Abstract
Immune evasion by Lyme spirochetes is a multifactorial process involving numerous mechanisms. The OspE protein family undergoes antigenic variation during infection and binds factor H (fH) and possibly FHL-1/reconectin. In Borrelia burgdorferi B31MI, the OspE family consists of three paralogs: BBL39 (ErpA), BBP38, and BBN38 (ErpP). BBL39 and BBP38 are identical and therefore are referred to here as BBL39. The goals of this study were to assess the specificity of the antibody (Ab) response to the OspE paralogs and to identify the domains or determinants of OspE that are required for the binding of fH and OspE-targeting Abs that develop during infection. Here we demonstrate that at least some of the anti-OspE Abs produced during infection are paralog specific and that Ab binding requires conformational determinants whose formation requires both the N- and C-terminal domains of OspE. The binding of fH to OspE was also found to be dependent on conformational determinants. It is also demonstrated here that all of the OspE paralogs expressed by B. burgdorferi B31MI are capable of binding fH. The binding of fH to members of the OspF protein family was also assessed. In contrast to an earlier report, no binding of BBO39 or BBR42 to human fH was detected. Lastly, a series of competitive binding enzyme-linked immunosorbent assay analyses, designed to determine if fH and infection serum Abs bind to the same sites on OspE, revealed that these ligands interact with different regions of OspE.
Lyme disease is a chronic infection caused by pathogenic species of the Borrelia burgdorferi sensu lato complex (31). The ability to maintain chronic infection indicates that the Lyme spirochetes are capable of immune evasion. The immune evasion strategies of the Lyme disease spirochetes are multifactorial, with several different contributing mechanisms (1, 6, 7, 16, 24, 25, 29, 33, 34, 37). The OspE paralogs contribute to immune evasion in two distinct ways: by antigenic variation (34) and through the binding of the complement regulatory protein, factor H (fH) (3, 17, 19; this work). The potential for OspE to be involved in these processes is supported by several studies that have demonstrated it to be surface exposed (11, 15, 21), to be expressed in both ticks and mammals, and to elicit a strong antibody (Ab) response (14, 16, 22, 27, 34). Analyses of OspE sequences have revealed that the protein is organized into conserved regions interspersed by two hypervariable domains (23, 34, 35). Analyses of the specificity of the Ab response to different OspE variants suggest that it is the hypervariable regions that are targeted by the Ab response during infection in mice (34). In contrast, the ability of divergent OspE proteins to bind fH (26) suggests that it is the conserved regions of OspE that are involved in fH binding.
Some Lyme spirochete isolates are serum resistant (20, 36), and as indicated above, some isolates can bind fH and fH-like protein 1/reconectin (FHL-1) (2, 18). fH and FHL-1, which are derived from the same transcript through processing events (13), serve as cofactors for factor I-mediated degradation of C3b. C3b degradation results in decreased levels of the C3 convertase complex, which facilitates complement evasion. Borreliae express several fH binding proteins (FHBPs) that were originally referred to as CRASPs (complement regulator-acquiring surface proteins) (19). OspE is the only Borrelia FHBP that has been identified at the sequence level. OspE interacts with the C terminus of fH (17). Neither the OspE determinants required for fH binding nor those required for the binding of Abs generated during infection have been identified.
In this report we assess the specificity of the Ab response to the OspE paralogs, localize OspE epitopes that are exposed during infection, and identify OspE determinants that are required for the binding of fH and Abs elicited during infection. These analyses demonstrate that all OspE paralogs of B. burgdorferi B31MI can bind fH, that the variable regions of the OspE proteins are immunodominant, and that the formation and proper presentation of these epitopes, as well as the fH binding site, involve conformational determinants. Using competitive binding assays, we demonstrate that the fH and infection serum Ab binding sites on OspE are separate and distinct. In addition, B. burgdorferi B31MI was also found to express a dominant 27-kDa protein that may specifically bind human fH (hfH) or FHL-1 protein and play an important role in fH-mediated evasion of complement attack.
MATERIALS AND METHODS
Protein nomenclature, bacterial cultivation, and generation of recombinant proteins (r proteins).
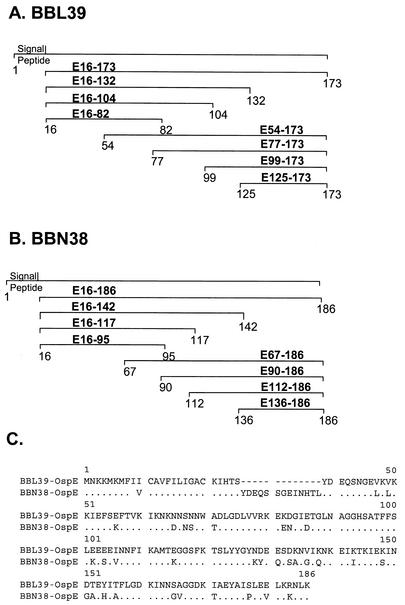
This analysis focuses on B. burgdorferi B31MI and its OspE and OspF proteins. The complete genome sequence for B31MI has been determined by The Institute for Genomic Research (TIGR) (12). Several different nomenclature strategies have been applied to the OspE proteins. In this report we collectively refer to these proteins as the OspE proteins and employ the paralog designations assigned by TIGR, with cross-references to other nomenclatures indicated where appropriate. The OspE protein family of B. burgdorferi B31MIpc (a clone derived from B31MI by subsurface plating) is made up of BBP38 (ErpA), BBL39 (ErpA), and BBN38 (ErpP) (12). BBP38 and BBL39 are identical and are collectively referred to below as BBL39. The amino acid sequences for these proteins are presented in Fig. 1. In isolate B31MI the OspF protein family is composed of three proteins: BBM38, BBO39, and BBR42.
FIG. 1.
Schematics of OspE (BBL39 and BBN38) constructs employed in this study and alignment of BBL39 and BBN38 amino acid sequences. (A and B) Schematics depict the BBL39 and BBN38 truncations that were generated and the nomenclature assigned to each. All constructs were generated as N-terminal S-Tag fusion proteins in E. coli as described in the text. Numbering is based on amino acid positions of the unprocessed proteins. (C) Alignment of the amino acid sequences of BBL39 and BBN38 (12).
B. burgdorferi B31MIpc was cultivated in BSK-H complete medium (Sigma) supplemented to 6% with rabbit sera (Sigma) at 33°C. To obtain recombinant BBL39 (rBBL39), rBBR42, and rBBO39 (or subfragments thereof), paralog-specific PCR amplicons were generated by using boiled cell lysates as the template. Primers (Table 1) were designed with tail sequences to allow for ligase-independent cloning (LIC) of the amplicons into the pET-32 LIC expression vector and subsequent expression as S-Tag fusion proteins (25). The S-Tag fusion adds approximately 17 kDa to the molecular mass of each r protein. Schematics of the constructs are given in Fig. 1. The sequence of each construct was verified by DNA sequencing (MWG Biotech). Expression of the r proteins was carried out as described by the vector manufacturer (Novagen) and confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting using the S-Tag protein for detection (24). Throughout this report, the term “full-length protein” refers to the unlipidated form of each protein minus its leader peptide (∼16 amino acids [aa]). To specifically PCR amplify, clone, and express full-length BBN38, a different PCR strategy was necessary. A 1,620-bp segment of cp32-9 (plasmid N) was amplified by using the BBN38-UHB (+) and BBN39-R2 (−) primers and TA cloned as previously described (34). The r plasmid was then used in a second round of PCR using the LIC-tailed primers UHB-N38 (+) and BBN39-R1 (−) to allow for the amplification of BBN38. The amplicon was then annealed into the pET-32 LIC vector (Novagen) and expressed as described above.
TABLE 1.
Oligonucleotide primers
| Primer | Sequence (5′ to 3′) |
|---|---|
| UHB(+) | GTTGTTTAAAAATTACATTTGCG |
| L40-R(−) | TATTGTTTTCTTCTACACCCTT |
| N38-UHB(+) | GTAGTCTTTTGTAATGAGCCGG |
| N39-R2(−) | CATTTAATTTAGTTCTTAGCGCGT |
| UHB-N38(+) | GTTGGTTAAAATTACATTTGAG |
| N39-R1(−) | CTTCTTTTTCTTCTTCTTG |
| L39-1(+) | GACGACGACAAGATGGAGAAATTTATGAATAAG |
| L39-16(+) | GACGACGACAAGATGCTTATAGGTGCTTGCAAG |
| L39-54(+) | GACGACGACAAGATGAGTAATAACTGGGCAGAC |
| L39-77(+) | GACGACGACAAGATGAACGCTGGGGGACATTC |
| L39-99(+) | GACGACGACAAGATGGCAATGACTGAAGGTGGA |
| L39-125(+) | GACGACGACAAGATGAAGAATAAAGAGATAAAAACAAAG |
| L39-82(−) | GAGGAGAAGCCCGGTTTACGAATGTCCCCCAGCG |
| L39-104(−) | GAGGAGAAGCCCGGTTTATCCACCTTCAGTCATTGC |
| L39-132(−) | GAGGAGAAGCCCGGTTTACTTTGTTTTTATCTCTTTATTC |
| L39-173(−) | GAGGAGAAGCCCGGTTTATTTTAAATTTCTTTTAAGCTC |
| N38-16(+) | GACGACGACAAGATGCTTATAGGTGCTTGCAAAATTC |
| N38-67(+) | GACGACGACAAGATGAATAGTAACTGGACAGAC |
| N38-90(+) | GACGACGACAAGATGAACGCTGGGGGACATTC |
| N38-112(+) | GACGACGACAAGATGGCAATGACTAAAGGCGG |
| N38-136(+) | GACGACGACAAGATGGGTATCCAAAACAAAGAGATC |
| N38-186(−) | GAGGAGAAGCCCGGTTTATTTTAAATTTTTTTTAAGCAC |
| N38-142(−) | GAGGAGAAGCCCGGTTTAGATCTCTTTGTTTTGGATACC |
| N38-117(−) | GAGGAGAAGCCCGGTTTATCCGCCTTTAGTCATTGC |
| N38-95(−) | GAGGAGAAGCCCGGTTTACGAATGTCCCCCAGCG |
| O39-F1(+) | GACGACGACAAGATTTATGCAAGTGGTGAAAATCTA |
| O39-R3(−) | GAGGAGAAGCCCGGTTGTTATTCTTTTTTATCTTCTTCTATTCC |
| R42-F1(+) | GACGACGACAAGATTGATGTAACTAGTAAAGATTTA |
| R42-R3(−) | GAGGAGAAGCCCGGTCTTTATTCTTTTTTACCTTCTACAG |
Underlining indicates the tail sequences incorporated into each primer to allow for annealing with LIC vectors as described in the text.
Infection of mice with B. burgdorferi B31MIpc and generation of infection sera.
C3H/HeJ mice were infected with B. burgdorferi B31MIpc by intradermal needle inoculation of 103 spirochetes between the shoulder blades. Blood was collected at weeks 4, 8, and 12, and sera were obtained as previously described (34). The mice were sacrificed at week 12, a time point at which full dissemination of the spirochetes had occurred.
Peptide synthesis and generation of anti-OspE peptide antisera.
Two 25-aa peptides that span the C-terminal 46 aa of BBL39 (with a short overlap) were synthesized as previously described (30). For the purpose of generating antipeptide antisera, a T-cell epitope (OVA323-339 T, ISQAVHAAHAEINEAGR) was incorporated at the N terminus of each peptide. Peptide sequences were as follows: OspE-pep1, INNSAGGDKIAEYAISLEELK RNLK; OspE-pep2, IKTKIEKINDTEYITFLGDKINNSA. To generate antisera, each peptide (50 μg) was injected into C3H/HeJ mice (three per set) in combination with either incomplete Freund's adjuvant and alum or montanide as previously described (30). Two boosters were administered at 2-week intervals. Blood was collected by tail snip, sera were obtained, and Ab titers were assessed by enzyme-linked immunosorbent assay (ELISA) (30). The specificities of the antisera were assessed by immunoblot analysis of rBBL39, rBBN38, and B. burgdorferi B31MIpc cell lysates as described below.
Immunoblot analysis and preabsorption of infection sera.
Cell lysates of B. burgdorferi B31MIpc or Escherichia coli that had been induced to express r proteins were fractionated by SDS-PAGE in 12% Bio-Rad Ready Gels and immunoblotted as previously described (25). In some cases, infection sera were preabsorbed prior to screening of the immunoblots. To preabsorb infection sera, E. coli cells expressing the appropriate r protein were first boiled in phosphate-buffered saline (PBS) for 15 min and then an aliquot of the cell lysate was incubated with the diluted antisera (in blocking buffer, consisting of PBS with 5% nonfat dry milk and 0.2% Tween) overnight at 4°C. The sample was centrifuged to pellet cellular debris, and the supernatant was recovered. Another aliquot of the E. coli lysate was added, and samples were incubated for 1 h at room temperature. These absorbed sera were then employed in the immunoblot analyses at a dilution of 1:500 with goat anti-mouse immunoglobulin G (IgG) as the secondary Ab. Other Ab preparations and antisera were employed in the immunoblot analyses at the following dilutions (with horseradish peroxidase-conjugated secondary Abs given in parentheses): anti-rOspE antisera (21), 1:1,000 (secondary Ab, goat anti-rabbit IgG); anti-fH antisera, 1:800 (secondary Ab, rabbit anti-goat IgG); anti-OspE pep1 and anti-OspE pep2 antisera, 1:250 (secondary Ab, goat anti-mouse IgG); mouse anti-hfH monoclonal Ab (clone OX23, Accurate Chemical and Scientific Corporation), 1:500 (secondary Ab, goat anti-mouse IgG). Immunoblots were generated by electroblotting as previously described (24) by using Immobilon P membranes (Millipore). All secondary Abs were from Pierce and were used at a 1:40,000 dilution. In all cases the Pierce Super Signal substrate was employed for chemiluminescent detection of Ab binding.
Nondenaturing PAGE.
To allow for immunoblot analysis of the various proteins in a state more closely approximating their native conformation, cell lysates expressing the r proteins were fractionated by electrophoresis under nondenaturing conditions. The separating gel (12% polyacrylamide) was cast by using 1.5 M Tris-HCl (pH 8.8), and the stacking gel (4% polyacrylamide) was cast by using 1.0 M Tris-HCl (pH 6.8). The running buffer consisted of 0.19 M glycine and 0.04 M Tris base, and the loading buffer consisted of 0.125 M Tris-HCl (pH 6.8), 20% glycerol, and 1% bromophenol blue. Samples were electrophoresed under constant voltage (200 V) and electroblotted as described above.
Affinity ligand binding immmunoblot assays: fH binding assays.
The fH used in affinity ligand binding immunoblot assays was purchased from Calbiochem. The manufacturer purified the protein from human sera. It is estimated to be 95% pure. Data below suggest that it may also contain trace amounts of FHL-1. The abilities of membrane-immobilized r proteins to bind fH were assessed as follows. r proteins, cell lysates of B. burgdorferi, and hfH (Calbiochem) were fractionated by SDS-PAGE and immunoblotted onto Immobilon P membranes as detailed above. After the initial blocking of the membranes, hfH was incubated with the blots at 10 ng/μl in blocking buffer for 1 h. Membranes were washed extensively prior to addition of either goat anti-hfH antisera (1:800) (Calbiochem) or an anti-hfH monoclonal Ab (1:500). A goat-anti-human IgG Ab (1:40,000) served as the secondary Ab in both cases. As a control, a second set of blots was treated as above except that hfH was not added.
ELISAs: fH and infection serum Ab binding assays.
In all ELISAs, r proteins, purified by using the S-Tag rEK Purification Kit as instructed by the manufacturer (Novagen), were used. The concentration of each purified r protein was determined by bicinchoninic acid assay (Pierce). The wells of the 96-well microtiter plates were coated by incubating the protein or peptide of interest at a concentration of 10 ng/μl in 20 mM Tris-HCl (pH 8.5) at room temperature for 3 h. Nonspecific binding was blocked by use of blocking buffer (PBS-Tween [PBST] with 5% nonfat dry milk). To assess fH binding, hfH was added (10 ng/μl in 100 μl of Tris-buffered saline with 20 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 0.05% Tween 20) for 3 h at room temperature. When normal human serum served as the source of fH, it was used at a 1:800 dilution. Wells were washed five times with 400 μl of PBST. The appropriate antisera (as indicated in each figure) were added in a total volume of 100 μl (PBST with 5% nonfat dry milk) and incubated for 3 h at room temperature. Antisera were removed, and wells were washed as above. The appropriate secondary Ab (dilution, 1:40,000) was added in 100 μl of blocking buffer for 1 h at room temperature, followed by washing as above. For detection, the 1-Step Slow TMB ELISA kit was used as directed by the manufacturer (Pierce). Plates were read by using a Spectromax 250 plate reader (Molecular Devices) at 450 nm. Data presented are average readings from three different plates, and standard deviations are given.
RESULTS
Demonstration that OspE paralog-specific Abs develop during infection in mice.
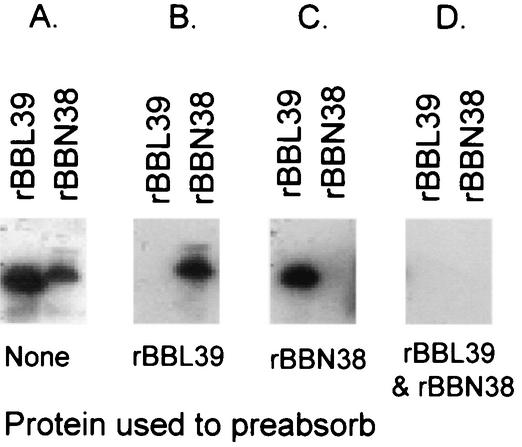
To determine if OspE paralog-specific Abs are elicited during infection of mice with B. burgdorferi B31MIpc, an immunoblot with repeated sets of cell lysates of E. coli that had been induced to express rBBL39 and rBBN38 was generated. Expression of the r proteins was confirmed by screening with S-Tag protein, and screening with anti-rOspE antisera confirmed that the proteins induced were OspE (data not shown). When screened with infection sera, both proteins reacted strongly (Fig. 2). To determine if paralog-specific Abs develop during infection, infection sera were absorbed with BBL39, BBN38, or both prior to immunoblot analysis. Absorption with BBL39 completely eliminated detection of BBL39 but had no effect on detection of BBN38. Similarly, absorption of infection sera with BBN38 completely eliminated detection of BBN38 but had no discernible effect on detection of BBL39. Absorption of antisera with both proteins completely eliminated detection of both BBL39 and BBN38. Hence, while BBN38 and BBL39 share regions of high sequence identity, it is evident that it is the paralog-specific sequences that are immunodominant and are the primary targets of Abs that develop during infection. These data also demonstrate that both paralogs are expressed during infection in mice.
FIG. 2.
Demonstration that a component of the anti-OspE Ab response elicited during infection in mice is paralog specific. E. coli harboring the full-length BBL39 or BBN38 construct was induced to express the r protein, and cell lysates were fractionated by SDS-PAGE and immunoblotted. Membranes were screened with sera collected from a mouse infected for 12 weeks with B. burgdorferi B31MIpc (A) or with infection sera preabsorbed with either BBL39 (B) or BBN38 (C) or both (D). All methods were as described in the text.
Demonstration that the binding of OspE-targeting Abs elicited during infection in mice requires conformational or discontinuous epitopes for optimal binding.
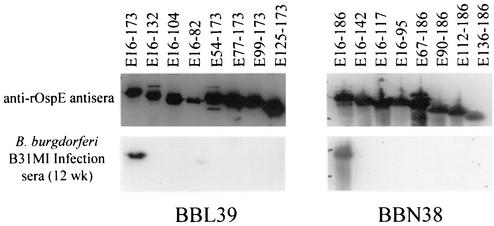
To identify the determinants of BBL39 and BBN38 that are involved in the binding of infection serum Abs, OspE r proteins and subfragments were fractionated by SDS-PAGE and immunoblotted. All of the OspE r proteins were immunoreactive with S-Tag (data not shown) and anti-rOspE polyclonal antisera (Fig. 3). In contrast, infection sera, which harbor Abs elicited to the more natural and native forms of the OspE proteins, reacted only with full-length forms of BBL39 and BBN38 and not with the C- or N-terminal truncations. The fact that independent deletions at either end of the protein completely eliminated reactivity suggests that the epitopes recognized during infection are not solely linear in nature. The requirement for both N- and C-terminal domains for Ab binding indicates that conformational or discontinuous epitopes are the primary target of Abs.
FIG. 3.
Analysis of the BBL39 and BBN38 determinants necessary for binding of anti-OspE Abs elicited during infection in mice. Subfragments of BBL39 and BBN38 were generated as S-Tag fusion proteins, fractionated by SDS-PAGE, immunoblotted, and screened with anti-rOspE antisera or 12-week infection sera. All methods were as described in the text. Note that addition of the S-Tag to each OspE subfragment adds approximately 17 kDa to the molecular mass.
Demonstration of infection serum Ab binding to OspE truncations fractionated under nondenaturing conditions.
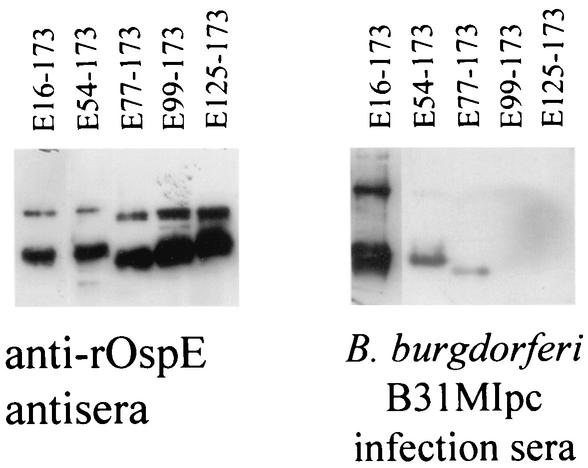
To further assess the postulate that conformational determinants are involved in the binding of Abs, a subset of the BBL39 subfragments was fractionated under nondenaturing conditions and immunoblotted. BBN38 and some of the BBL39 subfragments could not be analyzed by this approach because they could not be effectively resolved under native electrophoresis conditions. Screening of the immunoblots with anti-rOspE antisera verified that the fragments were efficiently resolved (Fig. 4). Interestingly, in this gel system, a larger immunoreactive protein was observed that may represent a dimerized form of OspE or a heterodimer of OspE and a yet-to-be-identified protein. Screening with infection sera revealed that full-length BBL39 and two of the N-terminally truncated fragments (E54-173 and E77-173) were immunoreactive with infection sera. These two truncations were not immunoreactive with infection sera when the proteins were fractionated in denaturing gels. The restoration of Ab binding to proteins fractionated under nondenaturing conditions provides direct evidence that conformational determinants are necessary for optimal Ab binding.
FIG. 4.
Demonstration of the binding of infection-induced anti-OspE Abs to BBL39 subfragments fractionated by nondenaturing PAGE. Cell lysates of E. coli induced to express recombinant forms of BBL39 were fractionated by nondenaturing PAGE and immunoblotted as described in the text. Immunoblots were screened with anti-rOspE antisera or with infection sera as indicated.
Optimal binding of hfH requires full-length protein.
By using the OspE r proteins described above, several identical immunoblots were generated. A cell lysate of B. burgdorferi B31MIpc was included as a positive control for fH binding, and hfH was included as a positive control for anti-fH Ab binding. In addition, BBR42 and BBO39 (OspF proteins) were included in order to assess their abilities to bind fH. After blocking, one immunoblot was incubated with hfH in blocking buffer while a second blot was incubated with blocking buffer only. Both blots were then incubated with an anti-fH Ab. Several important observations were made. Only the full-length forms of OspE were capable of binding fH (Fig. 5). No binding was detected to the OspF paralogs BBO39 and BBR42 or with any of the OspE truncations. In addition, the full-length rOspE proteins and the lipidated form of the protein expressed by B. burgdorferi B31MIpc were immunoreactive with the anti-fH antisera even when exogenous hfH was not added. While it is conceivable that this observation could indicate cross-immunoreactivity of the anti-fH Ab directly with the OspE paralogs, several pieces of evidence argue against this. First, even though the anti-fH antisera were used at a 1:800 dilution, it is likely that sufficient levels of endogenous fH remained in the goat antisera to allow for OspE-fH complexes to form even without addition of exogenous fH. Second, binding assays described below demonstrate that when rBBL39 and BBN38 were incubated with hfH, they were immunoreactive with an anti-hfH monoclonal Ab preparation. This monoclonal Ab is strictly specific for hfH and is devoid of endogenous fH. In addition, recent studies by Alitalo and colleagues have demonstrated that in the absence of any endogenous fH, radiolabeled rfH binds directly to both BBL39 and BBN38 (2). The significance of the binding of the endogenous fH present in the anti-fH antisera and its relevance to the interpretation of earlier reports is discussed in detail below.
FIG. 5.
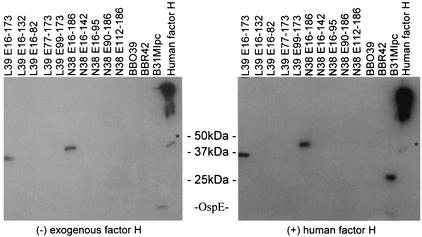
Analysis of the binding of hfH to OspE and OspF proteins by use of an affinity ligand binding immunoblot format. r proteins were fractionated by SDS-PAGE and immunoblotted. As indicated, the blots were then incubated either with purified hfH or with blocking buffer prior to the addition of goat anti-fH antisera. All methods are described in the text. Asterisks indicate the migration position of FHL-1.
The binding characteristics of individual FHBPs expressed by B. burgdorferi B31MIpc were found to differ. In contrast to the OspE proteins, a pair of ∼27-kDa FHBPs were observed only when exogenous hfH was included in the binding assay. These proteins did not interact with the endogenous fH present in the goat anti-fH antisera. This observation may indicate that these proteins can bind only fH derived from certain mammalian species.
ELISA analysis of Ab binding to OspE paralogs.
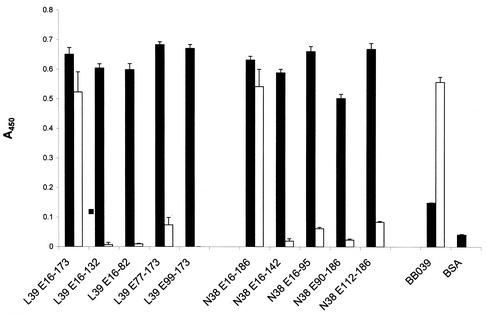
To identify OspE epitopes that are exposed during infection, ELISAs were performed using BBL39 and BBN38 subfragments as the immobilized antigen and 12-week infection sera as the detecting Ab. Several controls were performed, and all assays were performed in triplicate. Screening of the immobilized antigens with the rabbit anti-rOspE antisera verified that all antigens bound efficiently to the wells of the plates (Fig. 6). As a control, normal rabbit serum was also used to screen a set of wells, and with this serum no Ab binding was detected (data not shown). The specificity of the anti-OspE antisera was further demonstrated by screening wells containing immobilized BBO39 or BBR42 (OspF proteins) or bovine serum albumin (BSA) (negative control). These proteins did not react to any significant degree with the antisera (data not shown for BBR42). The binding of the OspF proteins to the wells was confirmed by using anti-rOspF antisera (data not shown). When infection sera were employed as the detecting antisera, BBO39, BBR42, and the full-length forms of BBL39 and BBN38 bound Abs, in agreement with reports demonstrating a robust IgG response to these proteins (25, 34). While the full-length forms of BBL39 and BBN38 bound Abs in the infection sera, the truncated proteins did not or did so at greatly reduced levels. The C-terminal 41-aa truncation of BBL39 and the 44-aa truncation of BBN38 completely abolished infection serum Ab binding. In contrast, some N-terminal truncations (BBL39 E77-173) still bound Abs but at reduced levels relative to that of the full-length protein. This is consistent with the data presented in Fig. 4. These results indicate that the optimal presentation of the epitopes is dependent upon higher-order structure or conformational determinants that involve the N- and C-terminal domains of the protein.
FIG. 6.
ELISA analysis of the abilities of recombinant BBL39 and BBN38 proteins or subfragments to bind polyclonal anti-OspE antisera and infection sera. Wells of ELISA plates were coated with OspE r proteins or subfragments. This and all other procedures are described in detail in the text. BBO39 served as a positive control for infection serum Ab binding, and BSA served as a negative control for both anti-rOspE antiserum and infection serum Ab binding. In one set of triplicate wells, anti-rOspE antisera were added (solid bars). Infection sera obtained from a mouse infected for 12 weeks with B. burgdorferi B31MIpc were added to another set of triplicate wells (open bars). Ab binding was measured as described in the text. Data are averages of three separate determinations. Error bars, standard deviations.
Demonstration of fH binding to full-length OspE paralogs but not to OspE truncations or OspF proteins.
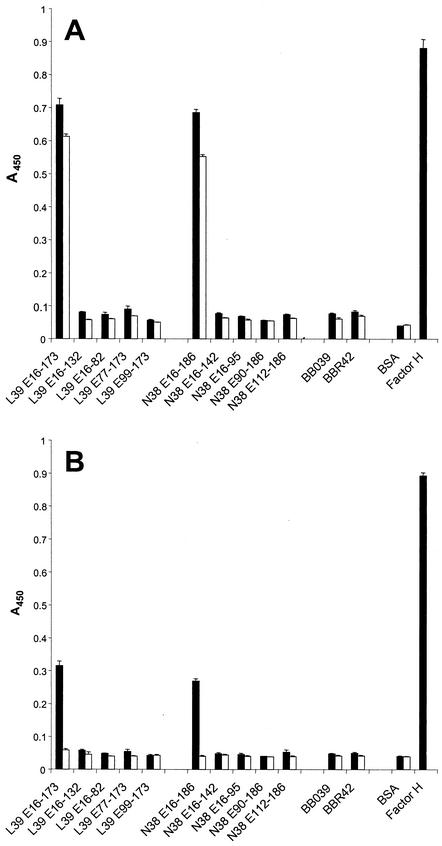
To determine if BBL39 protein subfragments can bind hfH, ELISAs were performed using either polyclonal anti-hfH antisera (Fig. 7, A) or a monoclonal Ab to hfH (Fig. 7B). Wells were coated with the BBL39 or BBN38 full-length protein or subfragments, incubated with or without fH, and then anti-hfH was added. Consistent with the immunoblot analyses, only the full-length forms of BBL39 (L39 E16-173) and BBN38 (N38 E16-186) bound fH. Binding was not observed with OspE truncations or with either of the OspF proteins, BBO39 and BBR42. As with infection serum Ab binding, the fact that independent truncations at the N and C termini of the protein eliminated fH binding indicates that fH does not recognize a solely linear determinant and that higher-order protein structure is required for efficient fH binding. Similar results were obtained with the monoclonal Ab, further confirming that the OspE proteins can bind hfH as well as goat fH.
FIG. 7.
Analysis of the fH binding abilities of BBL39 and BBN38 subfragments. All methods were as described in the text. r proteins were immobilized in wells, and then one set of triplicate wells was incubated with a preparation of purified fH (solid bars) while a second set was incubated with blocking buffer (open bars). After a wash, either polyclonal anti-hfH antisera (A) or a monoclonal anti-hfH Ab (B) was added. Binding was measured as described in Materials and Methods and was expressed as the average of three determinations. Error bars, standard deviations.
Analysis of binding of fH to BBL39 peptides.
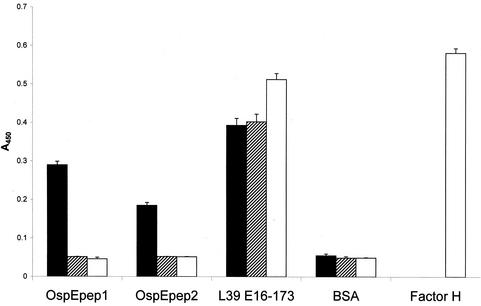
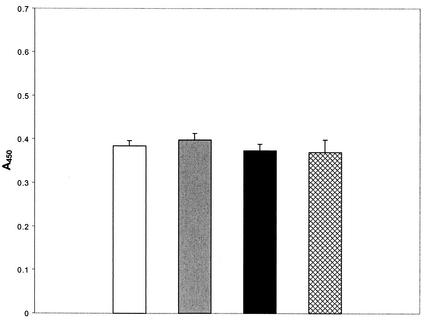
The data presented above demonstrate that N- or C-terminal deletions of BBL39 and BBN38 eliminate fH and infection serum Ab binding. In light of the absolute requirement for the C termini of these proteins for fH and infection serum Ab binding, we further explored the potential role of the C terminus in binding. Two peptides with a 5-aa overlap spanning the last 45 aa of BBL39 were synthesized, antisera were generated to each in mice, and a series of competitive binding ELISAs was performed. In the first set of assays, the peptides were immobilized in the wells of ELISA plates and then screened with the anti-rOspE antisera. As expected, both peptides and the positive control (L39 E16-173) reacted strongly with these antisera (A450, >0.18) while the negative controls (BBO39 and BSA) reacted only weakly (A450, <0.1) (Fig. 8). When screened with the 12-week infection sera, the peptides were nonreactive while all positive and negative controls behaved as predicted. Similarly, no binding of fH to the immobilized peptides was observed (A450, <0.05). These analyses provide evidence that linear determinants located within the C terminus of OspE are not in and of themselves sufficient for efficient binding of infection serum Abs or fH.
FIG. 8.
ELISA analysis of binding of fH to BBL39 and BBN38 peptides. Wells of a 96-well microtiter plate were coated with peptides or appropriate controls as indicated. All procedures are described in the text. Data are averages of three separate determinations. Binding of peptides and r proteins to the wells was assessed by using anti-OspE antisera (solid bars). The abilities of the peptides to bind infection serum Abs were assessed by using anti-B. burgdorferi B31MIpc infection sera (hatched bars). Abilities to bind hfH were assessed by using anti-fH antisera (open bars).
Competitive binding ELISAs: demonstration that fH and infection Abs bind to separate domains.
To determine if anti-OspE Abs in infection sera bind to the same site as fH, competitive binding ELISAs were performed. rBBL39 and control proteins were immobilized to a series of wells (in triplicate). One set of wells was preincubated with normal human sera, which served as the source of hfH and FHL-1. After allowing for binding of the fH and FHL-1, sera were removed, wells were washed, and infection sera were added. As demonstrated above, infection sera harbor Abs to BBL39 and BBN38. Preincubation with the human sera did not affect the binding of anti-OspE Abs in infection sera, indicating that fH and Abs are binding to separate domains (Fig. 9). Similarly, when purified hfH was preincubated with rBBL39 prior to addition of infection sera, the purified fH did not block the binding of anti-OspE Abs elicited during infection. As a control for these assays, the binding of fH from the purified preparation and from the human sera to BBL39 was verified by using goat anti-fH antisera.
FIG. 9.
Competitive binding ELISA analyses of fH and Ab binding to BBL39. All methods were as described in the text. In brief, BBL39 was immobilized in the wells of a 96-well microtiter plate in triplicate. Normal human serum (open bar) or purified hfH (shaded bar) was added, wells were washed, and infection sera (mouse) were added. Binding of mouse IgG to BBL39 was assessed by using rabbit anti-mouse IgG antisera. As a control for fH binding to immobilized BBL39, one set of wells was incubated with fH (solid bar) and washed, goat anti-fH antisera were added, wells were washed, and a rabbit anti-goat secondary Ab was added. To verify that anti-OspE IgG was able to bind to the immobilized BBL39, infection sera were added to one set of wells (crosshatched bar), wells were washed, and IgG binding was detected by using rabbit anti-mouse IgG. Detection methods were as described in the text. Data are average readings from three wells. Error bars, standard deviations.
It has been suggested that fH binds within the C terminus of OspE (3). To assess this further, competitive binding ELISAs were performed. Antisera against peptides spanning the C-terminal 46 aa of BBL39 were generated and incubated in the wells of ELISA plates coated with full-length BBL39. After removal of the antiserum, fH was added. The antipeptide antisera, which bound efficiently to the immobilized protein in control wells, did not inhibit or decrease the binding of fH (data not shown). The converse experiment also demonstrated that there was no competition for binding. These analyses indicate that linear determinants in the C terminus of OspE are not directly involved in the binding of fH.
DISCUSSION
The OspE proteins of the Lyme spirochetes are postulated to contribute to immune evasion through antigenic variation and to circumvent complement attack through the binding of fH (17-19, 34). Upon initiation of this study, little was known about the structure of OspE or its interaction with Abs and fH. Sequence analyses have revealed that variation among OspE proteins is localized to specific variable domains that are predicted to be surface exposed and antigenic (23). However, surface exposure of the variable domains had not been directly demonstrated, and the binding site for fH had not been localized. The goals of this study were to address these questions.
To identify the OspE immunodominant epitopes that are exposed during infection, immunoblots of rBBN38 and rBBL39 were screened with infection sera absorbed with each r protein. The specificity of the Ab response to these proteins during infection depends on whether the variable or conserved regions are presented on the cell surface. In spite of a high degree of sequence identity between BBN38 and BBL39, absorption selectively removed only the Ab to the protein used to absorb and had no impact on the immunoreactivity of the heterologous protein. These data demonstrate that a paralog-specific Ab response develops during infection in mice. The temporally expressed OspF proteins of B. burgdorferi B31MI (BBO39, BBR42, and BBM38) also elicit a paralog-specific Ab response during infection (25). Hence, differential expression of OspE paralogs and the development of sequence variation within the variable, surface-exposed domains could alter the antigenic profile of the bacteria and thus contribute to immune evasion.
To identify the domains of BBL39 and BBN38 that bind to fH or Ab, immunoblot and ELISA analyses were performed using truncated OspE proteins. Removal of a 41- to 45-aa domain from the C termini of these proteins eliminated infection serum Ab and fH binding but did not affect the binding of polyclonal anti-rOspE antisera. This suggested that the Ab and fH binding determinants reside within the C termini of these proteins. However, subsequent analyses revealed that N-terminal truncations also abolished Ab and fH binding. The requirement for both the N and C termini of these proteins for fH and Ab binding suggests that conformational or discontinuous binding domains are involved. While it might be expected that SDS-PAGE conditions would have eliminated conformational determinants in the full-length protein, it has been demonstrated that some proteins, such as herpes simplex virus protein and glycoprotein D, can retain some degree of native conformation upon SDS-PAGE (5).
To further assess the possible existence of conformational binding determinants, BBL39 subfragments were fractionated under nondenaturing conditions, immunoblotted, and either screened with infection sera or tested for their abilities to bind fH. While fH did not bind to the truncated fragments (data not shown), binding of infection serum Abs was restored to the E54-173 and E77-173 BBL39 subfragments. These results are in agreement with the ELISA analyses, in which the E77-173 fragment bound infection-induced Abs at a low level but did not bind fH. Unfortunately, the BBN38 subfragments could not be analyzed by this approach because they could not be resolved by native PAGE. Collectively, these analyses demonstrate that both the C- and N-terminal domains of BBL39 are required for optimal binding of Abs and fH, and they suggest that long-range interactions between domains may be required to allow for the formation or proper presentation of infection-exposed epitopes and fH binding sites.
Based on a recent study employing surface plasma resonance and OspE peptides, Alitalo et al. concluded that fH binds to C-terminal linear determinants of OspE (3). Our initial analyses of the C-terminal OspE truncations appeared to support this conclusion. However, since N-terminal deletions of OspE had the same effect, it is evident that determinants other than solely linear ones are involved. We interpret our data to indicate that both C- and N-terminal deletions prevent the formation of long-range conformational binding determinants. To further test the potential binding of fH to the C terminus of OspE, ELISAs were performed in which OspE C-terminal peptides were immobilized in the wells. No binding of fH to peptides spanning the terminal 46 residues of BBL39 was observed. The plasma resonance peptide-scanning approach employed by Alitalo et al. (3) does not allow for an assessment of the potential contribution of conformational determinants in fH binding. It is possible that the high sensitivity of the surface plasma resonance approach may lead to the detection of weak binding sites that may or may not be of importance in vivo. The data presented here indicate that optimal binding of fH by OspE requires the maintenance of native conformation. It should be noted that infection sera also did not bind to the peptides, further supporting the postulate that OspE conformational determinants are also involved in infection serum Ab binding.
While the analyses conducted here did not identify the specific amino acid residues necessary for Ab and fH binding, competitive ELISA analyses revealed that these ligands do not compete for the same binding site(s). In light of the ability of divergent OspE proteins to bind fH (26), it is likely that the fH binding domain resides within or involves conserved regions or structural elements of the protein. In contrast, the data presented above indicate that the Ab response to OspE is directed at conformational determinants involving variable regions of the proteins. This suggests that the binding domains for Abs and fH are separable. Since the fH binding domain must be surface exposed during infection, the absence of an Ab in infection sera targeting the fH binding site (as evidenced by the lack of competition between infection serum Abs and fH) may appear paradoxical. However, since fH is part of the innate immune response and is highly abundant, the fH binding site of OspE most likely becomes quickly saturated with fH upon infection and thus may not be exposed and targeted by Abs during infection.
An unexpected observation noted in these analyses was the immunoreactivity of full-length BBL39 and BBN38 with the anti-hfH antisera when hfH was omitted from the fH binding assay. Cross-reactivity of these proteins with the antisera is unlikely, because they exhibit no homology to fH. In addition, none of the OspE truncations were immunoreactive with the anti-fH antisera. A possible explanation for the interaction of anti-fH antisera with OspE in the absence of exogenous hfH is that endogenous goat fH naturally present in the goat anti-hfH antisera is bound by OspE and is thus detected by the anti-fH Ab. In human sera the concentrations of fH and FHL-1 are 300 to 500 and 45 μg ml−1, respectively (28). To test for the presence of detectable fH in the goat anti-hfH antisera, 1 μl of the antisera was fractionated by SDS-PAGE, immunoblotted, and screened with the goat anti-hfH antisera. Goat fH was readily detected (data not shown). Similarly, others have shown that fH can be detected in the antisera of several mammals by using the same anti-fH antisera used here (32). The presence of endogenous fH in the antisera was not recognized in earlier studies and impacts on the interpretation of earlier data (32). For example, Stevenson et al. concluded that OspE and other Borrelia proteins bind fH from different mammals in a species-specific fashion (32). However, the absence of essential controls, coupled with the presence of endogenous fH and FHL-1 in those assays at estimated concentrations of 65 and 8 μg ml−1, respectively, renders conclusions reached about binding specificity potentially invalid.
The Lyme spirochetes have been demonstrated to express several FHBPs (19, 26). While it is clear that OspE binds fH, there have been conflicting reports regarding fH binding to the OspF and Elp proteins (3, 32). Consistent with the findings reported by Alitalo et al. (3), the analyses presented here indicate that OspE binds fH while the OspF proteins (BBO39 and BBR42) do not. The weak binding of fH to some of the OspF and Elp proteins observed by Stevenson et al. is most likely an artifact that resulted from the exceptionally high antiserum concentration used (a 1:8 dilution) in their fH binding assays (32). The affinity ligand binding immunoblot assays conducted here also revealed that while OspE clearly binds fH, a pair of 27-kDa FHBPs of B. burgdorferi also bind fH. These proteins specifically bind hfH (or possibly hFHL-1) and do not bind goat fH. Kraiczy et al. (18) detected an FHBP of similar size in Borrelia afzelii, designated CRASP-1, that preferentially binds hFHL-1. In a separate comprehensive study of the FHBP profiles of diverse Lyme spirochete species and isolates, McDowell et al. demonstrate that the 27-kDa proteins are the dominant FHBPs expressed by these bacteria during in vitro cultivation (26). The sequence and specific identity of these proteins are the subject of ongoing analyses.
In summary, in this study we demonstrate that the immunodominant regions of BBN38 and BBL39 reside within the variable regions of these proteins and elicit a paralog-specific Ab response. Consistent with an earlier report from our laboratory (34), this finding indicates that OspE sequence variation influences the antigenic properties of the protein and that OspE sequence changes could play a role in immune evasion. We also demonstrate that the binding of Abs and fH requires conformational or discontinuous epitopes. Analyses of the BBL39 sequence revealed the existence of potential coiled-coil structural motifs. Coiled-coil motifs are important determinants of higher-order structure in some proteins. While the presence of putative coiled-coil motifs in BBL39 is worthy of mention, it is important to point out that BBN38 is not predicted to possess coiled-coil domains. Regarding fH binding, this may represent an important virulence mechanism for the Lyme disease spirochetes and play a key role in ensuring population maintenance in nature. At least some of the FHBPs are encoded by the 32-kb circular plasmids (4) which have been demonstrated by Samuels and colleagues to be bacteriophage genomes that have evolved to become integral parts of the B. burgdorferi genome (8-10). The presence of FHBP genes on potentially transferable genetic elements may allow for the transfer of the fH binding phenotype. In closing, the analyses presented here provide further insight into the structure-function relationships of the OspE proteins and the potential mechanisms of immune evasion used by the Lyme spirochetes.
ADDENDUM IN PROOF
In a recent study, Kraiczy et al. (P. Kraiczy, J. Hellwage, C. Skerka, M. Kirschfink, V. Brade, P. F. Zipfel, and R. Wallich, Eur. J. Immunol. 33:697-707, 2003) concluded that fH binds to the C terminus of OspE, but these investigators did not study N-terminal truncations of OspE and therefore could draw no conclusion about the influence of the N terminus or of protein folding on fH binding.
Acknowledgments
We thank Erol Fikrig for kindly providing anti-rOspE antisera. We are indebted to Darrin Akins, Merri Seppo, and their colleagues for freely sharing data regarding the binding of fH to OspE prior to their publication while this study was ongoing.
These studies were supported in part by a grant from the NIH (RO1AI37787-06). J. V. McDowell was supported in part by a Molecular Pathogenesis training grant from NIAID and by a predoctoral award from NINDS (F31NS43088).
Editor: V. J. DiRita
REFERENCES
- 1.Akins, D., S. F. Porcella, T. G. Popova, D. Shevchenko, S. I. Baker, M. Li, M. V. Norgard, and J. D. Radolf. 1995. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Mol. Microbiol. 18:507-520. [DOI] [PubMed] [Google Scholar]
- 2.Alitalo, A., T. Meri, L. Rämö, T. S. Jokiranta, T. Heikkilä, I. J. T. Seppälä, J. Oksi, M. Viljanen, and S. Meri. 2001. Complement evasion by Borrelia burgdorferi: serum-resistant strains promote C3b inactivation. Infect. Immun. 69:3685-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alitalo, A., T. Meri, H. Lankinen, I. Seppala, P. Lahdenne, P. S. Hefty, D. Akins, and S. Meri. 2002. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 169:3847-3853. [DOI] [PubMed] [Google Scholar]
- 4.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, G. H., V. J. Isola, J. Kuhns, P. W. Berman, and R. J. Eisenberg. 1986. Localization of discontinuous epitopes of herpes simplex virus glycoprotein D: use of a nondenaturing (“native” gel) system of polyacrylamide gel electrophoresis coupled with Western blotting. J. Virol. 60:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Silva, A. M., and E. Fikrig. 1997. Borrelia burgdorferi genes selectively expressed in ticks and mammals. Parasitol. Today 13:267-270. [DOI] [PubMed] [Google Scholar]
- 7.de Silva, A. M., E. Fikrig, E. Hodzic, F. S. Kantor, S. R. Telford III, and S. W. Barthold. 1998. Immune evasion by tickborne and host-adapted Borrelia burgdorferi. J. Infect. Dis. 177:395-400. [DOI] [PubMed] [Google Scholar]
- 8.Eggers, C., and D. S. Samuels. 1999. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J. Bacteriol. 181:7308-7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eggers, C. H., S. Casjens, S. F. Hayes, C. F. Garon, C. J. Damman, D. B. Oliver, and D. S. Samuels. 2000. Bacteriophages of spirochetes. J. Mol. Microbiol. Biotechnol. 4:365-373. [PubMed] [Google Scholar]
- 10.Eggers, C. H., B. J. Kimmel, J. L. Bono, A. F. Elias, P. Rosa, and D. S. Samuels. 2001. Transduction by φBB-1, a bacteriophage of Borrelia burgdorferi. J. Bacteriol. 183:4771-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Hage, N. E., L. D. Lieto, and B. Stevenson. 1999. Stability of erp loci during Borrelia burgdorferi infection: recombination is not required for chronic infection of immunocompetent mice. Infect. Immun. 67:3146-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser, C., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischman, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 13.Friese, M. A., J. Hellwage, T. S. Jokiranta, S. Meri, H. H. Peter, H. Eibel, and P. F. Zipfel. 1999. FHL-1/reconectin and factor H: two human complement regulators which are encoded by the same gene are differently expressed and regulated. Mol. Immunol. 36:809-818. [DOI] [PubMed] [Google Scholar]
- 14.Hefty, P. S., C. S. Brooks, A. M. Jett, G. L. White, S. K. Wikel, R. C. Kennedy, and D. R. Akins. 2002. OspE-related, OspF-related, and Elp lipoproteins are immunogenic in baboons experimentally infected with Borrelia burgdorferi and in human Lyme disease patients. J. Clin. Microbiol. 40:4256-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, and D. R. Akins. 2002. Changes in temporal and spatial patterns of outer surface lipoprotein expression generate population heterogeneity and antigenic diversity in the Lyme disease spirochete Borrelia burgdorferi. Infect. Immun. 70:3468-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, J. D. Radolf, and J. R. Akins. 2001. Regulation of the OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 69:3618-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hellwage, J., T. Meri, T. Heikkila, A. Alitalo, J. Panelius, P. Lahdenne, I. J. T. Seppala, and S. Meri. 2001. The complement regulator factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427-8435. [DOI] [PubMed] [Google Scholar]
- 18.Kraiczy, P., C. Skerka, M. Kirschfink, V. Brade, and P. F. Zipfel. 2001. Immune evasion of Borrelia burgdorferi by acquisition of human complement regulators FHL-1/reconectin and factor H. Eur. J. Immunol. 31:1674-1684. [DOI] [PubMed] [Google Scholar]
- 19.Kraiczy, P., C. Skerka, and P. F. Zipfel. 2001. Further characterization of complement regulator-acquiring surface proteins of Borrelia burgdorferi. Infect. Immun. 69:7800-7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurtenbach, K., H.-S. Sewell, N. H. Ogden, S. E. Randolph, and P. A. Nuttall. 1998. Serum complement sensitivity as a key factor in Lyme disease ecology. Infect. Immun. 66:1248-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam, T. T., T.-P. K. Nguyen, R. R. Montgomery, F. S. Kantor, E. Fikrig, and R. A. Flavell. 1994. Outer surface proteins E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect. Immun. 62:290-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magnarelli, L. A., J. W. Ijdo, S. J. Padula, R. A. Flavell, and E. Fikrig. 2000. Serologic diagnosis of Lyme borreliosis by using enzyme-linked immunosorbent assays with recombinant antigens. J. Clin. Microbiol. 38:1735-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marconi, R. T., S. Y. Sung, C. N. Hughes, and J. A. Carlyon. 1996. Molecular and evolutionary analyses of a variable series of genes in Borrelia burgdorferi that are related to ospE and ospF, constitute a gene family, and share a common upstream homology box. J. Bacteriol. 178:5615-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDowell, J. V., S. Y. Sung, L. T. Hu, and R. T. Marconi. 2002. Evidence that the variable regions of the central domain of VlsE are antigenic during infection with the Lyme disease spirochetes. Infect. Immun. 70:4196-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDowell, J. V., S.-Y. Sung, G. Price, and R. T. Marconi. 2001. Demonstration of the genetic stability and temporal expression of select members of the Lyme disease spirochete OspF protein family during infection in mice. Infect. Immun. 69:4831-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDowell, J. V., J. Wolfgang, E. Tran, M. S. Metts, D. Hamilton, and R. T. Marconi. 2003. Comprehensive analysis of the factor H binding capabilities of Borrelia species associated with Lyme disease: delineation of two distinct classes of factor H binding proteins. Infect. Immun. 71:3597-3602. [DOI] [PMC free article] [PubMed]
- 27.Nguyen, T.-P. K., T. T. Lam, S. W. Barthold, S. R. Telford III, R. A. Flavell, and E. Fikrig. 1994. Partial destruction of Borrelia burgdorferi within ticks that engorged on OspE- or OspF- immunized mice. Infect. Immun. 62:2079-2084. [DOI] [PMC free article] [PubMed]
- 28.Pangburn, M. K. 2000. Host recognition and target differentiation by factor H, a regulator of the alternate pathway of complement. Immunopharmacology 49:149-157. [DOI] [PubMed] [Google Scholar]
- 29.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 99:1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts, D., M. Caimano, J. McDowell, M. Theisen, A. Holm, S. Alban, E. Orff, D. Nelson, S. Wikel, J. Radolf, and R. Marconi. 2002. Environmental regulation and differential expression of members of the Bdr protein family of Borrelia burgdorferi. Infect. Immun. 70:7033-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steere, A. C., R. L. Grodzicki, A. N. Kornblatt, J. E. Craft, A. G. Barbour, W. Burgdorfer, G. P. Schmid, E. Johnson, and S. E. Malawista. 1983. The spirochetal etiology of Lyme disease. N. Engl. J. Med. 308:733-740. [DOI] [PubMed] [Google Scholar]
- 32.Stevenson, B., N. El-Hage, M. A. Hines, J. C. Miller, and K. Babb. 2002. Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 70:491-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suk, K., S. Das, W. Sun, B. Jwang, S. W. Barthold, R. A. Flavell, and E. Fikrig. 1995. Borrelia burgdorferi genes selectively expressed in the infected host. Proc. Natl. Acad. Sci. USA 92:4269-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung, S. Y., J. McDowell, J. A. Carlyon, and R. T. Marconi. 2000. Mutation and recombination in the upstream homology box-flanked ospE-related genes of the Lyme disease spirochetes results in the development of new antigenic variants during infection. Infect. Immun. 68:1319-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sung, S.-Y., C. LaVoie, J. A. Carlyon, and R. T. Marconi. 1998. Genetic divergence and evolutionary instability in ospE-related members of the upstream homology box gene family in Borrelia burgdorferi sensu lato complex isolates. Infect. Immun. 66:4656-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Dam, A. P., A. Oei, R. Jaspars, C. Fijen, B. Wilske, L. Spanjaard, and J. Dankert. 1997. Complement-mediated serum sensitivity among spirochetes that cause Lyme disease. Infect. Immun. 65:1228-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, J.-R., J. M. Hardham, A. G. Barbour, and A. G. Norris. 1997. Antigenic variation in Lyme disease Borreliae by promiscuous recombination of vmp like sequence cassettes. Cell 89:275-285. [DOI] [PubMed] [Google Scholar]