Abstract
The Bordetella BvgAS signal transduction system controls the expression of at least three phenotypic phases, the Bvg+ or virulent phase, the Bvg− or avirulent phase, and the Bvgi or Bvg intermediate phase, which has been hypothesized to be important for transmission. bipA, the first identified Bvgi-phase gene, encodes a protein with similarity to the well-characterized bacterial adhesins intimin and invasin. Proteins encoded by the bipA genes present in Bordetella pertussis Tohama I and Bordetella bronchiseptica RB50 differ in the number of 90-amino-acid repeats which they possess and in the sequence of the C-terminal domain. To investigate the possibility that bipA alleles segregate according to host specificity and to gain insight into the role of BipA and the Bvgi phase in the Bordetella infectious cycle, we compared bipA alleles across members of the B. bronchiseptica cluster, which includes both human-infective (B. pertussis and B. parapertussishu) and non-human-infective (B. bronchiseptica and B. parapertussisov) strains. bipA genes were present in most, but not all, strains. All bipA genes present in B. bronchiseptica strains were identical to bipA of RB50 (at least with regard to the DNA sequence of the 3′ C-terminal-domain-encoding region, the number of 90-amino-acid repeats encoded, and expression patterns). Although all bipA genes present in the other Bordetella strains were identical in the 3′ C-terminal-domain-encoding region to bipA of B. pertussis Tohama I, they varied in the number of 90-amino-acid repeats that they encoded and in expression level. Notably, the genes present in B. parapertussishu strains were pseudogenes, and the genes present in B. parapertussisov strains were expressed at significantly reduced levels compared with the levels in B. pertussis and B. bronchiseptica strains. Our results indicate that there is a correlation between specific bipA alleles and specific hosts. They also support the hypothesis that both horizontal gene transfer and fine-tuning of gene expression patterns contribute to the evolution of host adaptation in lineages of the B. bronchiseptica cluster.
Bordetellae are gram-negative coccobacilli that cause respiratory infections in mammals, birds, and reptiles. The classical bordetellae (19), Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica, are so closely related that they are now considered subspecies or strains of a single species (5, 27, 37, 39, 40, 57). Because the progenitor organism of this group appears to have been B. bronchiseptica, with B. pertussis and B. parapertussis strains diverging at different times and from different branches of the phylogenetic tree, these bacteria have been referred to as the B. bronchiseptica cluster (19). The organisms identified as B. bronchiseptica have a broad host range, infecting nearly all four-legged mammals, and cause infections that range from asymptomatic colonization to tracheobronchitis and pneumonia (22). In contrast, B. pertussis strains uniformly cause the acute and severe childhood illness known as whooping cough or pertussis and are adapted exclusively to humans (8). B. parapertussis strains comprise two separate lineages. B. parapertussishu strains have been isolated only from humans and cause a respiratory illness that is very similar to that caused by B. pertussis (24, 60), while B. parapertussisov strains were isolated from the respiratory tracts of healthy sheep, as well as sheep with chronic nonprogressive pneumonia (13, 43).
Members of the B. bronchiseptica cluster express overlapping sets of highly related virulence factors. These include putative adhesins, such as filamentous hemagglutinin (45), fimbriae (Fim) (36), pertactin (Prn) (46), tracheal colonization factor (TcfA) (17), and BrkA (Bordetella resistance to killing) (16), and toxins, such as adenylate cyclase toxin (CyaA) (25), dermonecrotic toxin (Dnt) (33), tracheal cytotoxin (20, 21), pertussis toxin (Ptx) (38, 42), and proteins secreted via a type III secretion system encoded by the bsc locus (62, 63). With the exception of tracheal cytotoxin, all of these factors are positively regulated by BvgAS sensory transduction systems that are nearly identical and functionally interchangeable (34, 51, 56, 59); hence, expression of these molecules characterizes a phenotypic state designated the Bvg+ phase. Experiments with phase-locked and ectopic expression mutants have shown that the Bvg+ phase is necessary and sufficient for the development of respiratory infection (1, 11, 35).
Comparisons of Bvg+-phase factors and their expression patterns across the Bordetella species have provided useful information for understanding phylogenetic and evolutionary relationships among these bacteria (5-7, 31, 48). Such information has also proven to be valuable for formulating hypotheses regarding the roles of the factors in the Bordetella infectious cycle. For example, since Ptx is expressed only by B. pertussis strains (5), it can be concluded that Ptx is not absolutely required for respiratory infection. However, since only B. pertussis strains induce leukocytosis (24, 58), it is likely that Ptx plays a significant role in causing this specific parameter of disease. Reciprocally, as type III secretion systems appear to be functional only in B. bronchiseptica and B. parapertussisov strains, which commonly cause chronic subclinical infections, it has been hypothesized that type III secreted proteins may function to down-regulate the host immune response and thereby contribute to long-term persistence (62).
Comparative analyses have similarly provided insight into Bvg-repressed phenotypes and the role of the Bvg− or avirulent phase. Bvg-repressed phenotypes in B. bronchiseptica include the expression of flagella, motility, chemotaxis, and the ability to grow under nutrient-limiting conditions, and it has been hypothesized that the role of the Bvg− phase is to allow the bacteria to survive for extended periods of time in the environment while they are between mammalian hosts (1, 9, 10). B. pertussis, B. parapertussishu, and B. parapertussisov strains, which do not express these phenotypes, are thought to be restricted to transmission via direct contact or aerosol droplets (9, 10). The Bvg-repressed genes expressed exclusively by B. pertussis include vrg and vra loci (28, 50). Although the functions of the products of these genes are unknown, determination of these functions should allow formulation of hypotheses regarding the role of the Bvg− phase in this species.
A third phenotypic phase, induced by growth in the presence of semimodulating concentrations of nicotinic acid or MgSO4 (chemicals that down-regulate BvgAS activity) or by a specific mutation in bvgS, has been described for B. bronchiseptica strain RB50 (12). This Bvg intermediate (Bvgi) phase, characterized by expression of a subset of Bvg+-phase factors, lack of expression of Bvg−-phase factors, and expression of phenotypes that are maximally if not exclusively expressed in this phase, is hypothesized to be important for aerosol transmission (12). The Bvgi-specific phenotypes identified so far include autoaggregation (12) and expression of the recently identified outer membrane protein BipA (14, 54). The function of BipA is unknown; however, the predicted similarity of this protein to intimin of enteropathogenic and enterohemorrhagic Escherichia coli and invasin of Yersinia spp., including a similar C-terminal exposed topology, suggests that it may play a role in adherence (54). Initial studies revealed the presence of a BipA cross-reactive protein in whole-cell lysates of B. pertussis strain GMT1 (54), and DNA sequence data from the Sanger Centre's B. pertussis genome project (http://www.sanger.ac.uk/Projects/B_pertussis/) indicate that there is a bipA homolog in B. pertussis Tohama I. The majority of the predicted amino acid differences between the BipA proteins produced by B. bronchiseptica RB50 and B. pertussis Tohama I are located at the extreme C terminus. This observation suggests the intriguing possibility that if BipA functions in adherence like intimin and invasin, it could be involved in host specificity since the C-terminal domains of intimin and invasin are directly involved in binding to receptor proteins on host cells (26, 30, 32, 61) and variability in this region among intimin family members has been hypothesized to play a role in tissue tropism (18, 41, 44). The conservation of bipA and the Bvgi phase in bordetellae is unknown. To study the role of BipA and the Bvgi phase in the Bordetella infectious cycle, we conducted a comparative analysis of bipA homologs and their expression patterns within and across strains of the B. bronchiseptica cluster.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. Bordetella strains were grown on Bordet-Gengou agar supplemented with 15% defibrinated sheep blood or in Stainer-Scholte (SS) broth (49). E. coli strains were grown in L broth or on L agar. When appropriate, media were supplemented with ampicillin (100 μg ml−1), gentamicin (30 μg ml−1), streptomycin (70 μg ml−1), rifampin (20 μg ml−1), kanamycin (50 μg ml−1), or sucrose (10%). B. parapertussisov strain Fr107i and B. parapertussishu strain 12822i were constructed by transferring the bvgS-I1 mutation to the chromosomes of Fr107 and 12822, respectively, by allelic exchange with plasmid pEG129, as described previously (12).
TABLE 1.
Strains used in this study
| Species | Strain | Electrophoretic type | Host | Geographic origin | Reference or source |
|---|---|---|---|---|---|
| B. bronchiseptica | RB50 | Rabbit | United States | 11 | |
| 548 | 6 | Pig | The Netherlands | 57 | |
| 590 | 6 | Dog | United States | 57 | |
| 595 | 4 | Dog | United States | 57 | |
| 625 | 16 | Rat | United States | 57 | |
| 675 | 14 | Human | Germany | 57 | |
| 676 | 3 | Pig | Australia | 57 | |
| 704 | 1 | Rabbit | United States | 58 | |
| Cb2 | Dog | United States | This study | ||
| JC100 | Human | United States | This study | ||
| RB53 | 11 | ||||
| RB53i | 11 | ||||
| B. pertussis | Tohama I | 37 | Human | Japan | 27 |
| 18323 | 38 | Human | United States | ATCC 9797 | |
| CS | Human | United States | This study | ||
| 6235 | Human | United States | This study | ||
| 6068 | Human | United States | This study | ||
| GMT1 | Human | United States | 34 | ||
| GMT1i | 12 | ||||
| B. parapertussishu | 12822 | 28a | Human | Germany | 24 |
| Ccug38844 | 28a | Human | Germany | 24 | |
| No7 | 28a | Human | France | 24 | |
| 840994 | 28a | Human | Finland | 24 | |
| 803 | 28a | Human | Italy | 24 | |
| H789 | 28a | Human | The Netherlands | 24 | |
| B. parapertussisov | Fr107 | Ovine | New Zealand | 13 | |
| J1 | Ovine | Scotland | 43 | ||
| C | Ovine | Scotland | 43 | ||
| H1 | Ovine | Scotland | 43 | ||
| Fr107i | This study | ||||
| E. coli | DH5α(pEG129) | 12 | |||
| DH5α(pTEN34) | This study |
The electrophoretic type was not determined, but it was almost definitely electrophoretic type 28 as all B. parapertussishu strains tested by van der Zee et al. were electrophoretic type 28 strains.
Recombinant DNA techniques.
Standard methods were used for preparation of plasmid DNA, restriction endonuclease digestion, agarose gel electrophoresis, DNA ligation, and other DNA manipulations (47). Nucleotide sequences were determined by Laragen, Inc. (Los Angeles, Calif.).
Plasmid rescue.
The 5′ end of bipA from B. parapertussishu strain 12822 was cloned by plasmid rescue. Briefly, a suicide plasmid derivative of plasmid pEG7 carrying a 621-bp PCR product corresponding to the C-terminal 307 amino acids of BipA from B. parapertussishu strain 12822 was introduced into 12822 by conjugation, and cointegrates were selected on Bordet-Gengou-gentamicin agar. Chromosomal DNA was then prepared, digested with FseI (which does not cut within the pEG7 derivative), self-ligated, and transformed into E. coli DH5α. Plasmid DNA recovered from the transformants carried chromosomal DNA from the bipA locus flanking the 621-bp region on the plasmid that provided homology for recombination. This DNA was characterized by restriction endonuclease digestion and nucleotide sequence analysis.
β-Galactosidase assays.
To measure bipA expression, plasmid pTEN34, which contains a 321-bp fragment of bipA corresponding to nucleotides encoding amino acids 70 to 177 of BipA fused to the promoterless lacZ gene on plasmid pEGZ (34), was constructed. Integration of this suicide plasmid into the chromosomes of the various strains at the bipA locus resulted in bipA-lacZ fusions. PCR was used to confirm that plasmids had integrated at the bipA locus as intended. β-Galactosidase assays were performed as previously described (34).
PCR and Southern blotting.
Primers used for PCR and Southern blotting are listed in Table 2. PCR was performed by using total cellular DNA templates. The cycling parameters were 94°C for 4 min and 30 cycles of 94°C for 30 s, 55 or 60°C for 45 s, and 72°C for 1 min, with a final extension step of 72°C for 5 min. Southern blotting was performed as previously described (2).
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | DNA sequence |
|---|---|
| QCPCRbipF | 5′AATTGCTTGGACAGCGGTTC3′ |
| QCPCRbipR | 5′GCGACTATCTCAAGCGTGAA3′ |
| bBipRepF1 | 5′GTCGCGGATTATCGCCG3′ |
| bBipRepR1 | 5′GGCACGGCGGATCCGGACAAC3′ |
| BipCtFwd2 | 5′CACGGCGGATCCGGACAACGAT3′ |
| pBipCtRvs3 | 5′CGCCTGCTCGCCAGACAGTG3′ |
| BipCtFwd3 | 5′CGGACAACGATGTGACGGT3′ |
| pBipCtRvs1 | 5′GTCTTACGGCGCTTAGTA3′ |
| KB1For | 5′CGCGGTACCCCGGCCTGTTCGACGTGCCG3′ |
| LMBipNtc | 5′GGGGATCCGGTTCCGAGAGATTGATGTTCAGG3′ |
| LM7BipNt | 5′CCTATGCTGCATGTATTGGGTTCC3′ |
Western immunoblotting.
Immunoblotting was performed as previously described (11, 12). Polyclonal anti-BipA antibody was used at a 1:5,000 dilution, and horseradish peroxidase-conjugated anti-rabbit immunoglobulin (secondary antibody) was used at a dilution of 1:5,000. Antigen-antibody complexes were detected by enhanced chemiluminescence (Amersham).
RESULTS
Prevalence of bipA homologs among Bordetella strains that cause respiratory infections in mammals.
We used PCR to investigate the prevalence of the bipA gene within and across the Bordetella species that cause respiratory infections in mammals. The collection of strains tested (Table 1) included members of all of the main subdivisions of the B. pertussis, B. bronchiseptica, B. parapertussishu, and B. parapertussisov species as determined by van der Zee et al. (57) except the B. bronchiseptica lineage represented by ET21, which was unavailable. A few recent clinical isolates of B. pertussis (strains CS, 6235, and 6068) and B. bronchiseptica (strains Cb2 and JC100) were also included. The primers used, QCPCRBipF and QCPCRBipR (Table 2 and primers 1 and 2 in Fig. 1), amplified a 727-bp fragment from B. bronchiseptica and B. pertussis genomic DNA corresponding to the region encoding amino acids 115 to 355 of BipA. Products that were approximately 727 bp long, and no other products, were amplified from all strains tested except B. bronchiseptica strains 548, 590, and 675 (data not shown), suggesting that most, but not all, bordetellae that infect mammals contain bipA homologs.
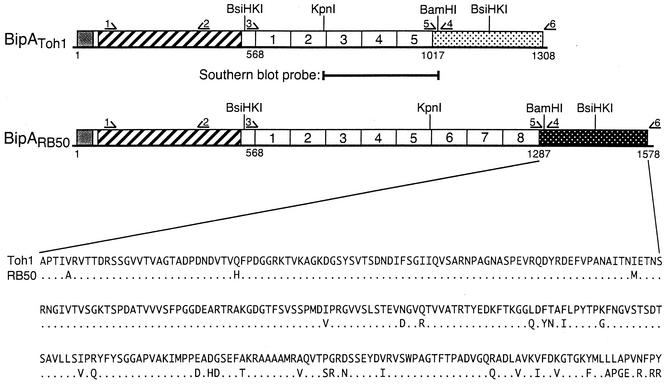
FIG. 1.
Schematic diagram of the bipA loci of B. bronchiseptica RB50 and B. pertussis Tohama I. The open reading frames of B. bronchiseptica RB50 and B. pertussis Tohama I code for N-terminal signal sequences (shaded boxes), followed by ∼410-amino-acid regions with similarity to intimin and invasin (cross-hatched boxes) and then by five (B. pertussis) or eight (B. bronchiseptica) 90-amino-acid repeated domains (indicated by the numbers 1 to 5 and 1 to 8, respectively) and finally by 291-amino-acid C-terminal domains. The amino acid sequences of the C-terminal domains, as predicted from their nucleotide sequences, are shown. Restriction endonuclease sites and the locations of primers (arrows 1 to 6) used in this study are indicated. The fragment used as a probe for Southern blotting is also indicated.
The number of 90-amino-acid repeats varies across, but not within (except for B. pertussis strain 18323), Bordetella species.
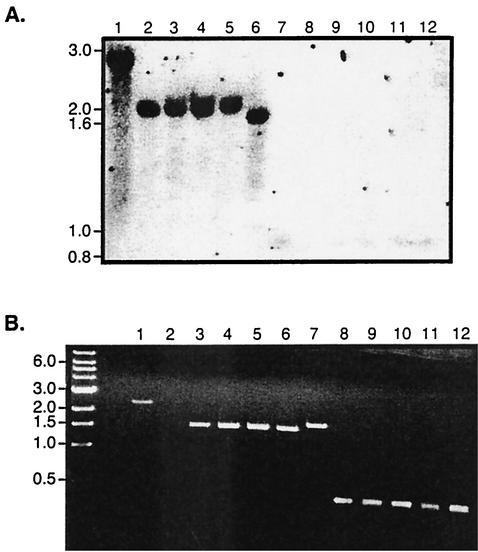
The bipA gene of B. pertussis Tohama I is predicted to encode a 1,308-amino-acid protein containing five highly conserved 90-amino-acid repeats extending from amino acid 568 to amino acid 1017, while the bipA gene of B. bronchiseptica RB50 is predicted to encode a 1,578-amino-acid protein containing eight of these highly conserved 90-amino-acid repeats (54) (Fig. 1). To estimate the number of 90-amino-acid repeat domains encoded by the bipA genes in the various strains included in this study, we performed Southern blot analyses. Genomic DNA digested with BsiHKI was probed with an 870-bp KpnI-BamHI fragment corresponding to the region encoding amino acids 1018 to 1310 of BipA of B. bronchiseptica (Fig. 1 and 2A). This probe hybridized to ∼2.75-kb fragments in all B. bronchiseptica strains tested except strains 548, 590, and 675. These data confirm the PCR results, indicating that all B. bronchiseptica strains tested except 548, 590, and 675 contain bipA homologs. They further suggest that all B. bronchiseptica strains containing bipA genes have the potential to encode BipA proteins containing eight 90-amino-acid repeats. With the exception of strain 18323, the probe hybridized to a ∼1.95-kb fragment in all B. pertussis strains tested (Fig. 2A and data not shown), suggesting that the bipA genes in these strains encode proteins containing five 90-amino-acid repeats. The probe hybridized to a ∼870-bp fragment in strain 18323, suggesting that this strain encodes a protein containing only one of the 90-amino-acid domains. The probe also recognized a ∼870-bp fragment in all B. parapertussishu strains, suggesting that these strains also encode only one 90-amino-acid domain. A ∼2.2-kb fragment was recognized by the probe in all B. parapertussisov strains tested, suggesting that these strains encode proteins containing six 90-amino-acid repeats. These results were confirmed by PCR by using primers bBipRepF1 and bBipRepR1 (primers 3 and 4 in Fig. 1), which anneal to regions flanking the DNA region encoding the 90-amino-acid repeat (Fig. 2B). These primers amplified 2.2-kb fragments in B. bronchiseptica strains, 1.35-kb fragments in B. pertussis strains, 1.6-kb fragments in B. parapertussisov strains, and 270-bp fragments in B. parapertussishu strains and B. pertussis 18323.
FIG. 2.
Comparison of the 90-amino-acid repeat domain-containing regions of bipA within and across Bordetella species. (A) Southern blot. BsiHKI-digested genomic DNA was probed with the 870-bp KpnI-BamHI fragment shown in Fig. 1. Lane 1, B. bronchiseptica RB50; lane 2, B. parapertussisov Fr107; lane 3, B. parapertussisov JI; lane 4, B. parapertussisov C; lane 5, B. parapertussisov HI; lane 6, B. pertussis Tohama I; lane 7, B. parapertussishu 12822; lane 8, B. parapertussishu No7; lane 9, B. parapertussishu 840994; lane 10, B. parapertussishu 803; lane 11, B. parapertussishu 789; lane 12, B. pertussis 18323. The positions of molecular weight markers are indicated on the left. (B) PCR. DNA fragments were amplified from genomic DNA by using primers bBipRepF1 and bBipRepR1 (primers 3 and 4 in Fig. 1). Lane 1, B. bronchiseptica strain RB50; lane 2, B. bronchiseptica strain 590; lanes 3 to 6, B. pertussis strains Tohama I, GMT1, 6068, and CS, respectively; lane 7, B. parapertussisov Fr107; lane 8, B. pertussis strain 18323; lanes 9 to 12, B. parapertussishu strains 12822, No7, 840994, and 803, respectively. The sizes of the 1-kb-marker fragments (in kilobase pairs) are indicated on the left.
The nucleotide sequences encoding the BipA C-terminal domains of all B. bronchiseptica strains are identical to those of RB50, and the nucleotide sequences encoding the BipA C-terminal domains of all other strains are identical to those of Tohama I.
Nearly all (34 of 39) of the predicted amino acid differences encoded by the bipA genes of B. pertussis Tohama I and B. bronchiseptica RB50 (exclusive of the additional 90-amino-acid repeats in BipA of RB50) are located in the C-terminal domain, and most are located at the extreme C terminus (Fig. 1) (54). To investigate the variability in this region within and across the Bordetella species, we used primers BipCtFwd2 and pBipCtRvs3 (primers 5 and 6 in Fig. 1) to PCR amplify 875-bp DNA fragments corresponding to the C-terminal domains of bipA and had their nucleotide sequences determined. The nucleotide sequences of these fragments from all B. bronchiseptica strains listed in Table 1 were identical to the nucleotide sequence of RB50. The nucleotide sequences of all other Bordetella strains listed in Table 1 were identical to the nucleotide sequence of Tohama I. These data suggest that all B. bronchiseptica strains that contain bipA encode BipA proteins with the same amino acid sequence in their C-terminal domains and that all non-B. bronchiseptica strains encode BipA proteins with C-terminal domains identical to the C-terminal domain of B. pertussis Tohama I BipA.
BipA protein can be detected in B. bronchiseptica and B. pertussis strains but not in B. parapertussishu or B. parapertussisov strains.
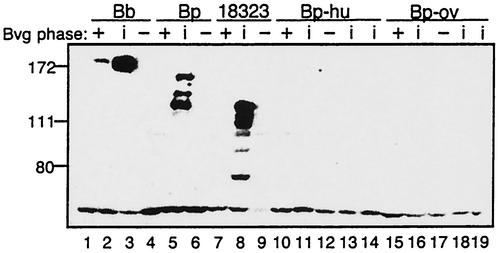
To investigate BipA protein expression in the various strains, we performed Western blot analyses with whole-cell lysates prepared from cultures grown under Bvg+-, Bvgi-, and Bvg−-phase conditions (i.e., in medium containing various concentrations of nicotinic acid or MgSO4 or both). Polyclonal antibodies raised against the C terminus of BipA (54) recognized polypeptides of the expected sizes in lysates of all B. bronchiseptica and B. pertussis strains tested (except B. bronchiseptica strains 548, 590, and 675) but failed to recognize polypeptides in lysates made from B. parapertussishu or B. parapertussisov strains (data not shown). Because the conditions under which the Bvgi phase is expressed can vary within and across species (34), we constructed B. parapertussishu and B. parapertussisov strains containing the bvgS-I1 mutation (12). In both B. bronchiseptica RB50 and B. pertussis GMT1, this mutation causes the bacteria to express Bvgi-phase phenotypes when they are grown under Bvg+-phase conditions (12, 54). Western blot analysis of whole-cell lysates of these strains confirmed that the BipA protein could be detected in B. pertussis and B. bronchiseptica but not in B. parapertussishu or B. parapertussisov (Fig. 3). Our inability to detect BipA in lysates of B. parapertussishu strain 12822 contradicts our previous report (54). We repeated this experiment with all B. parapertussishu isolates included in this study grown with various concentrations of the chemical modulators nicotinic acid and MgSO4 and also with selected strains in which the bvgS-I1 mutation had been introduced, and we still failed to detect the BipA protein (data not shown). Together with the results shown below, these data indicate conclusively that B. parapertussishu strains do not express the BipA protein, indicating that our previously reported result was in error.
FIG. 3.
BipA protein expression as determined by Western blotting. Whole-cell lysates of B. bronchiseptica RB50 (lanes 1 to 3), B. pertussis Tohama I (lanes 4 to 6), B. pertussis 18323 (18323) (lanes 7 to 9), B. parapertussishu 12822 (lanes 10 to 12), B. parapertussishu No7 (Bvgi phase only) (lane 13), B. parapertussishu 803 (Bvgi phase only) (lane 14), B. parapertussisov Fr107 (Fr107) (lanes 15 to 17), B. parapertussisov JI (Bvgi phase only) (lane 18), and B. parapertussisov HI (Bvgi phase only) (lane 19) were analyzed by Western blotting with anti-BipA (CT4) antibody. The positions of molecular size markers (in kilodaltons) are indicated on the left. Bb, B. bronchiseptica RB50; Bp, B. pertussis Tohama I; Bp-hu, B. parapertussishu; Bp-ov, B. parapertussisov.
The 5′ ends of the bipA loci of B. parapertussishu and B. parapertussisov strains differ from those of B. pertussis and B. bronchiseptica.
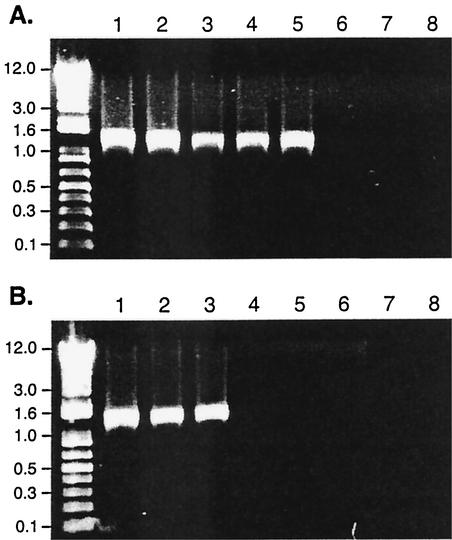
Because we did not detect BipA expression in the B. parapertussishu and B. parapertussisov strains by Western blotting (Fig. 3) despite the presence of bipA hybridizing sequences in the genomes (Fig. 2), we investigated the 5′ ends of the bipA loci in these strains by PCR. Primers designed to amplify 1.36-kb fragments from the 5′ ends of the bipA loci of B. bronchiseptica and B. pertussis (KB1For and LMBipNtc [Table 2]) amplified fragments of the expected sizes from B. bronchiseptica, B. pertussis, and B. parapertussisov strains but failed to amplify fragments from B. parapertussishu strains (Fig. 4A). Other primers designed to amplify DNA fragments within the same region also failed to yield products from B. parapertussishu strains (data not shown). These results suggested that the nucleotide sequence at the 5′ end of bipA in B. parapertussishu strains differed from the nucleotide sequences of the other bordetellae. We therefore cloned the bipA 5′ region from the chromosome of B. parapertussishu strain 12822 by plasmid rescue (see Materials and Methods) and determined the nucleotide sequence of the resulting clone. The sequence obtained matched exactly DNA sequence data newly released by the Sanger Centre (http://www.sanger.ac.uk/Projects/B_parapertussis/) and indicated that the homology between bipA of B. parapertussishu strain 12822 and the bipA genes of B. bronchiseptica RB50 and B. pertussis Tohama I begins with nucleotides encoding amino acid 67 (a glycine residue) of the BipA protein. Strain 12822 sequences immediately 5′ to this position exhibited no similarity to the 5′ ends of the bipA loci of RB50 and Tohama I and lacked discernible translation initiation elements. Promoter elements and putative BvgA binding sites were also not recognizable. Also, 793 bp 5′ to the beginning of the truncated ′bipA gene, an IS1001 element was found with its TnpA-encoding gene oriented in the opposite direction relative to the orientation of ′bipA. By searching the Sanger Centre's Bordetella genome databases, we found sequences identical to the 793-bp sequence that intervenes between IS1001 and ′bipA in 12822 in the RB50 genome (at a site unlinked to bipA) but not in the Tohama I genome. Sequences immediately 3′ to ′bipA in 12822 matched the sequences 3′ to bipA in Tohama I (99.1% identity) and RB50 (94.9% identity) for 700 bp, at which point there was a 450-bp gap in the 12822 sequence. Beyond that gap, the 12822, RB50, and Tohama I sequences were nearly identical for 690 bp, and then the Tohama I sequences diverged significantly while the RB50 and 12822 sequences remained nearly identical for at least another 3 kb. The region of DNA containing ′bipA and ∼700 bp 3′ to ′bipA in 12822, therefore, appears to be of B. pertussis origin, while the flanking sequences are more similar to B. bronchiseptica sequences, as expected for B. parapertussishu DNA in general due to its closer phylogenetic relationship with B. bronchiseptica DNA than with B. pertussis DNA. To determine if the genetic organization of the bipA region in 12822 was conserved in other B. parapertussishu isolates, we designed primers to amplify a 1.58-kb fragment from the 5′ end of bipA from B. parapertussishu 12822 (LM7BipNt and LMBipNtc [Table 2]). LM7BipNt annealed immediately 3′ to the IS1001 element, within the 793-bp intervening sequences. These primers amplified 1.58-kb fragments from all B. parapertussishu strains tested and did not yield PCR products from B. bronchiseptica, B. pertussis, or B. parapertussisov strains (Fig. 4B). The bipA homolog in all B. parapertussishu strains tested, therefore, appears to be a pseudogene.
FIG. 4.
Comparison of the 5′ regions of bipA homologs by PCR. (A) Genomic DNA amplified by PCR by using primers KB1For and LMBipNtc. Lane 1, B. bronchiseptica RB50; lane 2, B. pertussis Tohama I; lane 3, B. parapertussisov Fr107; lane 4, B. parapertussisov JI; lane 5, B. parapertussisov HI; lane 6, B. parapertussishu 12822; lane 7, B. parapertussishu No7; lane 8, B. parapertussishu 803. (B) Genomic DNA amplified by PCR by using primers LM7BipNt and LMBipNtc. Lane 1, B. parapertussishu 12822; lane 2, B. parapertussishu No7; lane 3, B. parapertussishu 803; lane 4, B. bronchiseptica RB50; lane 5, B. pertussis Tohama I; lane 6, B. parapertussisov Fr107; lane 7, B. parapertussisov JI; lane 8, B. parapertussisov HI.
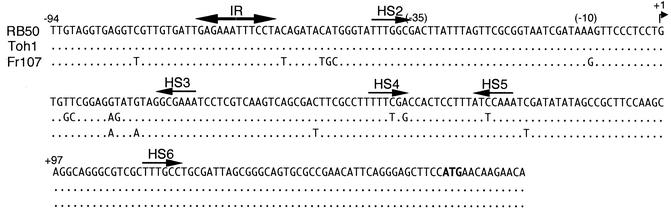
Our inability to detect BipA in B. parapertussisov strains by Western blotting was not due to gross differences in the 5′ regions of the bipA loci of these strains, as primers designed to amplify the promoter regions of bipA in RB50 and Tohama I amplified fragments of the same size in B. parapertussisov strains (Fig. 4A). We therefore determined the nucleotide sequences of the 5′ ends of the bipA genes of all of the B. parapertussisov strains used in this study. All of the sequences obtained were identical to each other, and although they were nearly identical to the sequences of B. pertussis and B. bronchiseptica, there were notable differences in the region between the BvgA binding sites designated IR1 and HS2 and at position −11 relative to the transcriptional start site that were identified in B. bronchiseptica (Fig. 5).
FIG. 5.
Comparison of bipA promoter regions. The nucleotide sequences of the promoter regions of B. bronchiseptica RB50, B. pertussis Tohama I, and B. parapertussisov Fr107 are shown. BvgA binding sites identified for RB50 and hypothesized to be required for transcriptional activation of bipA are indicated by arrows and are labeled IR1 and HS2 (14). BvgA binding sites hypothesized to be required for repression of bipA under Bvg+-phase conditions are also indicated by arrows and are labeled HS3, HS4, HS5, and HS6. The translation initiation codon is indicated by boldface type.
bipA transcription is maximal under Bvgi-phase conditions in B. pertussis, B. bronchiseptica, and B. parapertussisov, but the levels are different in different species.
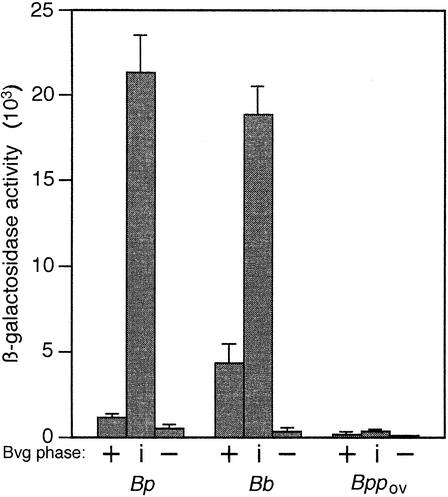
To compare bipA transcription levels in the various species, we created bipA-lacZ fusions in representative strains (B. bronchiseptica strain RB50, B. pertussis strain GMT1, and B. parapertussisov strain Fr107) using plasmid pTEN34, which contained a 321-bp fragment of bipA corresponding to nucleotides encoding amino acids 70 to 177 of BipA fused to a promoterless lacZ gene. Integration of this suicide plasmid into the chromosome resulted in the formation of a bipA′-′lacZ fusion. To determine expression levels under Bvg+- and Bvg−-phase conditions, we used wild-type strains (RB50::pTEN34, GMT1::pTEN34, and Fr107::pTEN34) grown in SS medium and in SS medium containing 20 mM MgSO4, respectively. To determine Bvgi-phase expression levels, we used strains containing the bvgS-I1 mutation (RB50i::pTEN34, GMT1i::pTEN34, and Fr107i::pTEN34) grown in SS medium. Consistent with previous reports (14, 54), the levels of bipA expression in B. bronchiseptica were maximal in the Bvgi phase (Fig. 6). The bipA-lacZ expression levels in B. pertussis were nearly the same as those in B. bronchiseptica in the Bvgi and Bvg− phases. The level of bipA-lacZ expression in B. pertussis grown under Bvg+-phase conditions, however, was only about 15% of that of B. bronchiseptica (∼500 β-galactosidase units, compared with ∼4,200 β-galactosidase units) (Fig. 6). This result is consistent with the fact that the BipA protein can be detected by Western blot analysis in lysates of Bvg+-phase B. bronchiseptica but not in lysates of Bvg+-phase B. pertussis (Fig. 3). bipA expression in B. parapertussisov strain Fr107 was significantly decreased under all conditions compared with bipA expression in B. bronchiseptica and B. pertussis. The overall expression pattern (maximal in the Bvgi phase), however, was the same as that in the other species (Fig. 6). Our inability to detect the BipA protein in lysates of B. parapertussisov strains by Western blotting was therefore most likely due to a decreased level of transcription compared with that in B. bronchiseptica and B. pertussis.
FIG. 6.
bipA expression patterns: β-galactosidase activity in B. bronchiseptica RB50 (Bb), B. pertussis GMT1 (Bp), and B. parapertussisov Fr107 (Bppov) in the Bvg+, Bvgi, and Bvg− phases. The units are nanomoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per milligram of protein. The error bars indicate one standard deviation.
DISCUSSION
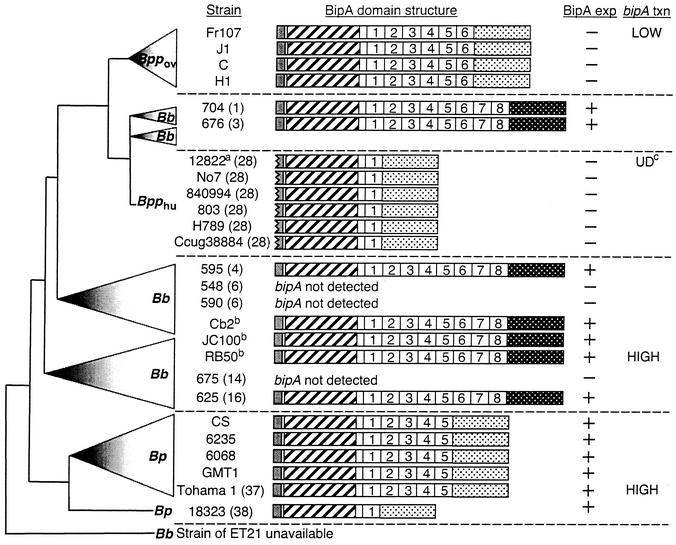
We have been studying bipA and its product, BipA, to determine its role and the role of the Bvgi phase in the Bordetella infectious cycle. Initial DNA sequence analyses revealed significant similarity between the predicted amino acid sequence of BipA and the amino acid sequences of intimin and invasin, suggesting that BipA represents a new member of this class of bacterial adhesins (54). The fact that most of the amino acid differences between the BipA homologs of B. bronchiseptica RB50 and B. pertussis Tohama I are located within the C-terminal domain, the region analogous to the regions of intimin and invasin that participate in direct contact with receptors on host cells, suggested further that if BipA does function as an adhesin, it might play a role in determining host specificity. To explore this possibility by using a genomic approach, we determined the prevalence, genetic organization, and expression pattern of bipA alleles in strains representing various clades of the B. bronchiseptica cluster. Our results are summarized in Fig. 7.
FIG. 7.
Schematic diagram of bipA alleles in strains of the B. bronchiseptica cluster. The phylogenetic relationships shown are based on the work of van der Zee et al. (58), and the dendrogram was adapted from the study of Gerlach et al. (19). The strains used in this study are included, and where known, electrophoretic types (ET) are indicated in parentheses after the strain designations. The cross-hatched area represents the region of BipA that exhibits amino acid sequence similarity with intimin and invasin, the numbered boxes represent the 90-amino-acid repeats, and the distribution of the two different C-terminal sequences are indicated (black dots on a white background, Tohama I-like; white dots on a black background, RB50-like). Although the electrophoretic types of the B. parapertussishu strains used in this study were not determined, they are almost definitely electrophoretic type 28 as all B. parapertussishu strains tested by van der Zee et al. were electrophoretic type 28 strains. The electrophoretic types of strains RB50, Cb2, and JC100 are unknown so these strains have been placed between the two large clusters of B. bronchiseptica strains. For RB50, this is consistent with its lack of IS elements. The ability to detect BipA by Western blotting (BipA exp) and the relative level of bipA expression under Bvgi-phase conditions (bipA txn) are indicated. Bppov, B. parapertussisov; Bb, B. bronchiseptica; Bpphu, B. parapertussishu; Bp, B. pertussis; UD, undetectable.
PCR and Southern blot analyses showed that most, but not all, strains contain bipA genes. In B. parapertussishu strains, these genes appear to be pseudogenes. Although B. parapertussisov strains contain intact bipA loci, the genes were expressed at very low levels compared with the levels of expression in B. bronchiseptica and B. pertussis strains. These low levels of expression were probably due to nucleotide differences at positions −11, −51 to −53, and −59 relative to the predicted transcriptional start site as these positions are predicted to be involved in BvgA-mediated transcriptional activation in B. bronchiseptica (14). The fact that bipA is absent from some strains and either is not expressed or is expressed at only a very low level in other strains suggests that if BipA plays a role in the Bordetella infectious cycle, the role is either not essential or can be compensated for by another gene(s) in some strains. Alternatively, BipA could play an important role in a behavior that is unique to those strains in which it is expressed. We and other workers have shown that the Bvg+ phase is necessary and sufficient for respiratory colonization and that failure to repress Bvg−-phase phenotypes is detrimental to the development of infection (1, 11, 12, 35). Based on these and other data, we hypothesized that the Bvgi phase is important for transmission (9, 10, 12). As transmission routes may include survival in one or more in vivo niches or ex vivo environments and as the various species, or even lineages within species, may be differentially restricted in their modes of transmission, it is conceivable that factors required for specific routes could be absolutely required in some strains and dispensable in others. BipA's limited distribution, therefore, may reflect a specialized role in this process.
In our analyses we identified five bipA alleles, and in most cases strains of a single species contained the same allele; the only exception was B. pertussis strain 18323, which contained its own unique allele. With regard to the C-terminus-encoding region, however, only two types were identified, those identical to the RB50 region (present in all B. bronchiseptica strains containing bipA) and those identical to the Tohama I region (present in all non-B. bronchiseptica strains). If only strains expressing high levels of BipA are considered, the two types segregate according to host specificity; all human-infective strains contain alleles encoding Tohama I-like C termini, while all non-human-infective strains contain alleles encoding RB50-like C termini. Host specificity is likely, although not necessarily, controlled by specific interactions that occur when the bacterium first encounters a potential host (i.e., at the level of adherence). Although similarity to intimin and invasin suggests that BipA plays a role in adherence, we were unable to detect BipA-dependent adherence in any of multiple cell lines, including those of respiratory, endothelial, and macrophage origin (54; unpublished observations). It is still possible, however, that BipA mediates adherence to specific respiratory cells that are either unavailable as cell lines or are available but lack a specific receptor(s) when they are cultured in vitro or that BipA mediates adherence to noncellular components of the respiratory tract, such as mucus. We are currently performing experiments to test these possibilities.
In addition to nucleotide sequence differences at their 3′ ends, bipA alleles were distinguished by differences in the number of 90-amino-acid repeats which they encoded. It is possible that these differences reflect variability in surface characteristics displayed by the different species. It is known that the lipopolysaccharide structures of B. pertussis, B. parapertussishu, and B. bronchiseptica differ; B. pertussis strains express only lipid A and a branched-chain core oligosaccharide, while B. bronchiseptica and B. parapertussishu strains add O-antigen-like homopolymers to their core structures under Bvg−-phase conditions (3, 4, 15, 23, 29, 55). Although predominance of O antigens or other large surface molecules in the Bvgi phase is unknown, our results suggest that the surfaces of B. bronchiseptica and B. pertussis differ such that eight 90-amino-acid domains are required for exposure of the C terminus of BipA in B. bronchiseptica, while only five 90-amino-acid domains may be required for exposure of the C terminus of BipA in B. pertussis. It is curious that bipA of B. pertussis 18323 encodes only one 90-amino-acid domain. In many respects, 18323 appears to be more similar to B. parapertussishu and B. bronchiseptica strains than to B. pertussis strains (5, 19, 40, 52). The presence of only one 90-amino-acid domain in BipA of 18323, like the data obtained for the B. parapertussishu strains, provides more evidence for the unique position of this B. pertussis strain on the Bordetella phylogenetic tree. The fact that the bipA allele in 18323 is expressed in a pattern similar to the pattern in B. bronchiseptica and other B. pertussis strains, however, provides support for the hypothesis that BipA confers a selective advantage to members of these species. If BipA does provide some selective advantage for 18323, however, the surface characteristics of 18323 must be such that the C terminus of a BipA protein with only one 90-amino-acid domain is exposed.
With regard to phylogenetic and evolutionary implications, our results are consistent with observations which indicate that there are significant genomic rearrangements within, and potentially across, Bordetella species (52, 53). As the progenitor organism for the B. bronchiseptica cluster is proposed to have been B. bronchiseptica (19, 57) and our analyses indicate that all B. bronchiseptica strains have the same bipA allele, vertical inheritance of bipA would require the occurrence and selection of the same set of mutations independently in each of three lineages in order for B. pertussis, B. parapertussishu, and B. parapertussisov strains to encode the same BipA C-terminal domain, which is significantly different from that of B. bronchiseptica. Vertical inheritance of bipA is even less probable given the lack of bipA expression in B. parapertussishu strains and the low level of expression of bipA in B. parapertussisov strains. Moreover, a sequence comparison of bipA and flanking regions suggests strongly that the truncated ′bipA gene present in B. parapertussishu strain 12822 was acquired horizontally from B. pertussis. Although little is known about the genome of B. parapertussisov strains, it seems likely that bipA was acquired horizontally in these strains as well, with subsequent mutations giving rise to variation in the number of regions encoding the 90-amino-acid repeat and nucleotide sequence differences at the promoter. We can only speculate about the selective advantages of these gene-swapping events. With regard to B. parapertussishu strains, an interesting possibility is that acquisition of ′bipA from B. pertussis resulted in replacement of a B. bronchiseptica-like bipA allele that conferred a selective disadvantage to B. parapertussishu strains during their adaptation to human hosts. Similarly, one can imagine that expression of a B. pertussis-like bipA allele in B. parapertussisov strains might be disadvantageous, leading to accumulation of promoter mutations that decrease transcription levels. The possibility that the wrong bipA allele could be detrimental at some point in the infectious cycle is an interesting one that we are investigating using chimeric strains.
Our results are consistent with the hypothesis that BipA plays a role in a specific mode of transmission that is used by a subset of Bordetella strains. They also contribute to the growing body of information indicating the highly fluid nature of the Bordetella genome and high degree of gene transfer and recombination within and among strains of the B. bronchiseptica cluster. The anticipated publication of the comparative genome sequences of B. pertussis Tohama I, B. parapertussishu 12822, and B. bronchiseptica RB50 will undoubtedly provide considerably more insight into the complex phylogenetic and evolutionary relationships among these bacteria.
Acknowledgments
We thank the Sanger Centre for releasing Bordetella genome sequence data to the public domain prior to publication and members of our laboratory for helpful discussions.
This work was supported by NIH grant AI43986.
Editor: V. J. DiRita
REFERENCES
- 1.Akerley, B. J., P. A. Cotter, and J. F. Miller. 1995. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80:611-620. [DOI] [PubMed] [Google Scholar]
- 2.Akerley, B. J., and J. F. Miller. 1993. Flagellin gene transcription in Bordetella bronchiseptica is regulated by the BvgAS virulence control system. J. Bacteriol. 175:3468-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, A., and D. Maskell. 1996. The identification, cloning and mutagenesis of a genetic locus required for lipopolysaccharide biosynthesis in Bordetella pertussis. Mol. Microbiol. 19:37-52. [DOI] [PubMed] [Google Scholar]
- 4.Allen, A. G., T. Isobe, and D. J. Maskell. 1998. Identification and cloning of waaF (rfaF) from Bordetella pertussis and use to generate mutants of Bordetella spp. with deep rough lipopolysaccharide. J. Bacteriol. 180:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arico, B., R. Gross, J. Smida, and R. Rappuoli. 1987. Evolutionary relationships in the genus Bordetella. Mol. Microbiol. 1:301-308. [DOI] [PubMed] [Google Scholar]
- 6.Arico, B., V. Scarlato, D. M. Monack, S. Falkow, and R. Rappuoli. 1991. Structural and genetic analysis of the bvg locus in Bordetella species. Mol. Microbiol. 5:2481-2491. [DOI] [PubMed] [Google Scholar]
- 7.Betsou, F., O. Sismeiro, A. Danchin, and N. Guiso. 1995. Cloning and sequence of the Bordetella bronchiseptica adenylate cyclase-hemolysin-encoding gene: comparison with the Bordetella pertussis gene. Gene 162:165-166. [DOI] [PubMed] [Google Scholar]
- 8.Cherry, J. D., and U. Heininger. 1998. Pertussis, p. 1423-1440. In R. D. Feigin and J. D. Cherry (ed.), Textbook of pediatric infectious diseases, 4th ed. Saunders, Philadelphia, Pa.
- 9.Cotter, P. A., and V. J. DiRita. 2000. Bacterial virulence gene regulation: an evolutionary perspective. Annu. Rev. Microbiol. 54:519-565. [DOI] [PubMed] [Google Scholar]
- 10.Cotter, P. A., and J. F. Miller. 2001. Bordetella, p. 619-674. In E. A. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, San Diego, Calif.
- 11.Cotter, P. A., and J. F. Miller. 1994. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 62:3381-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotter, P. A., and J. F. Miller. 1997. A mutation in the Bordetella bronchiseptica bvgS gene results in reduced virulence and increased resistance to starvation, and identifies a new class of Bvg-regulated antigens. Mol. Microbiol. 24:671-685. [DOI] [PubMed] [Google Scholar]
- 13.Cullinane, L. C., M. R. Alley, R. B. Marshall, and B. W. Manktelow. 1987. B. parapertussis from lambs. N. Z. Vet. J. 35:175.. [DOI] [PubMed] [Google Scholar]
- 14.Deora, R., H. J. Bootsma, J. F. Miller, and P. A. Cotter. 2001. Diversity in the Bordetella virulence regulon: transcriptional control of a Bvg-intermediate phase gene. Mol. Microbiol. 40:669-683. [DOI] [PubMed] [Google Scholar]
- 15.Di Fabio, J. L., M. Caroff, D. Karibian, J. C. Richards, and M. B. Perry. 1992. Characterization of the common antigenic lipopolysaccharide O-chains produced by Bordetella bronchiseptica and Bordetella parapertussis. FEMS Microbiol. Lett. 76:275-281. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez, R. C., and A. A. Weiss. 1994. Cloning and sequencing of a Bordetella pertussis serum resistance locus. Infect. Immun. 62:4727-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finn, T. M., and L. A. Stevens. 1995. Tracheal colonization factor: a Bordetella pertussis secreted virulence determinant. Mol. Microbiol. 16:625-634. [DOI] [PubMed] [Google Scholar]
- 18.Fitzhenry, R. J., D. J. Pickard, E. L. Hartland, S. Reece, G. Dougan, A. D. Phillips, and G. Frankel. 2002. Intimin type influences the site of human intestinal mucosal colonisation by enterohaemorrhagic Escherichia coli O157:H7. Gut 50:180-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlach, G., F. von Wintzingerode, B. Middendorf, and R. Gross. 2001. Evolutionary trends in the genus Bordetella. Microbes Infect. 3:61-72. [DOI] [PubMed] [Google Scholar]
- 20.Goldman, W. E., and B. T. Cookson. 1988. Structure and functions of the Bordetella tracheal cytotoxin. Tokai J. Exp. Clin. Med. 13(Suppl.):187-191. [PubMed] [Google Scholar]
- 21.Goldman, W. E., and L. A. Herwaldt. 1985. Bordetella pertussis tracheal cytotoxin. Dev. Biol. Stand. 61:103-111. [PubMed] [Google Scholar]
- 22.Goodnow, R. A. 1980. Biology of Bordetella bronchiseptica. Microbiol. Rev. 44:722-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harvill, E. T., A. Preston, P. A. Cotter, A. G. Allen, D. J. Maskell, and J. F. Miller. 2000. Multiple roles for Bordetella lipopolysaccharide molecules during respiratory tract infection. Infect. Immun. 68:6720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heininger, U., K. Stehr, S. Schmitt-Grohe, C. Lorenz, R. Rost, P. D. Christenson, M. Uberall, and J. D. Cherry. 1994. Clinical characteristics of illness caused by Bordetella parapertussis compared with illness caused by Bordetella pertussis. Pediatr. Infect. Dis. J. 13:306-309. [DOI] [PubMed] [Google Scholar]
- 25.Hewlett, E. L., V. M. Gordon, J. D. McCaffery, W. M. Sutherland, and M. C. Gray. 1989. Adenylate cyclase toxin from Bordetella pertussis. Identification and purification of the holotoxin molecule. J. Biol. Chem. 264:19379-19384. [PubMed] [Google Scholar]
- 26.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 27.Kloos, W. E., N. Mohapatra, W. J. Dobrogosz, J. W. Ezzel, and C. R. Manclark. 1981. Deoxyribonucleotide sequence relationships among Bordetella species. J. Int. Syst. Bacteriol. 31:173-176. [Google Scholar]
- 28.Knapp, S., and J. J. Mekalanos. 1988. Two trans-acting regulatory genes (vir and mod) control antigenic modulation in Bordetella pertussis. J. Bacteriol. 170:5059-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Blay, K., P. Gueirard, N. Guiso, and R. Chaby. 1997. Antigenic polymorphism of the lipopolysaccharides from human and animal isolates of Bordetella bronchiseptica. Microbiology 143:1433-1441. [DOI] [PubMed] [Google Scholar]
- 30.Leong, J. M., R. S. Fournier, and R. R. Isberg. 1990. Identification of the integrin binding domain of the Yersinia pseudotuberculosis invasin protein. EMBO J 9:1979-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, J., N. F. Fairweather, P. Novotny, G. Dougan, and I. G. Charles. 1992. Cloning, nucleotide sequence and heterologous expression of the protective outer-membrane protein P.68 pertactin from Bordetella bronchiseptica. J. Gen. Microbiol. 138:1697-1705. [DOI] [PubMed] [Google Scholar]
- 32.Liu, H., L. Magoun, S. Luperchio, D. B. Schauer, and J. M. Leong. 1999. The Tir-binding region of enterohaemorrhagic Escherichia coli intimin is sufficient to trigger actin condensation after bacterial-induced host cell signalling. Mol. Microbiol. 34:67-81. [DOI] [PubMed] [Google Scholar]
- 33.Livey, I., and A. Wardlaw. 1984. Production and properties of Bordetella pertussis heat-labile toxin. J. Med. Microbiol. 17:91-103. [DOI] [PubMed] [Google Scholar]
- 34.Martinez de Tejada, G., J. F. Miller, and P. A. Cotter. 1996. Comparative analysis of the virulence control systems of Bordetella pertussis and Bordetella bronchiseptica. Mol. Microbiol. 22:895-908. [DOI] [PubMed] [Google Scholar]
- 35.Merkel, T. J., S. Stibitz, J. M. Keith, M. Leef, and R. Shahin. 1998. Contribution of regulation by the bvg locus to respiratory infection of mice by Bordetella pertussis. Infect. Immun. 66:4367-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mooi, F. R., W. H. Jansen, H. Brunings, H. Gielen, H. G. J. van der Heide, H. C. Walvoort, and P. A. M. Guinee. 1992. Construction and analysis of Bordetella pertussis mutants defective in the production of fimbriae. Microbiol. Pathol. 12:127-135. [DOI] [PubMed] [Google Scholar]
- 37.Muller, M., and A. Hildebrandt. 1993. Nucleotide sequences of the 23S rRNA genes from Bordetella pertussis, B. parapertussis, B. bronchiseptica and B. avium, and their implications for phylogenetic analysis. Nucleic Acids Res. 21:3320.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munoz, J. J. 1988. Action of pertussigen (pertussis toxin) on the host immune system in pathogenesis and immunity in pertussis, p. 173-192. In A. C. Wardlaw and R. Parton (ed.), Pathogenesis and immunity in pertussis. Wiley and Sons, Chichester, United Kingdom.
- 39.Musser, J. M., D. A. Bemis, H. Ishikawa, and R. K. Selander. 1987. Clonal diversity and host distribution in Bordetella bronchiseptica. J. Bacteriol. 169:2793-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Musser, J. M., E. L. Hewlett, M. S. Peppler, and R. K. Selander. 1986. Genetic diversity and relationships in populations of Bordetella spp. J. Bacteriol. 166:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phillips, A. D., and G. Frankel. 2000. Intimin-mediated tissue specificity in enteropathogenic Escherichia coli interaction with human intestinal organ cultures. J. Infect. Dis. 181:1496-1500. [DOI] [PubMed] [Google Scholar]
- 42.Pittman, M. 1979. Pertussis toxin: the cause of the harmful effects and prolonged immunity of whooping cough. A hypothesis. Rev. Infect. Dis. 1:401-412. [DOI] [PubMed] [Google Scholar]
- 43.Porter, J. F., K. Connor, and W. Donachie. 1994. Isolation and characterization of Bordetella parapertussis-like bacteria from ovine lungs. Microbiology 140:255-261. [DOI] [PubMed] [Google Scholar]
- 44.Reece, S., C. P. Simmons, R. J. Fitzhenry, S. Matthews, A. D. Phillips, G. Dougan, and G. Frankel. 2001. Site-directed mutagenesis of intimin alpha modulates intimin-mediated tissue tropism and host specificity. Mol. Microbiol. 40:86-98. [DOI] [PubMed] [Google Scholar]
- 45.Relman, D. A., M. Domenighini, E. T. Tuomanen, R. Rappuoli, and S. Falkow. 1989. Filamentous hemagglutinin of Bordetella pertussis: nucleotide sequence and crucial role in adherence. Proc. Natl. Acad. Sci. USA 86:2634-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts, M., N. F. Fairweather, E. Leininger, D. Pickard, E. L. Hewlett, A. Robinson, C. Hayward, G. Dougan, and I. G. Charles. 1991. Construction and characterization of Bordetella pertussis mutants lacking the vir-regulated P.69 outer membrane protein. Mol. Microbiol. 5:1393-1404. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Savelkoul, P. H., D. P. de Kerf, R. J. Willems, F. R. Mooi, B. A. van der Zeijst, and W. Gaastra. 1996. Characterization of the fim2 and fim3 fimbrial subunit genes of Bordetella bronchiseptica: roles of Fim2 and Fim3 fimbriae and flagella in adhesion. Infect. Immun. 64:5098-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stainer, D. W., and M. J. Scholte. 1970. A simple chemically defined medium for the production of phase I Bordetella pertussis. J. Gen. Microbiol. 63:211-220. [DOI] [PubMed] [Google Scholar]
- 50.Stenson, T. H., and M. S. Peppler. 1995. Identification of two bvg-repressed surface proteins of Bordetella pertussis. Infect. Immun. 63:3780-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stibitz, S., and J. F. Miller. 1994. Coordinate regulation of virulence in Bordetella pertussis mediated by the vir (bvg) locus, p. 407-422. In V. L. Miller, J. B. Kaper, D. A. Portnoy, and R. R. Isberg (ed.), The molecular biology of bacterial pathogenesis. ASM Press, Washington, D.C.
- 52.Stibitz, S., and M. S. Yang. 1997. Genomic fluidity of Bordetella pertussis assessed by a new method for chromosomal mapping. J. Bacteriol. 179:5820-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stibitz, S., and M. S. Yang. 2001. Genomic plasticity in natural populations of Bordetella pertussis. J. Bacteriol. 181:5512-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stockbauer, K. E., B. Fuchslocher, J. F. Miller, and P. A. Cotter. 2001. Identification and characterization of BipA, a Bordetella Bvg-intermediate phase protein. Mol. Microbiol. 39:65-78. [DOI] [PubMed] [Google Scholar]
- 55.Turcotte, M. L., D. Martin, B. R. Brodeur, and M. S. Peppler. 1997. Tn5-induced lipopolysaccharide mutations in Bordetella pertussis that affect outer membrane function. Microbiology 143:2381-2394. [DOI] [PubMed] [Google Scholar]
- 56.Uhl, M. A., and J. F. Miller. 1995. Bordetella pertussis BvgAS virulence control system, p. 333-349. In J. A. Hoch and T. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 57.van der Zee, A., F. Mooi, J. Van Embden, and J. Musser. 1997. Molecular evolution and host adaptation of Bordetella spp.: phylogenetic analysis using multilocus enzyme electrophoresis and typing with three insertion sequences. J. Bacteriol. 179:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verghese, M. W., C. D. Smith, and R. Snyderman. 1985. Potential role for a guanine nucleotide regulatory protein in chemoattractant receptor mediated polyphosphoinositide metabolism, Calif.++ mobilization and cellular responses by leukocytes. Biochem. Biophys. Res. Commun. 127:450-457. [DOI] [PubMed] [Google Scholar]
- 59.Weiss, A. A., and S. Falkow. 1984. Genetic analysis of phase change in Bordetella pertussis. Infect. Immun. 43:263-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wirsing von Konig, C. H., and H. Finger. 1994. Role of pertussis toxin in causing symptoms of Bordetella parapertussis infection. Eur. J. Clin. Microbiol. Infect. Dis. 13:455-458. [DOI] [PubMed] [Google Scholar]
- 61.Worley, M. J., I. Stojiljkovic, and F. Heffron. 1998. The identification of exported proteins with gene fusions to invasin. Mol. Microbiol. 29:1471-1480. [DOI] [PubMed] [Google Scholar]
- 62.Yuk, M. H., E. T. Harvill, P. A. Cotter, and J. F. Miller. 2000. Modulation of host immune responses, induction of apoptosis and inhibition of NF-KB activation by the Bordetella type III secretion system. Mol. Microbiol. 35:991-1104. [DOI] [PubMed] [Google Scholar]
- 63.Yuk, M. H., E. T. Harvill, and J. F. Miller. 1998. The bvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Mol. Microbiol. 28:945-959. [DOI] [PubMed] [Google Scholar]