Abstract
This study investigated whether the recently recognized emergence of canine streptococcal toxic shock syndrome (STSS) and necrotizing fasciitis (NF) might be partly attributed to the use of fluoroquinolones to treat Streptococcus canis infections in dogs. Both mitomycin and the fluoroquinolone enrofloxacin caused bacteriophage-induced lysis of S. canis strain 34, an isolate from a case of canine STSS and NF. Fluoroquinolone-evoked, bacteriophage-induced lysis occurred over a range of concentrations similar to those that would occur after treatment of dogs with these agents. To search for a possible bacteriophage-encoded streptococcal superantigen gene(s), a library of the 36.5 (±1.1)-kb bacteriophage, designated φsc1, was made by ligating 3- to 7-kb Tsp5091-digested φsc1 fragments into an EcoRI-digested λZapII vector. Recombinants were screened for mitogenic activity by using canine peripheral blood lymphocytes. Of 800 recombinants screened, 11 recombinants with mitogenic effects were identified, and their inserts were sequenced. The highest homology of 11.6 kb of sequenced φsc1 DNA was to the completely sequenced Streptococcus pneumoniae bacteriophage MM1. Seven of the 11 φsc1 sequenced inserts contained a 552-bp open reading frame, scm, with 27% amino acid similarity to pokeweed (Phytolacca americana) mitogen. PCR showed this gene to be present in 22 of 23 S. canis isolates tested. Quantitative reverse transcription-PCR showed that bacteriophage induction was associated with a 58-fold enhancement of expression of this gene relative to that in a noninduced culture of a similar age. The presence of this gene on a fluoroquinolone-induced bacteriophage may explain the association observed between fluoroquinolone use in dogs and the development of canine STTS and NF.
Streptococcus canis is a commensal bacterium of dogs and some other animal species, in which it may cause a variety of opportunistic infections (10). These infections include streptococcal toxic shock syndrome (STSS) and necrotizing fasciitis (NF) (20, 22), diseases similar to STSS and NF caused by Streptococcus pyogenes in humans. In humans, STSS and NF are associated with S. pyogenes isolates possessing lysogenic bacteriophage-encoded superantigen genes, which are thought to be crucial in the development of the disease (4, 9, 12, 19, 25, 30). Powerful inducers of T-cell proliferation, superantigens cause the release of massive amounts of host cytokines with sometimes lethal effects (7, 16). Investigators attempting to identify superantigen genes in S. canis by DNA hybridization analysis using various S. pyogenes superantigen genes found that S. canis lacked genes with significant homology to the probes (5). However, DNA homology to other virulence genes from S. pyogenes, including M proteins, was observed.
Reported cases of canine STSS and NF in Ontario, Canada, have increased steadily since 1995, shortly after the introduction of a fluoroquinolone antimicrobial drug, enrofloxacin, into canine veterinary medicine (20, 22). For example, in 1996 Miller and colleagues described seven STSS and/or NF cases where four of seven dogs had been treated with the fluoroquinolone in the early stages of S. canis infection (20). The treatment was ineffective, and it was speculated that the antimicrobial drug might have been associated with progression of the infection. Shiga-toxigenic Escherichia coli encodes the Shiga toxin gene on bacteriophages (21), and Shiga toxin expression is increased by fluoroquinolones (14, 18). Fluoroquinolones have been shown to induce lysogenic bacteriophages, causing the expression of bacteriophage-encoded Shiga toxin genes as a result of induction (26, 31). Mice colonized by Shiga-toxigenic E. coli and treated with the fluoroquinolone ciprofloxacin showed increased Shiga toxin levels in their feces and died, whereas a colonized but untreated control group survived (26). Fluoroquinolone induction of a bacteriophage-encoded superantigen gene(s) might therefore provide a mechanism by which clinical use of this antimicrobial drug could result in STSS and/or NF in dogs infected with S. canis.
In this study, we report the identification of a lysogenic bacteriophage in S. canis that can be induced by a fluoroquinolone and the identification of a novel bacteriophage-encoded putative mitogen gene with significant homology to a group of mitogens identified in the pokeweed plant, Phytolacca americana.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
S. canis isolates used were obtained from veterinary hospitals across Ontario, Canada. Virulent isolates were defined as isolates acquired from dogs with STSS and/or NF; 10 of these 15 isolates were from dogs treated with enrofloxacin prior to development of severe disease. Commensal isolates (n = 12) were recovered from the mucosae or skin of healthy dogs. All S. canis strains were stored at −70°C in skim milk-sucrose solution and then grown in Todd-Hewitt broth (THB) (Oxoid, Nepean, Ontario, Canada) and on blood agar plates (Columbia agar base [Oxoid] and 5% defibrinated sheep blood). THB was supplemented with 10 mM glycine to weaken the cell wall of S. canis for more efficient lysis of the cells. Both S. canis and λZAPII (Stratagene, La Jolla, Calif.) bacteriophages were resuspended in SM buffer and stored in TM buffer (24). E. coli XL1-Blue MRF′ (Stratagene), used for expression of cloned S. canis DNA, was cultured by using Luria-Bertani (LB) broth supplemented with 0.2% maltose and 10 mM MgSO4 and LB agar plates. S. canis bacteriophage DNA was cloned by using the pGEM-3Zf(+) vector (Promega, Madison, Wis.), as well as the λZAPII vector.
Induction of bacteriophage by mitomycin C or enrofloxacin, and electron microscopic examination.
MICs of enrofloxacin (Bayer Inc., Toronto, Ontario, Canada) for STSS and/or NF isolates of S. canis were determined. Virulent S. canis strains were grown in THB at 37°C with gentle shaking, and growth was monitored by hourly measuring the optical density at 600 nm (OD600). Mitomycin C (Sigma, St Louis, Mo.) or enrofloxacin was added to the cultures to final concentrations in the range of 0.06 to 16 μg/ml during the early-, mid-, or late-logarithmic-growth phase. A decrease of at least 0.3 in the OD600 was taken as indicating bacteriophage induction. A negative-control culture, to which no mitomycin C or enrofloxacin was added, was included in each experiment.
To quantify the bacteriophage produced, two cultures of S. canis strain 34 of equal volume were grown at 37°C in THB until the OD600 reached 0.2, at which time enrofloxacin was added to one of the cultures to a final concentration of 16 μg/ml. Both cultures were reincubated until the OD600 of the culture containing enrofloxacin had decreased by approximately 0.3. Bacteriophage and bacteriophage DNA were isolated from both cultures by using a protocol from Sambrook and Russell (24). The DNA in both samples was then quantified spectrophotometrically by using a GeneQuant II RNA/DNA calculator (Pharmacia Biotech, Baie d'Urfé, Quebec, Canada).
A purified and concentrated bacteriophage suspension on a Formvar-coated grid was negatively stained with 1% phosphotungstic acid and examined by transmission electron microscopy using a Philips EM 300 electron microscope at 60 kV.
Cloning of S. canis bacteriophage DNA into λZapII and screening for mitogenicity.
S. canis bacteriophage DNA was partially digested with Tsp5091 (New England Biolabs, Mississauga, Ontario, Canada) into 3- to 7-kb fragments. The fragments were ligated into the λZapII vector and packaged into the GigaPack III Gold extract as described by the manufacturer (Stratagene) and described briefly below. An overnight culture of E. coli XL1-Blue was resuspended in 10 mM MgSO4 to an OD600 of 0.5. A 10−2 and a 10−4 dilution of the packaging reaction mixture were prepared in SM buffer, and 10 μl of these dilutions was mixed with 200 μl of the diluted E. coli and incubated for 15 min at 37°C. A 3-ml aliquot of LB broth supplemented with 0.7% agarose was added to each tube, and the mixture was plated onto LB agar plates, which were then incubated at 37°C overnight. Individual plaques were selected and washed into individual tubes containing 1 ml of SM buffer and 50 μl of chloroform. Eight hundred individual plaques were randomly selected for screening of mitogenic activity.
The method used for screening the bacteriophage λZAPII library for mitogenic activity was adapted from that used by Artuishen and colleagues (2). Briefly, recombinant λZAPII bacteriophage recovered from individual plaques were added to 200 μl of E. coli and resuspended in 10 mM MgSO4 (OD600 = 0.5) to a final concentration of about 3 × 104 PFU/ml. After incubation at 37°C for 15 min, 3 ml of LB broth supplemented with 0.7% agarose was added to each tube. One hundred microliters of this mixture was added to triplicate wells in a 96-well plate. The plates were incubated at 37°C overnight until plaques were observed. Then 100 μl of SM buffer was added to each well, and the plates were agitated at 4°C for 2 h. The SM buffer, containing proteins expressed by the recombinant bacteriophage, was removed and transferred to the wells of a new 96-well plate, to each well of which 100 μl of RPMI 1640 supplemented with 10% fetal calf serum (FCS) and 25 μg of gentamicin/ml was added. The plates were incubated for 5 h at 37°C under 5% CO2.
To determine the mitogenic activity of the λZAPII library phage supernatants, 100-μl aliquots of canine peripheral blood mononuclear cells (PBMCs) at a concentration of 106 per ml were added to each well of a 96-well plate and cultured in RPMI 1640 medium containing 10% heat-inactivated FCS and supplemented with 100 U of penicillin per ml and 20 μg of streptomycin per ml. A 100-μl aliquot of λZAPII library phage supernatant was added to each well, and plates were incubated at 37°C under 5% CO2 for 48 h. The PBMCs were then pulsed for 18 h with [3H]thymidine (ICN Biochemicals, Costa Mesa, Calif.) at 1 μCi/well. Finally, cells were harvested onto glass fiber filter mats, and counts per minute, resulting from [3H]thymidine incorporation into DNA of proliferating cells, were determined. The mean counts per minute for triplicate wells was calculated for each λZAPII recombinant. Counts from untreated cells were considered background. A stimulation index (SI) was calculated by dividing the mean counts per minute of the test clone by the mean counts per minute of a Streptococcus equi-negative λZAPII clone control; an SI score of ≥1.5 was regarded as positive. The positive control used was a λZAPII clone with an S. equi superantigen gene (sePE-H) insert, and the negative control was a λZAPII clone of S. equi containing a nonmitogenic DNA insert (both supplied by J. F. Timoney, University of Kentucky) (2).
DNA sequence determination and analysis of cloned S. canis bacteriophage fragments.
Phagemids were isolated from all positive mitogenic clones with a mean SI of ≥1.5 by following the single-clone excision protocol described by the manufacturer for λZAPII. Phagemids containing bacteriophage DNA inserts were selected, and the inserts were sequenced in both directions (Guelph Molecular Supercentre, University of Guelph). The DNA sequences of the 11 mitogenic λZapII clones were aligned with each other to generate four larger fragments of S. canis bacteriophage DNA by using the VectorNTI program (InforMax, Bethesda, Md.). Putative open reading frames (ORFs) were identified by using the ORF finder graphical analysis tool at the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov/gorf/gorf.html), in combination with the homology results obtained by doing a BLASTX search of the four fragments in GenBank (1). To find conserved domains in ORFs of interest, the Conserved Domain Database in GenBank was searched. Multiple alignment of amino acid sequences was performed by using both CLUSTALW (www.ebi.ac.uk/clustalw/) and Genebee (www.genebee.msu.su/services/malign_reduced.html).
Southern blotting and PCR of S. canis genomic DNA.
Genomic DNA from 11 virulent and 12 commensal S. canis isolates was hybridized with a probe of bacteriophage DNA isolated from S. canis 34. A 1.06-kb probe was synthesized by PCR using primers which encompassed part of the putative large terminase subunit and part of the putative minor capsid protein 1. The PCR product was labeled with digoxigenin (DIG) by random-primed labeling (Boehringer Mannheim, Laval, Quebec, Canada). The yield of DIG-labeled probe was determined by a spot test. Approximately 3 μg of chromosomal DNA of each S. canis isolate was double-digested overnight with XbaI and PstI (Pharmacia Biotech), separated on a 0.8% agarose gel run overnight at 20 V, and then stained with ethidium bromide and photographed. Southern transfers, hybridization, and chemiluminescent detection were performed under low-stringency conditions according to the manufacturer's instructions.
To establish the presence of the scm gene in the virulent and commensal isolates mentioned above, PCR amplification was performed using the forward primer scmF (5′ GGTGCTGCAATGCTTATGGT 3′) and the reverse primer scmR (5′ GCAGCAATAGCAGCAGATAC 3′), which encompass 500 bp of the gene. Touchdown PCR was performed as follows: cycles were of 90°C for 30 s, 60°C for 30 s, and 72°C for 2 min, with the annealing temperature declining by 1°C for 15 cycles until it reached 45°C, which was then maintained for 15 more cycles.
Quantitation of mitogen gene expression.
Quantitative reverse transcription-PCR (RT-PCR) was conducted by combined use of the ThermoScript RT-PCR system (Invitrogen, Carlsbad, Calif.) and LightCycler-FastStart DNA Master SVBR Green I (Roche, Indianapolis, Ind.) kits in conjunction with a LightCycler (Roche) real-time PCR instrument. RNA was extracted, by using the Qiagen RNeasy Midi-Kit, from S. canis strain 34 grown in THB containing 10 mM MgSO4 at 37°C, with gentle shaking, in the presence or absence of enrofloxacin. Enrofloxacin was added at a final concentration of 24 μg/ml to one broth culture when the OD600 was 0.26. The culture was then grown to an OD600 of 0.76 before lysis started to occur; RNA was extracted from the nonlysed bacteria when the OD600 fell to 0.64. The OD600 of a similarly treated culture fell over the next 3 h to 0.32. The noninduced control culture was also grown to an OD600 of 0.76, and RNA was then extracted. Equal amounts (500 ng) of total bacterial RNA from fluoroquinolone-induced and noninduced S. canis cultures were used in parallel in reverse transcription reactions, which were set up by following the manufacturer's instructions and using the random hexamers provided (Invitrogen). The reverse transcription products were diluted 10 times in Tris-EDTA (TE) buffer; then 1 μl was used as a template in each quantitative PCR.
Serial 10-fold dilutions of the scm gene PCR products in TE buffer (1 pg/μl to 0.01 fg/μl) were used with primers scmF and scmR in quantitative PCR to create a standard curve. The quantitative PCR mixtures were prepared by following the manufacturer's instructions (Roche) with the primer pair described above and Mg2+ at a final concentration of 2.0 mM. Quantitative PCRs were carried out in the LightCycler with 40 cycles of 95°C for 10 s for denaturing, 67°C for 10 s for annealing, and 72°C for 20 s for elongation and signal detection. The data were analyzed using the software provided with the LightCycler instrument.
Nucleotide sequence accession number.
The GenBank accession number for the scm gene with flanking bacteriophage DNA is AF522293.
RESULTS
Bacteriophage induction of virulent S. canis isolates.
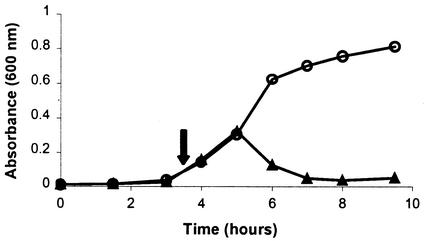
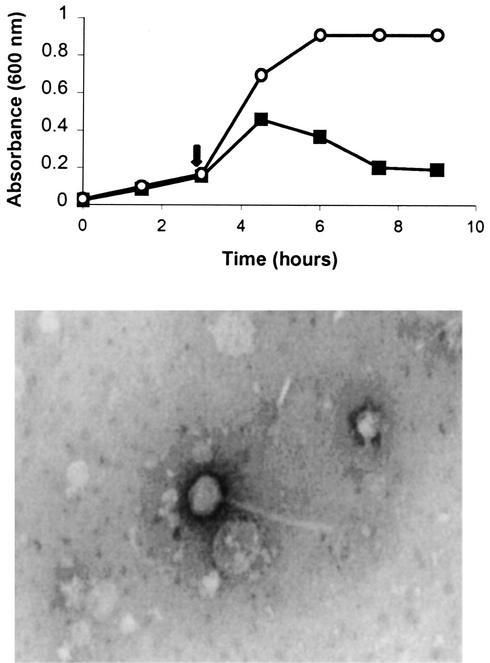
Fifteen virulent isolates of S. canis were tested for bacteriophage induction during growth in broth after the addition of either mitomycin C, an antibiotic commonly used to isolate bacteriophage, or enrofloxacin. The addition of mitomycin C at concentrations of 0.5 to 1.0 μg/ml during early-logarithmic growth demonstrated a lytic effect on the growth of 3 of 15 isolates tested, with a decrease in OD600 of up to 0.3 (Fig. 1). Addition of enrofloxacin at concentrations of 4.0 to 8.0 μg/ml to S. canis broth cultures in early-log-phase growth (OD600, 0.17 to 0.20) demonstrated a decrease in OD600 for only 1 of the 15 isolates; this isolate (strain 34) was 1 of the 3 isolates induced by mitomycin C (Fig. 2, top). Induction attempted at mid- or late-logarithmic growth failed to produce lysis.
FIG. 1.
Identification of bacteriophage induction during growth with mitomycin C. S. canis strain 39126 was grown alone or in the presence of mitomycin C (added at the early-logarithmic-growth phase) at a final concentration in broth of 0.5 μg/ml. The arrow indicates the time of addition of mitomycin C. At timed intervals, the culture medium was examined for induction by determining the optical densities of the control culture (○) and the culture with added mitomycin C (▴). The values plotted are mean concentrations of triplicate wells.
FIG. 2.
Identification of bacteriophage induction during growth with enrofloxacin. (Top) S. canis strain 34 was grown alone or in the presence of enrofloxacin (added at the early-logarithmic-growth phase) at a final concentration in broth of 4.0 μg/ml. The arrow indicates the time of addition of enrofloxacin. At timed intervals, the culture medium was examined for induction by determining the optical densities of the control culture (○) and the culture with added enrofloxacin (▪). The values plotted are mean concentrations of triplicate wells. (Bottom) Electron micrograph (with direct negative stain) of the bacteriophage found in the S. canis 34 lysed culture containing enrofloxacin.
Transmission electron microscopy confirmed the presence of a single morphological type of bacteriophage in an enrofloxacin-lysed culture of S. canis 34 (Fig. 2, bottom). Each bacteriophage had an icosahedral capsid of approximately 50.5 nm in diameter and a tail of approximately 216 nm. To further confirm the presence of bacteriophage, as well as the ability to isolate bacteriophage DNA for further cloning experiments, DNA was isolated from the bacteriophage particles and cloned into a plasmid, and the cloned insert was sequenced (details and data not shown). The DNA sequence obtained showed high (about 75%) amino acid identity to three genes of Streptococcus pneumoniae bacteriophage MM1. Therefore, a unique bacteriophage was identified in S. canis and designated φsc1. Approximately 18 times more φsc1 DNA was isolated in the presence of enrofloxacin than in its absence, confirming the bacteriophage-inducing effect of enrofloxacin. φsc1 DNA was digested singularly with a variety of restriction enzymes, and restriction fragments were examined in an agarose gel beside a 1-kb DNA ladder; the genome size was found to be 36.5 ± 1.1 kb.
Screening λZapII containing cloned φsc1DNA for mitogenicity.
Given an average insert size of 1 kb (superantigen gene and upstream promoter) in a genome of 36.5 ± 1.1 kb (F = gene size/genome size), the minimum number of clones requiring screening to ensure a 99% chance (P) of identifying the hypothetical superantigen gene was between 163 and 168, assessed by the Poisson distribution equation. Since random screening of 45 phagemids isolated from λZapII potential recombinant clones showed that only 10 phagemids contained inserts, 800 clones were screened for mitogenic activity to encompass all genes encoded by the bacteriophage genome of S. canis. SI scores were calculated by dividing the mean counts per minute of the test clone by the mean counts per minute of an S. equi negative λZAPII clone control; an SI score of ≥1.5 was regarded as positive. Of 23 clones that had SI scores of >1.5 at least once during screening, 11 clones contained bacteriophage DNA inserts.
Alignment of the 11 sequences resulting from the positive mitogenic clones selected by blastogenesis assays identified four bacteriophage fragments, designated A, B, C, and D. The total size of φsc1 DNA sequenced was 11.6 kb, approximately 32% of the bacteriophage genome. The highest homology (42 to 83% amino acid homology) of 10 of the 14 complete or partial genes identified was to putative or hypothetical proteins of the S. pneumoniae bacteriophage MM1. One putative 381-bp ORF had no homology to any known or hypothetical genes, and the other is described below.
Fragment C was 2.909 kb in length and represents the alignment of the sequences of 7 out of the 11 λZapII clones that expressed mitogenic activity in the blastogenesis assay. Of particular interest, an ORF was found to be present in all seven of the C fragment recombinants, in the middle of a gene with highest amino acid homology to an S. pyogenes bacteriophage putative minor tail protein (GenBank accession number AAK33897), partial sequence for which was obtained (bp 1 to 2475). This 552-bp ORF lies between 650 and 1,202 bp from the beginning of fragment C, is in a different reading frame from the putative minor tail protein gene into which it is inserted, and contains a start codon (ATG), a putative Shine-Dalgarno sequence (GGCAAG), putative −10 (GTATAT) and −35 (TTGATA) promoter sites, and the stop codon TAA. It has been deposited (with flanking bacteriophage DNA) in GenBank as accession number AF522293. The 16- to 444-bp section of the ORF has 27% amino acid identity to amino acids 102 to 258 of the 361-amino-acid protein mitogen lectin-B (PL-B) from P. americana (common name, Virginia pokeweed). This ORF is referred to below as scm, for a putative S. canis mitogen gene.
The G+C content of the 11.6 kb of sequenced bacteriophage DNA was 41.1 mol%, whereas that of the scm gene was 53.9 mol%.
The positive-control λZAPII clone containing the S. equi superantigen gene (sePE-H) insert resulted in an SI score of ≥1.5 81% of the time (n = 26), with an average SI score of 8.2 ± 2.2 (mean ± standard error [SE]) (range, 0.6 to 19.3); the λZAPII clones containing the scm gene resulted in an SI score of ≥1.5 23% of the time (n = 80), with an average SI score of 1.2 ± 0.2 (range, 0.6 to 3.2).
Clustal analysis of pokeweed mitogen PL-B and the φsc1 Scm homologue.
The amino acid sequence of φsc1 scm was compared with that of the 295 amino acids that compose the pokeweed mitogen protein. Forty-five of 142 amino acids between bp 16 and 444 on scm aligned with the same amino acids in the pokeweed mitogen lectin PL-B. Seventeen of these 45 amino acids are cysteine residues, and 7 are phenylalanine residues. The complete protein of PL-B is composed of seven sequential, repetitive domains with high homology. Therefore, scm was divided into domains based on alignment of scm with the PL-B domains, to determine by alignment analysis whether the hypothetical domains had homology with any of the seven PL-B domains (Fig. 3). Three hypothetical domains were identified, since the complete sequence of scm encompassed three complete domains from PL-B (domains 3, 4, and 5). scm has no homology to PL-B domains 1 and 7 but is homologous from the end of domain 2 to the beginning of domain 6. Twelve amino acid residues including 8 cysteine residues in PL-B are absolutely conserved in chitin-binding domains. Domain 1 of φsc1 contains 8 of these residues (6 are cysteines), domain 2 contains 4 of these residues (3 are cysteines), and domain 3 contains 8 of these residues (8 are cysteines). Scm domain 3 had the highest amino acid identity, 30.2%, with domain 5 of PL-B, because it contains the 8 cysteine residues of the conserved chitin-binding domain (Fig. 3). Scm domain 2 has 21.4% amino acid identity with domain 4 of PL-B, with 4 of the amino acid residues corresponding to chitin-binding domain residues. Scm domain 1 has 18% amino acid identity with domain 7 of PL-B, with 5 of the amino acid residues corresponding to conserved chitin-binding domains. Scm domains 1, 2, and 3 have approximately 25% amino acid identity with each other.
FIG. 3.
Alignment of the amino acids of three hypothetical domains in Scm, the gene of which is located in the φsc1 genome (domain 1, domain 2, domain 3), with the seven domains in pokeweed mitogen PL-B (PL-B1 to PL-B7). Twelve amino acid residues are conserved in chitin-binding domains and are boldfaced in the PL-B domains. The amino acid residues that are homologous to these conserved amino acids and are present in the three hypothetical domains of Scm are also boldfaced. Several gaps (indicated by dashes) are inserted to obtain maximal homology among these domains.
Southern blotting and PCR of S. canis genomic DNA for bacteriophage homologous DNA.
In Southern hybridization analysis of genomic DNA from virulent and commensal S. canis isolates, only genomic DNA from one commensal S. canis isolate hybridized to the probe that encompassed part of the putative large terminase subunit and part of the minor capsid protein 1 (data not shown). Both positive controls, bacteriophage and genomic DNA isolated from S. canis 34, showed strong hybridization bands. The DNA from the virulent S. canis isolates that demonstrated mitomycin C-induced bacteriophage lysis (isolates 98-1065A and 39126) did not hybridize to the probe. In the PCR amplification of genomic DNA, all 11 virulent isolates and 11 of 12 commensal isolates showed a 500-bp product (data not shown), confirming the presence of the scm gene in these isolates.
Quantitation of mitogen gene expression.
Quantitative RT-PCR showed that 0.11-fg-equivalent scm templates were detected in 1 μl of reverse transcription product originating from the noninduced S. canis culture, whereas 6.55-fg-equivalent scm templates were detected in 1 μl of reverse transcription product originating from the fluoroquinolone-induced S. canis culture. Therefore, expression of scm during fluoroquinolone induction was about 58 times higher than that in the noninduced culture.
DISCUSSION
This is the first demonstration that a fluoroquinolone can induce bacteriophage lysis in a pyogenic Streptococcus. The enrofloxacin-induced bacteriophage-lysis likely acts by inducing the SOS response, similarly to fluoroquinolone-induced bacteriophage lysis and enhanced Shiga toxin production in E. coli (26, 31). In the SOS response, the RecA protein acts indirectly as a coprotease to stimulate autodigestion of the phage repressor protein, LexA, triggering the bacteriophage to become lytic (17). Although bacteriophages in three S. canis isolates could be induced by mitomycin, a well-established bacteriophage inducer, only one of these isolates had bacteriophage that could be induced by the fluoroquinolone. The concentration at which induction occurred was similar to that which could be achieved during normal dosage with enrofloxacin, but the time at which the antimicrobial drug was added during logarithmic growth determined whether bacteriophage induction occurred. Thus, a combination of appropriate growth phase, growth rate, and fluoroquinolone concentration may determine whether or not bacteriophage induction and mitogen expression in S. canis in dogs will result from fluoroquinolone use, so that, if it is clinically important, this effect might not often be observed. Although mitomycin induced bacteriophage lysis in only three isolates, and although specific φsc1 DNA was detected in only two isolates, bacteriophages may be more widespread in S. canis than this might suggest, since the scm gene was identified in almost all isolates examined by PCR. Further work is needed to demonstrate if this gene is phage associated in all cases. Since at least two superantigen genes are located on lysogenic bacteriophages in many S. pyogenes isolates (9, 19, 30), it would be important to determine whether fluoroquinolone use might trigger bacteriophage induction and superantigen expression in this species. So far, there has been no association between fluoroquinolone use and development of STSS and/or NF in humans, but such an association may not have been looked for.
Induction of prophage by enrofloxacin from strain 34 appeared to be associated with transcription of the scm gene and production of mitogenic activity, since 7 of the 11 cloned φsc1 fragments contained the scm gene, showing that cloning of this gene was unlikely to have been by chance. Quantitative RT-PCR using equal amounts of starting RNA also showed that the scm gene was markedly expressed when enrofloxacin caused phage-induced lysis compared to a similarly grown but noninduced culture. Although Scm had no homology to identified superantigens, it has significant amino acid identity to domains of the pokeweed mitogen PL-B, so that Scm may stimulate T lymphocytes in a nonspecific manner, thus behaving like a superantigen. Many mitogenic plant lectins have been described (23, 27). Lectins bind to specific carbohydrate receptors on the surfaces of lymphocytes, in many cases the major histocompatability antigens (HLA), and trigger the mitogenic process (13). Lectin-induced membrane reactions lead to generation of second messengers as well as the synthesis of immunoregulatory lymphokines. Under appropriate conditions, over 80% of the total lymphocyte population may be induced to undergo mitosis with some lectins (13). Several pokeweed mitogens have been identified and characterized (15, 28, 29). PL-B has the highest mitogenicity of the pokeweed lectins (15). PL-B activates both T and B lymphocytes and also binds to the HLA of antigen-presenting cells (3). Although most mitogens, including superantigens, cause maximal incorporation of [3H]thymidine after 3 days of culture, PL-B reaches maximal incorporation on the 5th day (15). We did not examine [3H]thymidine after 5 days, because we were expecting to find a superantigen. Another complication in detecting the mitogen is that some lectins may be mitogenic at low concentrations but antimitogenic at high concentration (23). Based on the mitogenic activities of the Scm protein relative to that of the streptococcal superantigen control, Scm is about 7 times less potent than a streptococcal superantigen. However, the relatively low mitogenicity of Scm compared to the superantigen control may have been the result of the variables discussed. Further work is needed to address these aspects.
PL-B is a glycoprotein composed of 7 sequential chitin-binding domains, which have 48 to 79% homology. The high mitogenic activity of PL-B may be ascribed to its 7-domain structure, since other pokeweed lectins with fewer domains are less mitogenic (28). The presence of an unusually large number of intrachain disulfide bridges suggests that the pokeweed mitogen molecule, or at least some of its regions, is relatively inflexible. A significant proportion of the amino acids in Scm are cysteine residues, which are likely essential for the mitogenic activity of the protein. Besides possible methodological effects, the weak mitogenic effect on canine lymphocytes in the presence of Scm may be attributed to the presence of only three hypothetical domains in the protein. Whether expression of the scm gene would produce superantigen-like effects awaits further study, but our study suggests that there would be a superantigen-like effect, although it would be relatively weak.
The widespread nature of the scm gene in S. canis was a surprising finding, since it was anticipated that the virulent rather than the commensal isolates would possess this gene. The gene, which on the basis of its different G+C content must have been acquired from a nonstreptococcal source, appears to be important for some function in S. canis; otherwise one might expect that it would be lost. Defining its role in virulence will have to await analysis of the virulence of strains with deletions of scm. This gene appears to be unique to S. canis, since it has not been recognized in the sequenced genomes of other pathogenic streptococci, such as S. pyogenes (6).
S. canis bacteriophage φsc1 appears to be a typical lysogenic bacteriophage. For example, the bacteriophage resulted in a small degree of spontaneous lysis of the bacterium even in the absence of an inducer, although lysis increased in the presence of enrofloxacin, producing ≥17.8 times more bacteriophage DNA. The vast majority of double-stranded DNA-tailed bacteriophages have a common ancestry but have undergone extensive exchange of functional genetic elements obtained from a large shared pool (11). This was apparent for φsc1. Partial sequencing suggested that φsc1 is most closely related to bacteriophage MM1 of S. pneumoniae (8; GenBank accession number AJ302074). Sequence analysis suggested that only one type of bacteriophage was induced by enrofloxacin in S. canis 34, since cloning of the bacteriophage DNA yielded four fragments, of which 10 out of 14 of the hypothetical ORFs identified had high percentages of amino acid identity and similar gene order to the structural genes of S. pneumoniae bacteriophage MM1. It is therefore highly likely that all cloned DNA resulted from the same bacteriophage. Bacteriophage MM1 is a 40.3-kb lysogenic bacteriophage found in about 75% of natural isolates of S. pneumoniae (8). The complete genome of bacteriophage MM1 has been sequenced, and all hypothetical ORFs have been identified, with their corresponding functions, if known (GenBank accession number AJ302074). This sequence was used to assign positions for the sequence obtained from φsc1. Bacteriophage MM1 does not carry the scm gene, which, on the basis of differences in G+C content, must have been acquired by φsc1 from an unknown source. The fact that 100% amino acid identity was not observed for any gene shows that φsc1 is different from MM1. Many of the hypothetical ORFs identified in φsc1 also had high amino acid identities to the same structural proteins of bacteriophage found in other streptococcal species, including S. pyogenes and Streptococcus thermophilus. This indicates that φsc1 may have evolved from streptococcal bacteriophages. Since in Southern blotting only one other of the S. canis isolates examined showed the presence of phage DNA with homology to that of φsc1, it can be concluded that φsc1 is not widespread in S. canis. Whether the widespread scm gene is phage encoded in the other S. canis isolates in which it is present remains to be determined.
This study raises the question of the role that antimicrobial agents may play in the dissemination of virulence genes between different bacterial species and genus. This possibility has been raised for the use of fluoroquinolones in spreading Shiga toxin-encoding bacteriophage in E. coli, in which the toxin gene is fairly widespread among different serovars (26). Bacteriophage induction in S. canis by enrofloxacin might result in the spread of the bacteriophage and/or the scm gene into S. canis or other bacteria. Since bacteriophage may encode virulence genes in many bacterial pathogens, antimicrobial drugs such as the fluoroquinolones that evoke the SOS response may inadvertently enhance the spread of virulence genes and contribute to the emergence of new pathogens.
Acknowledgments
We thank Ontario Veterinary College's Pet Trust for funding.
We are grateful to Durda Slavic for invaluable assistance with some of the experiments and to Vivian Nicholson and Paul Huber for technical assistance. We thank Carlton Gyles and Janet MacInnes for critical comments, John Timoney for the S. equi controls, and David Acheson for sharing unpublished data relating to fluoroquinolone use in Shiga-toxigenic E. coli early in the course of this work.
Editor: V. J. DiRita
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artiushin, S. C., J. F. Timoney, A. S. Sheoran, and S. K. Muthupalani. 2002. Characterization and immunogenicity of pyrogenic mitogens SePE-H and SePE-I of Streptococcus equi. Microb. Pathog. 32:71-85. [DOI] [PubMed] [Google Scholar]
- 3.Basham, T. Y., and M. J. Waxdal. 1975. The stimulation of immunoglobulin production in murine spleen cells by the pokeweed mitogens. J. Immunol. 114:715-716. [PubMed] [Google Scholar]
- 4.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeWinter, L. M., D. E. Low, and J. F. Prescott. 1999. Virulence of Streptococcus canis from canine streptococcal toxic shock syndrome and necrotizing fasciitis. Vet. Microbiol. 70:95-110. [DOI] [PubMed] [Google Scholar]
- 6.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleischer, B. 1995. Bacterial superantigens. Rev. Med. Microbiol. 6:49-57. [Google Scholar]
- 8.Gindreau, E., R. Lopez, and P. Garcia. 2000. MM1, a temperate bacteriophage of the type 23F Spanish/USA multiresistant epidemic clone of Streptococcus pneumoniae: structural analysis of the site-specific integration system. J. Virol. 74:7803-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goshorn, S. C., and P. M. Schlievert. 1989. Bacteriophage association of streptococcal pyrogenic exotoxin type C. J. Bacteriol. 171:3068-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greene, C. E., and J. F. Prescott. 1998. Streptococcal and other Gram-positive bacterial infections, p. 205-214. In C. E. Greene (ed.), Infectious diseases of the dog and cat, 2nd ed. W. B. Saunders, Philadelphia, Pa.
- 11.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaul, R., A. McGeer, D. E. Low, K. Green, B. Schwartz, and A. E. Simor. 1997. Population-based surveillance for group A streptococcal necrotizing fasciitis: clinical features, prognostic indicators, and microbiology analysis of seventy-seven cases. Am. J. Med. 103:18-24. [DOI] [PubMed] [Google Scholar]
- 13.Kilpatrick, D. C. 1999. Mechanisms and assessment of lectin-mediated mitogenesis. Mol. Biotechnol. 11:55-65. [DOI] [PubMed] [Google Scholar]
- 14.Kimmitt, P. T., C. R. Harwood, and M. R. Barer. 1999. Induction of type 2 Shiga toxin synthesis in Escherichia coli 0157 by 4-quinolones. Lancet 353:1588-1589. [DOI] [PubMed] [Google Scholar]
- 15.Kino, M., K. Yamaguchi, H. Umekawa, and G. Funatsu. 1995. Purification and characterization of three mitogenic lectins from the roots of pokeweed (Phytolacca americana). Biosci. Biotechnol. Biochem. 59:683-688. [DOI] [PubMed] [Google Scholar]
- 16.Kotb, M. 1995. Bacterial pyrogenic exotoxins as superantigens. Clin. Microbiol. Rev. 8:11-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Little, J. W. 1991. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie 73:411-421. [DOI] [PubMed] [Google Scholar]
- 18.Matsushiro, A., K. Sato, H. Miyamoto, T. Yamamura, and T. Honda. 1999. Induction of prophages of enterohemorrhagic Escherichia coli O157:H7 with norfloxacin. J. Bacteriol. 181:2257-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McShan, W. M. 2000. The bacteriophages of group A streptococci, p. 105-116. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 20.Miller, C. W., J. F. Prescott, K. A. Mathews, S. D. Betschel, J. A. Yager, V. Guru, L. DeWinter, and D. E. Low. 1996. Streptococcal toxic shock syndrome in dogs. J. Am. Vet. Med. Assoc. 209:1421-1426. [PubMed] [Google Scholar]
- 21.O'Brien, A. D., L. R. Marques, C. F. Kerry, J. W. Newland, and R. K. Holmes. 1989. Shiga-like toxin converting phage of enterohemorrhagic Escherichia coli strain 933. Microb. Pathog. 6:381-390. [DOI] [PubMed] [Google Scholar]
- 22.Prescott, J. F., C. W. Miller, K. A. Mathews, J. A. Yager, and L. DeWinter. 1997. Update on canine streptococcal toxic shock syndrome and necrotizing fasciitis. Can. Vet. J. 38:241-242. [PMC free article] [PubMed] [Google Scholar]
- 23.Pusztai, A. 1991. Plant lectins. Cambridge University Press, Cambridge, United Kingdom.
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Stevens, D. L., M. H. Tanner, J. Winship, R. Swarts, K. M. Ries, P. M. Schlievert, and E. Kapland. 1989. Severe group A streptococcal infections associated with a toxic shock-like syndrome and scarlet fever toxin. N. Engl. J. Med. 321:1-7. [DOI] [PubMed] [Google Scholar]
- 26.Wagner, P. L., M. N. Neely, X. Zhang, D. W. Acheson, M. K. Waldor, and M. K. Friedman. 2001. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J. Bacteriol. 183:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waxdal, M. J. 1974. Isolation, characterization, and biological activities of five mitogens from pokeweed. Biochemistry 13:3671-3677. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi, K., N. Yurino, M. Kino, M. Ishiguro, and G. Funatso. 1997. The amino acid sequence of mitogenic lectin-B from the roots of pokeweed (Phytolacca americana). Biosci. Biotechnol. Biochem. 61:690-698. [DOI] [PubMed] [Google Scholar]
- 29.Yokoyama, K., T. Terao, and T. Osawa. 1976. Purification and biological activities of pokeweed (Phytolacca americana). Biochim. Biophys. Acta 538:384-396. [DOI] [PubMed] [Google Scholar]
- 30.Yu, C. E., and J. J. Ferretti. 1991. Molecular characterization of new group A streptococcal bacteriophages containing the gene for streptococcal erythrogenic toxin A (speA). Mol. Gen. Genet. 231:161-168. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, X., A. D. McDaniel, L. E. Wolf, G. T. Keusch, M. K. Waldor, and D. W. K. Acheson. 2000. Quinolone production induces Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664-670. [DOI] [PubMed] [Google Scholar]