Abstract
Correlations between FimH mutations and virulence were established by studying a collection of human commensal and extraintestinal pathogenic Escherichia coli natural isolates. Pathoadaptive (A27V and, to a lesser extent, A119V) and “commensal-adaptive” (A202V) mutations were evidenced in B2 phylogenetic group strains. fimH phylogenetic analysis indicates that these pathoadaptive mutations occurred several times.
One of the mechanisms in bacterial evolution towards virulence is the allelic variation of existing genes that has been called pathoadaptive (19). One of the most studied pathoadaptive mutations is the allelic variation within the adhesive subunit of type I fimbriae, FimH, of Escherichia coli (15, 16, 18). This bacterium is a normal inhabitant of the intestinal tract of vertebrates but can also be responsible for intestinal and extraintestinal diseases (mainly urinary tract infections [UTI] and septicemia). Indeed, all E. coli isolates are capable of binding to trimannose receptors, allowing bacteria to bind buccal epithelial cells. In addition, 70% of UTI isolates bind to both trimannose and monomannose residues (17), allowing them to adhere strongly to uroepithelial cells. This change in tissue tropism is due to a small number of amino acid changes in FimH, an otherwise highly conserved molecule (1, 7, 14, 15). In vitro mutagenesis and analysis of the FimH sequences of a small number of natural isolates had allowed the identification of the amino acids responsible for an increase in binding monomannose residues (13, 17, 18). Moreover, previous studies have shown a link between long-term evolutionary history (strain phylogeny) and virulence in E. coli, as extraintestinal E. coli pathogens (including UTI strains) belonged preferentially to one of the four main E. coli phylogenetic groups, i.e., the B2 phylogenetic group (9, 12). Thus, the simultaneous action on virulence of both single-mutation effects and the accumulated divergence in genetic background over millions of years led us to study more carefully the diversity and evolutionary history of the fimH gene from natural strains belonging to the various E. coli phylogenetic groups (6). We sequenced the fimH gene from the first postsignal peptide codon to codon 232 in an epidemiologic collection encompassing both 44 commensal and 69 pathogenic strains. The strains were isolated mainly in France and in Benin and included two previously published collections (11, 12). fimH amplification was performed by standard PCR from bacterial lysate using primer fimHF (5′-TGATGGGCTGGTCGGTAAATG-3′) and fimH3R (5′-CGATACCGTGTTATTCGCTGG-3′). PCR products were directly sequenced on both strands without interim cloning. In addition, the phylogenetic group (A, B1, D, and B2 [6]) to which the strains belong was determined by PCR as described by Clermont et al. (2), and the intrinsic extraintestinal virulence of the strains was tested by inoculating 10 mice per strain followed by scoring of deaths, as described by Picard et al. (12).
The Pearson correlation coefficients between the different FimH amino acid mutations and the characteristics of the strains, i.e., their origin of isolation (normal feces, urine, blood, miscellaneous infections), their phylogenetic groups, and the lethality in mice (0 to 1 mouse, 2 to 8 mice, and 9 to 10 mice killed of 10 mice inoculated), were calculated, and the statistical significance was evaluated by a Fisher test. Among the coding sequence, 33 amino acids were found to be variable, but only 7 of those, which were found variable in more than seven strains, were considered in the statistical analyses (Table 1). Except for A202V, all amino acid changes considered here (A27V, G66S/C, N70S, S78N, A119V, V163A) were previously described as polymorphic and potentially pathoadaptive for E. coli (15). Fifteen different FimH alleles (I to XV) were found in our collection of natural isolates (Table 1). While most amino acid mutations were widespread among the 15 alleles, the V163A mutation was found only in allele V in combination with both N70S and S78N mutations. Moreover, mutations N70S and S78N were almost always found together (0.865; P < 0.0001) and were the most common amino acid mutations. These two mutations are localized within β-sheet 6 in the three-dimensional structure of FimH (1).
TABLE 1.
FimH alleles of the studied natural isolates
| Allelea | No. of strains (%) | Mutation at amino acid residueb:
|
In vitro binding affinity to yeast mannan cells (strain)c | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 27 | 66 | 70 | 78 | 119 | 163 | 202 | |||
| I | 32 (26.23) | A | G | N | S | A | V | A | Low (KB21) |
| II | 23 (18.85) | V | High (PC31) | ||||||
| III | 23 (18.85) | S | N | Low (F18) | |||||
| IV | 11 (9.01) | V | Low (CI3) | ||||||
| V | 9 (7.34) | S | N | A | Low (MI11-2) | ||||
| VI | 8 (6.56) | V | |||||||
| VII | 4 (3.28) | S | V | ||||||
| VIII | 3 (2.46) | C | |||||||
| IX | 3 (2.46) | V | S | N | V | ||||
| X | 2 (1.64) | H | |||||||
| XI | 1 (0.82) | T | |||||||
| XII | 1 (0.82) | S | |||||||
| XIII | 1 (0.82) | N | |||||||
| XIV | 1 (0.82) | S | N | V | |||||
| XV | 1 (0.82) | S | S | N | |||||
Alleles are defined as combinations of amino acid mutations. Only mutations retreived more than seven times were used to define the alleles. The wild-type allele (allele I) is arbitrarily defined as the most frequently isolated.
Blank entries indicate identity with the wild type, whereas mutations are indicated.
As determined in reference 17.
When all strains were considered, significant correlations were found only between N70S/S78N mutations, the B2 phylogenetic group (0.467; P < 0.0001), and lethality in mice (0.316; P < 0.0009). In addition, a link between virulence in mice and B2 phylogenetic group was observed in our data (0.529; P < 0.0001), in accordance with previous works (8, 12). To know whether N70S and S78N mutations were correlated with lethality in mice independently from phylogenetic groups, we looked for correlations between FimH amino acid mutations, origin, and virulence in B2 group strains (68 strains, of which 28 are commensal and 40 are pathogenic). No significant correlation was observed for the N70S/S78N mutations, indicating that the previously observed correlation with lethality in mice was due to the B2 specificity of these mutations and not to the pathogenic nature of the strain.
Among the B2 strains, a highly significant correlation was found between the A27V mutation and UTI strains (0.378; P < 0.002). To a lesser extent, significant correlations were also found between the A119V mutation and UTI strains (0.273; P < 0.03) and between the A202V mutation and strains isolated from feces (0.307; P < 0.016). Because of the small sample size used to calculate these last two coefficients, additional B2 strains should be tested to confirm these data. In addition, a correlation was found between the A27V mutation and virulence in mice (0.330; P < 0.009). This mutation is localized in a loop between β-sheets 3 and 4 (1), and previous in vitro binding assays had shown a potential role of A27V mutation in the capacity of the strain to bind to monomannose (17) (Table 1). No correlation was found when non-B2 group strains were analyzed (data not shown). Taken together, these data strongly suggest a pathoadaptive role of the A27V mutation in the natural environment.
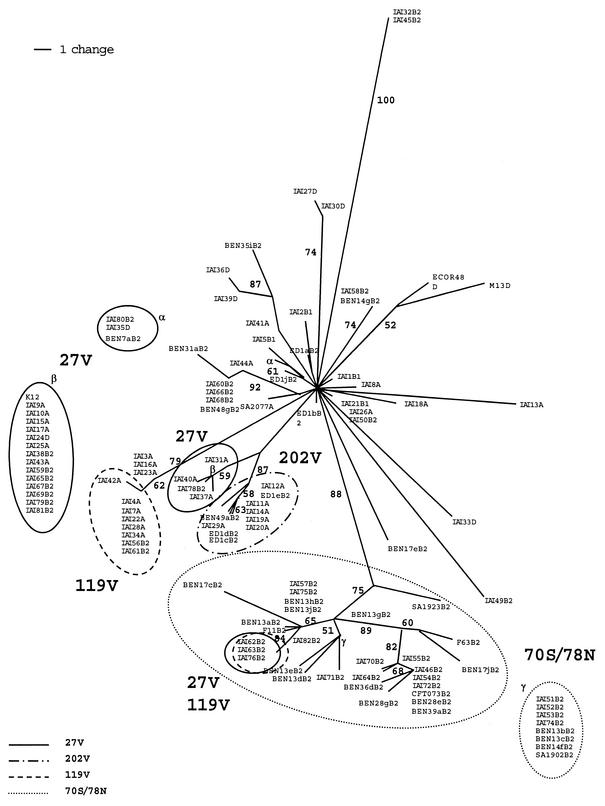
Having demonstrated that FimH has both mutations that are linked to the global phylogeny of the strains (N70S/S78N) and mutations that are linked to the environment (A27V, A119V, and A202V), we then compared the fimH gene and the strain phylogenies to understand the fimH evolutionary history. The unrooted tree reconstructed from the fimH sequences showed a grouping that is incongruent with the phylogenetic groups to which the strains belong (Fig. 1), an argument for the occurrence of horizontal gene transfers during fimH evolution (3, 5). The mutations separating the monophyletic group of B2 strains supported by a bootstrap value of 88% from the remaining strains, which also encompass some B2 strains (Fig. 1), were determined with MacClade (10). They are not scattered all over the sequence but are clustered between nucleotides 183 and 351 (data not shown), a feature typical of gene transfer (3). It could be assumed that the monophyletic group of B2 strains which possesses the N70S/S78N mutations corresponds to the true B2, whereas the remaining B2 strains have suffered from horizontal gene transfers. In addition, the phylogenetic tree (Fig. 1) indicated that the nucleotide changes corresponding to the A27V and A119V mutations (C80T and C356T, respectively) were each flanked by different sequences (three and two sequences, respectively). Thus, pathoadaptive mutations had arisen several times during the evolution.
FIG. 1.
Unrooted semistrict consensus tree reconstructed from the fimH gene sequences by using parsimony in PAUP* (20); 696 nucleotides, of which 62 are informative for parsimony (l = 241; consistency index, 0.456; retention index, 0.877). The numbers at branches are the bootstrap proportions obtained from 100 replicates. Only bootstraps with proportions above 50% are given. The phylogenetic groups to which the strains belong, determined as described in reference 2, are indicated: A, B1, D, and B2. Similar topology was obtained with a neighbor-joining approach (data not shown). Strains within circles correspond to the strains with the A27V, N70S/S78N, A119V, and A202V mutations. K12 and CFT073 sequences have been extracted from GenBank.
This study is the first large-scale analysis of FimH pathoadaptive mutations from a collection of well-characterized natural isolates. Altogether, our data indicated that pathoadaptive mutations are selected for in nature in B2 phylogenetic group strains. Since B2 strains have numerous extraintestinal virulence determinants, virulence can be considered a multifactorial trait as previously suggested (8), with a probably essential effect of the genetic background of the strain in the rise of particular mutations such as pathoadaptive ones. Furthermore, the concept of commensal-adaptive mutation could be proposed for the A202V mutation, which was preferentially present in B2 group strains isolated from feces. It would be interesting to test whether FimH A202V strains adhere better to gut epithelium. Moreover, one could propose that the balancing selection for these patho- and commensal-adaptive mutations could explain the presence of B2 group strains in normal feces (4, 21).
Acknowledgments
This study was partially supported by a grant from the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires (MENRT).
We are grateful to Olivier Pradillon for technical help and to Patricia Escobar-Páramo and Olivier Tenaillon for their comments on the manuscript.
Editor: V. J. DiRita
REFERENCES
- 1.Choudhury, D., A. Thompson, V. Stojanoff, S. Langermann, J. Pinkner, S. J. Hultgren, and S. D. Knignt. 1999. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285:1061-1066. [DOI] [PubMed] [Google Scholar]
- 2.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic groups. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denamur, E., G. Lecointre, P. Darlu, O. Tenaillon, C. Acquaviva, C. Sayada, I. Sunjevaric, R. Rothstein, J. Elion, F. Taddei, M. Radman, and I. Matic. 2000. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell 103:711-721. [DOI] [PubMed] [Google Scholar]
- 4.Duriez, P., O. Clermont, S. Bonacorsi, E. Bingen, A. Chaventré, J. Elion, B. Picard, and E. Denamur. 2001. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 147:1671-1676. [DOI] [PubMed] [Google Scholar]
- 5.Dykhuizen, D. E., and L. Green. 1991. Recombination in Escherichia coli and the definition of biological species. J. Bacteriol. 173:7257-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittman. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hung, C. S., J. Bouckaert, D. Hung, J. Pinkner, C. Widberg, A. DeFusco, C. G. Auguste, R. Strouse, S. Langermann, G. Walsman, and S. J. Hultgren. 2002. Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol. Microbiol. 44:903-915. [DOI] [PubMed] [Google Scholar]
- 8.Johnson, J. R., M. Kuskowski, E. Denamur, J. Elion, and B. Picard. 2000. Clonal origin, virulence factors, and virulence. Infect. Immun. 68:424-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 10.Maddison, D. R., and W. P. Maddison. 2000. MacClade 4: analysis of phylogeny and character evolution, version 4.0. Sinauer Associates, Sunderland, Mass.
- 11.Picard, B., P. Duriez, S. Gouriou, I. Matic, E. Denamur, and F. Taddei. 2001. Mutator natural Escherichia coli isolates have an unusual virulence phenotype. Infect. Immun. 69:9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Picard, B., J. S. Garcia, S. Gouriou, P. Duriez, N. Brahimi, E. Bingen, J. Elion, and E. Denamur. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schembri, M. A., E. V. Sokurenko, and P. Klemm. 2000. Functional flexibility of the FimH adhesin: insights from a random mutant library. Infect. Immun. 68:2638-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sokurenko, E. V., V. Chesnokova, R. J. Doyle, and D. L. Hasty. 1997. Diversity of the Escherichia coli type 1 fimbrial lectin. J. Biol. Chem. 272:17880-17886. [DOI] [PubMed] [Google Scholar]
- 15.Sokurenko, E. V., V. Chesnokova, D. E. Dykhuizen, I. Ofek, X. R. Wu, K. A. Krogfelt, C. Struve, M. A. Schembri, and D. L. Hasty. 1998. Pathogenic adaptation of Escherichia coli by natural variation of FimH adhesin Proc. Natl. Acad. Sci. USA 95:8922-8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sokurenko, E. V., H. S. Courtney, S. N. Abraham, P. Klemm, and D. L. Hasty. 1992. Functional heterogeneity of type 1 fimbriae of Escherichia coli. Infect. Immun. 60:4709-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sokurenko, E. V., H. S. Courtney, J. Maslow, A. Siitonen, and D. L. Hasty. 1995. Quantitative differences in adhesiveness to type 1 fimbriated Escherichia coli due to structural differences in fimH genes. J. Bacteriol. 177:3680-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokurenko, E. V., H. S. Courtney, D. E. Ohman, P. Klemm, and D. L. Hasty. 1994. FimH family of type 1 adhesins: functional heterogeneity due to minor sequence variations among fimH genes. J. Bacteriol. 176:748-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sokurenko, E. V., D. L. Hasty, and D. E. Dykhuizen. 1999. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 7:191-195. [DOI] [PubMed] [Google Scholar]
- 20.Swofford, D. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0. Sinauer Associates, Sunderland, Mass.
- 21.Zhang, L., B. Foxman, and C. Marrs. 2002. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J. Clin. Microbiol. 40:3951-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]