Abstract
The major virulence factors of toxigenic Vibrio cholerae are cholera toxin, which is encoded by a lysogenic filamentous bacteriophage (CTXΦ), and toxin-coregulated pilus (TCP), an essential colonization factor that is also the receptor for CTXΦ. The genes involved in the biosynthesis of TCP reside in a pathogenicity island, which has been reported to correspond to the genome of another filamentous phage (designated VPIΦ) and to encode functions necessary for the production of infectious VPIΦ particles. We examined 46 V. cholerae strains having diverse origins and carrying different genetic variants of the TCP island for the production of the VPIΦ and CTXΦ in different culture conditions, including induction of prophages with mitomycin C and UV irradiation. Although 9 of 10 V. cholerae O139 strains and 12 of 15 toxigenic El Tor strains tested produced extracellular CTXΦ, none of the 46 TCP-positive strains produced detectable VPIΦ in repeated assays, which detected as few as 10 particles of a control CTX phage per ml. These results contradict the previous report regarding VPIΦ-mediated horizontal transfer of the TCP genes and suggest that the TCP island is unable to support the production of phage particles. Further studies are necessary to understand the mechanism of horizontal transfer of the TCP island.
Cholera is a severe dehydrating diarrhea caused by toxigenic strains of the gram-negative bacterium Vibrio cholerae. The profuse watery diarrhea is mainly due to an enterotoxin (cholera toxin [CT]) produced by V. cholerae (10, 22) when it colonizes the small intestine. Colonization of the brush border of the small intestine is assumed to be mediated by a rigid pilus colonization factor designated the toxin-coregulated pilus (TCP), the expression of which is coordinately regulated with expression of cholera toxin (20, 45, 46).
In V. cholerae, the major virulence genes are in clusters. The ctxAB genes encoding cholera toxin reside in the genome of a lysogenic filamentous phage (CTXΦ) (47), whereas the genes encoding the major colonization factor, TCP, are encoded by the TCP gene cluster or island (7, 30, 34, 40, 43, 44). Since the genes of the TCP cluster are not present in many strains of V. cholerae, this region was clearly recently acquired by V. cholerae through horizontal gene transfer (30, 34, 43, 44).
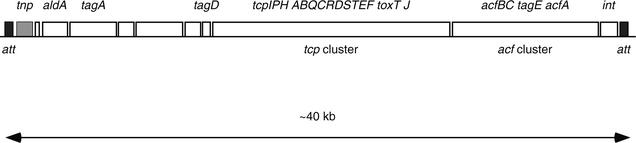
The major subunit of TCP is encoded by the tcpA gene, but pilus formation and function require the products of several other genes located on the chromosome adjacent to the tcpA gene. Immediately adjacent to and downstream of the tcp cluster is the acf gene cluster (8), which is assumed to encode an accessory colonization factor, although the exact nature of this colonization factor is unclear. A putative integrase gene (int) is located downstream of and adjacent to the tcp-acf gene cluster (18, 30). A nearly 40-kb region, including the tcp-acf gene cluster, is flanked on both sides by putative 20-bp att-like attachment sequences (Fig. 1) (26, 30). This structural feature, together with the cluster's association with pathogenic strains of V. cholerae, prompted Karaolis and colleagues to rename the region the Vibrio pathogenicity island (VPI) (26). The VPI is absent from most nontoxigenic environmental strains, but it has recently been found in a variety of environmental strains (26, 36-38, 44).
FIG. 1.
Genetic organization of the TCP island of V. cholerae. att sites are at both ends, and the putative integrase gene is near the right end of the island. The transposase sequence is near the left end of the island (grey box). To improve clarity, the att sites are not drawn to scale.
Remarkably, the cholera toxin-converting bacteriophage CTXΦ uses TCP as its receptor for infecting cells (47). Since CTXΦ uses TCP as its receptor, it appears that acquisition of the TCP island is the initial genetic element acquisition required for origination of epidemic strains. However, the mechanism of horizontal transfer of the TCP island is controversial (34, 43, 47). In a previous report the authors suggested that the TCP island corresponds to the genome of another filamentous phage, designated VPIΦ (25). However, this finding has not been verified independently and has not been supported by any other investigation. In the present study, we reexamined V. cholerae strains having diverse origins and carrying genetic variants of the TCP island for the production of VPIΦ. Our results do not support the conclusion that VPIΦ particles are produced by TCP island-positive V. cholerae.
MATERIALS AND METHODS
Bacterial strains.
A total of 46 TCP island-positive V. cholerae strains were analyzed in the present study. These strains included 6 classical biotype strains and 18 El Tor biotype strains of V. cholerae O1, 10 strains of V. cholerae O139, and 12 V. cholerae strains belonging to six different non-O1, non-O139 serogroups. The clinical O1 and O139 strains were either obtained from our culture collection or freshly isolated from diarrhea patients at the treatment center of the International Centre for Diarrheal Disease Research, Bangladesh (ICDDR,B) located in Dhaka, Bangladesh. Environmental isolates of O1 and O139 vibrios were isolated from surface water samples collected in Dhaka, whereas the non-O1, non-O139 strains were initially cultured from three different freshwater lakes and ponds in the eastern part of Calcutta, India (3). These strains carry genetic variants of the TCP island with new alleles of several TCP island genes, including tcpA and toxT (36). The strains analyzed in this study are described in Table 1. Strains were stored either in lyophilized form or in sealed deep nutrient agar at room temperature. Before use, the identities of the cultures were verified by biochemical and serological methods (48). All strains were also tested for the presence of the ctx and tcp gene clusters by using specific DNA probes or PCR assays as described below.
TABLE 1.
Analysis of 46 strains of TCP-positive V. cholerae belonging to different serogroups for production of CTXΦ and VPIΦ particles
| Strain(s) | No. of strains analyzed | Serogroup and/or biotype | Presence of genes
|
Production of phages in supernatantb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| tcpAa | tcpI | toxTa | acfB | rstR | ctxA | zot | VPIϕ | CTXΦc | |||
| Clinical isolates | |||||||||||
| S-224, S-262, L-351, L-262, O395, AE-2882 | 6 | O1 classical | C | + | + | + | + | + | + | − | − |
| SA317, SA406, SA508 | 3 | O1 El Tor | E | + | + | + | − | − | − | − | − |
| G-755, N-16961, P-27457, AM-13860, AM-11726, AF-1471, AP-13547, AP-13530, AM-11706 | 9 | O1 El Tor | E | + | + | + | + | + | + | − | + |
| P-28108, P-27821, AL-12890 | 3 | O1 El Tor | E | + | + | + | + | + | + | − | − |
| AI-1836 | 1 | O139 | E | + | + | + | + | + | + | − | − |
| AL-11089, AP-32541, AK-17334, AR-1432, AR-1468, AL-12077 | 6 | O139 | E | + | + | + | + | + | + | − | + |
| Surface water isolates | |||||||||||
| EV-02, EV-11, 102V138 | 3 | O1 El Tor | E | + | + | + | + | + | + | − | + |
| SB-Lake, 224V541, Env-91 | 3 | O139 | E | + | + | + | + | + | + | − | + |
| SCE4, SCE359 | 2 | O8 | C | + | + | + | − | − | − | − | − |
| SCE223, SCE354 | 2 | O27 | Env | + | + | + | + | + | + | − | − |
| SCE225, SCE226, SCE227 | 3 | O35 | C | + | Env | + | + | − | − | − | |
| SCE256, SCE264 | 2 | O42 | C | + | Env | + | + | − | − | − | − |
| SCE263 | 1 | O10 | C | + | Env | + | + | + | − | − | − |
| SCE340, SCE341 | 2 | O69 | E | + | Env | − | − | − | − | − | − |
Env,+, present; −, not present; new alleles of the tcpA or toxT genes, which are different from those normally found in epidemic strains; C, classical tcpA type gene present; E, El Tor type tcpA gene present.
Most strains carried other unidentified phage genomes, as shown by analysis of the total nucleic acids extracted from phage preparations of the strains, but these nucleic acids did not hybridize with probes for CTXΦ or VPIΦ. Strains were grown in the presence of mitomycin C (20 ng /ml) for phage induction (see text for details).
CTXΦ was produced irrespective of whether strains were grown at 30 or 37°C.
Induction of lysogens.
V. cholerae strains were either treated with mitomycin C or subjected to UV irradiation to induce possible prophages carried by the strains. Strains were grown in Luria-Bertani broth (LB) at 30°C to an absorbance at 540 nm of 0.2. The cells were collected by centrifugation, washed, and resuspended in fresh LB. Each suspension was divided into aliquots to which mitomycin C (Sigma Chemical Company, St. Louis, Mo.) was added at a concentration of 20 ng/ml. To analyze induction by UV irradiation, 2-ml aliquots of cultures containing 2 × 105 cells were spread on a UV-transparent tray, placed on a UV transilluminator (model T-2201; Sigma), and exposed for 2 to 3 min. The treated cells were washed with LB and inoculated into test flasks containing 500 ml of fresh LB. Different sets of cultures were grown at 30 and 37°C and for 6 and 16 h (overnight) with shaking. The culture supernatants were analyzed for extracellular phage particles as described below. Strains grown in a similar way but without mitomycin C or UV irradiation were also analyzed for phage production.
Isolation of phage DNA.
Phage preparations were made by using 500-ml LB cultures and the method described by Karaolis et al. (25), and total nucleic acids were extracted from these preparations. In brief, cultures were centrifuged at 10,000 × g, and the supernatants were sterilized by filtration. To confirm that the filtrates from the culture supernatants did not contain bacterial cells, aliquots of the filtrates were streaked on Luria agar plates and incubated overnight at 37°C. Pancreatic DNase I (100 U/ml) and RNase A (50 μg/ml) were added to the filtrates and incubated at room temperature for 3 h. NaCl and polyethylene glycol 8000 were added to final concentrations of 1 M and 10% (wt/vol), respectively, and the mixtures were left to precipitate overnight at 4°C. The supernatants were centrifuged at 11,000 × g for 20 min, and the pellets were resuspended in 2 ml of SM buffer. Each solution was extracted twice with an equal volume of phenol-chloroform and then once with chloroform to disrupt possible phage particles, and the total nucleic acid was precipitated with ethanol. The pellet was washed with 80% ethanol, vacuum dried, and resuspended in a final volume of 50 μl. Preparations containing phage DNA were identified by PCR and Southern blot hybridization. For each PCR assay, 2 to 5 μl of a solution was used, whereas 10 μl was used for Southern blot analysis. Serial dilutions in LB of CTX-KmΦ (10 to 104 particles/ml) isolated from a mitomycin C-induced culture of strain SM44 (10) were used as positive controls for the phage assay. CTX-KmΦ derived from SM44 was quantified as described previously by incubating serially diluted filter-sterilized culture supernatants with strain RV508 and then determining the number of Kmr colonies (11, 47).
Probes and PCR assays.
The gene probes used in this study to analyze different V. cholerae strains included a 0.5-kb EcoRI fragment of pCVD27 encoding part of the ctxA gene (23) and an 840-bp region internal to the zot gene, amplified by PCR from the recombinant plasmid pBB241 as described previously (14). The El Tor and classical biotype-specific rstR probes were SacI-XbaI fragments of pHK1 and pHK2, respectively (28). PCR assays specific for the tcpA, tcpI,acfB, and toxT genes of the TCP pathogenicity island, as well as the ctxA and zot genes of CTX prophage, have been described previously (3, 9, 14, 15, 27). Phage nucleic acid preparations were tested both by PCR assays and by Southern blot hybridization with specific probes. The multiplex PCR assay for classical or El Tor biotype-specific tcpA genes (27) was used to detect the presence of possible VPIΦ genomes. To detect possible VPIΦ produced by environmental non-O1, non-O139 strains carrying the env variant of tcpA, a PCR assay with primers flanking tcpA (tcpI-F [5′GCCGTCTCCGCATTAAGCTCTGCAC] and tcpQ-R [5′ GAGGACTGTTCTGCAATCTGCTCAT]) was performed as described previously (36). The 5.4-kb PCR-generated amplicon corresponding to the tcpI-Q region and the tcpA amplicon were also used as probes to screen for possible VPIΦ by Southern blot hybridization of phage nucleic acids. For PCR-based detection of the CTXΦ genome the ctxA PCR was used, whereas for detection of the CTX-KmΦ genome a PCR assay for the zot gene was used (14). A strand-specific oligonucleotide probe (5′TCTATCTCTGTAGCCCCTATTACG) was also used to specifically identify the (+) strand of the single-stranded genome of CTXΦ.
Colony or Southern blots were prepared by using nylon filters (Hybond; Amersham Biosciences, Uppsala, Sweden) and were processed by standard methods (33). The polynucleotide probes were labeled by random priming (16) by using a random primer DNA labeling kit (Bethesda Research Laboratories, Gaithersburg, Md.) and [α-32P]dCTP (3,000 Ci/mmol; Amersham). Oligonucleotide probes were labeled by 3′ tailing by using terminal deoxynucleotide transferase and [α-32P]dCTP (Amersham). Southern blots and colony blots were hybridized with the labeled probes and autoradiographed as described previously (11).
Construction of Kmr-oriR6K insertions in the TCP island.
Internal fragments of tcpI (VC0825), int (VC0847; encodes a putative integrase), tnp (VC0817; encodes a putative transposase), and aldA (VC0819; encodes an aldehyde dehydrogenase) were cloned into pNJ16, which contains a Kmr gene and an origin of replication (oriR6K) that is not functional in Vibrio (21). The resulting plasmids were inserted into the chromosome of El Tor strain N16961 or classical strain O395 by homologous recombination.
Infant mouse infections.
Infant mouse colonization assays were performed as previously described (17). In brief, 6-day-old infant CD-1 mice were inoculated intragastrically with a mixture of overnight cultures of N16961 tcpI::Kmr-oriR6K (Kmr Smr) and Vc857 (Nalr TCP−) at a ratio of approximately 1:5. After 24 h of incubation, the mice were sacrificed, and the small intestines were removed and homogenized. The homogenates were plated on Luria agar with appropriate antibiotics to determine the number of kanamycin-resistant N16961 or Vc857 bacteria present.
RESULTS AND DISCUSSION
The only previous study (25) to demonstrate production of VPIΦ and transfer of the TCP island to a recipient strain raised a number of intriguing questions regarding the biology and ecology of VPIΦ (31). However, there has been no other confirmation of the existence of VPIΦ, despite the important implications of the discovery of VPIΦ for understanding the role of vibriophages in the emergence and evolution of epidemic V. cholerae strains. We therefore decided to reexamine the ability of diverse V. cholerae strains to produce VPIΦ by using the same culture conditions that were used in the original report describing VPIΦ (25). Of the 46 V. cholerae strains analyzed in this study, 44 were positive in PCR assays for the tcpA, tcpI, toxT, and acfB genes, indicating that these genes and presumably the entire TCP island are present in these strains (Table 1). Two strains were negative for the acfB gene but were positive for the tcpA, tcpI, and toxT genes. Except for three El Tor biotype strains and nine non-O1, non-O139 strains, all other strains were also positive for the CTX prophage, as shown by the presence of the ctxA and zot genes.
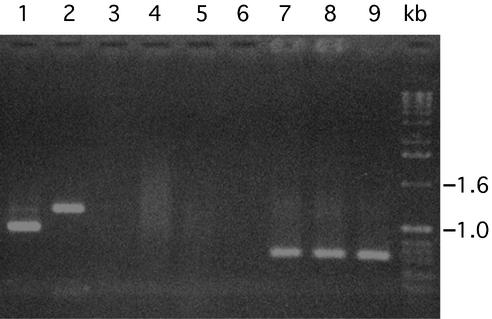
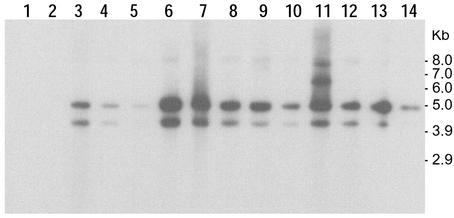
PCR analysis of total nucleic acid extracted from phage preparations indicated that all phage preparations were negative for the tcpA gene. Southern blot analysis of phage nucleic acid with probes for different TCP island genes was also negative (data not shown). The same nucleic acid extracts were tested for the CTXΦ-specific gene ctxA by using both PCR assays and DNA probe assays; representative data obtained in these experiments are presented in Fig. 2 and 3. Phage preparations from all toxigenic classical biotype strains were negative for the CTXΦ genome, while phage preparations from 12 of 15 toxigenic El Tor strains and 9 of 10 O139 strains were positive for CTXΦ production. These findings are consistent with previous studies, which showed that the classical biotype strains of V. cholerae O1 do not produce infectious CTXΦ, which has been attributed to a dysfunctional CTX prophage (6). It should be emphasized that the ctxA hybridizing sequences or the PCR templates that gave positive PCR results for ctxA were evidently present in phage particles, since these DNAs should have been the only DNAs protected from nuclease digestion during phage preparation. The same preparations did not give positive reactions in the PCR assay for tcpA. Thus, in the present study, the presence of CTXΦ and the absence of VPIΦ served as reciprocal controls and ruled out the possibility of chromosomal DNA contamination of the phage nucleic acid preparations.
FIG. 2.
PCR analysis of phage nucleic acid preparations of V. cholerae strains carrying the TCP island. Preparations from various strains were tested by PCR for tcpA (lanes 4 to 6) and ctxA (lanes 7 to 9). The following V. cholerae strains were tested: lanes 4 and 7, AL-11089; lanes 5 and 8, AL-12077; lanes 6 and 9, P-27457. As positive controls, lanes 1 and 2 contained PCR assay mixtures from assays for tcpA in which chromosomal DNA templates from El Tor strain G-755 and classical strain 569B, respectively, were used. Lane 3 contained the tcpA PCR product of the chromosomal DNA template from strain Env-37, which lacks the tcpA gene. The numbers on the right indicate the positions of bands derived from a 1-kb DNA ladder (Invitrogen) used as molecular size markers.
FIG. 3.
Southern blot hybridization analysis of total phage nucleic acids derived from different V. cholerae strains by using a ctxA gene probe. The V. cholerae strains used for phage preparation were AL-12890, P-28108, AM-11726, AM-11706, EV-02, AL-11089, AL-12077, P-27457, N-16961, AP-32541, AF-1471, AR-1402, AR-1432, and G-755 (lanes 1 through 14, respectively). The numbers on the right indicate the positions of bands derived from a supercoiled DNA ladder (Invitrogen) used as molecular size markers.
In the case of LB mixed with exogenous CTX-KmΦ and used as a positive control, a minimum of 5 × 104 phage particles added to 500 ml of medium (102 particles/ml) could be distinctly detected by Southern blot hybridization of 10 μl of the ultimate phage nucleic acid preparation by using a zot probe. Positive PCR results were obtained with less than 1 μl of the phage nucleic acid preparation (data not shown), which is theoretically equivalent to detecting the phage in a solution containing fewer than 10 particles/ml.
DNA-damaging agents, such as mitomycin C and UV irradiation, are known to induce lysogenic phages. We therefore used these agents to encourage phage production. Despite treatment with either mitomycin C or UV irradiation, none of the strains produced VPIΦ. The production of CTXΦ, however, was markedly influenced by both mitomycin C and UV irradiation. In the absence of any induction, only 5 strains produced CTXΦ, but with induction 21 of 25 toxigenic O1 El Tor and O139 strains produced extracellular CTXΦ particles. In view of the report that hemagglutinin protease produced during the late log phase of a culture can inactivate CTXΦ (29), in the present study we included precautions to ensure that any VPIΦ present in the culture supernatants was not degraded by proteases (thus escaping detection). We therefore tested culture supernatants for the presence of phage after 6 h of growth, as well as after 16 h of growth. It should be noted that the presence of CTXΦ served as an indicator, and since CTXΦ was clearly detected in cultures grown under either of these conditions, it is highly unlikely that VPIΦ was produced and degraded by extracellular proteases produced by the bacteria.
Another method for detecting phage is to monitor the transfer of genetic material by transduction. This method is commonly used for detecting and quantifying CTXΦ transfer (47). We therefore constructed several Kmr-marked versions of the TCP island and examined their ability to move into Escherichia coli or TCP-negative V. cholerae strains. Kmr-oriR6K was inserted into tcpI, aldA, tnp, and int of N16961 and O395, and the resulting strains were used as donors for transduction of E. coli DH5αλpir or Vc857, a V. cholerae environmental isolate that lacks the entire TCP island. No TCP island transfer was detected by standard conjugation and transduction techniques (data not shown). We repeated these assays under various conditions, such as toxT overproduction (which induces expression of most TCP genes), overproduction of the putative phage integrase, starvation, or conditions favoring biofilm formation. Again, not a single TCP transfer event was detected.
It has been reported that the efficiency of CTXΦ transduction in El Tor strains is greatly increased during host infection (47). We therefore coinoculated N16961 tcpI::Km and the recipient strain Vc857 into 6-day-old infant mice and recovered bacteria after 24 h of incubation. For all six mice examined, we did not recover a single Kmr Vc857 bacterium, while an average of 2.4 × 106 N16961 tcpI::Km bacteria were recovered per mouse. Therefore, we concluded that TCP islands are not transferred between bacteria under such in vivo conditions.
The putative VPIΦ is presumed to have a TCP-like morphology and to be composed of TcpA subunits, but expression of TCP is known to vary under different growth conditions (35). We therefore used two different temperatures (30 and 37°C) to examine the production of VPIΦ. Karaolis et al. observed VPIΦ production at 30°C in the classical biotype strain O395 and at 37°C in the El Tor biotype strain N16961 (25). We included both these strains in our study, but in our assays neither of these strains was positive for the production of VPIΦ, although strain N16961 produced CTXΦ under all conditions tested (Table 1).
Karaolis et al. reported that classical strain O395 produced VPIΦ, as shown by PCR detection of VPI DNA within phage particles that were produced at 30°C and that were immunoprecipitable with anti-TCP antibody (25). However, these particles appeared to be morphologically identical to the TCP bundles that are known to be produced by O395 (45). TCP production in O395 is tightly linked to autoagglutination (4, 45). Phage preparations that we derived from highly autoagglutinated cultures of O395 grown at 30°C consistently failed to show the presence of VPI DNA, as assessed by PCR or Southern blotting, nor did these phage preparations transduce the TCP island. We therefore concluded that the TCP made by O395 do not correspond to VPI phage particles.
In El Tor strains like N16961, TCP production occurs at a low level in vitro, but it can be detected by CTXΦ transduction either in vitro under the conditions described here or in vivo (13, 47). This supports the hypothesis that TCP were indeed expressed by the El Tor strains but that they also did not correspond to VPIΦ as assessed by PCR, Southern blotting, or (in the case of N16961) the ability to transduce the TCP island.
Since our marked TCP islands carried the aphA-3 gene at locations different from that used by Karaolis et al. (25), there is a formal possibility that our insertions might have inactivated the phage. However, since our PCR and hybridization analyses of phage DNA preparations from unmarked wild-type strains did not detect VPIΦ either, we believe that our inability to detect VPIΦ or to mobilize the TCP island was not due to insertional inactivation of phage function.
Many V. cholerae strains are also lysogenized with a phage designated phage K139 (42). To determine whether this phage can facilitate transduction of the TCP island, we constructed donor strains containing K139 phage (marked with Apr in the glo gene) and Kmr-oriR6K inserted at various locations within the tcp region and used them as donor strains. K139 phage were transferred at high frequencies to recipients RV79 and Vc857 (10−1 and 10−4, respectively), while TCP markers were not transferred at all (Table 2). Since it is possible that the recipients that we tested were not able to receive the TCP island, we also examined phage preparations directly by PCR. As shown in Fig. 4, the phage K139 gene glo was readily detected in the phage preparation, while the tcpJ gene was not. VC0815, which lies adjacent to the tcp genes but still within the TCP island, also was not detected in phage preparations. These data further indicated that under these conditions the TCP island is not transferred and is not detectable in a phage preparation.
TABLE 2.
Transduction efficiency of phage K139 and the TCP island
| Supernatanta | Transduction efficiency with the following recipients:
|
|||
|---|---|---|---|---|
| RV79 (TCP+)
|
Vc857 (TCP−)b
|
|||
| Apr | Kmr | Apr | Kmr | |
| N16961ΩK139 (glo::bla), tcp::Kmc | 5 × 10−1 | <1 × 10−6d | 5 × 10−4 | <1 × 10−6d |
| O395ΩK139 (glo::bla), tcp::Kmc | 6 × 10−1 | <1 × 10−6d | 5 × 10−4 | <1 × 10−6d |
There was 106 PFU of phage K139 per ml in each supernatant.
A number of TCP− strains were tested as recipients.
Constructs with Kmr-oriR6K inserted in tcpI, aldA, tnp, and int were tested.
No Kmr colonies were found in 106 recipient cells.
FIG. 4.
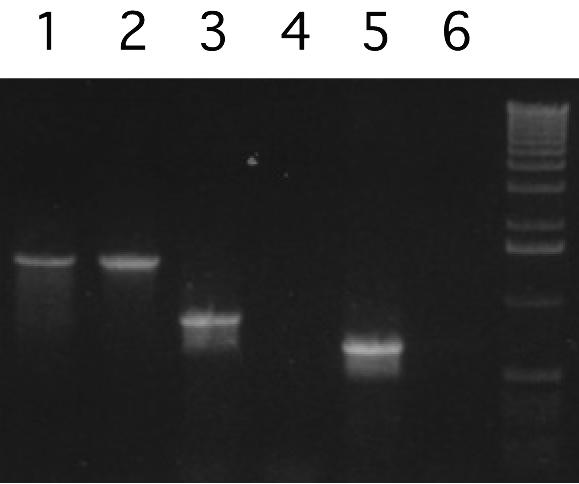
PCR detection of genes in K139 phage and genes in and adjacent to the TCP island. Whole cells were used as templates in lanes 1, 3, and 5, while phage preparations were used in lanes 2, 4, and 6. Primers for the K139 gene glo were used in lanes 1 and 2; primers for tcpJ were used in lanes 3 and 4; and primers for VC0815 were used in lanes 5 and 6.
A recent study found that genetic variants of the TCP island were carried by a number of environmental V. cholerae strains belonging to nonepidemic serogroups (1, 24, 36-38). Since environmental strains of V. cholerae are assumed to have evolved into pathogenic clones by acquisition of virulence genes, the TCP gene clusters in these environmental strains could be progenitors of the TCP island found in pathogenic clones and therefore genetically more closely related to an ancestral TCP phage that might have invaded a TCP-negative strain and converted it to a TCP-positive strain. Using this assumption, we tested environmental non-O1, non-O139 strains for production of VPIΦ. However, analysis of 12 strains, including 2 strains carrying a new allele of the tcpA gene, 8 strains carrying a new allele of the toxT gene, and 2 strains carrying conventional tcpA and toxT genes, showed that none of these strains produced VPIΦ (Table 1).
In summary, although CTXΦ is produced by most toxigenic strains, none of the TCP island-positive strains analyzed here produced the putative VPIΦ. The detection limit of the PCR assays was less than 10 phage particles/ml. Assuming that 1 ml of an overnight culture contained ∼109 bacterial cells, the phage would have been detectable even if 1 phage particle was produced per 108 cells. Since Karaolis et al. reported that VPIΦ donor strains produced more than 104 transducing particles per 3 × 107 donor cells, we estimated that our level of sensitivity was more than 106 times that needed to detect VPIΦ. Therefore, the results of the present study contradict the previous report demonstrating the production of VPIΦ and emphasize that further studies are needed to understand the mechanism involved in horizontal transfer of the TCP island.
The mechanism that significantly limits distribution of TCP genes to mostly strains of the O1 and O139 serogroups out of the more than 200 serogroups of V. cholerae should also be investigated. In a recent study, O'Shea and Boyd (41) demonstrated that the VPI was transferred to a recipient strain by the generalized transducing phage CP-T1 (19, 39). Since these authors observed transfer of the VPI only at low frequencies (10−8 to 10−9 per PFU), we concluded that the mechanism is simply generalized transduction of a chromosomal marker rather than a specific, VPI-mediated process. Therefore, we favor the hypothesis that the VPI corresponds to a satellite element similar to TSST element (32), P4 (2), or RS1 (5, 12). Such elements can be packaged efficiently into phage particles by a helper phage but are not viable phages themselves. We are currently exploring this hypothesis by isolating diverse vibriophages and testing them for efficient packaging and transfer of the VPI.
Acknowledgments
This research was funded in part by the United States Agency for International Development under grant HRN-5986-A-00-6005-00 with the ICDDR,B. The ICDDR,B is supported by countries and agencies that share its concern for the health problems of developing countries. Current donors providing unrestricted support include the aid agencies of the governments of Australia, Bangladesh, Belgium, Canada, Japan, the Kingdom of Saudi Arabia, The Netherlands, Sweden, Sri Lanka, Switzerland, and the United States. Work in the laboratory of J.J.M. was supported by National Institutes of Health grant AI18045 (to J.J.M.) and by a National Research Service Award (to J.Z.).
We thank Matthew Waldor (New England Medical Center, Boston, Mass.) for providing the rstR gene probes and Su Chiang for critiquing and editing the manuscript.
Editor: V. J. DiRita
REFERENCES
- 1.Boyd, E. F., and M. K. Waldor. 2002. Evolutionary and functional analyses of variants of the toxin- coregulated pilus protein TcpA from toxigenic Vibrio cholerae non-O1/non-O139 serogroup isolates. Microbiology 148:1655-1666. [DOI] [PubMed] [Google Scholar]
- 2.Briani, F., G. Deho, F. Forti, and D. Ghisotti. 2001. The plasmid status of satellite bacteriophage P4. Plasmid 45:1-17. [DOI] [PubMed] [Google Scholar]
- 3.Chakraborty, S., A. K. Mukhopadhyay, R. K. Bhadra, A. N. Ghosh, R. Mitra, T. Shimada, S. Yamasaki, S. M. Faruque, Y. Takeda, R. R. Colwell, and G. B. Nair. 2000. Virulence genes in environmental strains of Vibrio cholerae. Appl. Environ. Microbiol. 66:4022-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang, S. L., R. K. Taylor, M. Koomey, and J. J. Mekalanos. 1995. Single amino acid substitutions in the N-terminus of Vibrio cholerae TcpA affect colonization, autoagglutination, and serum resistance. Mol. Microbiol. 17:1133-1142. [DOI] [PubMed] [Google Scholar]
- 5.Davis, B. M., H. H. Kimsey, A. V. Kane, and M. K. Waldor. 2002. A satellite phage-encoded antirepressor induces repressor aggregation and cholera toxin gene transfer. EMBO J. 21:4240-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, B. M., K. E. Moyer, E. F. Boyd, and M. K. Waldor. 2000. CTX prophages in classical biotype Vibrio cholerae: functional phage genes but dysfunctional phage genomes. J. Bacteriol. 182:6992-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everiss, K. D., K. J. Hughes, M. E. Kovach, and K. M. Peterson. 1994. The Vibrio cholerae acfB colonization determinant encodes an inner membrane protein that is related to a family of signal-transducing proteins. Infect. Immun. 62:3289-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everiss, K. D., K. J. Hughes, and K. M. Peterson. 1996. The accessory colonization factor and the toxin-coregulated pilus gene clusters are physically linked in the Vibrio cholerae O395 genome. DNA Sequence 5:51-55. [DOI] [PubMed] [Google Scholar]
- 9.Faruque, S. M., K. M. Ahmed, A. R. Abdul Alim, F. Qadri, A. K. Siddique, and M. J. Albert. 1997. Emergence of a new clone of toxigenic Vibrio cholerae O1 biotype El Tor displacing V. cholerae O139 Bengal in Bangladesh. J. Clin. Microbiol. 35:624-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faruque, S. M., Asadulghani, A. R. Alim, M. J. Albert, K. M. Islam, and J. J. Mekalanos. 1998. Induction of the lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic Vibrio cholerae O1 and O139. Infect. Immun. 66:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faruque, S. M., Asadulghani, M. Kamruzzaman, R. K. Nandi, A. N. Ghosh, G. B. Nair, J. J. Mekalanos, and D. A. Sack. 2002. RS1 element of Vibrio cholerae can propagate horizontally as a filamentous phage exploiting the morphogenesis genes of CTXΦ. Infect. Immun. 70:163-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faruque, S. M., Asadulghani, M. N. Saha, A. R. Alim, M. J. Albert, K. M. Islam, and J. J. Mekalanos. 1998. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXΦ: molecular basis for origination of new strains with epidemic potential. Infect. Immun. 66:5819-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faruque, S. M., L. Comstock, J. B. Kaper, and M. J. Albert. 1994. Distribution of Zonula occludens toxin (zot) gene among clinical isolates of Vibrio cholerae O1 from Bangladesh and Africa. J. Diarrhoeal Dis. Res. 12:222-224. [PubMed] [Google Scholar]
- 15.Faruque, S. M., A. K. Siddique, M. N. Saha, Asadulghani, M. M. Rahman, K. Zaman, M. J. Albert, D. A. Sack, and R. B. Sack. 1999. Molecular characterization of a new ribotype of Vibrio cholerae O139 Bengal associated with an outbreak of cholera in Bangladesh. J. Clin. Microbiol. 37:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinberg, A. P., and B. Vogelstein. 1984. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Addendum. Anal. Biochem. 137:266-267. [DOI] [PubMed] [Google Scholar]
- 17.Gardel, C., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64:2246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harkey, C. W., K. D. Everiss, and K. M. Peterson. 1994. The Vibrio cholerae toxin-coregulated-pilus gene tcpI encodes a homolog of methyl-accepting chemotaxis proteins. Infect. Immun. 62:2669-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hava, D. L., and A. Camilli. 2001. Isolation and characterization of a temperature-sensitive generalized transducing bacteriophage for Vibrio cholerae. J. Microbiol. Methods 46:217-225. [DOI] [PubMed] [Google Scholar]
- 20.Herrington, D. A., R. H. Hall, G. Losonsky, J. J. Mekalanos, R. K. Taylor, and M. M. Levine. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168:1487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Judson, N., and J. J. Mekalanos. 2000. TnAraOut, a transposon-based approach to identify and characterize essential bacterial genes. Nat. Biotechnol. 18:740-745. [DOI] [PubMed] [Google Scholar]
- 22.Kaper, J. B., J. G. Morris, and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaper, J. B., J. G. J. Morris, and M. Nishibuchi. 1988. DNA probes for pathogenic Vibrio species, p. 65-67. In F. C. Tenover (ed.), DNA probes for infectious diseases. CRC Press, Inc., Boca Raton, Fla.
- 24.Karaolis, D. K., R. Lan, J. B. Kaper, and P. R. Reeves. 2001. Comparison of Vibrio cholerae pathogenicity islands in sixth and seventh pandemic strains. Infect. Immun. 69:1947-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karaolis, D. K., S. Somara, D. R. Maneval, Jr., J. A. Johnson, and J. B. Kaper. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375-379. [DOI] [PubMed] [Google Scholar]
- 26.Karaolis, D. K. R., J. A. Johnson, C. C. Bailey, E. C. Boedeker, J. B. Kaper, and P. R. Reeves. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA 95:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keasler, S. P., and R. H. Hall. 1993. Detecting and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet 341:1661.. [DOI] [PubMed] [Google Scholar]
- 28.Kimsey, H. H., and M. K. Waldor. 1998. CTXφ immunity: application in the development of cholera vaccines. Proc. Natl. Acad. Sci. USA 95:7035-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimsey, H. H., and M. K. Waldor. 1998. Vibrio cholerae hemagglutinin/protease inactivates CTXφ. Infect. Immun. 66:4025-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovach, M. E., M. D. Shaffer, and K. M. Peterson. 1996. A putative integrase gene defines the distal end of large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology 142:2165-2174. [DOI] [PubMed] [Google Scholar]
- 31.Lee, C. A. 1999. Vibrio cholerae TCP: a trifunctional virulence factor? Trends Microbiol. 7:391-392. (Discussion, 7:393.) [DOI] [PubMed]
- 32.Lindsay, J. A., A. Ruzin, H. F. Ross, N. Kurepina, and R. P. Novick. 1998. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 29:527-543. [DOI] [PubMed] [Google Scholar]
- 33.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Mel, S. F., and J. J. Mekalanos. 1996. Modulation of horizontal gene transfer in pathogenic bacteria by in vivo signals. Cell 87:795-798. [DOI] [PubMed] [Google Scholar]
- 35.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukhopadhyay, A. K., S. Chakraborty, Y. Takeda, G. B. Nair, and D. E. Berg. 2001. Characterization of VPI pathogenicity island and CTXφ prophage in environmental strains of Vibrio cholerae. J. Bacteriol. 183:4737-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nandi, B., R. K. Nandy, A. C. Vicente, and A. C. Ghose. 2000. Molecular characterization of a new variant of toxin-coregulated pilus protein (TcpA) in a toxigenic non-O1/non-O139 strain of Vibrio cholerae. Infect. Immun. 68:948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novais, R. C., A. Coelho, C. A. Salles, and A. C. Vicente. 1999. Toxin-co-regulated pilus cluster in non-O1, non-toxigenic Vibrio cholerae: evidence of a third allele of pilin gene. FEMS Microbiol. Lett. 171:49-55. [DOI] [PubMed] [Google Scholar]
- 39.Ogg, J. E., T. L. Timme, and M. M. Alemohammad. 1981. General transduction in Vibrio cholerae. Infect. Immun. 31:737-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogierman, M. A., S. Zabihi, L. Mourtzios, and P. A. Manning. 1993. Genetic organization and sequence of the promoter-distal region of the tcp gene cluster of Vibrio cholerae. Gene 126:51-60. [DOI] [PubMed] [Google Scholar]
- 41.O'Shea, Y., and E. F. Boyd. 2002. Mobilization of the Vibrio pathogenicity island between Vibrio cholerae isolates mediated by CP-T1 generalized transduction. FEMS Microbiol. Lett. 214:153-157. [DOI] [PubMed] [Google Scholar]
- 42.Reidl, J., and J. J. Mekalanos. 1995. Characterization of Vibrio cholerae bacteriophage K139 and use of a novel mini-transposon to identify a phage-encoded virulence factor. Mol. Microbiol. 18:685-701. [DOI] [PubMed] [Google Scholar]
- 43.Rubin, E. J., M. K. Waldor, and J. J. Mekalanos. 1998. Mobile genetic elements and the evolution of new epidemic strains of Vibrio cholerae, p. 147-161. In R. A. Krause (ed.), Emerging infections. Academic Press, New York, N.Y.
- 44.Taylor, R., C. Shaw, K. Peterson, P. Spears, and J. Mekalanos. 1988. Safe, live Vibrio cholerae vaccines? Vaccine 6:151-154. [DOI] [PubMed] [Google Scholar]
- 45.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thelin, K. H., and R. K. Taylor. 1996. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 64:2853-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 48.World Health Organization. 1974. World Health Organization guidelines for the laboratory diagnosis of cholera. World Health. Organization, Geneva, Switzerland.