for those that were also expressing GFP. Following overnight growth on chocolate agar, bacteria were screened for those not expressing GFP, and these clones were used to infect additional animals for the library's second in vivo pass. Following the final round of sorting, single-colony isolates were screened in vitro for lack of fluorescence. Those clones not exhibiting fluorescence were prepared for plasmid isolation and identification of the insert DNA sequence. Thus, clones containing insert DNA in the correct orientation and sequence 5′ to a predicted ORF containing a putative promoter that was preferentially active when the organisms were in the chinchilla middle ear and was down-regulated when the clones were grown in vitro.
FIG. 5.
Strategy for isolating clones containing promoters driving gfp expression in vivo. (Step 1) A portion of the promoter trap library was obtained by overnight growth on chocolate agar. (Step 2) Clones containing promoters induced in vitro were removed by FACS. (Step 3) The remaining population was used for in vivo inoculation. (Step 4) Following 24 and 48 h, bacterium-containing effusions were recovered, labeled with an R-PE-conjugated antibody, and sorted for clones containing promoters induced in vivo. (Step 5) Clones with down-regulated expression in vitro were used in a subsequent round of infection to enrich for promoters induced in vivo.
Identification of promoter activity in vivo in NTHI-induced AOM.
Entire insert sequences obtained from clones isolated from middle ear fluids by FACS and screened for the absence of fluorescence in vitro were compared with the complete genome sequence of H. influenzae strain Rd. In this study, a total of 16,000 clones were screened in the chinchilla for promoter activity induced early in the course of OM. By DFI we isolated 52 clones with putative promoters that were regulated in vivo. These candidates showed homology to genes necessary for general metabolic processes, environmental informational processing, or membrane transport and to genes encoding membrane proteins or hypothetical proteins. Eight of these clones contained sequences unique to NTHI strain 86-028NP, a strain whose genome has been sequenced to threefold coverage (http://www.microbial-pathogenesis.org).
In order to confirm the induction of putative promoter candidates in vivo, we compared the relative amounts of mRNA produced for each gene when NTHI strain 86-028NP was grown in vitro to mid-log phase or in vivo for 48 h. Transcription levels were assessed by real-time RT-PCR using SYBR Green. Known amounts of bacterial genomic DNA (102 to 107 genomic copies of DNA) were used to generate a standard curve for RT-PCR quantitation by amplifying a fragment of the gyrase gene (gyr). Gyrase is constitutively expressed in vitro under various growth conditions (our unpublished data) and was therefore used to normalize total bacterial RNA levels in different samples. Total RNA was isolated from NTHI strain 86-028NP grown in vitro to mid-log phase and from effusions recovered 48 h after inoculation of the chinchilla middle ear. By using primers designed to amplify a portion of each ORF downstream of the putative in vivo-induced promoters as identified by FACS analysis, gene-specific mRNA was reverse transcribed and amplified by RT-PCR. The amount of product was calculated using the standard curve and normalized relative to gyr expression. Relative gene expression in vivo was compared to gene expression in vitro, and data were expressed as the fold increase. Of the 44 candidates with homology to ORFs in H. influenzae strain Rd, 26 were confirmed to be regulated in vivo, displaying a minimum twofold increase in relative gene expression compared to expression in vitro by RT-PCR (Table 1). Three genes, lppB, lolA, and mukF, were isolated from two independent library screens in different animals and demonstrated 2.6-, 2.4-, and 2.0-fold increases in relative gene expression, respectively. Five genes were expressed at ≥5-fold-higher levels (rbsC, purE, ribB, arcB, and the gene encoding hypothetical protein HI0094).
TABLE 1.
H. influenzae genes identified as expressed in vivo
| Category | Gene or ORF | Fold inductiona | Product of function |
|---|---|---|---|
| Amino acid metabolism | hisB | 2.9 | Histidine biosynthesis bifunctional protein |
| Lipoprotein | lppB | 2.6 | Lipoprotein B homologue |
| Membrane transport | sapA | 2.8 | Peptide ABC transporter; periplasmic SapA precursor |
| lolA | 2.4 | Outer membrane lipoprotein carrier protein precursor | |
| rbsC | 5.1 | Ribose transport system permease protein | |
| Purine synthesis | purE | 51.7 | Phosphoribosylaminoimidazole carboxylase catalytic subunit; PurE |
| Biosynthetic and metabolic functions | ribB | 8.3 | 3,4-Dihydroxy-2-butanone 4-phosphate synthase; riboflavin biosynthesis |
| argF (arcB) | 10 | Ornithine carbamoyl transferase; arginine degradation | |
| uxuA | 3.1 | Mannonate dehydratase; production of glyceraldehyde 3-phosphate | |
| dsbB | 2.6 | Disulfide oxidoreductase; disulfide bond formation protein B | |
| ureH | 3.9 | Urease accessory protein | |
| licC | 2.3 | Phosphocholine (ChoP) cytidylyltransferase | |
| HI1647 | 2.0 | Putative pyridoxine biosynthesis protein; singlet oxygen resistance protein | |
| DNA replication, repair | ispZ | 2.5 | Probable intracellular septation protein |
| radC | 2.1 | DNA repair protein | |
| mukF | 2.0 | MukF protein homologue; remodeling of nucleoid structure | |
| Gene regulation | glpR | 2.8 | Glycerol-3-phosphate regulon repressor |
| ihfB | 2.5 | Integration host factor beta subunit | |
| argR | 2.7 | Arginine repressor | |
| cspD | 2.1 | Cold shock-like protein; stress response protein | |
| Hypothetical or unknown proteins | HI0094 | 8.3 | Hypothetical protein |
| HI1163 | 2.3 | Conserved hypothetical protein; putative oxidase | |
| HI1063 | 2.7 | Hypothetical protein | |
| HI0665 | 2.8 | Hypothetical protein | |
| HI1292 | 2.6 | Hypothetical protein | |
| HI1064 | 2.6 | Hypothetical protein |
Fold increase in relative gene expression in RNA samples obtained in vivo over that in samples obtained in vitro as determined by quantitative RT-PCR.
DISCUSSION
Whereas DFI has been successfully used to identify promoters active in cell culture models of infection or under in vitro conditions designed to simulate an in vivo environment, few have applied DFI to identify promoters regulated in a specific biological niche within the whole animal. This is likely due to the numerous challenges associated with sorting from an in vivo environment. The host inflammatory response, dissemination and/or clearance of bacterial cells from the site of infection, and adherence of bacteria to epithelial cells, possibly via biofilm formation, can make bacteria inaccessible for retrieval from the living animal. These factors, among others, contribute to the complexity of the microenvironment and the heterogeneity of gene expression as the bacteria sense and respond to these changes. Recently, DFI has been used to identify promoters expressed in S. pneumoniae when the bacteria were screened in a mouse model of respiratory tract infection and a gerbil infection model of OM (32, 33). In the present study, we applied DFI technology to a relevant animal model to identify NTHI gene products involved in early events of OM as determined by up-regulation of their levels of expression.
In order to identify differentially regulated promoters, a promoter trap library was constructed and sorting parameters were defined. A portion of the promoter trap library was inoculated directly into the chinchilla middle ear, and OM development was monitored by video otoscopy and tympanometry at 24 and 48 h. This bacterial adaptation to the host environment results in an inflammatory response, indicated by erythema, vessel dilation and bulging of the tympanic membrane, infiltration of polymorphonuclear cells, and accumulation of fluid in the middle ear cavity as observed by otoscopy and microscopic examination of recovered effusions. At 24 and 48 h after infection, the middle ear fluids were recovered. It should be noted that our analysis is limited to those bacteria recoverable from the middle ear fluid. In some cases it was necessary to lavage the middle ear cavity to collect the bacteria for FACS analysis. Thus, our data likely include genes up-regulated when NTHI bacteria are loosely adherent to mucosae. Recently, NTHI has been observed to form a biofilm in the middle ear cavity in a chinchilla model of OM (18). Since our protocol selects for clones recovered from the planktonic population, we would not expect to recover those clones in which genes are up-regulated when the bacteria are associated with mucosal biofilms. Homogenization of middle ear mucosae and subsequent bacterial cell isolation, however, would enable us to recover these clones. It is also possible that some GFP-expressing clones were recovered in the effusion yet were adherent to eukaryotic cells present in the effusion as exfoliated cells or in aggregates. We therefore treated the middle ear fluids with a mucolytic agent and then by centrifugation to remove large aggregates and eukaryotic cells, and we subsequently labeled the bacteria with an R-PE-conjugated antibody. By two-color FACS analysis, we isolated bacteria that were expressing GFP from other cells and debris associated with the effusion. Following isolation, DNA sequences of the Haemophilus inserts 5′ of the gfpmut3 gene were determined and analyzed. In this manner, we identified genes that are up-regulated as NTHI bacteria sense and respond to the environment of the chinchilla middle ear during AOM.
Following our DFI procedure, described above (Fig. 5), and subsequent FACS analysis of GFP-expressing clones, we isolated 52 candidate clones containing potential in vivo-regulated promoters. The genes they control were categorized based on their general description and function within the cell and include genes involved in general metabolic processes, environmental informational processing, or membrane transport and genes encoding membrane proteins or hypothetical proteins. Eight of these 52 clones contain sequences that are unique to NTHI strain 86-028NP, a clinical OM isolate. Importantly, three clones were isolated from independent screens in more than one animal, thereby verifying our method of isolation.
In order to independently confirm the FACS data, we determined the relative expression of candidate genes by quantitative RT-PCR. The parent strain, 86-028NP, was used for these studies. Thus, we analyzed wild-type gene expression without the influence of plasmid copy number on gene regulation, which could result in GFP production and false-positive clone identification by FACS. Among the 44 candidate clones containing sequences similar to that identified in H. influenzae strain Rd, quantitative comparison of gene expression in vitro and in vivo confirmed up-regulation of expression of 26 genes (60%) when NTHI bacteria respond to environmental cues present in the chinchilla middle ear (Table 1).
The bacteria multiplied in the middle ear cavity, reaching a concentration 500 times the inoculum dose by 48 h (5). The bacteria thus must sense and respond to the environment, acquiring or synthesizing the necessary nutrients for growth and survival. This is reflected in the strong induction of expression of rbsC, the gene encoding the membrane component of the ribose transporter. This implies that free ribose, a fermentable carbohydrate in Haemophilus (31), is present in the middle ear effusion. In contrast, the strong induction of the purE and ribB genes, required for purine and riboflavin synthesis, respectively, implies that purine and riboflavin concentrations in the effusion are insufficient for the growth observed. This is consistent with observations in other experimental infection systems, where purine and riboflavin concentrations are too low to sustain an experimental infection (22, 49). Additional genes encoding proteins involved in metabolic processes that show increased expression include argF, encoding ornithine carbamoyltransferase, involved in arginine degradation via the urea cycle; uxuA, encoding mannonate hydrolase, required for d-glucuronate transformation into glyceraldehyde 3-phosphate; glpR, encoding the glycerol-3-phosphate regulon repressor; and hisB, encoding imidazole glycerol-phosphate dehydratase, which is required for histidine biosynthesis.
NTHI adaptation to the middle ear environment results in twofold induction of sapA expression. In other organisms, the products of the sap operon have homology to a peptide uptake transport system and confer resistance to antimicrobial peptides (29), suggesting that NTHI may be encountering antimicrobial peptides in the middle ear environment. Consistent with this hypothesis is the increased expression of the licC gene, encoding phosphocholine cytidyltransferase. Phosphorylcholine (ChoP) has been implicated in the pathogenesis of NTHI (51). NTHI modulates ChoP expression by phase variation, decorating the lipooligosaccharide on the cell surface. ChoP may contribute to NTHI persistence in the respiratory tract via decreased susceptibility to antimicrobial peptides (30) and may alter the sensitivity to serum killing mediated by C-reactive protein (50). Tong and coworkers suggest that the microenvironment of the nasopharynx and middle ear cavity may select for the ChoP+ phenotype, since ChoP+ strains show greater colonization of the chinchilla nasopharynx (44).
In the middle ear, NTHI increases gene expression (fourfold) of ureH, a homologue of a gene required for expression of active urease in Helicobacter. Urease has been shown to be involved in acid tolerance in Helicobacter (54) and A. pleuropneumoniae infection models (8, 11). It is notable that, in one study, 87% of NTHI isolates from middle ear effusions were urease positive (15). Similarly, we observed an increase in expression of numerous other genes involved in replication, growth, and transcriptional regulation in response to the middle ear environment. Disulfide bond formation is important for folding and assembly of many secreted proteins in bacteria. We demonstrated an increase of approximately threefold in dsbB gene transcription, illuminating an important role for disulfide interchange in NTHI growing in the middle ear environment.
Recently, many innovative molecular strategies utilizing genome sequence information have been developed, significantly contributing to our understanding of how Haemophilus adapts and responds to the surrounding environment. Akerley and coworkers utilized a high-density transposon mutagenesis strategy to identify H. influenzae genes essential for growth on rich medium (1). We identified six genes in our screen that are included in Akerley's essential gene set (hisB, lppB, lolA, ispZ, mukF, and the gene encoding the unknown protein HI0665) which were up-regulated during infection. In recent work, van Ulsen and coworkers identified genes of NTHI that are expressed upon interaction with two human respiratory tract-derived epithelial cell lines (48). In our work, we have extended this line of investigation to a relevant model of OM using a low-passage clinical isolate of NTHI. Included among the gene set identified by van Ulsen et al. are the stress response gene cspD, genes involved in purine and riboflavin biosynthesis, and a gene encoding a protein of unknown function, vapA. Similarily, the cspD gene was identified in our system. We also demonstrated a twofold increase in vapA gene expression in vivo but did not include vapA on Table 1 due to its detectable level of expression in vitro.
In summary, we have identified, by DFI, a set of genes whose expression is up-regulated when we inoculate NTHI clones directly into the chinchilla middle ear. These data advance our understanding of Haemophilus pathogenesis. In another, more relevant model, NTHI bacteria are administered to the nasopharynx, where they colonize and ascend the eustachian tube, infecting the middle ear. We will utilize the latter model to study site-specific gene expression during the entire course of disease progression as well as to identify other genes that are up-regulated in experimental OM. In addition, we will use defined mutants to allow us to elucidate the function of each gene product, particularly with regard to its role in virulence.
Acknowledgments
We thank Susan West for plasmid pGZRS-39A and Anthea Lee for gfpmut3. We also thank Cindy McAllister for technical assistance with flow cytometry and FACS, Huachun Zhong for sequencing, Linda Johnson and Laurie Tarantino for technical assistance with plasmid construction and transformations, and Brooke Johnson for the chinchilla illustration. We thank Jennifer Neelans for manuscript preparation.
The DNA sequence was determined by the Core Facility at Columbus Children's Research Institute; the Core Facility was supported in part by National Institutes of Health grant HD34615. This work was supported by grant R01-DC0391 to L.O.B. from NIDCD/NIH.
Editor: D. L. Burns
REFERENCES
- 1.Akerley, B. J., E. J. Rubin, V. L. Novick, K. Amaya, N. Judson, and J. J. Mekalanos. 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 99:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badger, J. L., C. A. Wass, and K. S. Kim. 2000. Identification of Escherichia coli K1 genes contributing to human brain microvascular endothelial cell invasion by differential fluorescence induction. Mol. Microbiol. 36:174-182. [DOI] [PubMed] [Google Scholar]
- 3.Bakaletz, L. O., R. L. Daniels, and D. J. Lim. 1993. Modeling adenovirus type 1-induced otitis media in the chinchilla: effect on ciliary activity and fluid transport function of eustachian tube mucosal epithelium. J. Infect. Dis. 168:865-872. [DOI] [PubMed] [Google Scholar]
- 4.Bakaletz, L. O., and K. A. Holmes. 1997. Evidence for transudation of specific antibody into the middle ears of parenterally immunized chinchillas after an upper respiratory tract infection with adenovirus. Clin. Diagn. Lab. Immunol. 4:223-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakaletz, L. O., B. J. Kennedy, L. A. Novotny, G. Duquesne, J. Cohen, and Y. Lobet. 1999. Protection against development of otitis media induced by nontypeable Haemophilus influenzae by both active and passive immunization in a chinchilla model of virus-bacterium superinfection. Infect. Immun. 67:2746-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakaletz, L. O., E. R. Leake, J. M. Billy, and P. T. Kaumaya. 1997. Relative immunogenicity and efficacy of two synthetic chimeric peptides of fimbrin as vaccinogens against nasopharyngeal colonization by nontypeable Haemophilus influenzae in the chinchilla. Vaccine 15:955-961. [DOI] [PubMed] [Google Scholar]
- 7.Baldwin, R. L. 1993. Effects of otitis media on child development. Am. J. Otol. 14:601-604. [PubMed] [Google Scholar]
- 8.Baltes, N., W. Tonpitak, G. F. Gerlach, I. Hennig-Pauka, A. Hoffmann-Moujahid, M. Ganter, and H. J. Rothkotter. 2001. Actinobacillus pleuropneumoniae iron transport and urease activity: effects on bacterial virulence and host immune response. Infect. Immun. 69:472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartilson, M., A. Marra, J. Christine, J. S. Asundi, W. P. Schneider, and A. E. Hromockyj. 2001. Differential fluorescence induction reveals Streptococcus pneumoniae loci regulated by competence stimulatory peptide. Mol. Microbiol. 39:126-135. [DOI] [PubMed] [Google Scholar]
- 10.Black, S., H. Shinefield, B. Fireman, E. Lewis, P. Ray, J. R. Hansen, L. Elvin, K. M. Ensor, J. Hackell, G. Siber, F. Malinoski, D. Madore, I. Chang, R. Kohberger, W. Watson, R. Austrian, and K. Edwards. 2000. Efficacy, safety and immunogenicity of heptavalent pneumococcal conjugate vaccine in children. Pediatr. Infect. Dis. J. 19:187-195. [DOI] [PubMed] [Google Scholar]
- 11.Bosse, J. T., and J. I. MacInnes. 2000. Urease activity may contribute to the ability of Actinobacillus pleuropneumoniae to establish infection. Can. J. Vet. Res. 64:145-150. [PMC free article] [PubMed] [Google Scholar]
- 12.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher. 1994. Green fluorescent protein as a marker for gene expression. Science 263:802-805. [DOI] [PubMed] [Google Scholar]
- 13.Clarke, L., and J. Carbon. 1976. A colony bank containing synthetic ColE1 hybrid plasmids representative of the entire E. coli genome. Cell 9:91-99. [DOI] [PubMed] [Google Scholar]
- 14.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 15.DeMaria, T. F., D. J. Lim, J. Barnishan, L. W. Ayers, and H. G. Birck. 1984. Biotypes of serologically nontypeable Haemophilus influenzae isolated from the middle ears and nasopharynges of patients with otitis media with effusion. J. Clin. Microbiol. 20:1102-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhandayuthapani, S., L. E. Via, C. A. Thomas, P. M. Horowitz, D. Deretic, and V. Deretic. 1995. Green fluorescent protein as a marker for gene expression and cell biology of mycobacterial interactions with macrophages. Mol. Microbiol. 17:901-912. [DOI] [PubMed] [Google Scholar]
- 17.Dunn, A. K., and J. Handelsman. 1999. A vector for promoter trapping in Bacillus cereus. Gene 226:297-305. [DOI] [PubMed] [Google Scholar]
- 18.Ehrlich, G. D., R. Veeh, X. Wang, J. W. Costerton, J. D. Hayes, F. Z. Hu, B. J. Daigle, M. D. Ehrlich, and J. C. Post. 2002. Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. JAMA 287:1710-1715. [DOI] [PubMed] [Google Scholar]
- 19.Eskola, J., and T. Kilpi. 2000. Potential of bacterial vaccines in the prevention of acute otitis media. Pediatr. Infect. Dis. J. 19:S72-S78. [DOI] [PubMed] [Google Scholar]
- 20.Eskola, J., T. Kilpi, A. Palmu, J. Jokinen, J. Haapakoski, E. Herva, A. Takala, H. Kayhty, P. Karma, R. Kohberger, G. Siber, and P. H. Makela. 2001. Efficacy of a pneumococcal conjugate vaccine against acute otitis media. N. Engl. J. Med. 344:403-409. [DOI] [PubMed] [Google Scholar]
- 21.Eskra, L., A. Canavessi, M. Carey, and G. Splitter. 2001. Brucella abortus genes identified following constitutive growth and macrophage infection. Infect. Immun. 69:7736-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuller, T. E., R. J. Shea, B. J. Thacker, and M. H. Mulks. 1999. Identification of in vivo induced genes in Actinobacillus pleuropneumoniae. Microb. Pathog. 27:311-327. [DOI] [PubMed] [Google Scholar]
- 23.Hunter, L. L., R. H. Margolis, and G. S. Giebink. 1994. Identification of hearing loss in children with otitis media. Ann. Otol. Rhinol. Laryngol. Suppl. 163:59-61. [DOI] [PubMed] [Google Scholar]
- 24.Infante-Rivard, C., and A. Fernandez. 1993. Otitis media in children: frequency, risk factors, and research avenues. Epidemiol. Rev. 15:444-465. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan, B., T. L. Wandstrat, and J. R. Cunningham. 1997. Overall cost in the treatment of otitis media. Pediatr. Infect. Dis. J. 16:S9-S11. [DOI] [PubMed] [Google Scholar]
- 26.Kennedy, B. J., L. A. Novotny, J. A. Jurcisek, Y. Lobet, and L. O. Bakaletz. 2000. Passive transfer of antiserum specific for immunogens derived from a nontypeable Haemophilus influenzae adhesin and lipoprotein D prevents otitis media after heterologous challenge. Infect. Immun. 68:2756-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein, J. O. 1997. Role of nontypeable Haemophilus influenzae in pediatric respiratory tract infections. Pediatr. Infect. Dis. J. 16:S5-S8. [DOI] [PubMed] [Google Scholar]
- 28.Lee, A. K., and S. Falkow. 1998. Constitutive and inducible green fluorescent protein expression in Bartonella henselae. Infect. Immun. 66:3964-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez-Solanilla, E., F. Garcia-Olmedo, and P. Rodriguez-Palenzuela. 1998. Inactivation of the sapA to sapF locus of Erwinia chrysanthemi reveals common features in plant and animal bacterial pathogenesis. Plant Cell 10:917-924. [PMC free article] [PubMed] [Google Scholar]
- 30.Lysenko, E. S., J. Gould, R. Bals, J. M. Wilson, and J. N. Weiser. 2000. Bacterial phosphorylcholine decreases susceptibility to the antimicrobial peptide LL-37/hCAP18 expressed in the upper respiratory tract. Infect. Immun. 68:1664-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macfadyen, L. P., I. R. Dorocicz, J. Reizer, M. H. Saier, Jr., and R. J. Redfield. 1996. Regulation of competence development and sugar utilization in Haemophilus influenzae Rd by a phosphoenolpyruvate:fructose phosphotransferase system. Mol. Microbiol. 21:941-952. [DOI] [PubMed] [Google Scholar]
- 32.Marra, A., J. Asundi, M. Bartilson, S. Lawson, F. Fang, J. Christine, C. Wiesner, D. Brigham, W. P. Schneider, and A. E. Hromockyj. 2002. Differential fluorescence induction analysis of Streptococcus pneumoniae identifies genes involved in pathogenesis. Infect. Immun. 70:1422-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marra, A., S. Lawson, J. S. Asundi, D. Brigham, and A. E. Hromockyj. 2002. In vivo characterization of the psa genes from Streptococcus pneumoniae in multiple models of infection. Microbiology 148:1483-1491. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell, M. A., K. Skowronek, L. Kauc, and S. H. Goodgal. 1991. Electroporation of Haemophilus influenzae is effective for transformation of plasmid but not chromosomal DNA. Nucleic Acids Res. 19:3625-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyamoto, N., and L. O. Bakaletz. 1996. Selective adherence of non-typeable Haemophilus influenzae (NTHi) to mucus or epithelial cells in the chinchilla eustachian tube and middle ear. Microb. Pathog. 21:343-356. [DOI] [PubMed] [Google Scholar]
- 36.Pelton, S. I., and J. O. Klein. 2002. The future of pneumococcal conjugate vaccines for prevention of pneumococcal diseases in infants and children. Pediatrics 110:805-814. [DOI] [PubMed] [Google Scholar]
- 37.Runyen-Janecky, L. J., and S. M. Payne. 2002. Identification of chromosomal Shigella flexneri genes induced by the eukaryotic intracellular environment. Infect. Immun. 70:4379-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Schneider, W. P., S. K. Ho, J. Christine, M. Yao, A. Marra, and A. E. Hromockyj. 2002. Virulence gene identification by differential fluorescence induction analysis of Staphylococcus aureus gene expression during infection-simulating culture. Infect. Immun. 70:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snow, J. B., Jr. 2002. Progress in the prevention of otitis media through immunization. Otol. Neurotol. 23:1-2. [DOI] [PubMed] [Google Scholar]
- 41.Spinola, S. M., J. Peacock, F. W. Denny, D. L. Smith, and J. G. Cannon. 1986. Epidemiology of colonization by nontypeable Haemophilus influenzae in children: a longitudinal study. J. Infect. Dis. 154:100-109. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki, K., and L. O. Bakaletz. 1994. Synergistic effect of adenovirus type 1 and nontypeable Haemophilus influenzae in a chinchilla model of experimental otitis media. Infect. Immun. 62:1710-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teele, D. W., J. O. Klein, C. Chase, P. Menyuk, and B. A. Rosner. 1990. Otitis media in infancy and intellectual ability, school achievement, speech, and language at age 7 years. J. Infect. Dis. 162:685-694. [DOI] [PubMed] [Google Scholar]
- 44.Tong, H. H., L. E. Blue, M. A. James, Y. P. Chen, and T. F. DeMaria. 2000. Evaluation of phase variation of nontypeable Haemophilus influenzae lipooligosaccharide during nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect. Immun. 68:4593-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valdivia, R. H., and S. Falkow. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22:367-378. [DOI] [PubMed] [Google Scholar]
- 46.Valdivia, R. H., and S. Falkow. 1998. Flow cytometry and bacterial pathogenesis. Curr. Opin. Microbiol. 1:359-363. [DOI] [PubMed] [Google Scholar]
- 47.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 48.van Ulsen, P., M. van Schilfgaarde, J. Dankert, H. Jansen, and L. van Alphen. 2002. Genes of non-typeable Haemophilus influenzae expressed during interaction with human epithelial cell lines. Mol. Microbiol. 45:485-500. [DOI] [PubMed] [Google Scholar]
- 49.Wang, J., A. Mushegian, S. Lory, and S. Jin. 1996. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc. Natl. Acad. Sci. USA 93:10434-10439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiser, J. N., N. Pan, K. L. McGowan, D. Musher, A. Martin, and J. Richards. 1998. Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J. Exp. Med. 187:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiser, J. N., M. Shchepetov, and S. T. Chong. 1997. Decoration of lipopolysaccharide with phosphorylcholine: a phase-variable characteristic of Haemophilus influenzae. Infect. Immun. 65:943-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.West, S. E., M. J. Romero, L. B. Regassa, N. A. Zielinski, and R. A. Welch. 1995. Construction of Actinobacillus pleuropneumoniae-Escherichia coli shuttle vectors: expression of antibiotic-resistance genes. Gene 160:81-86. [DOI] [PubMed] [Google Scholar]
- 53.Wilson, R. L., A. R. Tvinnereim, B. D. Jones, and J. T. Harty. 2001. Identification of Listeria monocytogenes in vivo-induced genes by fluorescence-activated cell sorting. Infect. Immun. 69:5016-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young, G. M., D. Amid, and V. L. Miller. 1996. A bifunctional urease enhances survival of pathogenic Yersinia enterocolitica and Morganella morganii at low pH. J. Bacteriol. 178:6487-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]