Abstract
Multiple epiphyseal dysplasia (MED) is a degenerative cartilage condition shown in some cases to be caused by mutations in genes encoding cartilage oligomeric matrix protein or type IX collagen. We studied a family with autosomal dominant MED affecting predominantly the knee joints and a mild proximal myopathy. Genetic linkage to the COL9A3 locus on chromosome 20q13.3 was established with a peak log10 odds ratio for linkage score of 3.87 for markers D20S93 and D20S164. Reverse transcription–PCR performed on the muscle biopsy revealed aberrant mRNA lacking exon 3, which predicted a protein lacking 12 amino acids from the COL3 domain of α3(IX) collagen. Direct sequencing of genomic DNA confirmed the presence of a splice acceptor mutation in intron 2 of the COL9A3 gene (intervening sequence 2, G-A, -1) only in affected family members. By electron microscopy, chondrocytes from epiphyseal cartilage exhibited dilated rough endoplasmic reticulum containing linear lamellae of alternating electron-dense and electron-lucent material, reflecting abnormal processing of mutant protein. Type IX collagen chains appeared normal in size and quantity but showed defective cross-linking by Western blotting. The novel phenotype of MED and mild myopathy is likely caused by a dominant-negative effect of the exon 3-skipping mutation in the COL9A3 gene. Patients with MED and a waddling gait but minimal radiographic hip involvement should be evaluated for a primary myopathy and a mutation in type IX collagen.
Multiple epiphyseal dysplasia (MED) is a clinically and genetically heterogeneous group of autosomal dominant skeletal dysplasias characterized by early-onset osteoarthritis, a waddling gait, and sometimes short stature. Both the more severe Fairbank-type MED (1), which overlaps clinically with pseudoachondroplasia (PSACH) (2), and the milder Ribbing-type MED (3) display significant hip involvement, but a much wider phenotypic spectrum is currently recognized (4). Childhood-onset joint pain and stiffness usually progress to osteoarthritic degeneration of hips and knees, often necessitating total joint arthroplasty.
At least three genetic defects underlie this phenotype. Mutations in the gene encoding cartilage oligomeric matrix protein (COMP) on chromosome 19p13.1 (5) cause MED [EDM 1; Mendelian Inheritance of Man (MIM) #132400] and the related phenotype PSACH (6). A milder hip-sparing MED (EDM 2; MIM #600204) is caused by mutations in the gene encoding the α2 chain of type IX collagen, COL9A2, on chromosome 1p32.3–33 (7, 8). Other families do not show linkage to either locus (9–11). While this manuscript was in preparation, a mutation in the COL9A3 gene encoding the α3(IX) chain defined a third locus (EDM 3) for MED (12).
Mutations in various collagen genes cause disorders affecting connective tissue, basement membranes, and muscle (13). Mutations in the three chains of type VI collagen cause Bethlem myopathy, an autosomal dominant and slowly progressive myopathy with early joint contractures (14). Although type VI collagen has widespread tissue expression, the phenotype is largely restricted to muscle and joint contractures (15). In muscle, type VI collagen may interact with type IV collagen, the major collagenous component of basement membranes (16).
Type IX collagen is a heterotrimer composed of α1(IX), α2(IX), and α3(IX) chains encoded by genes on chromosomes 6q12-q13 (COL9A1), 1p32.3–33 (COL9A2), and 20q13.3 (COL9A3), respectively. Type IX collagen belongs to the fibril-associated collagens with interrupted triple helices (17). Each chain contains four noncollagenous domains (NC1–4) separated by three triple helical collagenous domains (COL1–3) (Fig. 5c). NC4 of the α1 chain forms an additional large globular domain at the N terminus. In cartilage, type IX collagen molecules are bound to the surface of type II collagen fibrils (18–20) and to each other (20) via lysine-derived cross-links. Type IX collagen in cartilage is believed to play a role in the organization and spacing of type II collagen fibrils (21) by cross-linking and regulating the growth of type II collagen fibrils through its COL3 and α1(IX) NC4 domains, which extend laterally into the perifibrillary space (22). In chick cartilage, the NC3 domain of the α2(IX) chain can be modified by a single glycosaminoglycan (chondroitin/dermatan sulfate) side chain, thereby making it a proteoglycan (23). The cationic NC4 domain may interact with other proteoglycans in cartilage. Expression of type IX collagen is predominantly restricted to cartilage and eye vitreous with only minor expression in other tissues (17). Tissue-specific isoforms of the α1(IX) chain are found in cartilage and eye vitreous (24).
Figure 5.
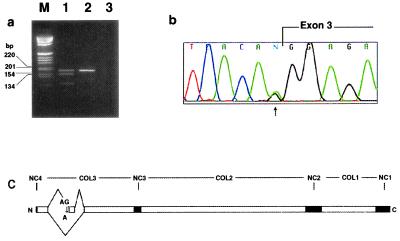
(a) Reverse transcription–PCR from skeletal muscle cDNA using primers flanking the mutation. Lane M, molecular size markers in bp; lane 1, patient sample showing a normal upper band and an abnormal lower band lacking the 36-bp exon 3; lane 2, normal control containing a single upper band; lane 3, negative control without DNA. (b) Chromatogram of genomic DNA sequence from the proband. The G-to-A mutation is present in a heterozygous state at position −1 with respect to the exon ( ). (c) Schematic of the splice defect relative to the domain structure of COL9A3. COL1–3, collagenous domains 1–3; and NC1–4, noncollagenous domains 1–4.
We describe an autosomal dominant disorder that combines clinical and radiographic features of MED with those of a mild myopathy. The genetic basis is a splice site mutation that causes skipping of exon-3 in the COL9A3 gene. We describe new morphologic findings in cartilage and muscle and present biochemical evidence for a dominant-negative effect of the mutation on type IX collagen interactions.
Clinical Study
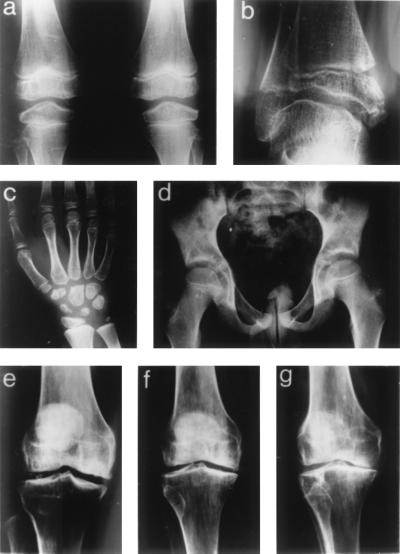
The proband is a 10-year-old boy (individual IV-5 in Fig. 1) referred to the Children's Hospital Neuromuscular Clinic for evaluation of proximal muscle weakness and mildly elevated serum creatine kinase (201 and 179 units/liter; normal <175 units/liter). He walked at 1 year of age but at age 3 was noted to have difficulty walking and climbing stairs. He always had difficulty rising from the floor and sometimes used a one-handed Gower maneuver. He tired easily, never ran well, and complained of knee pain. He had significant weakness of neck flexion, mild weakness of shoulder abduction and elbow extension, and proximal lower extremity weakness (hamstrings weaker than quadriceps). His height was 145.5 cm (90th percentile), and his hands appeared normal. The family history was notable for childhood-onset osteoarthritis and complaints of knee pain and stiffness in his mother and several relatives. Eleven members were affected across three generations (Fig. 1). The proband's x-rays revealed epiphyseal changes characteristic of MED (Fig. 2 a–c). The knee joints were affected the most (Fig. 2a), followed by the ankle joints (Fig. 2b). The distal ulnae were mildly shortened and the carpals and tarsals showed irregular ossification (Fig. 2c). The pelvis and hip joints (Fig. 2d), ribs, and spine appeared normal. X-rays of affected family members showed changes consistent with MED, predominantly affecting the knee joints and sparing the hips. Significant osteoarthritis developed with joint space narrowing, femoral condylar bony flattening, and development of osteophytes and subchondral cysts by mid-adulthood (Fig. 2 e–g). The proband's mother (III-7) had weak neck flexors and minimal weakness of the proximal extremities. His affected aunt (III-9) had minimal proximal lower extremity weakness, whereas her younger daughter (IV-7) with greater knee involvement had noticeable proximal lower and upper extremity weakness and weakness of neck flexion. Her older daughter (age 16, IV-6) was asymptomatic but had bilateral knee crepitus and radiographic changes of MED. A male second cousin (IV-3) had been diagnosed clinically with a proximal myopathy by a neurologist. His radiologic skeletal changes were more severe than female family members of comparable age. Both males were weaker than affected female relatives. In this family, the degree of muscle weakness correlated with the severity of the MED.
Figure 1.
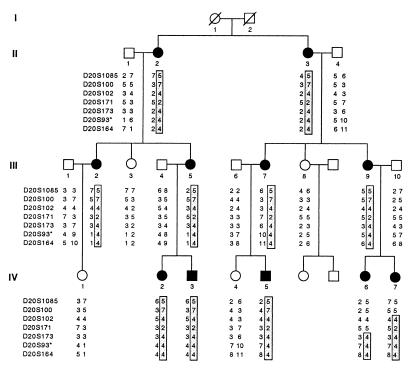
Pedigree of MED with mild myopathy genotyped at markers from chromosome 20q13.3. The disease haplotype is boxed. All affected family members share a common allele of markers D20S173, D20S93, and D20S164. D20S93* lies within an intron of the COL9A3 gene (34). Circles, female; squares, male; shaded symbols, affected; open symbols, unaffected.
Figure 2.
(a--d) Radiographs from the proband (individual IV-5) at 10 yr of age. (a) Anteroposterior view of both knees. Epiphyses are abnormally shaped especially at subchondral bone regions. (b) Abnormal ankle radiograph with irregular shaping of distal tibial epiphysis and a rounded talar surface. (c) Anteroposterior radiograph centered at the wrist shows irregular carpals and a mildly shortened distal ulna. (d) Normal hip and pelvic radiograph. Knee radiographs showing progressive degenerative osteoarthritis in (e) individual III-9 at 38 yr and her mother II-3 at 52 yr (f) and 63 yr (g).
Materials and Methods
Histology.
Iliac crest growth cartilage biopsy was processed immediately after removal of the outer fibrocartilaginous layer and spicules of metaphyseal bone. Epiphyseal and physeal cartilage was cut into 1 × 0.5 × 0.5 mm segments, fixed in modified Karnovsky solution (1% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4), and processed for light and electron microscopy as described (25, 26). The muscle biopsy was processed for histologic studies by standard frozen section histology, histochemistry, morphometry, and glutaraldehyde-fixed, Epon-embedded electron microscopy (27).
Genetic Linkage Analysis.
Genomic DNA was purified from blood by using a Puregene Kit (Gentra Systems). Microsatellite markers from the ABI Prism Linkage Mapping Set Version 1 (Applied Biosystems) or synthesized from published sequences (Life Technologies, Grand Island, NY; Research Genetics, Huntsville, AL) were amplified by PCR. They included D1S197, D2S220, and D1S209 (Col9A2); D6S257, D6S264, and D6S280 (Col9A1); D19S221, D19S226, and D19S414 (COMP); and D20S1085, D20S100, D20S102, D20S171, D20S173, D20S93, and D20S164 (COL9A3). Markers were genotyped by using an ABI Prism 377 DNA sequencer equipped with genescan and genotyper software. Two-point lod scores were calculated by using mlink (28). The disease was modeled as an autosomal dominant disorder with complete penetrance and an allele frequency of 0.0001.
Mutation Analysis.
Total RNA was isolated from the muscle biopsy by using a polytron homogenizer and Trizol Reagent (GIBCO/BRL). cDNA was synthesized with random hexonucleotides (Pharmacia) and superscript II (GIBCO/BRL) reverse transcriptase. Five oligonucleotide primer pairs were used to amplify overlapping fragments of the coding region of the cDNA by PCR, such that the first forward primer corresponded to bases 29–50 of the published cDNA sequence: COL9A3-P1F2 (5′-tcctgctcctgctcctcctcgg-3′) or COL9A3-P1F3′ (5′-cggccaggacggcattgac-3′) and COL9A3-P1R2 (5′-cttccccctgctcgcctttgtag-3′); COL9A3-P2F (5′-ctactgaccttcagtgcccaagt-3′) and COL9A3-P2R (5′-cgttctcccttctcgcctttg-3′); COL9A3-P3F2 (5′-agggagaggctggtcgcaacg-3′) and COL9A3-P3R (5′-ggtgccaaaggcttcctt aggtg-3′); and COL9A3-P4F (5′-gaacaaattgcacagttagccgc-3′) and COL9A3-P4R2 (5′-cttctctcaaatgctgggcttac-3′). COL9A3-P1F3′ and COL9A3-P1R9 (5′-ctggtttccccggctttcctg-3′) were used to confirm the aberrantly spliced product. PCR was performed by using Platinum Taq (GIBCO/BRL) at an annealing temperature of 60°C for 40 cycles in a PT-200 thermal cycler (MJ Research, Cambridge, MA).
PCR products were gel purified and sequenced automatically (Applied Biosystems). To identify the exon/intron structure surrounding putative exon 3, primer combinations located 300–400 bases from the 5′ end of the cDNA sequence were used to amplify and sequence the intervening intronic sequence from genomic DNA. Long-range PCR on genomic DNA was performed with a primer designed from intronic sequence, COL9A3-iR1 (5′-agactctccagctccccaaactc-3′) and the 5′ cDNA primer COL9A3-P1F2 (62°C annealing temperature, 35 cycles). The 4-kb product was subcloned into the TOPO vector (Invitrogen). Primers COL9A3-P1F5 (5′-ggtcctccaggtctgcct-3′) and COL9A3-P1R8 (5′-aggcagacctggaggacc-3′) from putative exon 3 were used to sequence the flanking introns, whose sequence was used to design primers COL9A3-e3F1 (5′-cttttgtctgctggggctgg-3′) and COL9A3-e3R (5′-ttccccctttctctccagactg-3′) from introns 3 and 4, respectively, for PCR amplification and sequencing of exon 3 from genomic DNA.
Collagen IX Analysis.
An aliquot of iliac crest growth cartilage biopsy was assayed for intact collagen IX α-chains in a denaturing solution and cross-linked peptides of the insoluble collagen IX pool after CNBr digestion of the tissue residue (19). Tissue was extracted in 4 M guanidine⋅HCl/0.05 M Tris⋅HCl (pH 7) at 4°C for 24 h. The extract was dialyzed to remove salt, digested with chondroitinase ABC (29) to destroy glycosaminoglycan chains, and analyzed by SDS/PAGE/Western blotting by using a polyclonal antiserum that recognizes all three chains of collagen type IX (26). The postextraction tissue residue was dried, digested with CNBr in 70% formic acid, freeze-dried, and analyzed by SDS/PAGE/Western blotting as above. Human fetal growth cartilage was treated similarly and used as a normal control.
Results
Biopsies.
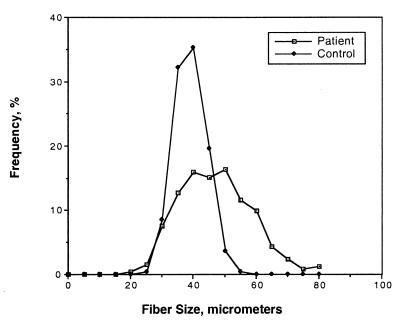
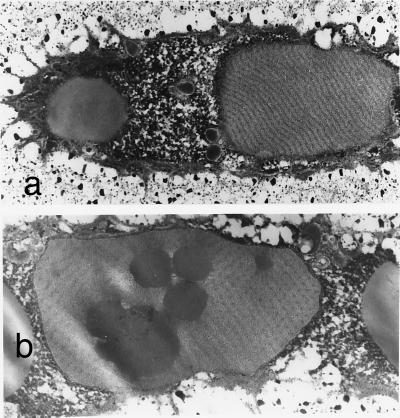
The proband's quadriceps muscle biopsy showed mild myopathic changes characterized by mild variability in fiber size (Fig. 3). Although the mean fiber diameter was 44.46 ± 11.42 μm, which is within the normal range for a 10-year-old (27, 30), the variability coefficient (standard deviation of the fiber diameter × 1,000 divided by the mean diameter of all the fibers) was increased to 260 (normal <250) (27, 31). Size variability was seen in both type 1 and type 2 fibers. Immunostaining for dystrophin, merosin, and sarcoglycans was normal. By light microscopy, virtually all iliac crest epiphyseal chondrocytes had extensive intracytoplasmic inclusions. The cartilage matrix was notable only for a more fibrillar appearance than normal in the longitudinal cartilage septae at the lower physis. The endochondral sequence appeared normal. By electron microscopy, the epiphyseal chondrocytes were strikingly abnormal with markedly dilated rough endoplasmic reticulum (RER) containing linear arrays of alternating electron-dense (60-nm wide) and electron-lucent (120-nm wide) material (Fig. 4a). Some sections of dilated RER showed a hexagonal array of punctate electron-dense material, 60-nm wide, corresponding to end-on profiles of the rod-like material (Fig. 4b). Occasional fat droplets were seen in the cytoplasm and RER. No whorled patterns of the RER material or degenerating cells were observed. Epiphyseal areas of cartilage were more abnormal than cartilage in the physeal regions. The pericellular matrix showed occasional collagen fibrils with focal widening and splitting on longitudinal sections and irregular outlines on transverse sections. The perimysial fibrillar collagen in muscle was normal by electron microscopy in cross-section with uniform diameters, round outlines, and equal spacing.
Figure 3.
Histogram of quadriceps muscle biopsy from the proband showing mild variability in fiber diameter compared with an age-matched control.
Figure 4.
Electron micrographs of epiphyseal chondrocytes with dilated RER containing (a) linear lamellar material in longitudinal section (×8,000) and (b) hexagonal arrays of punctate material in cross-section (×12,970). Fat inclusions are seen in both the cytoplasm and RER.
Genetic Linkage and Mutation Detection.
Several candidate loci for MED were evaluated for genetic linkage in this family by using markers flanking the COMP (9, 32), COL9A1 (33), COL9A2 (7, 10), and COL9A3 genes (34). Significant linkage was found only to COL9A3 gene on chromosome 20q13.3 with the maximal lod score, Zmax(θ = 0) = 3.87, at markers D20S93 and D20S164 (Table 1). The minimum haplotype shared by affected individuals extended 3.3 cM from D20S173 to the chromosome 20q telomere (Fig. 1). To determine whether the more severe phenotype seen in the two male second cousins was caused by a coinherited modifying locus on the X chromosome, X chromosome markers from the ABI Prism Linkage Mapping Set Version 1 were genotyped in all family members. The affected boys shared alleles identical by state at the adjacent markers DX986 and DXS990, separated by 7.7 cM, and DX1227 on distal Xq, limiting the possibility of a coinherited X-linked gene to these two small regions. Different haplotypes flanked the COL9A1, COL9A2, and COMP loci, excluding the coinheritance of a polymorphism in one of these genes.
Table 1.
lod scores of genetic markers near the COL9A3 locus at chromosome 20q13.3 over a range of recombination frequencies, θ, at 100% penetrance
| Locus | θ
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0.0 | 0.01 | 0.05 | 0.1 | 0.2 | 0.3 | 0.4 | Zmax | |
| D20S1085 | 3.83 | 3.77 | 3.51 | 3.16 | 2.43 | 1.62 | 0.76 | |
| D20S100 | −infini | −0.19 | 0.98 | 1.29 | 1.26 | 0.92 | 0.43 | |
| D20S102 | 2.97 | 2.91 | 2.70 | 2.43 | 1.85 | 1.21 | 0.54 | |
| D20S171 | −infini | 1.81 | 2.26 | 2.24 | 1.86 | 1.28 | 0.60 | |
| D20S173 | 2.66 | 2.62 | 2.42 | 2.18 | 1.64 | 1.07 | 0.46 | |
| D20S93* | 3.87 | 3.80 | 3.54 | 3.20 | 2.46 | 1.65 | 0.78 | 3.87 |
| D20S164 | 3.87 | 3.80 | 3.54 | 3.20 | 2.46 | 1.65 | 0.78 | 3.87 |
*Lies within an intron of the COL9A3 gene.
To search for a mutation in the COL9A3 gene, total RNA was isolated from the proband's muscle biopsy and α3(IX) chain cDNA was amplified in overlapping fragments by reverse transcription–PCR. Gel electrophoresis of one of the PCR products revealed two closely spaced bands in the patient but not in normal controls (Fig. 5a). DNA sequencing revealed an internal 36-bp deletion in the COL3 domain. Because only the cDNA sequence of COL9A3 was known, the intron/exon structure surrounding this deleted region was determined by genomic PCR by using primers predicted to lie within known exons of the homologous mouse col9a2 gene (35). The deleted 36 bp corresponded exactly to a single exon homologous to exon 3 from the mouse col9a2 (35) and human COL9A2 genes (36). PCR amplification of this region from the proband's genomic DNA did not reveal a deletion, suggesting a splice site mutation. Intronic primers were designed to PCR amplify and sequence the corresponding genomic region (Fig. 5b). A heterozygous G to A transition was found at position -1 in the consensus sequence for the splice acceptor site of intron 2 (IVS2, G-A, -1). Because this base is 100% invariant (37, 38), the mutation most likely prevents recognition of the splice acceptor site during mRNA processing and results in the skipping of exon 3 (39) (Fig. 5c). The mutation specifically segregated with the disease in the family and was not found in 100 control chromosomes.
Biochemical Studies.
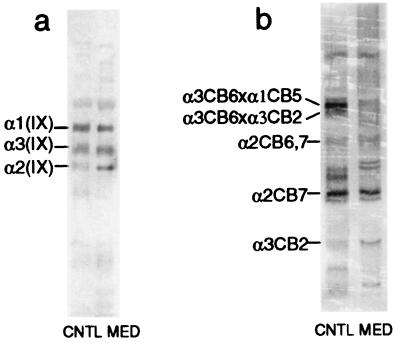
The small pool of type IX collagen extracted in 4 M guanidine⋅HCl presumably represents newly synthesized protein not yet cross-linked in the matrix. Western blot analysis of extract from the proband's iliac crest cartilage showed the same relative amounts and electrophoretic mobilities of α1(IX), α2(IX), and α3(IX) chains as in the normal control (Fig. 6a). The much larger pool of type IX collagen cross-linked in the matrix also appeared to be present in essentially normal amounts (Fig. 6b). However, the pattern of peptides was markedly deficient in cross-linked α3CB6, a band which is normally prominent and serves as a marker for the extent of collagen IX-to-IX covalent interactions in the matrix (19) (S. Ichimura, J. J. Wu, and D. R. Eyre, unpublished work). Nevertheless, other fragments of the α3(IX) collagen chain appeared to be present in normal amounts, implying defective cross-linking of type IX collagen rather than a deficiency of the α3(IX) chain in the matrix. Direct evidence that the α3(IX) product of the mutant allele was present in the matrix, such as by demonstration of a shortened CB-peptide from the COL3 domain, was beyond the limits of resolution.
Figure 6.
Collagen IX protein analysis. (a) Western blot analysis of denaturant-extracted intact collagen type IX chains showing no apparent quantitative difference between control (CNTL) and patient (MED) samples. (b) Western blot analysis of CNBr-extracted, cross-linked collagen IX fragments showing a clear deficiency in cross-linked α3CB6 fragments in patient (MED) compared with control (CNTL) samples.
Discussion
This family's early-onset degenerative joint disease with mild myopathy is a novel form of MED (40) lacking short stature and brachydactyly (4). The knee joints are most severely affected, similar to three families with analogous splice site mutations in the COL9A2 gene that result in the skipping of exon 3 (7, 8, 40). Sparing of hip joints distinguishes their conditions from the Ribbing and Fairbank types of MED and from PSACH. The effect of the mutation on COL9A3 mRNA splicing in the family reported here is similar to that described for a family with EDM 3 and a slightly different COL9A3 mutation (IVS2, G-A, -1 versus IVS2, A-T, -2) (12). However, several affected members of that family had radiographic hip involvement and no reported muscle weakness. Two mouse models of MED have been created by altering expression of the α1 chain of type IX collagen, either by overexpressing a transgene for an in-frame internally deleted α1(IX) cDNA (41) or by creating a homozygously disrupted col9a1 gene by homologous recombination (42). The phenotype in both consisted of early degenerative joint disease, particularly of the knees.
The phenotypic differences between the families with type IX collagen gene mutations are intriguing as the predicted consequence of all three mutations, a 12-amino acid deletion within the COL3 domain of the α2(IX) and α3 (IX) chains, is identical. The extent of hip involvement in patients with type IX collagen gene mutations may be influenced by differences in the degree of aberrant splicing, collagen polymorphisms, or other modifying loci. This phenomenon is reminiscent of nearly identical mutations in dysferlin that give rise to proximal muscle weakness in one form of autosomal recessive limb-girdle muscular dystrophy and distal muscle weakness in Myoshi distal muscular dystrophy (43, 44).
The variable muscle weakness in this family and the increased fiber-type variability in the proband's muscle biopsy appear to represent a genuine myopathy, not a secondary effect of MED. Variability in fiber size is a well-documented myopathic finding (30, 31, 45) and may be associated with normal or mildly elevated serum creatine kinase. Disuse atrophy secondary to joint discomfort causes selective type 2 fiber atrophy but both type 1 and type 2 fiber size variability were seen in the proband's biopsy, consistent with a true myopathy (45). The joint most involved was the knee; the hip was spared radiographically, precluding the latter as an explanation for proximal weakness. Neck flexor weakness, seen in some family members, also cannot be explained on the basis of skeletal changes. The only two affected males, related as second cousins, showed the most significant knee joint disease and weakness. Haplotype analysis excluded the possibility of coinherited polymorphisms in the COL9A1, COL9A2, and COMP genes and most of the X chromosome, but the possibility of modifying genes present in two small regions on the X chromosome or at autosomal loci cannot be ruled out. Family members with MED caused by a COL9A2 mutation showed variable severity but no sex bias (40). Clinical variability also was reported for the family harboring a similar COL9A3 mutation (12), but the most severely affected individual was female.
A role for type IX collagen in muscle has not been identified previously. Type IX collagen has been observed in tissues other than cartilage and eye vitreous, including notochord (46–50), inner ear (51), heart (52), brain, and skin in mice (53). In noncartilagenous tissues, gene expression of the three chains of collagen IX is not always precisely coordinated with each other or with collagen II (53). The expression of the α3 chain of collagen IX within muscle tissue permitted reverse transcription–PCR mutation analysis on COL9A3 cDNA. Type IX collagen recently has been shown to be expressed in the fibrocartilagenous region of tendinous insertions into bone (54). A myopathy that mainly affects myotendinous junctions histologically is caused by α7 integrin gene mutations in humans (55) and is observed in α7 integrin knockout mice (56). Patients with MED often have a waddling gait, even when hip radiographs appear structurally normal, as emphasized in the clinical description of the family with a COL9A2 mutation (40). It remains to be determined whether careful clinical assessment of others with MED caused by mutations in genes encoding type IX collagen will demonstrate myopathic weakness.
Light microscopic findings in iliac crest cartilage were similar to those described in tibial physial cartilage in a case of Fairbank MED with an unknown mutation (57). In both cases, most chondrocytes contained intracytoplasmic inclusions and cartilage septae in the lower part of the growth plate appeared more fibrillar than normal. Electron microscopy in our case revealed the inclusions as dilated RER packed with linear lamellar material, which are as striking as the whorled arrays seen in PSACH caused by COMP mutations (58, 59). The widths of the electron-dense and -lucent bands in the linear (60-nm and 120-nm) and whorled (45-nm and 90-nm) (58) arrays are similar. In cross-sections of dilated RER, a hexagonal array of fibrils was observed primarily in epiphyseal chondrocytes. Markedly less structured accumulations were described in a case of Fairbanks MED (57). The rod-like material likely represents accumulation of abnormally or incompletely processed matrix protein (60). Although triple helix formation of collagen proceeds from the C to N terminus in the RER (61), in this family, the synthesis and assembly of collagen IX likely is initiated normally given that the mutation affects the COL3 domain close to the N terminus (Fig. 5c). The morphologically distinct whorled precipitations seen in PSACH caused by COMP mutations (58, 59) may be because of different biophysical properties of the precipitated proteins. The abnormalities were more pronounced in epiphyseal than in physeal chondrocytes. Inclusions were not seen in articular cartilage biopsies from two patients with MED caused by an analogous COL9A2 mutation (62), perhaps because of differences in tissue sampling or regional gene expression.
Western blot analysis of growth cartilage revealed apparently normal amounts and sizes of the three type IX collagen α chains. However, by peptide mapping, we observed a gross deficiency of the cross-linked fragment α3CB6, a marker for collagen IX-to-IX covalent interactions (19). This finding suggests that the mutation interferes with type IX-to-IX collagen cross-linking, consistent with a transdominant mechanism of action of the internally deleted α3(IX) chain. In conjunction with the morphological alterations of the RER, multiple steps of collagen synthesis and assembly are apparently disturbed. These findings represent the first descriptions of the morphological and biochemical consequences of human type IX collagen mutations. MED is one of the most common skeletal dysplasias and the one requiring total joint arthroplasty most frequently. Abnormalities in type IX collagen in individuals without short stature leads us to believe that similar mutations may well underlie some cases of idiopathic osteoarthritis. The pathophysiology of premature osteoarthrosis caused by abnormalities of type IX collagen could be explained by reduced compressive stiffness of cartilage lacking normal type IX collagen. Disturbed interactions with other components of cartilage extracellular matrix may disrupt the arrangement and spacing of collagen II fibrils (63–66). Our finding of slightly thickened collagen II fibrils in the cartilage biopsy would be consistent with such a pathogenetic mechanism. We did not observe massive and irregular aggregates of collagen II fibrils as described in diastrophic dysplasia (25, 26).
The COL3 domain of type IX collagen is closest to the N terminus of the protein and is believed to project from the surface of the type II collagen fibrils, whereas the COL2 domain links type IX collagen molecules to the surface of type II collagen fibrils (17, 21, 22). The COL3 domain, the cationic NC4 domain and, if present, the glycosaminoglycan on the α2(IX) chain, likely interact with components of the extracellular matrix, such as other proteoglycans. Because the deleted exon maintains the (Gly-X-Y)n repeat of the collagen helix, the truncated protein is most likely translated and incorporated into a significant percentage of the forming heterotrimers of type IX collagen and exerts a dominant-negative effect through the misalignment of subunits. This mechanism also was postulated for the transgenic mouse model of MED expressing an internally deleted col9a1 gene as the severity of the phenotype correlated with the amount of transgene expression (41). It is not precisely known at which step in the synthesis of the type IX collagen trimer such a defective chain would exert a deleterious effect, e.g., during intracellular assembly in the RER, secretion, or incorporation into the extracellular matrix (67). The abnormal ultrastructure of the RER in the proband (Fig. 4 a--c) suggests that the earliest steps are affected. Further studies of type IX collagen and characterization of similar patients may yield insights into the pathophysiology of osteoarthritis and myopathy and suggest targets for therapeutic intervention.
Acknowledgments
We thank the family for their participation in this study. L.M.K. is an Investigator of the Howard Hughes Medical Institute. This work was supported by National Institutes of Health Grant P30-HD18655.
Abbreviations
- EDM
epiphyseal dysplasia multiplex
- MED
multiple epiphyseal dysplasia
- PSACH
pseudoachondroplasia
- COMP
cartilage oligomeric matrix protein
- lod
log10 odds ratio for linkage
- RER
rough endoplasmic reticulum
- NC
noncollagenous
References
- 1.Fairbank T. Br J Surg. 1947;34:225–232. doi: 10.1002/bjs.18003413502. [DOI] [PubMed] [Google Scholar]
- 2.Rimoin D L, Rasmussen I M, Briggs M D, Roughly P J, Gruber H E, Warman M L, Olsen B R, Hsia Y E, Yuen J, Reinker K, et al. Hum Genet. 1994;93:236–242. doi: 10.1007/BF00212015. [DOI] [PubMed] [Google Scholar]
- 3.Ribbing S. Acta Radiol Suppl. 1937;34:1–107. [Google Scholar]
- 4.International Working Group on Constitutional Disorders of Bone. Am J Med Genet. 1998;79:376–382. doi: 10.1002/(sici)1096-8628(19981012)79:5<376::aid-ajmg9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 5.Briggs M D, Hoffman S M, King L M, Olsen A S, Mohrenweiser H, Leroy J G, Mortier G R, Rimoin D L, Lachman R S, Gaines E S, et al. Nat Genet. 1995;10:330–336. doi: 10.1038/ng0795-330. [DOI] [PubMed] [Google Scholar]
- 6.Hecht J T, Nelson L D, Crowder E, Wang Y, Elder F F, Harrison W R, Francomano C A, Prange C K, Lennon G G, Deere M, et al. Nat Genet. 1995;10:325–329. doi: 10.1038/ng0795-325. [DOI] [PubMed] [Google Scholar]
- 7.Muragaki Y, Mariman E C, van Beersum S E, Perala M, van Mourik J B, Warman M L, Olsen B R, Hamel B C. Nat Genet. 1996;12:103–105. doi: 10.1038/ng0196-103. [DOI] [PubMed] [Google Scholar]
- 8.Holden P, Canty E G, Mortier G R, Zabel B, Spranger J, Carr A, Grant M E, Loughlin J A, Briggs M D. Am J Hum Genet. 1999;65:31–38. doi: 10.1086/302440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deere M, Blanton S H, Scott C I, Langer L O, Pauli R M, Hecht J T. Am J Hum Genet. 1995;56:698–704. [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs M D, Choi H, Warman M L, Loughlin J A, Wordsworth P, Sykes B C, Irven C M, Smith M, Wynne-Davies R, Lipson M H, et al. Am J Hum Genet. 1994;55:678–684. [PMC free article] [PubMed] [Google Scholar]
- 11.Oehlman R, Summerville G P, Yeh G, Weaver E J, Jiminez S A, Knowlton R G. Am J Hum Genet. 1994;54:3–10. [PMC free article] [PubMed] [Google Scholar]
- 12.Paassilta P, Lohiniva J, Annunen S, Bonaventure J, Le Merrer M, Pai L, Ala-Kokko L. Am J Hum Genet. 1999;64:1036–1044. doi: 10.1086/302328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuivaniemi H, Tromp G, Prockop D J. Hum Mutat. 1997;9:300–315. doi: 10.1002/(SICI)1098-1004(1997)9:4<300::AID-HUMU2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 14.Jöbsis G J, Keizers H, Vreijling J P, de Visser M, Speer M C, Woltermann R A, Baas F, Bolhuis P A. Nat Genet. 1996;14:113–115. doi: 10.1038/ng0996-113. [DOI] [PubMed] [Google Scholar]
- 15.Bethlem J, vanWijnaarden G K. Brain. 1976;99:91–100. [Google Scholar]
- 16.Kuo H J, Maslen C L, Keene D R, Glanville R W. J Biol Chem. 1997;272:26522–26529. doi: 10.1074/jbc.272.42.26522. [DOI] [PubMed] [Google Scholar]
- 17.Olsen B R. Int J Biochem Cell Biol. 1997;29:555–558. doi: 10.1016/s1357-2725(96)00100-8. [DOI] [PubMed] [Google Scholar]
- 18.van der Rest M, Mayne R. J Biol Chem. 1988;263:1615–1618. [PubMed] [Google Scholar]
- 19.Wu J J, Woods P E, Eyre D R. J Biol Chem. 1992;267:23007–23014. [PubMed] [Google Scholar]
- 20.Diab M, Wu J J, Eyre D R. Biochem J. 1996;314:327–332. doi: 10.1042/bj3140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eyre D R, Wu J J. J Rheumatol. 1995;43,Suppl.:82–85. [PubMed] [Google Scholar]
- 22.Vaughan L, Mendler M, Huber S, Bruckner P, Winterhalter K H, Irwin M I, Mayne R. J Cell Biol. 1988;106:991–997. doi: 10.1083/jcb.106.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konomi H, Seyer J M, Ninomiya Y, Olsen B R. J Biol Chem. 1986;261:6742–6746. [PubMed] [Google Scholar]
- 24.Nishimura I, Muragaki Y, Olsen B R. J Biol Chem. 1989;264:20033–20041. [PubMed] [Google Scholar]
- 25.Shapiro F. Calcif Tissue Int. 1992;51:324–331. doi: 10.1007/BF00334495. [DOI] [PubMed] [Google Scholar]
- 26.Diab M, Wu J J, Shapiro F, Eyre D. Am J Med Genet. 1994;49:402–409. doi: 10.1002/ajmg.1320490411. [DOI] [PubMed] [Google Scholar]
- 27.Dubowitz V. Pathological Changes in Muscle Biopsies. Eastbourne, U.K.: Baillière Tindall; 1985. pp. 86–90. [Google Scholar]
- 28.Lathrop G M, Lalouel J M, Julier C, Ott J. Proc Natl Acad Sci USA. 1984;81:3443–3446. doi: 10.1073/pnas.81.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J J, Eyre D R, Slayter S. Biochem J. 1987;248:373–381. doi: 10.1042/bj2480373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brooke M H, Engel W K. Neurology. 1969;19:591–605. doi: 10.1212/wnl.19.6.591. [DOI] [PubMed] [Google Scholar]
- 31.Banker B Q, Engel A G. In: Basic Reactions of Muscle. Engel A G, Franzini-Armstrong C, editors. Vol. 2. New York: McGraw–Hill; 1994. pp. 832–888. [Google Scholar]
- 32.Knowlton R G, Cekleniak J A, Cohn D H, Briggs M D, Hoffman S M G, Brandriff B F, Olsen A S. Genomics. 1995;28:513–519. doi: 10.1006/geno.1995.1183. [DOI] [PubMed] [Google Scholar]
- 33.Warman M L, Tiller G E, Polumbo P A, Seldin M F, Rochelle J M, Knoll J H, Cheng S D, Olsen B R. Genomics. 1993;17:694–698. doi: 10.1006/geno.1993.1391. [DOI] [PubMed] [Google Scholar]
- 34.Brewton R G, Wood B M, Ren Z X, Gong Y, Tiller G E, Warman M L, Lee B, Horton W A, Olsen B R, Baker J R, et al. Genomics. 1995;30:329–336. doi: 10.1006/geno.1995.9870. [DOI] [PubMed] [Google Scholar]
- 35.Perälä M, Elima K, Metsaranta M, Rosati R, de Crombrugghe B, Vuorio E. J Biol Chem. 1994;269:5064–5071. [PubMed] [Google Scholar]
- 36.Pihlajamaa T, Vuoristo M, Annunen S, Perälä M, Prockop D J, Ala-Kokko L. Matrix Biol. 1998;17:237–241. doi: 10.1016/s0945-053x(98)90063-4. [DOI] [PubMed] [Google Scholar]
- 37.Padget R A, Grabowski P J, Konarska M M, Seiler S, Sharp P A. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- 38.Shapiro M B, Senapathy P. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krawczak M, Reiss J, Cooper D N. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 40.van Mourik J B A, Hamel B C J, Mariman E C M. Am J Med Genet. 1998;77:234–240. doi: 10.1002/(sici)1096-8628(19980518)77:3<234::aid-ajmg9>3.3.co;2-#. [DOI] [PubMed] [Google Scholar]
- 41.Nakata K, Ono K, Miyazaki J, Olsen B R, Muragaki Y, Adachi E, Yamamura K, Kimura T. Proc Natl Acad Sci USA. 1993;90:2870–2874. doi: 10.1073/pnas.90.7.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fässler R, Schnegelsberg P N, Dausman J, Shinya T, Muragaki Y, McCarthy M T, Olsen B R, Jaenisch R. Proc Natl Acad Sci USA. 1994;91:5070–5074. doi: 10.1073/pnas.91.11.5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bashir R, Britton S, Strachan T, Keers S, Vafiadaki E, Lako M, Richard I, Marchand S, Bourg N, Argov Z, et al. Nat Genet. 1998;20:37–42. doi: 10.1038/1689. [DOI] [PubMed] [Google Scholar]
- 44.Liu J, Aoki M, Illa I, Wu C, Fardeau M, Angelini C, Serrano C, Urtizberea J A, Hentati F, Ben Hamida M, et al. Nat Genet. 1998;20:31–36. doi: 10.1038/1682. [DOI] [PubMed] [Google Scholar]
- 45.Dubowitz V. The Muscular Dystrophies. London: Baillière Tindall; 1985. pp. 289–404. [Google Scholar]
- 46.Ring C, Hassell J, Halfter W. Dev Biol. 1996;180:41–53. doi: 10.1006/dbio.1996.0283. [DOI] [PubMed] [Google Scholar]
- 47.Hayashi M, Hayashi K, Iyama K, Trelstad R L, Linsenmayer T F, Mayne R. Dev Dyn. 1992;194:169–176. doi: 10.1002/aja.1001940302. [DOI] [PubMed] [Google Scholar]
- 48.Swiderski R E, Solursh M. Dev Dyn. 1992;194:118–127. doi: 10.1002/aja.1001940205. [DOI] [PubMed] [Google Scholar]
- 49.Fitch J M, Mentzer A, Mayne R, Linsenmayer T F. Development (Cambridge, UK) 1989;105:85–95. doi: 10.1242/dev.105.1.85. [DOI] [PubMed] [Google Scholar]
- 50.Kosher R A, Solursh M. Dev Biol. 1989;131:558–566. doi: 10.1016/s0012-1606(89)80026-0. [DOI] [PubMed] [Google Scholar]
- 51.Slepecky N B, Savage J E, Yoo T J. Acta Oto-Laryngol. 1992;112:611–617. doi: 10.3109/00016489209137449. [DOI] [PubMed] [Google Scholar]
- 52.Liu C-Y, Olsen B R, Kao W W-Y. Dev Dyn. 1993;198:150–157. doi: 10.1002/aja.1001980208. [DOI] [PubMed] [Google Scholar]
- 53.Perälä M, Savontaus M, Metsaranta M, Vuorio E. Biochem J. 1997;324:209–216. doi: 10.1042/bj3240209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sagarriga Visconti C, Kavalkovich K, Wu J, Niyibizi C. Arch Biochem Biophys. 1996;328:135–142. doi: 10.1006/abbi.1996.0153. [DOI] [PubMed] [Google Scholar]
- 55.Hayashi Y K, Chou F-L, Engvall E, Ogawa M, Matsuda C, Hirabayashi S, Yokochi K, Ziober B L, Kramer R H, Kaufman S J, et al. Nat Genet. 1998;19:94–97. doi: 10.1038/ng0598-94. [DOI] [PubMed] [Google Scholar]
- 56.Mayer U, Saher G, Fassler R, Bornemann A, Echtermeyer F, von der Mark H, Miosge N, Poschl E, von der Mark K. Nat Genet. 1997;17:318–323. doi: 10.1038/ng1197-318. [DOI] [PubMed] [Google Scholar]
- 57.Stanescu R, Stanescu V, Muriel M-P, Maroteaux P. Am J Med Genet. 1993;45:501–507. doi: 10.1002/ajmg.1320450420. [DOI] [PubMed] [Google Scholar]
- 58.Cooper R R, Ponseti I V, Maynard J A. J Bone Jt Surg. 1973;55:475–484. [PubMed] [Google Scholar]
- 59.Cohn D H, Briggs M D, King L M, Rimoin D L, Wilcox W R, Lachman R S, Knowlton R G. Ann NY Acad Sci. 1996;785:188–194. doi: 10.1111/j.1749-6632.1996.tb56258.x. [DOI] [PubMed] [Google Scholar]
- 60.Horton W A, Campbell D, Machado M A, Aulthouse A L, Ahmed S, Ellard J T. Am J Med Genet. 1989;34:91–94. doi: 10.1002/ajmg.1320340116. [DOI] [PubMed] [Google Scholar]
- 61.Engel J, Prockop D J. Annu Rev Biophys Biophys Chem. 1991;20:137–152. doi: 10.1146/annurev.bb.20.060191.001033. [DOI] [PubMed] [Google Scholar]
- 62.van Mourik J B A, Buma P, Wilcox W R. Ultrastruct Pathol. 1998;22:249–251. doi: 10.3109/01913129809033476. [DOI] [PubMed] [Google Scholar]
- 63.Bader D L, Kempson G E, Egan J, Gilbey W, Barrett A J. Biochim Biophys Acta. 1992;1116:147–154. doi: 10.1016/0304-4165(92)90111-7. [DOI] [PubMed] [Google Scholar]
- 64.Diab M. Orthop Rev. 1993;22:165–170. [PubMed] [Google Scholar]
- 65.Bruckner P, van der Rest M. Microsc Res Tech. 1994;28:378–384. doi: 10.1002/jemt.1070280504. [DOI] [PubMed] [Google Scholar]
- 66.Hagg R, Hedbom E, Mollers U, Aszodi A, Fassler R, Bruckner P. J Biol Chem. 1997;272:20650–20654. doi: 10.1074/jbc.272.33.20650. [DOI] [PubMed] [Google Scholar]
- 67.Byers P H. In: Disorders of Collagen Biosynthesis and Structure. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. Vol. 3. New York: McGraw–Hill; 1995. pp. 4029–4077. [Google Scholar]