Abstract
The hormones that most directly control tissue activities in health and disease are delivered by two noninhibitory members of the serpin family of protease inhibitors, thyroxine-binding globulin (TBG) and corticosteroid-binding globulin. The structure of TBG bound to tetra-iodo thyroxine, solved here at 2.8 Å, shows how the thyroxine is carried in a surface pocket on the molecule. This unexpected binding site is confirmed by mutations associated with a loss of hormone binding in both TBG and also homologously in corticosteroid-binding globulin. TBG strikingly differs from other serpins in having the upper half of its main β-sheet fully opened, so its reactive center peptide loop can readily move in and out of the sheet to give an equilibrated binding and release of thyroxine. The entry of the loop triggers a conformational change, with a linked contraction of the binding pocket and release of the bound thyroxine. The ready reversibility of this change is due to the unique presence in the reactive loop of TBG of a proline that impedes the full and irreversible entry of the loop that occurs in other serpins. Thus, TBG has adapted the serpin inhibitory mechanism to give a reversible flip-flop transition, from a high-affinity to a low-affinity form. The complexity and ready triggering of this conformational mechanism strongly indicates that TBG has evolved to allow a modulated and targeted delivery of thyroxine to the tissues.
Keywords: corticosteroid-binding globulin, serpin, thyroxine-binding globulin, crystal structure
Thyroxine is the major hormone controlling cellular development as well as the rate of body metabolism. It is a small molecule formed by the linkage of two tyrosines, which are iodinated to give the alternative tri- or tetra-iodo forms of the hormone. Thyroxine is principally carried in the blood by thyroxine-binding globulin (TBG), which binds it with high affinity (Kd = 0.1 nM) in equilibrium with steady-state low levels of free thyroxine (1–3). Although TBG is not a protease inhibitor, it is otherwise a typical member of the serpin family of protease inhibitors. The alignment of its sequence shows that it retains the same framework structure as the archetypal inhibitory members of the family, α1-antitrypsin, antithrombin, and antichymotrypsin (4). It has similarly retained a typical reactive site loop, with a putative reactive center at the position denoted P1 and, with 17 residues before it, the hinge of the loop at P17 (see Fig. 1 and Table 1). In particular, TBG undergoes the profound and irreversible conformational change on cleavage of its reactive loop, which is characteristic of the serpins (5, 6). Such cleavage of the reactive loop of TBG by proteases does occur during sepsis to give a 3-fold reduction in its binding affinity (7–9). However, because only a minor proportion, <20%, of the circulating TBG is bound to thyroxine, even this relatively small decrease in affinity will result in an effective release of thyroxine (10), as demonstrably occurs at sites of inflammation (11). Normally, however, thyroxine is released without cleavage of TBG, and in the absence of an alternative mechanism it has been assumed that the release occurs because of passive diffusion. A reason for uncertainty about this assumption has been the previous placement of the binding site of thyroxine within a β-barrel of TBG. This presumed site has been consistently backed by a series of modeling, linkage, and domain-exchange studies (12–15), but it does not explain why thyroxine is released by the serpin conformational change or why mutations distant to the site result in a loss of binding. It also does not explain how a conformational change that is irreversible in other serpins has been adapted by TBG to give the reversible binding of thyroxine and to allow its tissue-targeted release. To answer these questions, we determined the crystal structure of TBG bound to thyroxine.
Fig. 1.
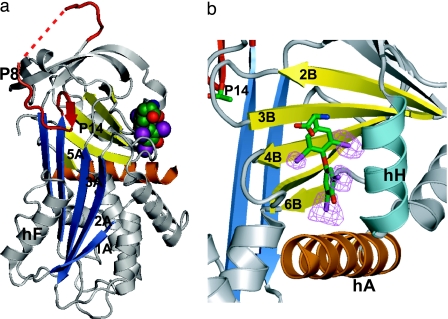
TBG and the thyroxine binding site. (a) Structure of TBG with thyroxine (space-filled). The upper half of the Aβ-sheet (blue) is opened, with initial insertion of the reactive loop (red) to P14 threonine, 14 residues before the reactive center P1. (b) Binding pocket showing thyroxine in stick form enclosed between strands 3–5 of the B-sheet and helices H and A and with iodine atoms, contoured at 5 times rms density in a log-likelihood gradient map for anomalous scattering (16).
Table 1.
Reactive loop sequences
| Serpin | P17 | P16 | P15 | P14 | P13 | P12 | P11 | P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 | P1’ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antichymo. | Glu | Thr | Gly | Thr | Glu | Ala | Ala | Ala | Ala | Thr | Gly | Val | Lys | Phe | Val | Pro | Met | Ser |
| TBG_Human | Glu | Lys | Gly | Thr | Glu | Ala | Ala | Ala | Val | Pro | Glu | Val | Glu | Leu | Ser | Asp | Gln | Pro |
| TBG_Chimp. | Glu | Lys | Gly | Thr | Glu | Ala | Ala | Ala | Val | Pro | Glu | Val | Glu | Leu | Ser | Asp | Gln | Pro |
| TBG_Bovine | Glu | Lys | Gly | Thr | Glu | Ala | Val | — | — | Pro | Glu | Val | Arg | Phe | Leu | Asn | Gln | Pro |
| TBG_Sheep | Glu | Lys | Gly | Thr | Glu | Ala | Ile | — | — | Pro | Glu | Val | Arg | Phe | Leu | Asn | Gln | Pro |
| TBG_Pig | Glu | Lys | Gly | Thr | Glu | Ala | Ile | — | — | Pro | Glu | Val | Thr | Phe | Leu | Asn | Gln | Pro |
| TBG_Mouse | Glu | Glu | Gly | Thr | Lys | Glu | Gly | Ala | Ser | Pro | Glu | Val | Gly | Ser | Leu | Asp | Gln | Gln |
| TBG_Rat | Glu | Glu | Gly | Thr | Lys | Glu | Gly | Ala | Ser | Pro | Glu | Ala | Gly | Ser | Leu | Asp | Gln | Gln |
TBG sequences are from GenBank, with the accession nos. in parentheses, as follows: human (P05543), chimpanzee (P61640), bovine (Q9TT36), sheep (P50450), pig (Q9TT35), mouse (P61939), and rat (P35577). Antichymotrypsin sequence is from Protein Data Bank ID code 1YXA. The reactive center loop (P17–P1) of human TBG corresponds to residues 339–355 of the mature protein. P8 are in bold type; prolines unique to TBG are in italic type.
Results and Discussion
Plasma-derived TBG is heterogeneous and resistant to crystallization, so the recombinant nonglycosylated form of the molecule was prepared by expression in Escherichia coli, with subsequent crystallization and x-ray diffraction as detailed in Materials and Methods. The resulting 2.8-Å structure of TBG complexed with tetra-iodo thyroxine is shown in Fig. 1. The molecule is well ordered in the structure with an R factor of 0.235 and Rfree of 0.284 and with good geometry (for statistics, see Table 2, which is published as supporting information on the PNAS web site) except for three residues at the N terminus, one at the C terminus, and eight in the mobile 350–357 segment of the reactive center loop. The molecule has five cysteines but no disulfide linkages.
Binding Site.
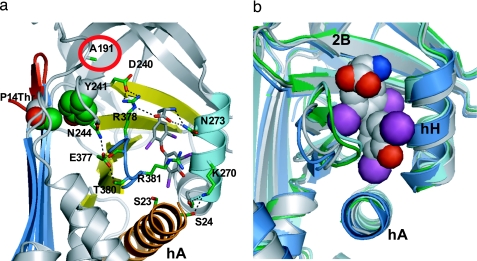
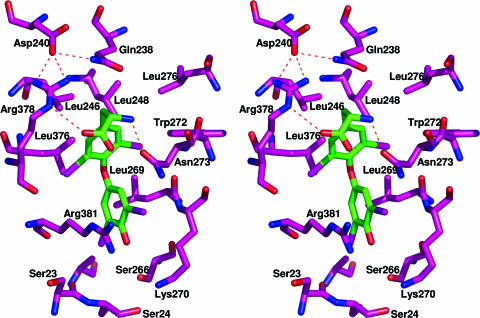
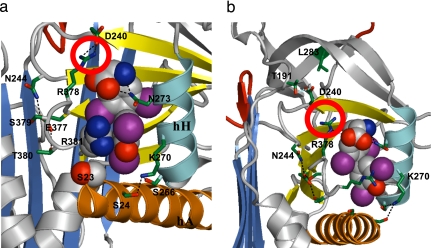
The thyroxine is clearly evident in the structure, bound in a pocket between helices H and A and strands 3–5 of the B-sheet on the opposite side of the molecule to the previously proposed site. This unexpected site is independently confirmed by the positioning of the four iodines in an anomalous dispersion gradient map (16), which shows (Fig. 1b) the thyroxine oriented in a cisoid conformation, with the aminopropionate and outer phenolic ring both on the same side of the inner ring of the molecule, as in the crystal structure of the albumin–thyroxine complex (16, 17). The thyroxine is held in the TBG pocket by a series of hydrophobic interactions with underlying residues and by hydrogen bonding (H-bonding) of the aminopropionate of the thyroxine with adjacent residues. These interactions, shown in Figs. 2a and 3, fulfill the stringent specifications defined by the previously determined binding affinities of 12 different thyroxine analogues, as collated and cited in ref. 13. The identification of the binding site immediately answers some existing puzzles. The mutation in humans (18) that most clearly affects thyroxine binding (Ser-23 → Thr) is now seen (Fig. 4a) to occur in direct proximity to the site, where the extra methyl group in the side chain of the replacement threonine will sterically hinder the binding of thyroxine. The structure also shows (Fig. 4b) how a mutation (Ala-191 → Thr) present in some 60% of Australian aborigines (19) will predictably perturb the H-bonds that normally stabilize the binding pocket and hence result in a decreased binding of thyroxine. The effects of these and other mutations are now apparent as detailed in Figs. 3 and 4, but most importantly, the structure also reveals how TBG reversibly binds thyroxine and allows its triggered release.
Fig. 2.
Binding and triggered release of thyroxine. (a) Interactions with adjacent side chains anchor thyroxine within the pocket. Thyroxine release will be triggered on full insertion of P14 threonine (space-filled upper left) displacing Tyr-241 and disrupting the H-bonds that anchor thyroxine and the flanking peptide loop between s4B and s5B (blue). This network will be similarly disrupted by the common presence (19) in Australian aborigines of a Thr at 191 (circled red; see also Fig. 4). (b) The triggered movement of the flanking s4B–5B loop is shown in the homologous pocket in antichymotrypsin. The open pocket of TBG before loop insertion (blue) matches that of active antichymotrypsin (gray). Transition to the fully inserted loop (cyan) or to the partially inserted δ-form (green) in antichymotrypsin both result in a 4- to 5-Å shift of the loop with a contraction of the binding pocket.
Fig. 3.
Stereoview of thyroxine-binding pocket showing hydrophobic and H-bonding interactions. The replacement of Leu-246 by Thr in the recombinant variant mD–TBG results in a marked reduction in binding affinity (14) with the replacement being compounded by a new glycosylation site at Asn-244, a residue that stabilizes the peptide loop flanking the binding site (see also Figs. 2a and 4a).
Fig. 4.
Thyroxine binding and mutations. (a) Space-filling depictions show how the side chain of Arg-381 stacks with the outer phenolic ring of the thyroxine and how the mutation of Ser-23 → Thr will sterically hinder the binding of thyroxine. Mutations in CBG causing a loss of hormone binding (at Trp-371 and Asp-367 in CBG) affect the structural equivalents of Arg-381 and Glu-377 in the TBG pocket. (b) The mutation Ala-191 → Thr is commonly present in West Australian aboriginals (TBG inheritance is X-linked: 56% of men were hemizygotes for the variant; 29% of women were homozygotes and 38% heterozygotes; ref. 19). As modeled here, Thr-191 will predictably compete for the H-bond network formed with Arg-378 (circled in a) that anchors thyroxine. A concomitant mutation in the aboriginal, Leu-283 → Phe, in an adjacent tightly packed region of the molecule, will exacerbate the perturbation of the binding site.
Reversible Release of Thyroxine.
As Fig. 1 shows, the reactive center loop of TBG has commenced the entry into the central fourth-strand position of the A-sheet that in inhibitory members of the family results in the entrapment of their target proteases (20). This initial insertion of the loop in TBG extends to a threonine situated 14 residues before the reactive center P1 (Table 1). As shown in Fig. 2a, further insertion of the side chain of this P14 threonine will displace an underlying conserved tyrosine with a consequent disruption of the H-bonds that stabilize the peptide loops flanking the binding pocket. The resultant changes in the conformation of the pocket are illustrated (Fig. 2b) by comparable structures of an inhibitory serpin that closely resembles TBG, antichymotrypsin (21). Antichymotrypsin shares with TBG not only the initial entry of the reactive loop into a relatively open A-sheet, but also the presence of extra residues in the distal portion of the loop that allow freer movement of the loop into and out of the sheet. Hence, antichymotrypsin readily undergoes a transition from the five- to six-stranded conformation. This transition usually results in the complete incorporation of its intact reactive loop into the A-sheet to give its irreversible latent form, but it can aberrantly result (22) in a partial insertion of just two residues, from P14 to P12, to give the δ-conformation (Fig. 5c). This small partial insertion is still sufficient to trigger the overall serpin conformational change in antichymotrypsin, including a 4- to 5-Å closure of the surface pocket that in TBG binds thyroxine (Fig. 2). Thus, a change to either the partial or fully six-stranded form in TBG will predictably give a closure of the thyroxine-binding pocket, as in Fig. 2b, with the consequent release of the bound thyroxine.
Fig. 5.
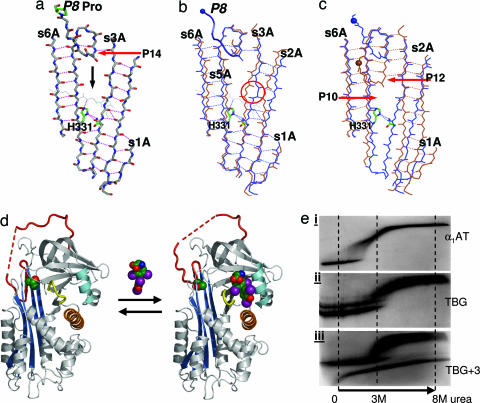
Reversibility of the conformational transition in TBG. (a) The A-sheet in TBG is blocked at the level of entry of the P8 residue of the reactive loop by a barrier centered on His-331 that cannot be readily displaced by the P8 proline uniquely present in TBG. (b) In other serpins, including antichymotrypsin (brown), the sheet is only partly opened (circled). (c) Entry of the loop to P12 in δ-antichymotrypsin (brown) requires a complete opening of its sheet. However, in TBG (blue), the sheet is already opened to allow ready movement of the loop to P10 without disruption of the His-331 barrier. (d) Schematic of flip-flop conformational equilibrium, with unbound TBG modeled on δ-antichymotrypsin on the left and the structure of thyroxine-bound TBG solved here on the right. (e) Transverse urea gradient gels confirm δ-equivalence of unbound TBG. (i) Typical serpin profile (α-1-antitrypsin), with unfolding near 1 M urea. (ii) TBG (without thyroxine) has a δ-conformation stabilized profile with unfolding at 3 M urea (22). (iii) Identical to that of the low-affinity TBG+3 mutant (25). The longer loop in the TBG+3 mutant also allows conversion in the urea gel with complete insertion of the loop to give the hyperstable latent form seen in iii as a second unbroken profile.
As shown in Fig. 5, the upper half of the A-sheet of TBG is fully opened, comparable with that of the aberrant δ-form of antichymotrypsin. However, whereas the complete opening of the A-sheet in the δ-conformation of antichymotrypsin allows the further irreversible entry of the loop, TBG has evolved adaptations that limit its loop insertion and enable it to reversibly bind and release thyroxine. The active five-stranded conformation of the serpins in general is maintained by the binding together of the lower halves of strands 3 and 5 of the A-sheet by a highly conserved histidine (23), equivalent to position 331 in TBG (Fig. 5a). In other serpins, the barrier formed by this histidine is broken on entry of the reactive center peptide loop into the sheet, by the side chain of a threonine, eight residues before the reactive center. However, in TBG the P8 threonine in the reactive loop is replaced by a proline, an iminoacid that cannot form interstrand H-bonds. The unique presence of this proline in all of the sequences of TBG from different species (Table 1) is clearly purposeful: to limit loop insertion to within the upper half of the A-sheet. The critical function of this limitation of loop insertion in maintaining the reversibility of the thyroxine-binding transition is demonstrated in a family with a mutation in TBG of His-331 to a Tyr (24). The loss of the histidine will allow unrestricted entry of the intact loop into the sheet and result in the affected family in markedly increased plasma levels of a “denatured” conformer of TBG together with an effective loss of thyroxine binding.
Triggering of Release.
The binding of thyroxine by TBG can be considered as an equilibrium between a high-affinity form, as in the structure here, with only an initial insertion of the loop to P14, and a low-affinity form of TBG as modeled in Fig. 5d, with a further insertion of the loop to P12. Direct experimental support for this two-conformation model comes from the findings of Grasberger et al. (25). They showed that the engineered deletion of four residues from the TBG reactive loop, which will hinder loop insertion, results in a stable form with an increased affinity for thyroxine. Conversely the insertion of three extra residues (TBG+3), which will allow ready insertion, results in a stable form with a reduction in affinity for thyroxine that matches that of the fully six-stranded, cleaved TBG. Their findings neatly fit with the structural results presented here, as summarized in Fig. 5d. Evidence that the uncomplexed TBG is in a δ-like partially inserted conformation as modeled in Fig. 5d is provided by transverse urea gradient electrophoresis (see Fig. 5e). Serpins typically have an unfolding transition at 1 M urea, whereas uncomplexed TBG has the characteristic profile of the δ-conformation of antichymotrypsin (22), with an unfolding transition at 3 M urea, identical to that of the low-affinity TBG+3 mutant of Grasberger et al. (25). Supportive evidence that the partial loop insertion in the low-affinity form of TBG is limited to just two further residues, as in δ-antichymotrypsin (Fig. 5c), comes from the finding that in the TBG of cattle, pigs, and sheep (26), in contrast to humans, the P8 proline in the reactive loop is preceded by two deletions (Table 1). This difference effectively limits loop entry in these species to just two further residues beyond P14. This additional evidence that a limited insertion to P12 is sufficient to cause the transition provides insight into the still-unsolved question of the events that trigger the serpin conformational change. We can see with TBG that, although the A-sheet is open to P10, the transition will result at an early stage of entry of the loop beyond P14. As shown in Fig. 2a, further entry of the loop into the A-sheet to P12 will necessarily result in the full insertion of the side chain of the P14 threonine into the body of the molecule. The resulting displacement by the side chain of a tyrosine within a closely packed region of the molecule explains the contraction of the binding pocket in TBG and will predictably also be an initiating factor in the overall serpin conformational change.
The concept of a flip-flop change in conformation linked to an on-and-off binding of thyroxine is well illustrated by the detailed structural findings with another closely related serpin, the coagulation-inhibitor antithrombin. Antithrombin has a surface site that avidly binds a heparin pentasaccharide, with the binding and release of the heparin being associated with movement of the reactive loop into and out of the A-sheet of the antithrombin (27). In a similar way, analogous to TBG, antithrombin predominantly circulates in a low-heparin-affinity form with a partially inserted reactive loop. The binding of heparin, however, causes an induced-fit change in its binding site on antithrombin, which is accompanied by an expulsion of the reactive loop from the A-sheet of the molecule. We can now see in video detail how this conformational transition in antithrombin allows an equilibrated shift from a low- to a high-affinity binding state (28). So although the carriage of thyroxine is discussed here in terms of the simple concept, that the transition in TBG results in the release of thyroxine, what is really happening is a transition of TBG from a high- to a low-affinity state with an accompanying change in the equilibrated binding and release of thyroxine (25).
Conclusions and Implications.
The central conclusion from our structural findings is that TBG has evolved to allow it to be an active rather than just a passive carrier of thyroxine. The deficiency of TBG is readily covered by the more simple transport of thyroxine by two other plasma proteins with a lesser affinity, transthyretin and albumin (3). The great advantage, however, of the serpin framework of TBG is that it can be readily adapted to allow a triggered and modulated release of thyroxine. There has long been inferential evidence of the targeted delivery of thyroxine to the placenta and other tissues (3), but further research has been hampered by the absence of a feasible mechanism for its selective release (3). We can now see how the opened sheet of TBG will readily allow the in-and-out movements of its reactive loop that are linked to the binding and release of thyroxine. Similar movements of the loop in other serpins are sensitive to changes in body temperature and pH as well as to peptide and other ligand binding. Is this sensitivity the case with TBG? What is the selective advantage to the Australian aboriginal of a mutation that results in a decreased binding of thyroxine? Could it provide a survival advantage in extreme heat exposure? Thyroxine is believed to be a major modulator of body thermogenesis (29), but just how, why, and where it does this modulation is still unclear. What other factors trigger the transition and the release of thyroxine? These are questions that need reexamination in the light of the new structural understandings. The questions are relevant not only to the release of thyroxine, the hormone that controls the daily activities of the body, but also to the delivery by corticosteroid-binding globulin (CBG) of the steroids that control the response of the body to disease. CBG is closely related to TBG, and its structure is unknown, but it had been assumed (8) that it shared the same hormone-binding site as that proposed for TBG. It can now be seen that the correct site in CBG is that modeled by Edgar and Stein in 1995 (30), homologous to that shown here in TBG. Confirmation of the equivalence and validity of each of the two sites is provided by two variants of CBG with ineffective hormone binding (31, 32), both involving replacements of residues, the structural equivalents of which are seen in TBG to be critical components of the hormone-binding pocket (Figs. 2a and 4a).
Materials and Methods
The human TBG cDNA IMAGE clone (GenBank accession no. BC020747) was purchased from Geneservice Ltd. (Cambridge, U.K.). The TBG coding sequence (covering residues 19–415 of mature TBG), together with the coding sequence for 6-histidine and a thrombin cleavage site, was amplified by PCR and cloned into expression vector pET16b (Novagen, San Diego, CA) by DNA restriction endonuclease sites NcoI and XhoI. Recombinant TBG was expressed in E. coli strain Bl21star (DE3) (Novagen) as a fusion protein with an N terminus of MDHHHHHHLVPRGS LYMSS. Protein expression was induced by 0.5 mM IPTG at 28°C in a 50-liter fermentor. TBG was purified from the supernatant of cell lysate by a nickel-chelating column (protein is eluted with a 0–0.2 M imidazole gradient in 10 mM phosphate buffer, pH 8) and subsequent ion exchange (protein eluted with 0–0.5 M NaCl gradient in 10 mM Tris·HCl/1 mM EDTA, pH 8) and gel-filtration chromatography. The N-terminal His-tag was removed by thrombin cleavage. The prepared recombinant TBG has an N terminus of GSLYKMSS … with the first two residues derived from the thrombin cleavage site. TBG–thyroxine complex was prepared by mixing TBG with excess thyroxine (T4) and separated from free T4 on a gel filtration column. Transverse urea gel electrophoresis was performed on 8% (wt/vol) polyacrylamide gels with a linear gradient of 0–8 M urea and a nondenaturing-PAGE buffer system (33, 34).
Recombinant wild-type TBG (termed native TBG) or its complexes with T4 were concentrated to ≈10 mg/ml in 10 mM Tris·HCl/50 mM NaCl/1 mM EDTA (pH 7.4) and screened for crystallization by vapor diffusion at room temperature. Crystallization of native TBG–T4 complexes was achieved in sitting drops consisting of 2 μl of the complexes and 2 μl of precipitant solution (20% PEG 3350/0.2 M NaF/5% glycerol). Crystals appeared overnight and grew to full size within a week, but under the same conditions, native uncomplexed TBG did not crystallize. Crystals of native TBG–T4 complexes were transferred into the precipitation solution with 20% glycerol and flash-cooled in liquid nitrogen. Diffraction data to 2.8-Å resolution were collected at Daresbury Synchrotron Station 14.2 (Daresbury, U.K.).
Data were processed by Mosflm (35) and scaled with Scala (36). The structure was solved by molecular replacement using Phaser (37) to find two copies in the asymmetric unit. The coordinates of the high-resolution structure of murine antichymotrypsin [Protein Data Bank (PDB) ID code 1YXA, with 44% sequence identity to human TBG], stripped of the reactive center loop, was used as the search model. An initial model of TBG was built in Coot (38) and refined with TLS (translation, libration, screw motion) parameters and tight NCS (noncrystallographic symmetry) restraints in Refmac5 (39) to a final R factor of 0.235 and Rfree of 0.284 (see Table 2). Figures were made with the open-source program Pymol (40), by using the following coordinate sets: native murine antichymotrypsin (21), PDB ID code 1YXA; human antichymotrypsin in the δ-conformation (22), PDB ID code 1QMN; and cleaved antichymotrypsin (41), PDB ID code 1AS4. The atomic coordinates and structure factors for TBG have been deposited in the PDB (PDB ID code 2ceo).
Supplementary Material
Acknowledgments
We thank our colleagues Penelope E. Stein for help in initiating this project, Beibei Du for help in preparing recombinant TBG, and James A. Huntington and David A. Lomas for comments on the paper. This work was supported by grants from the Alpha-1 Foundation, the British Heart Foundation, and the Wellcome Trust.
Abbreviations
- TBG
thyroxine-binding globulin
- CBG
corticosteroid-binding globulin.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors for human thyroxine-binding globulin have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2ceo).
References
- 1.Refetoff S., Murata Y., Mori Y., Janssen O. E., Takeda K., Hayashi Y. Horm. Res. 1996;45:128–138. doi: 10.1159/000184775. [DOI] [PubMed] [Google Scholar]
- 2.Schussler G. C. Thyroid. 2000;10:141–149. doi: 10.1089/thy.2000.10.141. [DOI] [PubMed] [Google Scholar]
- 3.Robbins J. J. Clin. Endocrinol. Metab. 2000;85:3994–3995. doi: 10.1210/jcem.85.11.7049. [DOI] [PubMed] [Google Scholar]
- 4.Huber R., Carrell R. W. Biochemistry. 1989;28:8951–8966. doi: 10.1021/bi00449a001. [DOI] [PubMed] [Google Scholar]
- 5.Loebermann H., Tokuoka R., Deisenhofer J., Huber R. J. Mol. Biol. 1984;177:531–556. [PubMed] [Google Scholar]
- 6.Pemberton P. A., Stein P. E., Pepys M. B., Potter J. M., Carrell R. W. Nature. 1988;336:257–258. doi: 10.1038/336257a0. [DOI] [PubMed] [Google Scholar]
- 7.Jirasakuldech B., Schussler G. C., Yap M. G., Drew H., Josephson A., Michl J. J. Clin. Endocrinol. Metab. 2000;85:3996–3999. doi: 10.1210/jcem.85.11.6966. [DOI] [PubMed] [Google Scholar]
- 8.Suda S. A., Gettins P. G., Patston P. A. Arch. Biochem. Biophys. 2000;384:31–36. doi: 10.1006/abbi.2000.2110. [DOI] [PubMed] [Google Scholar]
- 9.Janssen O. E., Golcher H. M., Grasberger H., Saller B., Mann K., Refetoff S. J. Clin. Endocrinol. Metab. 2002;87:1217–1222. doi: 10.1210/jcem.87.3.8332. [DOI] [PubMed] [Google Scholar]
- 10.Schreiber G. J. Endocrinol. 2002;175:61–73. doi: 10.1677/joe.0.1750061. [DOI] [PubMed] [Google Scholar]
- 11.Adelberg H. M., Siemsen J. K., Jung R. C., Nicoloff J. T. Radiology. 1971;99:141–146. doi: 10.1148/99.1.141. [DOI] [PubMed] [Google Scholar]
- 12.Terry C. J., Blake C. C. Protein Eng. 1992;5:505–510. doi: 10.1093/protein/5.6.505. [DOI] [PubMed] [Google Scholar]
- 13.Jarvis J. A., Munro S. L., Craik D. J. Protein Eng. 1992;5:61–67. doi: 10.1093/protein/5.1.61. [DOI] [PubMed] [Google Scholar]
- 14.Buettner C., Grasberger H., Hermansdorfer K., Chen B., Treske B., Janssen O. E. Mol. Endocrinol. 1999;13:1864–1872. doi: 10.1210/mend.13.11.0367. [DOI] [PubMed] [Google Scholar]
- 15.Grasberger H., Buettner C., Janssen O. E. J. Biol. Chem. 1999;274:15046–15051. doi: 10.1074/jbc.274.21.15046. [DOI] [PubMed] [Google Scholar]
- 16.McCoy A. J., Storoni L. C., Read R. J. Acta Crystallogr. D. 2004;60:1220–1228. doi: 10.1107/S0907444904009990. [DOI] [PubMed] [Google Scholar]
- 17.Petitpas I., Petersen C. E., Ha C. E., Bhattacharya A. A., Zunszain P. A., Ghuman J., Bhagavan N. V., Curry S. Proc. Natl. Acad. Sci. USA. 2003;100:6440–6445. doi: 10.1073/pnas.1137188100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janssen O. E., Astner S. T., Grasberger H., Gunn S. K., Refetoff S. J. Clin. Endocrinol. Metab. 2000;85:368–372. doi: 10.1210/jcem.85.1.6317. [DOI] [PubMed] [Google Scholar]
- 19.Takeda K., Mori Y., Sobieszczyk S., Seo H., Dick M., Watson F., Flink I. L., Seino S., Bell G. I., Refetoff S. J. Clin. Invest. 1989;83:1344–1348. doi: 10.1172/JCI114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huntington J. A., Read R. J., Carrell R. W. Nature. 2000;407:923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 21.Horvath A. J., Irving J. A., Rossjohn J., Law R. H., Bottomley S. P., Quinsey N. S., Pike R. N., Coughlin P. B., Whisstock J. C. J. Biol. Chem. 2005;280:43168–43178. doi: 10.1074/jbc.M505598200. [DOI] [PubMed] [Google Scholar]
- 22.Gooptu B., Hazes B., Chang W. S., Dafforn T. R., Carrell R. W., Read R. J., Lomas D. A. Proc. Natl. Acad. Sci. USA. 2000;97:67–72. doi: 10.1073/pnas.97.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou A., Stein P. E., Huntington J. A., Carrell R. W. J. Biol. Chem. 2003;278:15116–15122. doi: 10.1074/jbc.M211663200. [DOI] [PubMed] [Google Scholar]
- 24.Bertenshaw R., Takeda K., Refetoff S. Am. J. Hum. Genet. 1991;48:741–744. [PMC free article] [PubMed] [Google Scholar]
- 25.Grasberger H., Golcher H. M., Fingerhut A., Janssen O. E. Biochem. J. 2002;365:311–316. doi: 10.1042/BJ20020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janssen O. E., Lahner H., Grasberger H., Spring S. A., Saller B., Mann K., Refetoff S., Einspanier R. Mol. Cell. Endocrinol. 2002;186:27–35. doi: 10.1016/s0303-7207(01)00679-7. [DOI] [PubMed] [Google Scholar]
- 27.Jin L., Abrahams J. P., Skinner R., Petitou M., Pike R. N., Carrell R. W. Proc. Natl. Acad. Sci. USA. 1997;94:14683–14688. doi: 10.1073/pnas.94.26.14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huntington J. A. Trends Biochem. Sci. 2006;31:427–435. doi: 10.1016/j.tibs.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Silvestri E., Schiavo L., Lombardi A., Goglia F. Acta Physiol. Scand. 2005;184:265–283. doi: 10.1111/j.1365-201X.2005.01463.x. [DOI] [PubMed] [Google Scholar]
- 30.Edgar P., Stein P. Nat. Struct. Biol. 1995;2:196–197. doi: 10.1038/nsb0395-196. [DOI] [PubMed] [Google Scholar]
- 31.Emptoz-Bonneton A., Cousin P., Seguchi K., Avvakumov G. V., Bully C., Hammond G. L., Pugeat M. J. Clin. Endocrinol. Metab. 2000;85:361–367. doi: 10.1210/jcem.85.1.6315. [DOI] [PubMed] [Google Scholar]
- 32.Avvakumov G. V., Hammond G. L. J. Steroid Biochem. Mol. Biol. 1994;49:191–194. doi: 10.1016/0960-0760(94)90010-8. [DOI] [PubMed] [Google Scholar]
- 33.Goldenberg D. P. In: Protein Structure: A Practical Approach. Creighton T. E., editor. Oxford: IRL Press at Oxford Univ. Press; 1989. pp. 225–250. [Google Scholar]
- 34.Mast A. E., Enghild J. J., Salvesen G. Biochemistry. 1992;31:2720–2728. doi: 10.1021/bi00125a012. [DOI] [PubMed] [Google Scholar]
- 35.Leslie A. G. W. In: Crystallographic Computing 5: From Chemistry to Biology. Moras D., Podjarny A. D., Thierry J. C., editors. Oxford: Oxford Univ. Press; 1990. pp. 50–61. [Google Scholar]
- 36.Evans P. R. In: Proceedings of the CCP4 Study Weekend: Data Collection and Processing. Sawyer L., Isaacs N., Bailey S., editors. Daresbury, U.K.: Daresbury Laboratory; 1993. pp. 114–122. [Google Scholar]
- 37.McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. Acta Crystallogr. D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P., Cowtan K. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Winn M. D., Isupov M. N., Murshudov G. N. Acta Crystallogr. D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 40.Delano W. L. The PyMOL User’s Manual. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 41.Baumann U., Huber R., Bode W., Grosse D., Lesjak M., Laurell C. B. J. Mol. Biol. 1991;218:595–606. doi: 10.1016/0022-2836(91)90704-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.