Abstract
Protein p60 encoded by the iap gene is regarded as an essential gene product of Listeria monocytogenes. Here we report, however, the successful construction of a viable iap deletion mutant of L. monocytogenes EGD. The mutant, which produces no p60, shows abnormal septum formation and tends to form short filaments and hooked forms during logarithmic growth. These abnormal bacterial cells break into almost normal sized single bacteria in the late-stationary-growth phase. The iap mutant is strongly attenuated in a mouse model after intravenous injection, demonstrating the importance of p60 during infection, and the invasiveness of the Δiap mutant for 3T6 fibroblasts and Caco-2 epithelial cells is slightly reduced. Upon uptake by epithelial cells and macrophages, the iap mutant escapes from the phagosome into the cytosol with the same efficiency as the wild-type strain, and the mutant bacteria also grow intracellularly at a rate similar to that of the wild-type strain. Intracellular movement and cell-to-cell spread are drastically reduced in various cell lines, since the iap-negative bacteria fail to induce the formation of actin tails. However, the bacteria are covered with actin filaments. Most intracellular bacteria show a nonpolar and uneven distribution of ActA around the cell, in contrast to that for the wild-type strain, where ActA is concentrated at the old pole. In an iap+ revertant strain that produces wild-type levels of p60, intracellular movement, cell-to-cell spread, and polar distribution of ActA are fully restored. In vitro analysis of ActA distribution on the filaments of the Δiap strain shows that the loss of bacterial septum formation leads to ActA accumulation at the presumed division sites. In the light of data presented here and elswhere, we propose to rename iap (invasion-associated protein) cwhA (cell wall hydrolase A).
Listeria monocytogenes is a gram-positive, facultatively intracellular bacterium that can be isolated from the environment but is also a food-borne pathogen for humans and animals (41). Among the six characterized Listeria species (36), L. monocytogenes is the only human pathogen which causes severe infections with symptoms such as septicemia, meningitis, and encephalitis, mainly in immunocompromised individuals such as newborns and pregnant women (15, 40).
The virulence of L. monocytogenes depends on its capability of invading and multiplying in professional phagocytes as well as in normally nonphagocytic host cells (reviewed in references 6, 14, and 26).
A number of listerial virulence determinants involved in the intracellular life cycle of L. monocytogenes have been characterized (reviewed in references 6 and 45). A family of internalins was discovered in L. monocytogenes; at least two of these, called internalin A (InlA) and InlB, are necessary for triggering uptake by several cell types. A sulfhydryl-activated pore-forming cytolysin called listeriolysin is required, along with two phospholipases, for escape from the phagosome into the host cell cytosol. ActA, a listerial cell wall protein, promotes F-actin assembly, intracellular movement, and cell-to-cell spread (see below). Whereas the inlA and inlB genes form an operon, the other virulence factors mentioned above are located on the chromosome in the L. monocytogenes virulence gene cluster, and their expression is controlled in a complex manner by the positive regulatory factor PrfA (45).
L. monocytogenes has the ability to move within the host cell cytosol and also to spread from cell to cell without an extracellular phase, resulting in rapid spread of the bacteria within a layer of cultured mammalian cells and probably also within cells of the liver and spleen of infected hosts, which are the predominant target organs of this pathogen. Intracellular movement of L. monocytogenes inside the host cell cytoplasm, as well as intercellular spread, is mediated by actin polymerization (31, 43). Mutations in the actA gene, which codes for a proline-rich protein (ActA) of 639 amino acids, resulted in loss of virulence in mice, lack of intracellular actin polymerization around the bacteria, inability to move intracellularly, and formation of microcolonies inside the host cell (8, 21, 44). By use of immunofluorescence microscopy, data have been presented showing that ActA is not present at the new bacterial pole after cell division but seems to be concentrated at the old pole (22). Asymmetric distribution of the ActA protein was shown to be required and sufficient to direct actin-based motility in a cell-free system by studies in which either streptococci or latex beads were coated asymmetrically with genetically engineered ActA protein. (5, 42). During cell-to-cell spread, bacteria become transiently entrapped in double-membrane vacuoles, escape from which is mediated by listeriolysin together with a phospholipase and a metalloprotease (11, 30, 44). By this process L. monocytogenes facilitates propagation of the bacterial infection.
In 1989, Kuhn and Goebel identified (by sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) a 60-kDa extracellular protein of L. monocytogenes, termed p60, which is possibly involved in the invasion of mammalian cells (25; see also references 3 and 18) and which is obviously also expressed during growth of L. monocytogenes in human hosts, since antibodies directed against p60 are frequently present in sera of human listeriosis patients and healthy individuals (12, 24). Mutants of L. monocytogenes which have impaired synthesis of p60 show a rough-colony morphology (R mutants) and are strikingly attenuated in virulence in mice (34). These mutants have also lost the capability of invading 3T6 mouse fibroblasts. However, treatment of these mutants with partially purified p60 from wild-type L. monocytogenes restores their invasiveness (3, 25). One rough strain, L. monocytogenes RIII (SLCC 5779) (19), forms particularly long cell chains which disaggregate when they are exposed to purified p60 or ultrasonicated (25). Although this strain has no mutation in the gene encoding p60 (initially termed iap for invasion-associated protein), it produces greatly reduced amounts of p60, whereas iap-specific mRNA is present at the same level as in the wild-type strain (23). It was hence proposed that expression of the iap gene in L. monocytogenes is controlled on the posttranscriptional level.
Protein p60 is encoded by an open reading frame of 1,452 bp, which gives rise to a protein with a theoretical molecular size of 50.34 kDa and a theoretical pI of 9.75. The protein has a typical N-terminal signal sequence which is removed during secretion, and it is characterized by the presence of a series of threonine-asparagine repeats (23). The protein shows a clear domain structure, with two highly conserved regions at the N and C termini covering roughly 100 and 120 amino acids, respectively (3). The central part of the protein is constituted by a highly variable region including the threonine-asparagine repeats (3). An SH3 domain has been identified in the highly conserved N-terminal region of p60 (46); however, it is still unclear whether this domain is functional in any sense. The C-terminal region is homologous to a number of hydrolytic enzymes and hence is thought to confer the hydrolytic activity of p60 (48). Proteins highly related to p60 have been found in all six Listeria species (3). Overexpression of the iap gene in L. monocytogenes or in Bacillus subtilis results in bacteriolytic activity, suggesting that p60 acts as a murein hydrolase (48). This finding is in accordance with the putative function of p60 in a late step in cell division. Protein p60 also shows sequence homology to the repeat domain of an autolysin of Enterococcus faecium (10), further supporting the hypothesis that p60 acts as a hydrolase. Recently, Schubert et al. (39) identified a 45-kDa protein of L. monocytogenes with peptidoglycan lytic activity, which was termed p45 (encoded by the spl gene) and which shares high sequence similarity with p60 in its C terminus. Another two members of the p60 family were identified in the course of the L. monocytogenes sequencing project. Genes lmo0394 and lmo1104 encode putative proteins with homology to p60 (4, 13).
In order to characterize the role of p60 during infection in more detail, it would be desirable to work not with an L. monocytogenes strain that produces reduced amounts of p60, such as the rough strain L. monocytogenes SLCC 5779, but with a mutant that completely lacks p60. Earlier efforts to inactivate the iap gene in L. monocytogenes by homologous recombination either were unsuccessful (48) or resulted in a strain which produced reduced levels of p60 (1). In the present work, we report the construction of an L. monocytogenes deletion mutant that lacks the whole iap gene and hence is completely devoid of p60. This mutant has reduced virulence in mice and diminished actin-based intracellular motility. In light of recent data and the results presented here, we propose to rename the gene encoding p60, changing its designation from iap (invasion-associated protein) to cwhA, for cell wall hydrolase A, and hence to refer to p60 in the future as CwhA.
MATERIALS AND METHODS
Culture of bacterial strains.
L. monocytogenes strains (Table 1) were cultured in brain heart infusion (BHI) broth or on BHI agar plates (Difco) at 37°C. Bacteria grown for 36 h were used for infection of cell cultures and in animal experiments unless otherwise indicated. Bacteria were pelleted by centrifugation (at 4,000 ×g and 4°C), washed with phosphate-buffered saline, pH 7.4 (PBS), resuspended in 20% (vol/vol) glycerol-PBS, and stored at −70°C. Listeria strains harboring plasmid pLSVΔiap, pLSViap, or pLSViapR (Table 1) were grown in BHI supplemented with 5 μg of erythromycin (Sigma) ml−1. Escherichia coli DH10b was used as a host for DNA manipulations. E. coli strains harboring plasmid pLSVΔiap, pLSViap, or pLSViapR were grown at 37°C in Luria-Bertani (LB) broth supplemented with 600 μg of erythromycin ml−1.
TABLE 1.
Characteristics of bacterial strains and plamids
| Designation | Strain or plasmid | Relevant genotype | Description | Reference or source |
|---|---|---|---|---|
| L. monocytogenes strains | ||||
| EGD | Sv1/2a EGD | Wild type | Local strain collection | |
| WL-120 | Δiap | Δiap | iap deletion mutant | This work |
| WL-121 | Rev1 | iapΔA908 | iapΔA908 revertant | This work |
| WL-122 | iap+ | iap+ revertant | This work | |
| Plasmids | ||||
| pLSV1 | Emrori(Ts) | Mutagenesis plasmid | 47 | |
| pLSVΔiap | Emrori(Ts) ΔAB | iap knockout plasmid | This work | |
| pLSViap | Emrori(Ts) ΔA iap ΔB | iap reintegration plasmid | This work | |
| pLSViapR | Emrori(Ts) partial iap | iap repair plasmid | This work |
DNA manipulations.
Molecular cloning and recombinant DNA techniques followed standard protocols (38). For PCR, Pfu-polymerase (Promega) was used. Restriction enzymes (Amersham Pharmacia Biotech), calf alkaline phosphatase (Amersham Pharmacia Biotech), and T4 DNA ligase (Life Technologies) were utilized according to the manufacturer's instructions. Chromosomal DNA from L. monocytogenes was isolated by resuspending bacteria in Tris-EDTA (pH 8.2) containing 2 mg of lysozyme (Sigma) ml−1. After incubation at 37°C for 15 min, bacteria were harvested by centrifugation and further treated with DNAzol (Life Technologies) according to the manufacturer's instructions.
Construction of L. monocytogenes Δiap and the revertant iap+ strain.
The iap deletion mutant (called L. monocytogenes Δiap) was constructed by using L. monocytogenes Sv1/2a EGD as the parental strain. The iap+ revertant (L. monocytogenes iap+) was constructed by reintegration of the iap locus into the chromosome of L. monocytogenes Δiap. Deletion mutants were obtained by homologous recombination using constructs derived from the pLSV1 mutagenesis vector (47). Plasmid pLSV1 carries an erythromycin resistance gene (Emr) and a gram-negative and a gram-positive temperature-sensitive origin of replication [ori(Ts)] and was used to construct the iap knockout plasmid as follows. A 320-bp upstream fragment of the iap locus (ΔA) was amplified by using primers A1 (5′-TCATCAGGATCCTGTCTCATT-3′) and A2 (5′-TAACTCCCCGGGCTAAAGAGG-3′), and a second, 434-bp fragment (ΔB), which is localized downstream of the iap locus, was amplified with primers B1 (5′-ATGAAACCCGGGGTATGATGA-3′) and B2 (5′-AGCGTAGAATTCTAGACTGTA-3′) from chromosomal DNA derived from L. monocytogenes EGD. Both fragments were cut with PspAI and ligated, and a large fragment was then obtained by PCR amplification with primers A1 and B2 and with the ligation mixture as a template. This fragment (ΔAB) was cloned via the introduced BamHI and EcoRI sites into pLSV1, giving rise to pLSVΔiap. pLSVΔiap was transformed by electroporation into L. monocytogenes EGD, and erythromycin-resistant bacteria growing at 43°C (and hence harboring the chromosomally integrated knockout plasmid) were selected. These so-called integration mutants were grown in liquid culture at 30°C and screened for the loss of erythromycin resistance. Erythromycin-sensitive clones were screened by PCR to identify a mutant in which the iap gene was deleted.
A similar protocol was used to construct the iap+ revertant strain. With primers A1 and B2, a large fragment (2,373 bp) comprising fragment ΔA, the iap locus, and fragment ΔB was amplified from L. monocytogenes chromosomal DNA. This fragment was cloned into pLSV1 with the restriction enzymes mentioned above, resulting in pLSViap. This plasmid was transformed into L. monocytogenes Δiap, erythromycin-resistant bacteria growing at 43°C were selected, and after they were grown at 30°C, erythromycin-sensitive revertants were screened by PCR with appropriate primer pairs. Since it was not possible to obtain a functional, p60-expressing iap+ revertant by this method due to a frameshift mutation in the open reading frame of the reintegrated iap gene in strain L. monocytogenes Rev1, the iap locus was corrected as follows. A 571-bp fragment (partial iap) including the site where the frame shift had occurred was amplified from chromosomal DNA with primers R1 (5′-ATTTGCGGATCCAACAATCGCATCCGC-3′) and R2 (5′-TACGGAGAATTCCCAAATAGTGTCACC-3′) and cloned via BamHI and EcoRI into pLSV, giving rise to the so-called repair plamid pLSViapR. This plasmid was introduced into L. monocytogenes Rev1, and plasmid integration and excision were performed as described above. Since it was not possible to screen for clones with the corrected frame shift by PCR, a colony immunoblot assay with a monoclonal antibody against p60 (see below) was performed to identify a p60-expressing iap+ revertant strain, now called L. monocytogenes iap+. The sequences of the iap loci in L. monocytogenes Rev1 and L. monocytogenes iap+ were confirmed by nucleotide sequence analysis.
Isolation of supernatant and cellular proteins of L. monocytogenes.
Overnight cultures were diluted 1:100 and grown in BHI at 37°C to the late-exponential-growth phase (180 Klett units). Each culture was then centrifuged (at 4,000 ×g and 4°C). The supernatant was precipitated on ice with 10% trichloroacetic acid, pelleted by centrifugation (at 5,000 ×g and 4°C), washed in acetone, and resuspended in 2× Laemmli buffer to 1% of the original culture volume. The pellet of the culture was washed in PBS and resuspended in 2× Laemmli buffer to 10% of the original culture volume. The proteins were heated to 110°C for 10 min prior to being loaded onto SDS gels.
Antibodies, SDS-PAGE, and immunoblot analysis.
The antibodies used in this work were mouse monoclonal antibody K3A7, directed against protein p60 (37) (provided by A. Bubert, Darmstadt, Germany); mouse monoclonal antibody L2L2, directed against p45 of L. monocytogenes ATCC 19117 (39) (kindly provided by F. Fiedler, Munich, Germany); a rabbit polyclonal antiserum raised against ActA (33); a rabbit polyclonal antiserum directed against listeriolysin O (LLO) (provided by J. Kreft); a mouse monoclonal antibody (L7.7) against InlA (20) (kindly provided by P. Cossart, Paris, France); and a mouse polyclonal antiserum specific for listerial PlcB. This antiserum was generated by injecting mice three times with purified PlcB eluted from SDS gels which had been loaded with proteins from supernatants from overnight cultures of L. monocytogenes with pERL3, which expresses additional copies of PrfA (9) and hence results in increased PlcB expression. Affinity-purified rabbit polyclonal antibodies directed against InlA and InlB (32) were kindly provided by J. Wehland, Braunschweig, Germany. SDS-PAGE was performed according to the method of Laemmli (27a). In brief, 20 μl of samples was separated on SDS-10 or 15% polyacrylamide gels and transferred to a nitrocellulose membrane (Hybond C; Amersham Pharmacia Biotech) by using a semidry electrotransfer unit. Blots were blocked in 5% low-fat milk, washed, and incubated with dilutions of the different antibodies. Peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) (Fcγ) and peroxidase-conjugated goat anti-rabbit IgG (heavy plus light chains) (both from Dianova) were used as secondary antibodies. Bands were visualized by enhanced chemiluminescence according to the manufacturer's protocol (Amersham Pharmacia Biotech).
Mouse virulence assay.
Six- to 8-week-old female BALB/c mice(Harlan Winkelmann, Borchen, Germany) were used in groups of four to five mice for each bacterial strain. Experiments were performed twice. Mice were infected intravenously with 100 μl of a bacterial suspension. Mice were killed 3 days postinfection by cervical dislocation, and the livers and spleens were aseptically removed. Organs were placed in 5 ml of sterile distilled water and homogenized using glass homogenizers. Serial dilutions of the homogenates were then plated onto BHI agar and incubated at 37°C. CFU counts per organ were calculated for each mouse. The Student t test was used for statistical analysis. P values of <0.05 were considered statistically significant.
Cell culture and infection experiments.
3T6 mouse fibroblasts, Caco-2 human epithelial cells, P388.D1 mouse macrophages, and HepG2 human liver cells were obtained from the European Collection of Cell Cultures, Salisbury, United Kingdom. 3T6 fibroblasts, Caco-2 cells, and P388D.1 macrophages were grown in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum (FCS) at 37°C under a 5% CO2 atmosphere. HepG2 cells were grown in Eagle's minimal essential medium supplemented with 10% FCS, nonessential amino acids, 2 mM l-glutamine, 1 mM sodium pyruvate, and 0.15% sodium bicarbonate (all from Gibco). For invasion and intracellular growth assays, 2.5 × 105 cells were seeded in 12-well tissue culture plates 1 to 2 days prior to infection. Cell layers were washed with PBS containing 1 mM CaCl2 and 0.5 mM MgCl2 [PBS(Ca2+ Mg2+)] and infected in triplicate with 1 ml of bacteria suspended in medium without FCS. After 1 h of infection (1.5 h for HepG2 cells), cells were washed three times with PBS(Ca2+ Mg2+) and incubated for different periods of time with medium containing 100 μg of gentamicin (Sigma) ml−1. After they were washed twice with PBS(Ca2+ Mg2+), cells were harvested by lysis in ice-cold water and ultrasonication. To calculate infection rates, serial dilutions of the homogenized cell suspensions were plated on BHI agar. For statistical analysis of invasion assays, Student′s t test was used and P values of <0.05 were considered statistically significant.
Immunofluorescence assays.
For intracellular immunofluorescence staining according to previously described procedures (27), cells were grown on glass cover slides and infected as described above. Six hours postinfection, the cell layer was washed twice with PBS and once with 10 mM EGTA-1 mM MgCl2-60 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES)-23 mM HEPES (pH 6.9) (PHEM) and then permeabilized for 1 min in 0.25% Triton X-100 in PHEM. Fixation was carried out by a 3-min incubation in acetone at −20°C. The dried cell layer was then stained with an antiserum against ActA for 1 h at room temperature (RT), followed by a wash step with PBS, and afterwards stained with a lissamine rhodamine (LRSC)-labeled goat anti-rabbit IgG (Dianova) for another hour. For visualization of filamentous actin, cells were additionally incubated for 30 min at RT in fluorescein isothiocyanate (FITC)-labeled phalloidin (Sigma). Extracellular bacteria were stained as follows. Bacteria were grown to the exponential-growth phase, washed with PBS, and absorbed on polylysine-coated glass cover slides. After fixation with 4% paraformaldehyde for 30 min and quenching with 10 mM glycine for 15 min, bacteria were incubated for 1 h at RT with an anti-ActA antiserum or the monoclonal anti-internalin A antibody (both diluted 1:50 in PBS), washed extensively with PBS, and then incubated with a secondary FITC-labeled antiserum (Dianova) for another hour. The cover slides were then incubated in 1 mM 4′,6′-diamidino-2-phenylindole (DAPI) for 10 min at RT. Finally, the bacteria were embedded in 50% Vectashield mounting medium (Vector Laboratories)-1% low-melting-point agarose. Images of preparations were produced with a fluorescence-equipped microscope (Leica DMR HC) and an electronic camera (Diagnostic Instruments Inc.). Digital images were processed using META-MORPH software (Universal Imaging Corporation).
Heterologous plaque assay.
The heterologous plaque assay is a modification of the classical plaque assay and has been described in detail previously (16). P388.D1 macrophages were infected at a multiplicity of infection (MOI) of 20 for 1 h as described above. After macrophages were washed three times with PBS, a gentamicin-containing medium was added for 1.5 h to kill extracellular bacteria. The cells were then detached by addition of cold PBS(Ca2+ Mg2+) and were washed once with PBS(Ca2+ Mg2+) and once with a medium containing 10 μg of gentamicin ml−1. Cells were resuspended in a medium with gentamicin and counted. Various dilutions of the cell suspension were placed on the HepG2 cell layer and incubated for 2.5 h at 37°C, allowing the macrophages to settle down on the HepG2 cell layer. The medium was then removed, and the cells were overlaid with 0.5% agarose in HepG2 medium containing 10 μg of gentamicin ml−1. Twenty-four hours later, a second overlay was added which consisted of 0.01% neutral red, 0.5% agarose, and 10 μg of gentamicin ml−1 in HepG2 medium. Plaques were imaged after 3 days.
Scanning electron microscopy.
For ultrastructural studies, bacteria were grown to different growth phases, washed with PBS, and applied to polylysine-coated glass cover slides. The samples were fixed with 4% paraformaldehyde overnight at 4°C, dehydrated through a series of acetone dilutions (30 to 100%), and finally dehydrated by using a critical-point dryer (CPD 030; BAL-TEC, Witten, Germany). Samples were gold sputtered (SCD 005; BAL-TEC) to a thickness of 30 nm and were examined using a digital scanning microscope (DSM 962; Zeiss, Oberkochen, Germany).
Transmission electron microscopy.
Bacteria were grown to the exponential-growth phase, washed with PBS, incubated for 10 min on ice in 10 mM polylysine in PBS, and pelleted by centrifugation. The pellets were fixed for 3 h on ice with 2.5% glutaraldehyde and 2% formaldehyde in 100 mM cacodylate buffer (pH 7.2), washed with 50 mM cacodylate buffer, and further treated with 2% OsO4 for 2 h. After five wash steps in water, the pellets were incubated overnight with 0.5% uranyl acetate and washed again with water. The probes were dehydrated through a series of ethanol dilutions, embedded in Epon, cut with a diamond knife by using an ultramicrotome, and examined by using a Zeiss transmission microscope (EM 10). Caco-2 cells were grown on glass cover slides and infected as described above. Six hours postinfection, cells were fixed for 30 min with 0.05 M cacodylate buffer (pH 7.2) including 2.5% glutaraldehyde, washed with cacodylate buffer, and further treated with 2% OsO4 and 0.5% uranyl acetate as described above. Embedding and dehydrating steps were performed as described above.
RESULTS
Construction of the iap deletion mutant L. monocytogenes Δiap and its reversion by cis-complementation.
Based on the known sequence of the L. monocytogenes iap gene (GenBank accession no. X52268), a 1,655-bp deletion (positions 319 to 1973) comprising not only the open reading frame but also the promoter region (124 bp upstream of the transcription start site) and the terminator region (76 bp downstream of the transcription termination site) of the iap gene was introduced into the chromosome (all base pair numbering is according to the numbering in X52268). The knockout plasmid pLSVΔiap, which carries flanking sequences of the iap locus, was transformed into L. monocytogenes strain Sv1/2a EGD to produce an isogenic deletion mutant (L. monocytogenes Δiap) lacking the iap locus by a two-step integration-excision technique as described in detail in Materials and Methods. Immunoblot analysis confirmed that, as expected, no p60 was produced by this iap deletion mutant (Fig. 1). To prove that the resulting phenotype of the iap deletion mutant is exclusively caused by deletion of the iap locus, we generated an iap+ revertant strain by reintegrating the whole iap locus back into the chromosome of strain L. monocytogenes Δiap by use of plasmid pLSViap and the experimental procedure used for the generation of the Δiap strain. However, the first revertant strain which we obtained (L. monocytogenes Rev1) harbored a frameshift mutation in the open reading frame of the iap gene (a deletion of adenine at position 908), resulting in a total lack of p60 expression. This frameshift mutation was corrected by site-specific mutagenesis using plasmid pLSViapR, which carries a part of the iap gene (positions 508 to 1078) including the region of the frameshift mutation. The resulting corrected iap+ revertant strain, called L. monocytogenes iap+, secretes exactly the same amount of p60 as the wild-type strain (Fig. 1). Southern blot analyses (Fig. 1), PCR, and sequencing were used to confirm the structure of the iap deletion mutant and the iap+ revertant strain.
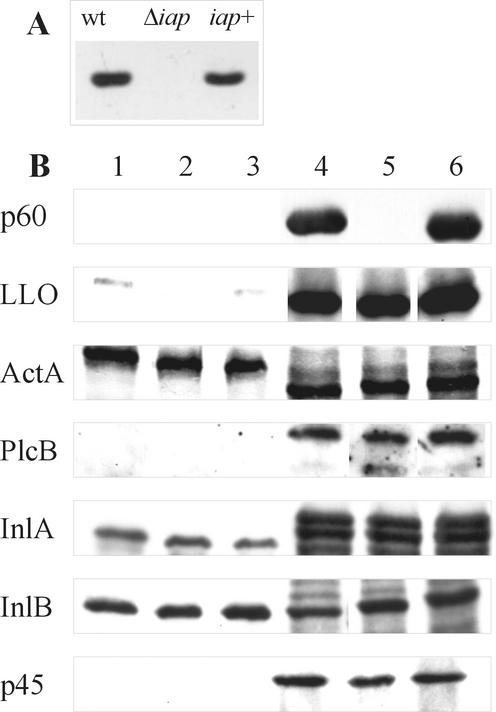
FIG. 1.
Characterization of wild-type (wt) L. monocytogenes, the Δiap mutant, and the iap+ revertant by Southern and Western blot analysis. (A) Southern blot analysis with HpaI-digested chromosomal DNA from wt L. monocytogenes, the Δiap mutant, and the iap+ revertant. An internal fragment of the iap gene was used as a probe. (B) Western blot analysis determines expression of p60 and other virulence proteins in wt L. monocytogenes (lanes 1 and 4), the Δiap mutant (lanes 2 and 5), and the iap+ revertant (lanes 3 and 6). Lanes 1 to 3, total cellular proteins; lanes 4 to 7, proteins of culture supernatants.
Expression of several virulence factors and lytic enzymes is unchanged in L. monocytogenes Δiap.
In order to test the effect of the deletion of the iap gene, we analyzed the expression of several cell surface-associated or secreted proteins of L. monocytogenes. For this purpose the wild-type strain EGD, the iap deletion mutant, and the iap+ revertant were grown to the late-exponential-growth phase, and total cellular proteins and proteins from the culture supernatant were isolated, separated by SDS-PAGE, and analyzed by immunoblotting using a set of polyclonal antisera against some of the known virulence factors of L. monocytogenes. As shown in Fig. 1, the amounts of the proteins LLO, ActA, PlcB, InlA, and InlB in the late-exponential-growth phase are not altered either by deletion of the iap gene or by cis-complementation of the Δiap mutant (iap+ revertant), demonstrating that lack of expression of p60 does not influence the expression of other L. monocytogenes virulence factors.
Since L. monocytogenes Δiap is viable, we suspected that the function of p60 as a murein hydrolase in L. monocytogenes can be taken over by other lytic enzymes. Using a monoclonal antibody (L2L2, provided by F. Fiedler) against the recently identified secreted lytic protein p45, we investigated whether expression of this protein was increased in the Δiap mutant. However, as shown in Fig. 1, immunoblot analysis revealed that the amount of p45 is the same in the wild-type strain, the Δiap mutant, and the iap+ revertant. By performing a zymogram assay using heat-killed L. monocytogenes as a substrate as described by Wuenscher et al. (48), we also attempted to detect other secreted lytic proteins which might be overexpressed in the Δiap strain compared to the wild-type strain. We were only able to visualize the activities of p60 and one other, unknown lytic protein with a high molecular mass (greater than 150 kDa). However, this high-molecular-weight protein was present in L. monocytogenes Δiap (completely lacking p60-specific bacteriolytic activity) in the same amount as in the wild-type strain (data not shown).
The iap deletion mutant leads to abnormal cell division.
When grown in liquid culture in BHI broth, L. monocytogenes Δiap grows at a rate almost indistinguishable from that of the wild-type strain when cell density is measured by light absorption at 590 nm (data not shown). However, light microscopy analysis reveals considerable differences in bacterial shape (Fig. 2A). In the exponential-growth phase the iap mutant tends to form longer filaments which are mostly hooked. In the stationary-growth phase and in a culture grown overnight, these filaments become shorter, and a culture of the Δiap mutant grown for 36 h shows predominantly short bacterial cells, which are on average only slightly longer than the wild-type cells. Scanning electron microscopy of L. monocytogenes Δiap (Fig. 2B) indicates that septum formation is initiated at several positions along the filaments, but there are only a few events which appear to be normal cell divisions. In most cases, the bacterial filaments seem to break at the positions of initiated septa. Transmission electron microscopy of L. monocytogenes Δiap also demonstrates the existence of septa between single bacterial cells that seem not to separate in a normal way but to break apart, since they lack normal constriction in the division area (Fig. 2C). The poles of such broken cells are often flatter than the poles of wild-type cells, and even most of the short cells from overnight cultures of the Δiap mutant exhibit irregular pole formation and often carry additional, incompletely built septa (Fig. 2B). The shape of the iap+ revertant is indistinguishable from that of the wild-type strain (Fig. 2A), suggesting that the abnormal cell division observed with the Δiap mutant is exclusively caused by deletion of the iap locus. In contrast to the extremely long filaments formed by the rough strain L. monocytogenes RIII (Fig. 2A) (26), the filaments of the Δiap mutant do not disaggregate upon ultrasonication (data not shown).
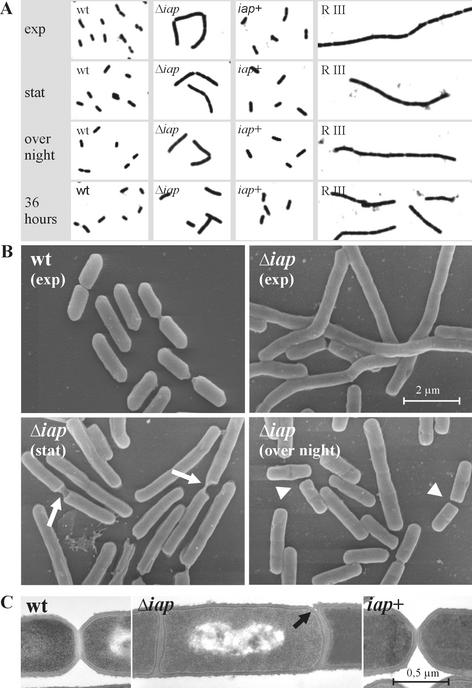
FIG. 2.
(A) Appearance of wild-type (wt) bacteria, the Δiap mutant, the iap+ revertant, and the rough (RIII) strain in phase-contrast microscopy. (B and C) Scanning electron microscopy (B) and transmission electron microscopy (C) images of the wt, Δiap, and iap+ strains. Pictures of the bacteria were taken in the exponential-growth phase (exp), in the stationary phase (stat), after overnight culture, and after 36 h of growth. Arrows indicate remarkable sites of abnormal cell division, where bacterial filaments seem to break; arrowheads indicate unusual flat poles present in the Δiap mutant. In panel C, the differences in the mode of septum formation among the three strains are clearly visible.
The virulence of L. monocytogenes Δiap is strongly attenuated in the mouse model.
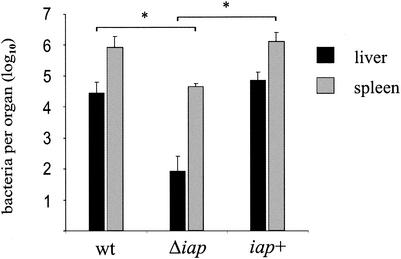
To test the virulence of the Δiap mutant, we infected groups of four to five BALB/c mice intravenously with a sublethal dose (5 × 103 CFU) of Δiap mutant bacteria or, as a control, with the same number of wild-type or iap+ revertant bacteria. The infection was monitored after 3 days by assessing bacterial loads in the livers and spleens. All bacteria used for infection were cultured to mid-log phase. While most mice infected with the wild-type or iap+ revertant bacteria showed signs of disease after 3 days, mice infected with L. monocytogenes Δiap had a normal, healthy appearance. As shown in Fig. 3, mice infected with the mutant strain carried bacterial loads more than 1 log unit lower in the spleen (P < 0.05) and more than 2 log units lower in the liver (P < 0.05) than those of the wild type at day 3 of infection. The iap+ revertant showed viable bacterial counts in the spleen and liver comparable to those of the wild-type strain. These results indicate that deletion of the iap gene leads to a strong attenuation of the virulence of L. monocytogenes upon intravenous infection.
FIG. 3.
Mouse virulence assay. BALB/c mice (n = 4 to 5) were infected intravenously with 5 × 103 CFU of wild-type (wt) L. monocytogenes, the Δiap mutant, or the iap+ revertant. Values are mean bacterial counts per liver or spleen at day 3 postinfection. Error bars, standard deviations. Asterisks indicate statistically significant differences (P < 0.05).
L. monocytogenes Δiap has a slightly reduced invasiveness.
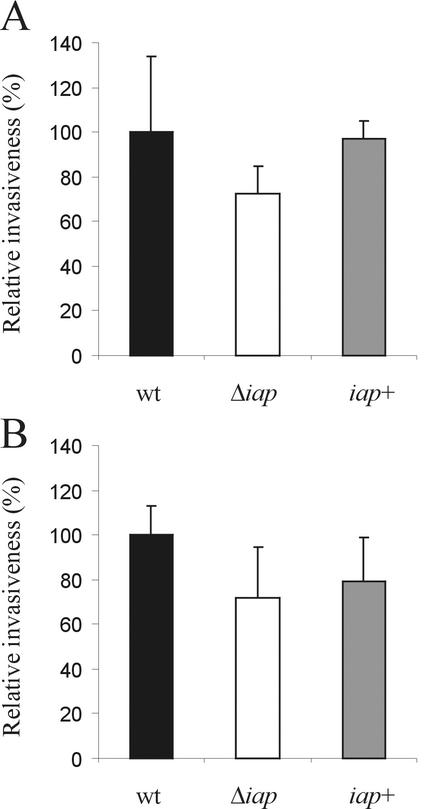
R mutants with drastically reduced expression of p60 (L. monocytogenes SLCC 5779) were found to have a strongly reduced ability to invade nonprofessional phagocytic fibroblasts of the 3T6 cell line (25). In order to confirm the role of p60 as an invasion-associated protein, we tested the invasion of wild-type L. monocytogenes, L. monocytogenes Δiap, and the iap+ revertant in 3T6 fibroblasts and Caco-2 epithelial cells. As shown in Fig. 4, the uptake of the Δiap mutant by 3T6 fibroblasts and Caco-2 epithelial cells was reduced to about 70% of uptake of the L. monocytogenes wild-type strain. This slight reduction in invasiveness was less pronounced than that described earlier for L. monocytogenes RIII (25). The uptake of the iap+ revertant was only insignificantly lower than that of the wild-type bacteria.
FIG. 4.
Invasiveness of wild-type (wt) L. monocytogenes, the Δiap mutant, and the iap+ revertant. Caco-2 epithelial cells (A) or 3T6 mouse fibroblasts (B) were infected for 1 h at an MOI of 5, and intracellular bacteria (grown to the late-exponential-growth phase) were quantified by determining the CFU per well. Each column represents the mean of one representative experiment performed in triplicate. Error bars, standard deviations.
The Δiap mutant is unable to induce actin tails within Caco-2 cells and exhibits impaired homologous and heterologous cell-to-cell spread.
L. monocytogenes Δiap was taken up by P388.D1 macrophages with the same efficiency as the wild-type strain. Release of the Δiap mutant into the host cell cytosol was also unaffected, as shown by the appearance of green fluorescent protein (GFP)-expressing bacteria when the mutant bacteria were equipped with a plasmid harboring the gfp gene under the control of the actA promoter (data not shown). Since GFP is expressed only when the bacteria are in the cytosol of the host cell (7), GFP expression is a marker for cytosolic localization. Upon release from the phagosome, extensive replication of the Δiap mutant occurred in the host cell cytosol, and the rate of replication was only slightly lower than that of the wild-type strain (Fig. 5). Identical results were obtained by directly counting Giemsa-stained intracellular bacteria (Fig. 5B) and by live-cell counts upon lysis of the macrophages and plating of the lysate (Fig. 5A). Immunofluorescence assays were used to monitor whether L. monocytogenes Δiap was able to induce polymerization of F-actin, to induce actin tails, and to move intracellularly in Caco-2 cells. Intracellular bacteria were stained with an antibody directed against ActA, and filamentous actin was visualized in parallel by FITC-phalloidin staining (Fig. 6A). It was striking that L. monocytogenes Δiap grew intracellularly in microcolonies, whereas the wild-type bacteria, as expected, colonized the whole cell. L. monocytogenes Δiap was still able to induce the polymerization of actin filaments on the surface, but formation of actin tails by these cytosolically replicating bacteria was strongly impaired. Whereas most of the wild-type bacteria induced the formation of actin tails, such actin tails were only very rarely found with the Δiap mutant. The strongly reduced capacity for actin tail formation hence nicely explains the formation of microcolonies due to a lack of intracellular movement. Transmission electron microscopy confirmed that L. monocytogenes Δiap growing in the host cell cytosol did not have typical actin tails. However, the bacteria were associated with a filamentous actin meshwork (Fig. 6B).
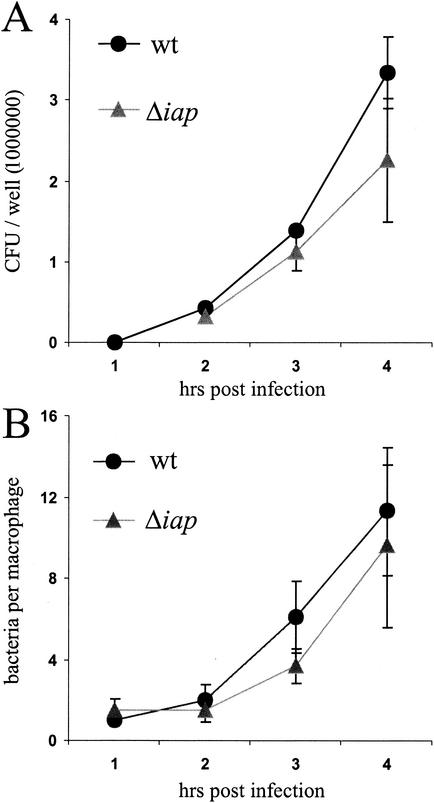
FIG. 5.
Comparison of intracellular replication rates of L. monocytogenes Δiap and wild-type (wt) L. monocytogenes. P388.D1 macrophages were infected with wt bacteria or the Δiap mutant, and at several time points postinfection, intracellular bacteria were quantified either by determining the CFU per well (A) or by microscopical analysis of bacteria which colocalize with the macrophages after Giemsa staining. Bacteria per macrophage were counted (B). Means and standard deviations from one representative experiment performed in triplicate are presented.
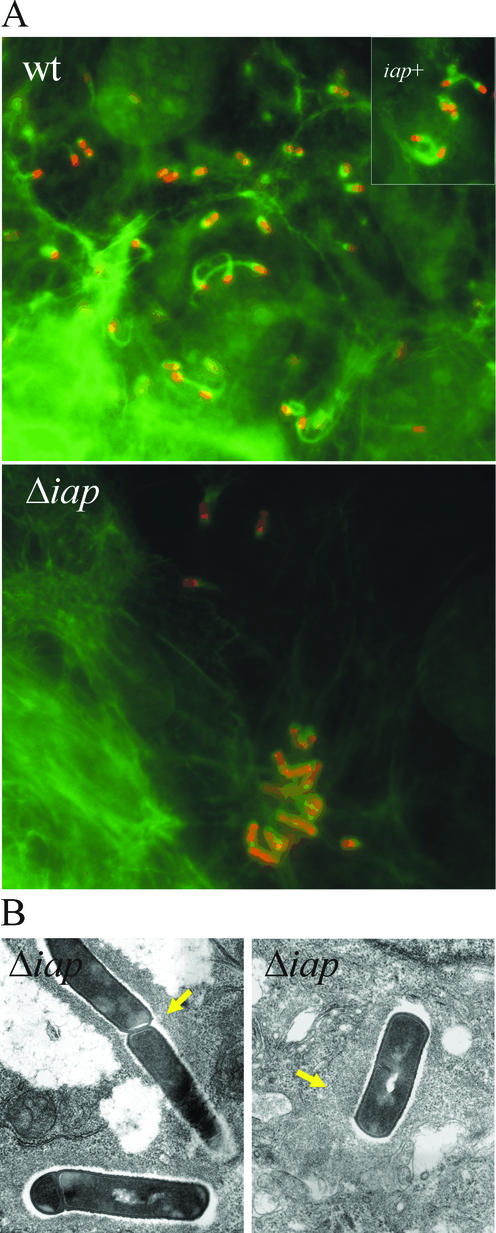
FIG. 6.
(A) Immunofluorescence analysis of Caco-2 cells infected with wild-type (wt) L. monocytogenes, the Δiap mutant, or the iap+ revertant for 6 h. Bacteria appear red due to labeling of surface ActA by the anti-ActA antiserum, and cellular F-actin appears green due to FITC-phalloidin. Filamentous actin is randomly distributed around the Δiap bacteria, in contrast to the F-actin tails visible at the poles of the wt and iap+ intracellular bacteria. (B) Transmission electron microscopy of the Δiap mutant in Caco-2 cells 6 h postinfection. Filamentous actin (arrows) is randomly distributed around the bacteria in both panels.
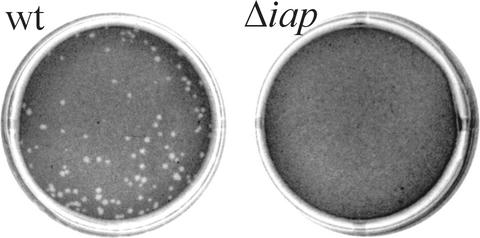
A heterologous plaque assay was used to test for cell-to-cell spread of the Δiap mutant. P388.D1 macrophages were infected with either wild-type or mutant bacteria and placed on a layer of HepG2 cells. Whereas the wild-type bacteria spread from the macrophages to the hepatocytes, resulting in plaque formation, the mutant bacteria were unable to spread from the infected macrophages into the hepatocytes, as shown by the absence of plaques (Fig. 7).
FIG. 7.
A heterologous plaque assay was performed to illustrate the spreading of L. monocytogenes from infected P388.D1 macrophages into a HepG2 cell layer. Plaques were imaged 3 days after infection. Whereas the wild-type (wt) bacteria produced plaques, L. monocytogenes Δiap was unable to spread from the macrophages to the hepatocytes and did not show plaque formation.
The iap+-revertant strain polymerizes actin filaments, induces actin tail formation, and moves intracellularly within the Caco-2 cells to a similar extent as the wild-type strain (see inset in Fig. 6A), suggesting that the loss of actin tail formation as well as intra- and intercellular movement is caused solely by the deletion of the iap gene.
ActA and InlA distribution in the L. monocytogenes Δiap mutant.
Actin polymerization by intracellular L. monocytogenes depends on the surface protein ActA (8, 21), and actin tail formation occurs at the old pole of the dividing listeria, where ActA appears to be concentrated (22). To test whether deletion of the iap gene, which—as shown above—leads to abnormal cell division, may affect the polar distribution of ActA, we determined the localization of ActA in intracellular replicating mutant and wild-type bacteria by staining them with ActA-specific polyclonal antibodies. As shown in Fig. 8, about 95% of the intracellular wild-type bacteria showed a polar distribution of ActA protein, which was in accord with previous reports (22). In contrast, only about 5% of the Δiap mutant bacteria exhibited a polar ActA accumulation, whereas the large majority of the bacterial cells showed an atypical ActA distribution (Fig. 8). That is, ActA neither accumulated polarly nor was evenly distributed around the entire cell surface; instead, it was concentrated at different positions of the bacterial cell surface. The iap+ revertant strain shows a polar ActA distribution (about 92%) similar to that of the wild-type strain (Fig. 8).
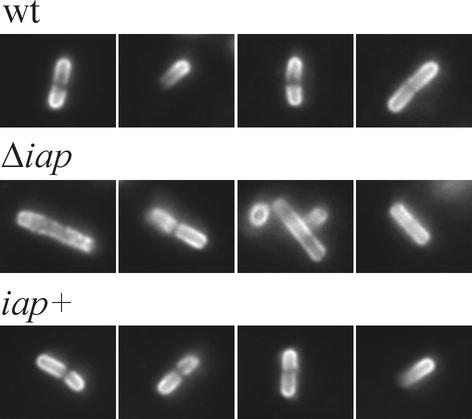
FIG. 8.
Distribution of ActA at the surface of intracellular L. monocytogenes. Caco-2 cells were infected for 6 h with either wild-type (wt) bacteria, the Δiap mutant, or the iap+ revertant, and intracellular bacteria were stained with an antiserum directed against ActA. The wt bacteria and the iap+ revertant show a typical polar localization of ActA, whereas ActA is unevenly arranged at the surface of L. monocytogenes Δiap.
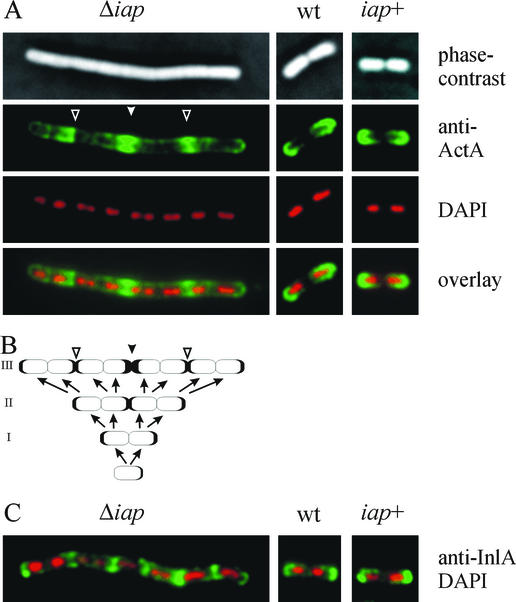
To clarify these data obtained with intracellular bacteria, we analyzed ActA distribution in bacteria grown in BHI broth. All three relevant strains were grown to mid-log phase, fixed on glass slides, and stained with the anti-ActA antiserum and in parallel with DAPI to mark the nucleoids of the individual bacterial cells within the filaments. As expected, wild-type and iap+ bacteria showed the typical polar distribution of ActA, with no detectable ActA at the site of septum formation (Fig. 9A). Filaments of the Δiap strain, however, showed a different distribution; several spots of high ActA concentration were visible along the filament (Fig. 9A). With the help of the corresponding phase-contrast images and especially the nucleoids marked by DAPI staining, we are able to present a rationale for the observed uneven ActA distribution along the filaments (Fig. 9B). The individual bacterial cells forming the filament send ActA on the surface—as usual—to the “old” cellular poles. However, starting from a single cell, after two rounds of replication, the two central bacteria in the four-cell chain send ActA to each other, since this central position would—in normally separated cells—represent the old poles of the two central cells (Fig. 9B). Hence, the center of the filament becomes enriched in ActA (Fig. 9A and B). A further round of replication should then result in two additional spots of ActA accumulation along the filament due to the same mechanism. In fact, these spots are clearly visible in the fluorescence image (Fig. 9A). Breaking of such filaments at different positions then leads to the uneven distribution of ActA apparent on the bacterial surface, as detected either at later stages of growth in broth (data not shown) or in intracellular bacteria (Fig. 8).
FIG. 9.
Distribution of ActA (A) and InlA (C) at the surfaces of L. monocytogenes bacteria grown in BHI. Wild-type (wt) bacteria, the Δiap mutant, and the iap+ revertant were stained with an antiserum directed against either ActA (A) or InlA (C) and with DAPI (A and C). Wild-type bacteria and the iap+ revertant show a typical polar localization of ActA and InlA, whereas both proteins are arranged differently at the surface of L. monocytogenes Δiap. (B) Model showing how ActA distribution along the filament is generated during growth of the filament from a single bacterial cell over three rounds of replication without complete division. Filled arrowheads, center of the filament enriched in ActA. Open arrowheads, spots of ActA accumulation along the filament.
In addition to ActA, the surface distibution of internalin A, another L. monocytogenes protein with known polar localization (28), was also analyzed in the Δiap mutant. As shown in Fig. 9C, internalin A is polarly located in the wild-type and iap+ strains, as expected, but it is unevenly distributed on the surface and localized mainly at the points of incomplete septum formation within the cell chains formed by the Δiap mutant. This finding clearly shows that the lack of p60 and in turn the lack of complete cell division interferes with the correct surface distribution of several virulence factors.
DISCUSSION
In this paper we report the construction and characterization of an L. monocytogenes mutant which carries a deletion of the entire iap gene including 124 bp of the 5′-upstream expression site. This deletion was introduced into the genome of L. monocytogenes EGD by homologous recombination and confirmed by sequence analysis of the deletion site. Additionally, an iap+ revertant strain was constructed by completely restoring the iap locus within the Δiap mutant. For this purpose we had to clone the whole iap gene, including its promoter region, into a shuttle vector with E. coli as the host. However, upon reintegration of the cloned iap gene into L. monocytogenes Δiap, we noticed that a frameshift mutation in the coding region of the iap gene had occurred during cloning or reintegration. Meanwhile, we knew that expression of p60 in E. coli often leads to mutations in the iap gene, most likely because p60 exhibits toxicity for E. coli (17). Due to the frameshift mutation in the iap gene of our initial revertant strain, we had to correct the mutation by site-specific mutagenesis in a second step. In the final iap+ revertant strain (L. monocytogenes iap+), p60 expression is fully restored and there is a correct iap locus as confirmed by DNA sequence analysis.
The successful construction of L. monocytogenes Δiap was unexpected in light of our previous failure to isolate an iap insertion mutant (48) by using a similar strategy (47). Indeed, most of the known naturally occurring mutations affecting synthesis of p60 (the product of iap gene) in L. monocytogenes change the level of expression of the iap gene rather than inactivating the iap gene itself (25, 37). Mutants of the latter type form bacterial colonies with a rough appearance; hence their designation R (for rough) mutants (19). It has been shown that the L. monocytogenes RIII mutant forms long filaments containing multiple double septa which are not properly separated (25). Recently, Bielecki reported the construction of two strains with insertions into the iap locus of L. monocytogenes strain 1043S (2) which express greatly reduced levels of the p60 protein and also show reductions in PlcA levels and hemolytic activity (1). These mutants are still poorly characterized on a molecular level, and since revertants of the insertion mutant were not obtained, the presence of additional mutations cannot be excluded.
The genetic basis of most R mutants is unknown. However, in a recent paper, Lenz and Portnoy (29) presented data showing that inactivation of the secA2 gene in L. monocytogenes results in an R phenotype. The secA2 gene is an additional copy of the secA gene present in all bacteria and, interestingly, is located directly upstream of the iap gene. Deletion or inactivation of secA2 also leads to reduced p60 secretion and hence to a phenotype resembling that of the RIII mutant described previously (25). However, not all R mutants tested so far have defects in secA2, showing that the R-phenotype may be caused by different mechanisms.
In contrast to the iap mutants described by Bielecki (1) and the secA2 mutants described by Lenz and Portnoy (29), the phenotype of our iap deletion mutant is different from that of the R mutants. The colonies formed by this mutant are smooth and almost indistinguishable from those formed by the wild-type strain. Clearly, the deletion of the iap gene also affects the cell division of L. monocytogenes, leading to abnormal cell forms. But instead of forming the extremely long filaments with double septa (cell chains) observed with the RIII mutant, the Δiap mutant forms shorter filaments, and septum formation is initiated at various points along the filament. These septa seem to be incomplete and obviously serve as break points for the disruption of the filaments and the formation of quasi-single cells similar in size to the wild-type cells. Filament formation by L. monocytogenes Δiap is mainly observed in fast, exponentially growing cultures, while in cultures grown into stationary phase and also in bacteria inside mammalian host cells, the predominant bacterial forms are short single cells. A role in cell division was recently also attributed to the PcsB protein from Streptococcus agalactiae (35), which is distantly related to p45 of L. monocytogenes. Lack of PcsB in S. agalactiae results in viable but irregularly shaped bacteria, since septum formation is initiated in different planes. Interestingly, with respect to cell division, the phenotype of the Streptococcus mutant is similar to the phenotype of the L. monocytogenes Δiap mutant described here.
L. monocytogenes Δiap is only slightly impaired in the invasion of 3T6 fibroblasts and Caco-2 epithelial cells. This minor reduction in invasion due to the lack of p60 expression is much smaller than the reduction in invasion by the RIII mutant (25). The extremely low level of p60 expression in the RIII mutant seems to be due to impaired translation of the iap transcript (23). Levels of other invasion-related proteins may hence also be reduced in the RIII mutant due to impaired translation, and their putatively reduced expression may contribute to the observed low level of invasion by the RIII mutant.
The virulence of L. monocytogenes Δiap is strongly attenuated in the mouse model of infection. Whereas mice infected intravenously with high doses of L. monocytogenes Δiap (5 × 103 bacteria per mouse) have a healthy appearance during the course of infection, most mice infected with the same dose of the wild-type strain look seriously ill. Numbers of viable bacteria in the spleens and livers of the infected mice are 1 to 2 log units lower than the numbers of wild-type bacteria at day 3 postinfection. The virulence of the iap+ revertant is completely restored. The reason for the dramatic reduction in virulence of L. monocytogenes Δiap is most likely the strongly impaired ability of the mutant to spread from cell to cell. This loss of intercellular spread is observed not only within a layer of homologous epithelial cells but also between macrophages and hepatocytes. In this respect the Δiap mutant resembles an actA insertion mutant which—due to its inability to polymerize actin within the infected host cells—is unable to move intracellularly and to spread from cell to cell (8). The virulence attenuation of the actA mutant in the mouse model is indeed in the same order as that of the Δiap mutant (8; our unpublished data). However, in contrast to the actA mutant, the Δiap mutant produces wild-type levels of ActA, and the expression and secretion of other known virulence factors are also not significantly altered. Despite the presence of normal amounts of ActA, formation of the typical tails consisting of polymerized actin filaments is only rarely observed upon infection of epithelial cells with the Δiap mutant. Additionally, in contrast to actA mutants, the Δiap mutant is still able to induce the polymerization of actin filaments around the bacteria. However, these actin filaments are not organized into the typical actin tails.
It is rather likely that the altered cell morphology of the Δiap mutant is the major reason for the observed lack of actin tail formation, the resulting inability to move intracellularly, and the subsequent attenuation in virulence. Actin polymerization occurs predominantly at the old poles of dividing wild-type L. monocytogenes cells (21). The ActA protein has been shown to accumulate at this pole, and this asymmetric distribution of ActA has been shown to be crucial for actin tail formation in heterologous systems in in vitro motility assays (5, 42). From the data presented here, we conclude that deletion of the iap gene initially does not impair the translocation of ActA to the old pole. However, the lack of cell division upon septum formation leads to mutant bacteria which exhibit polar ActA localization in less than 5% of all intracellularly localized bacteria. This low percentage of bacteria exhibiting polar ActA distribution correlates perfectly with the small number of bacteria inducing the formation of actin tails. Most of the intracellular Δiap bacteria display an uneven and clearly nonpolar ActA distribution, with spots of high ActA concentrations at several positions along the filaments. Such a distribution of ActA on the bacterial surface results in F-actin polymerization but not in the ordered formation of F-actin tails, which requires strictly polarly accumulated ActA. The precise reversion of the deletion of the iap gene by site-specific recombination, which restores expression of p60 to wild-type levels, also restores polar distribution of ActA as well as actin tail formation. Since normal cell division, leading to formation of single bacterial cells in exponentially growing cultures, is observed in the iap+ revertant strain, there seems to be a strict correlation between loss of p60, impaired cell division, elimination of polar ActA distribution, and actin tail formation.
Our analysis of ActA distribution in in vitro-grown L. monocytogenes clearly shows that lack of p60 expression does not directly result in lack of polar distribution of ActA. In contrast, the lack of cell division after septum formation leads to ActA accumulation at the points of septum formation along the filament. These spots of high ActA concentration are not at the ends (poles) of the filaments and hence cannot induce defined F-actin tails. Single mutant bacteria which are formed within host cells at low efficiency by the rupture of the filaments show polar ActA distribution, according to our model. These bacteria account for the rare cases in which Δiap mutant bacteria form F-actin tails. Since F-actin-dependent intra- and intercellular spread is a major virulence factor in L. monocytogenes, this lack of intracellular motility of the Δiap strain nicely explains the reduction in virulence of the strain completely devoid of p60.
Finaly, we propose to rename the gene encoding p60, changing the designation from iap (invasion-associated protein) to cwhA, for cell wall hydrolase A, and hence to refer to p60 in the future as CwhA.
Acknowledgments
We thank F. Engelbrecht and J. Kreft for helpful discussions, M. Beck for help with the animal experiments, I. Weiglein for the purification of PlcB for antiserum production, and B. Bergmann for support during the cell culture experiments. Many thanks also to A. Bubert, F. Fiedler, P. Cossart, and J. Wehland for providing antibodies.
This work was supported by the Deutsche Forschungsgemeinschaft through GK 520 (to S.P.), SFB 479-B1 (to W.G.), and SFB 479-B5 (to M.K.) and by grants from the Fonds der Chemischen Industrie (to W.G.) and through the Bundesministerium für Bildung und Forschung (AZ01 KS9603) (to A.K.-M.) within the scope of IZKF Würzburg.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Bielecki, J. 1994. Insertions within iap gene of Listeria monocytogenes generated by plasmid pLIV are not lethal. Acta Microbiol. Pol. 43:133-143. [PubMed] [Google Scholar]
- 2.Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. [PubMed] [Google Scholar]
- 3.Bubert, A., M. Kuhn, W. Goebel, and S. Köhler. 1992. Structural and functional properties of the p60 proteins from different Listeria species. J. Bacteriol. 174:8166-8171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabanes, D., P. Dehoux, O. Dussurget, L. Frangeul, and P. Cossart. 2002. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 10:238-245. [DOI] [PubMed] [Google Scholar]
- 5.Cameron, L. A., M. J. Footer, A. van Oudenaarden, and J. A. Theriot. 1999. Motility of ActA protein-coated microspheres driven by actin polymerization. Proc. Natl. Acad. Sci. USA 96:4908-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cossart. P., and M. Lecuit. 1998. Interactions of Listeria monocytogenes with mammalian cells during entry and actin-based movement: bacterial factors, cellular ligands and signaling. EMBO J. 17:3797-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dietrich, G., A. Bubert, I. Gentschev, Z. Sokolovic, A. Simm, A. Catic, S. H. E. Kaufmann, J. Hess, A. A. Szalay, and W. Goebel. 1998. Delivery of antigen-encoding plasmid DNA into the cytosol of macrophages by attenuated suicide Listeria monocytogenes. Nat. Biotechnol. 16:181-185. [DOI] [PubMed] [Google Scholar]
- 8.Domann, E., J. Wehland, M. Rohde, S. Pistor, M. Hartl, W. Goebel, M. Leimeister-Wächter, M. Wuenscher, and T. Chakraborty. 1992. A novel bacterial virulence gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 11:1981-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelbrecht, F., S. K. Chun, C. Ochs, J. Hess, F. Lottspeich, W. Goebel, and Z. Sokolovic. 1996. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol. Microbiol. 21:823-837. [DOI] [PubMed] [Google Scholar]
- 10.Fürst, P., H. U. Mosch, and M. Solioz. 1989. A protein of unusual composition from Enterococcus faecium. Nucleic Acids Res. 17:6724.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gedde, M. M., D. E. Higgins, L. G. Tilney, and D. A. Portnoy. 2000. Role of listeriolysin O in cell-to-cell spread of Listeria monocytogenes. Infect. Immun. 68:999-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gentschev, I., Z. Sokolovic, S. Köhler, G. F. Krohne, H. Hof, J. Wagner, and W. Goebel. 1992. Identification of p60 antibodies in human sera and presentation of this listerial antigen on the surface of attenuated salmonellae by the HlyB-HlyD secretion system. Infect. Immun. 60:5091-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chétouani, E. Couvé, A. de Daruvar, P. Dehoux, E. Domann, G. Domínguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K.-D. Entian, H. Fsihi, F. Garcia-Del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L.-M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. Mata Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J.-C. Pérez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J.-A. Vázquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 14.Goebel, W., and M. Kuhn. 2000. Bacterial replication in the host cell cytosol. Curr. Opin. Microbiol. 3:49-53. [DOI] [PubMed] [Google Scholar]
- 15.Gray, M. L., and A. H. Killinger. 1966. Listeria monocytogenes and listeric infections. Bacteriol. Rev. 30:309-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greiffenberg, L., W. Goebel, K. S. Kim, I. Weiglein, A. Bubert, F. Engelbrecht, M. Stins, and M. Kuhn. 1998. Interaction of Listeria monocytogenes with human brain microvascular endothelial cells: InlB-dependent invasion, long-term intracellular growth, and spread from macrophages to endothelial cells. Infect. Immun. 66:5260-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herler, M., A. Bubert, M. Goetz, Y. Vega, J.-A. Vazquez-Boland, and W. Goebel. 2001. Positive selection of mutations leading to loss or reduction of transcriptional activity of PrfA, the central regulator of Listeria monocytogenes virulence. J. Bacteriol. 183:5562-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hess, J., I. Gentschev, G. Szalay, C. Ladel, A. Bubert, W. Goebel, and S. H. E. Kaufmann. 1995. Listeria monocytogenes p60 supports host cell invasion by and in vivo survival of attenuated Salmonella typhimurium. Infect. Immun. 63:2047-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hof, H. 1984. Virulence of different strains of Listeria monocytogenes serovar 1/2a. Med. Microbiol. Immunol. 173:207-218. [DOI] [PubMed] [Google Scholar]
- 20.Jonqieres, R., H. Bierne, J. Mengaud, and P. Cossart. 1998. The inlA gene of Listeria monocytogenes LO28 harbors a nonsense mutation resulting in release of internalin. Infect. Immun. 66:3420-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kocks, C., E. Gouin, M. Tabouret, P. Berche, H. Ohayon, and P. Cossart. 1992. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell 68:521-531. [DOI] [PubMed] [Google Scholar]
- 22.Kocks, C., R. Hellio, P. Gounon, H. Ohayon, and P. Cossart. 1993. Polarized distribution of Listeria monocytogenes surface protein ActA at the site of directional actin assembly. J. Cell Sci. 105:699-710. [DOI] [PubMed] [Google Scholar]
- 23.Köhler, S., A. Bubert, M. Vogel, and W. Goebel. 1991. Expression of the iap gene coding for protein p60 of Listeria monocytogenes is controlled on the posttranscriptional level. J. Bacteriol. 173:4668-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolb-Mäurer, A., S. Pilgrim, E. Kämpgen, A. D. McLellan, E.-B. Bröcker, W. Goebel, and I. Gentschev. 2001. Antibodies against listerial protein 60 act as an opsonin for phagocytosis of Listeria monocytogenes by human dendritic cells. Infect. Immun. 69:3100-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn, M., and W. Goebel. 1989. Identification of an extracellular protein of Listeria monocytogenes possibly involved in intracellular uptake by mammalian cells. Infect. Immun. 57:55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn, M., and W. Goebel. 2000. Internalization of Listeria monocytogenes by nonprofessional and professional phagocytes. Subcell. Biochem. 33:411-436. [DOI] [PubMed] [Google Scholar]
- 27.Kuhn, M., M.-C. Prévost, J. Mounier, and P. J. Sansonetti. 1990. A nonvirulent mutant of Listeria monocytogenes does not move intracellularly but still induces polymerization of actin. Infect. Immun. 58:3477-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.Lebrun, M., J. Mengaud, H. Ohayon, F. Nato, and P. Cossart. 1996. Internalin must be on the bacterial surface to mediate entry of Listeria monocytogenes into epithelial cells. Mol. Microbiol. 21:579-592. [DOI] [PubMed] [Google Scholar]
- 29.Lenz, L. L., and D. A. Portnoy. 2002. Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol. Microbiol. 45:1043-1056. [DOI] [PubMed] [Google Scholar]
- 30.Marquis, H., H. Goldfine, and D. A. Portnoy. 1997. Proteolytic pathways of activation and degradation of a bacterial phospholipase C during intracellular infection by Listeria monocytogenes. J. Cell Biol. 137:1381-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mounier, J., A. Ryter, M. Coquis-Rondon, and P. J. Sansonetti. 1990. Intracellular and cell-to-cell spread of Listeria monocytogenes involves interaction with F-actin in the enterocytelike cell line Caco-2. Infect. Immun. 58:1048-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parida, S. K., E. Domann, M. Rohde, S. Müller, A. Darji, T. Hain, J. Wehland, and T. Chakraborty. 1998. Internalin B is essential for adhesion and mediates the invasion of Listeria monocytogenes into human endothelial cells. Mol. Microbiol. 28:81-93. [DOI] [PubMed] [Google Scholar]
- 33.Pfeuffer, T., W. Goebel, J. Laubinger, M. Bachmann, and M. Kuhn. 2000. LaXp180, a mammalian ActA-binding protein, identified with the two-hybrid system, co-localizes with intracellular Listeria monocytogenes. Cell. Microbiol. 2:101-114. [DOI] [PubMed] [Google Scholar]
- 34.Potel, J., and J. Schulze-Lammers. 1985. Listeria monocytogenes-vaccine: production and control. Zentbl. Bakteriol. Mikrobiol. Hyg. 259:331-340. [DOI] [PubMed] [Google Scholar]
- 35.Reinscheid, D. J., B. Gottschalk, A. Schubert, B. J. Eikmanns, and G. S. Chhatwal. 2001. Identification and molecular analysis of PcsB, a protein required for cell wall separation of group B streptococcus. J. Bacteriol. 183:1175-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rocourt, J. 1999. The genus Listeria and Listeria monocytogenes: phylogenetic position, taxonomy, and identification, p. 1-20. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis and food safety. Marcel Dekker, New York, N.Y.
- 37.Rowan, N. J., A. A. Candlish, A. Bubert, J. G. Anderson, K. Kramer, and J. McLauchlin. 2000. Virulent rough filaments of Listeria monocytogenes from clinical and food samples secreting wild-type levels of cell-free p60 protein. J. Clin. Microbiol. 38:2643-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Schubert, K., A. M. Bichlmaier, E. Mager, K. Wolff, G. Ruhland, and F. Fiedler. 2000. P45, an extracellular 45 kDa protein of Listeria monocytogenes with similarity to protein p60 and exhibiting peptidoglycan lytic activity. Arch. Microbiol. 173:21-28. [DOI] [PubMed] [Google Scholar]
- 40.Seeliger, H. P. R. 1961. Listeriosis. Hafner Press, New York, N.Y.
- 41.Seeliger, H. P. R. 1984. Modern taxonomy of the Listeria group relationship to its pathogenicity. Clin. Investig. Med. 7:217-221. [PubMed] [Google Scholar]
- 42.Smith, G. A., D. A. Portnoy, and J. A. Theriot. 1995. Asymmetric distribution of the Listeria monocytogenes ActA protein is required and sufficient to direct actin-based motility. Mol. Microbiol. 17:945-951. [DOI] [PubMed] [Google Scholar]
- 43.Tilney, L. G., and D. A. Portnoy. 1989. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 109:1597-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vazquez-Boland, J.-A., C. Kocks, S. Dramsi, H. Ohayon, C. Geoffroy, J. Mengaud, and P. Cossart. 1992. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect. Immun. 60:219-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vazquez-Boland, J.-A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whisstock, J. C., and A. M. Lesk. 1999. SH3 domains in prokaryotes. Trends Biochem. Sci. 24:132-133. [DOI] [PubMed] [Google Scholar]
- 47.Wuenscher, M. D., S. Köhler, W. Goebel, and T. Chakraborty. 1991. Gene disruption by plasmid integration in Listeria monocytogenes: insertional inactivation of the listeriolysin determinant lisA. Mol. Gen. Genet. 228:177-182. [DOI] [PubMed] [Google Scholar]
- 48.Wuenscher, M. D., S. Köhler, A. Bubert, U. Gerike, and W. Goebel. 1993. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J. Bacteriol. 175:3491-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]