Abstract
The endocannabinoid arachidonoyl ethanolamine (anandamide) is a lipid transmitter synthesized and released “on demand” by neurons in the brain. Anandamide is also generated by macrophages where its endotoxin (LPS)-induced synthesis has been implicated in the hypotension of septic shock and advanced liver cirrhosis. Anandamide can be generated from its membrane precursor, N-arachidonoyl phosphatidylethanolamine (NAPE) through cleavage by a phospholipase D (NAPE–PLD). Here we document a biosynthetic pathway for anandamide in mouse brain and RAW264.7 macrophages that involves the phospholipase C (PLC)-catalyzed cleavage of NAPE to generate a lipid, phosphoanandamide, which is subsequently dephosphorylated by phosphatases, including PTPN22, previously described as a protein tyrosine phosphatase. Bacterial endotoxin (LPS)-induced synthesis of anandamide in macrophages is mediated exclusively by the PLC/phosphatase pathway, which is up-regulated by LPS, whereas NAPE–PLD is down-regulated by LPS and functions as a salvage pathway of anandamide synthesis when the PLC/phosphatase pathway is compromised. Both PTPN22 and endocannabinoids have been implicated in autoimmune diseases, suggesting that the PLC/phosphatase pathway of anandamide synthesis may be a pharmacotherapeutic target.
Keywords: biosynthesis, phosphatase, phospholipase C, phosphoanandamide
The endocannabinoid N-arachidonoyl ethanolamine (anandamide, AEA) is a lipid transmitter synthesized and released “on demand” by neurons in the brain (1). AEA is also generated by macrophages (2), where its bacterial endotoxin (LPS)-induced synthesis has been implicated in the hypotension of septic shock (3, 4) and liver cirrhosis (5, 6). Macrophage-derived AEA has been also implicated in antiinflammatory effects both in the periphery (7) and in the central nervous system (8, 9). AEA is thought to be generated from its membrane precursor, N-arachidonoyl phosphatidylethanolamine (NAPE), through cleavage by a phospholipase D (NAPE–PLD) (10, 11), up-regulation of which can result in increased tissue levels of AEA (12). We earlier reported that LPS potently stimulates AEA synthesis in RAW264.7 mouse macrophages, in which it increases both the generation of NAPE from [14C]diarachidonoyl phosphatidylcholine and the conversion of NAPE to AEA (4). Because these effects could be prevented by inhibitors of RNA transcription or protein synthesis (4), we hypothesized that LPS induces the expression of proteins involved in the biosynthesis of AEA, and a subtraction cloning strategy using resting and LPS-treated macrophages may help identifying such proteins. Although a specific N-acyltransferase (NAT) involved in the generation of NAPE has not yet been discovered, a NAPE-specific PLD has been identified and its ability to generate AEA from NAPE has been established (11). The results presented here indicate that, unexpectedly, NAPE–PLD is not involved in the stimulated synthesis of AEA in RAW264.7 macrophages. Instead, we identified the lipid phosphoanandamide (pAEA), which is also present in brain, and is generated from NAPE by phospholipase C (PLC). We also identified PTPN22, previously described as a protein tyrosine phosphatase (13, 14), as one of the enzymes responsible for the generation of AEA from its phosphorylated precursor.
Results
NAPE–PLD Is Not Involved in LPS-Stimulated AEA Synthesis.
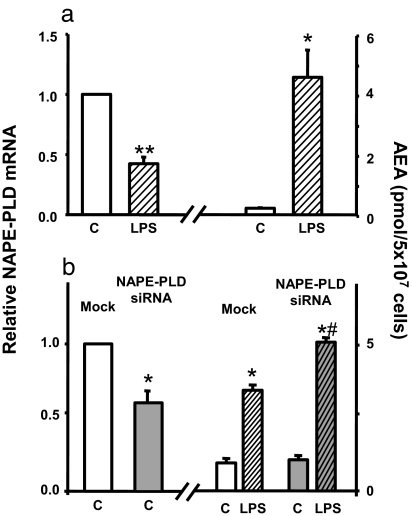
To confirm the feasibility of the subtraction cloning strategy mentioned above, we first tested whether NAPE–PLD expression in RAW264.7 cells is induced by LPS. Surprisingly, the dramatic increase in cellular AEA levels induced by LPS was associated by a marked decrease rather than increase in NAPE–PLD gene expression (Fig. 1a), suggesting that the increased conversion of NAPE to AEA may involve an alternative pathway. This was further indicated by the finding that siRNA knockdown of NAPE–PLD expression did not influence the basal level of AEA or its increase by LPS treatment, the latter being even greater than in mock-transfected controls (Fig. 1b).
Fig. 1.
Dissociation of NAPE–PLD from LPS-induced AEA synthesis. (a) LPS (10 ng/ml for 90 min) reduces NAPE–PLD mRNA and increases AEA levels in RAW264.7 cells. (b) siRNA knockdown of NAPE–PLD expression is associated with unchanged basal AEA and increased LPS-stimulated AEA levels. NAPE–PLD mRNA and AEA were quantified by real-time PCR and LC/MS, respectively, as described in Materials and Methods. Means ± SE from three to four experiments are shown. Asterisks indicate significant difference (∗, P < 0.05; ∗∗, P < 0.005) from values in control (C) cells or from value in mock-transfected, LPS-stimulated cells (#).
Identification of Proteins Involved in LPS-Induced AEA Synthesis.
To find proteins involved in the biosynthesis of AEA, we generated differentially expressed cDNAs in LPS-stimulated versus control RAW264.7 cells, using the PCR-Select method of cDNA subtraction. The differentially expressed cDNAs were labeled with [α-32P]dCTP and used as probes to screen a commercial cDNA library prepared from LPS-treated RAW264.7 cells. The DNA targets were sequenced and the identities of the corresponding genes were established by Blast search of the GenBank database. To find out which of these genes might be involved in the regulation of AEA synthesis, full-length cDNAs were transiently transfected into RAW264.7 cells, and the cellular levels of AEA were measured by liquid chromatography (LC)/MS. Transfection of 21 of the >150 LPS-induced genes resulted in a significant (>50%) increase in the cellular level of AEA, and RAW264.7 cell lines stably transfected with these genes have been developed.
LPS Induces the Expression of the Protein Tyrosine Phosphatase PTPN22 in RAW264.7 Cells.
One of the genes, whose stable overexpression resulted in a 2-fold increase in AEA levels in RAW264.7 cells, encodes PTPN22 (Lyp, PEP, PTPN8), a nonreceptor protein tyrosine phosphatase predominantly expressed in lymphoid tissues (13, 14). Exposure of wild-type RAW264.7 cells to 10 ng/ml LPS for 90 min resulted in a 3.2 ± 0.5-fold increase in the expression of PTPN22, as verified by real-time PCR (n = 3). The possibility suggested by these findings that AEA may arise by dephosphorylation of a precursor, generated from NAPE by a PLC, had been earlier contemplated (10, 14), but not proved because pAEA, once produced in the body, is quickly modified by phosphatases to AEA (15).
Identification of pAEA in RAW264.7 Cells and in Mouse Brain.
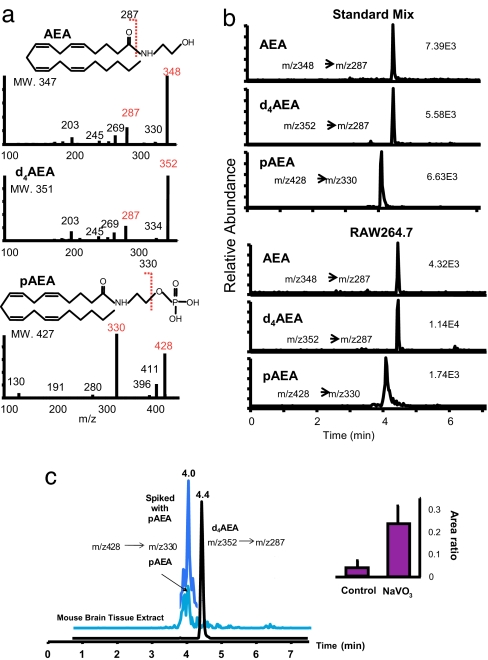
To test whether pAEA may be formed in vivo, we incubated homogenates of RAW264.7 cells with [14C]NAPE in the absence or presence of the nonselective tyrosine phosphatase inhibitor sodium orthovanadate (NaVO3, 1 mM), and analyzed the products by thin layer chromatography. A band comigrating with synthetic pAEA could be observed in the presence, but not in the absence, of NaVO3, in extracts of both control RAW264.7 cells and RAW264.7 cells stably transfected with PTPN22 (RAW264.7-Lyp cells, not shown). To definitively identify pAEA, extracts of both RAW264.7 cells and mouse brain incubated either with or without NaVO3 were analyzed by high-performance liquid chromatography/in line electrospray ionization tandem mass spectrometry (HPLC/ESI-MS/MS), using [2H4]AEA as internal standard. PAEA could be clearly identified along with AEA in both RAW264.7 cells and mouse brain extracts by selected reaction monitoring (SRM) using the transition of molecular ion at m/z 428 to m/z 330, the major fragment formed by dephosphorylation under collision-induced dissociation condition (Fig. 2b). In brain tissue incubated with NaVO3, the concentrations of pAEA increased up to 15-fold compared with controls (Fig. 2c).
Fig. 2.
pAEA is an intermediate of AEA synthesis. (a) MS/MS spectra of AEA, pAEA, and d4AEA, the internal standard. (b) Representative ion chromatograms for pAEA and AEA obtained from ≈400 fmol standards (Upper) and RAW 264.7 (Lower) cells treated with NaVO3 and LPS by using SRM. The transitions of m/z 348 → 287, m/z 352 → 287, and m/z 428 → 330 were selected for AEA, d4AEA, and pAEA, respectively. (c) Effect of NaVO3 on pAEA levels in brain tissue extract. (Left) Representative SRM ion chromatograms obtained from the control brain sample. The presence of pAEA peak at 4 min was further confirmed by spiking with standard pAEA. (Right) The peak area ratio of pAEA to d4AEA in brain samples incubated with or without NaVO3 (mean ± SE, n = 3).
Interaction of PAEA with CB1 Receptors.
The ability of pAEA to directly interact with CB1 receptors was tested in radioligand binding displacement assays using 0.5 nM [3H]CP-55,940 as the labeled ligand. PAEA bound to mouse brain CB1 receptors with an apparent Kd of 46.7 nM compared with a Kd of 13.2 nM for AEA, which is comparable to earlier findings (15). However, in the presence of NaVO3, the affinity of pAEA was reduced ≈10-fold to 432.1 nM, whereas the affinity of AEA remained unchanged (16.3 nM), indicating that pAEA itself is not functional at CB1 receptors and its apparent potency is due to its conversion to AEA in the assay mixture.
In Vivo Conversion of PAEA to AEA by Phosphatases.
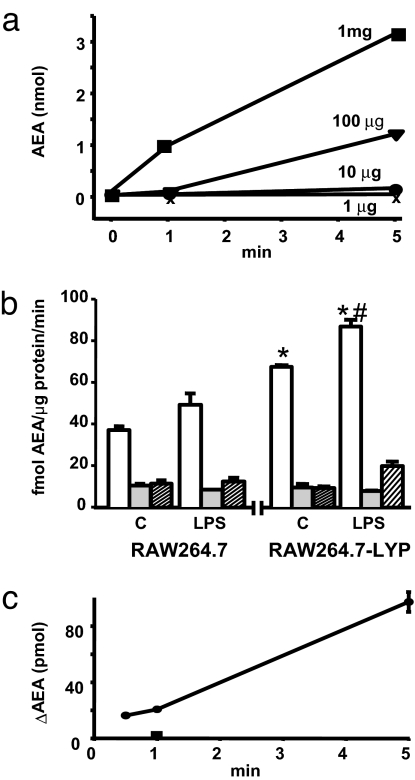
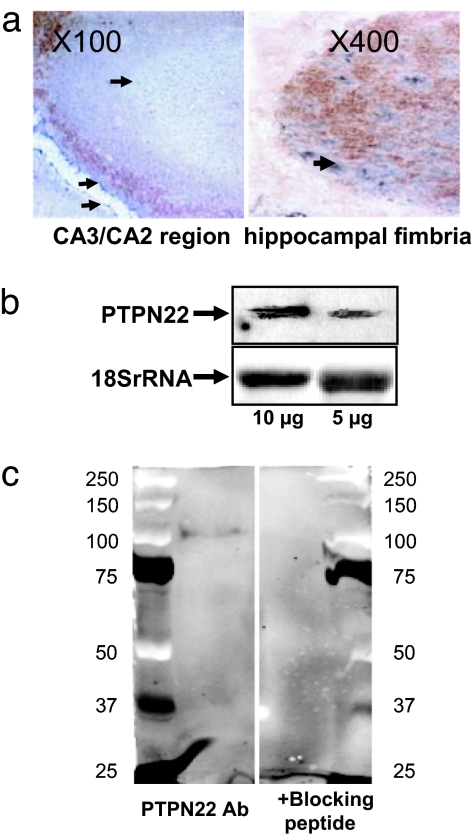
To determine whether pAEA is enzymatically converted to AEA in vivo, synthetic pAEA was incubated with extracts of control RAW264.7 cells or RAW264.7-Lyp cells in the presence of the fatty acid amidohydrolase inhibitor URB597 (16) to prevent AEA breakdown, and the amount of AEA generated determined by LC/MS. The formation of AEA was time and protein concentration-dependent (Fig. 3a), and it was about twice greater in RAW264.7-Lyp than in control RAW264.7 cells (Fig. 3b). Furthermore, preincubation of both cell types with 10 ng/ml LPS for 90 min significantly increased the conversion of pAEA to AEA, which could be blocked by boiling the cell extracts or by the presence of NaVO3 in the medium (Fig. 3b). Similar results were obtained by using mouse brain extracts (Fig. 4b), suggesting that PTPN22 is present in brain, which was then verified by Western and Northern blotting and immunohistochemistry (Fig. 5).
Fig. 3.
Phosphatase activity involved in conversion of pAEA to AEA. (a) Generation of AEA from synthetic pAEA is time- and protein concentration-dependent. Homogenates of RAW264.7 cells with different protein concentrations were incubated with 5 nmol of pAEA for the indicated times, and AEA was then determined in the lipid extracts by LC/MS. (b) Conversion of synthetic pAEA to AEA in homogenates of RAW264.7 cells (Left) and RAW264.7-Lyp cells (Right) is increased by LPS pretreatment and is heat- and NaVO3-sensitive. Cell homogenates (10 μg of protein) pretreated with vehicle (C) or 10 ng/ml LPS for 90 min (LPS) were incubated for 20 s with 5 pmol of synthetic pAEA (open columns). Parallel aliquots were tested after boiling for 5 min (shaded columns) or a preincubation with 1 mM NaVO3 for 1 h at 4°C before the initiation of enzymatic reaction (hatched columns). Means ± SE from three experiments are shown. Asterisk indicates significant difference (P < 0.01) from AEA levels in the same treatment group (C, control). Pound sign indicates significant difference (P < 0.05) from value in control cell homogenate. (c) Generation of AEA from synthetic pAEA by recombinant PTPN22 expressed in wheat germ extracts. Aliquots of wheat germ extract containing no construct (control) or the mPTPN22-pIVEX1.4WG construct were incubated with 5 nmol of synthetic pAEA for the indicated times, and the amount of AEA generated was measured by LC/MS. Values shown represent AEA generated over the amount detected in controls, which was abolished by boiling the extracts, as tested at 1 min (filled square). Points are means of two to three separate experiments.
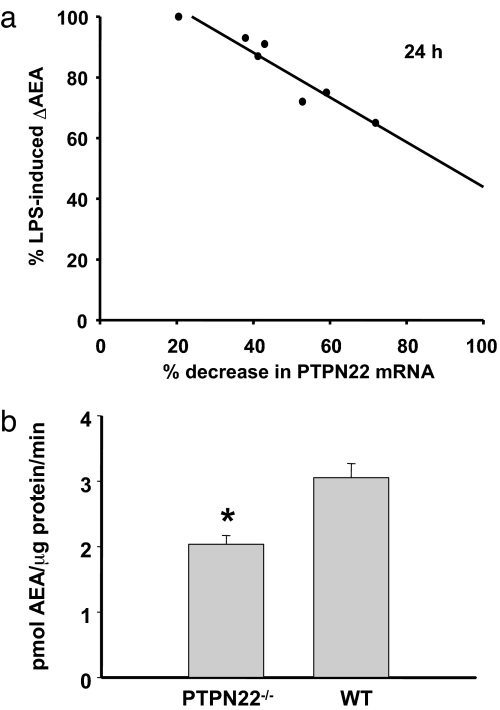
Fig. 4.
Knockdown or knockout of PTPN22 reduces LPS-induced AEA synthesis and the conversion of pAEA to AEA. (a) Graded siRNA knockdown of PTPN22 in RAW264.7 cells causes progressive but disproportional reduction of LPS-induced AEA synthesis. The degree of knockdown was verified by real-time PCR for each batch of cells. (b) Enzymatic conversion of pAEA to AEA by brain extracts from wild-type and PTPN22 knockout mice. Aliquots of brain extracts (10 μg of protein) were incubated with 5 nmol of synthetic pAEA for 1 min at 37°C, and the AEA generated was measured by LC/MS. Columns and bars represent means ± SE from three separate brains in each group.
Fig. 5.
PTPN22 is present in the mouse brain. (a) Immunohistochemical localization of PTPN22 in mouse hippocampus. PTPN22 is visualized by Ni-DAB double staining providing a blue color, whereas neuron-specific (Left) and glia-specific (Right) stains appear in brown. (b) Identification of PTPN22 mRNA in mouse brain by Northern blotting. (c) Western blot of 105-kDa PTPN22 protein in a mouse brain extract (Left) is eliminated by the presence of a blocking peptide (Right).
Contribution of PTPN22 to the Generation of AEA in Vivo.
To test whether PTPN22 is involved in the dephosphorylation of pAEA in vivo, we generated recombinant PTPN22 in a wheat germ cell-free protein translation system by using PTPN22 cDNA-pIVEX1.4WG construct as template. Possibly due to the relatively large size of the protein (105 kDa), only low level expression was achieved, which was verified by Western blotting. Nevertheless, when recombinant PTPN22 was incubated with 5 nmol of synthetic pAEA, AEA was generated in a time-dependent manner that could be abolished by boiling the extracts (Fig. 3c). The role of PTPN22 in the generation of AEA in vivo was further tested in two ways. First, graded siRNA knockdown of PTPN22 in RAW264.7 cells caused a progressive reduction in LPS-induced increase in AEA production, although the degree of the reduction was less than the reduction in PTPN22 message at any level of knockdown (Fig. 4a). Second, heat- and NaVO3-sensitive conversion of synthetic pAEA to AEA was 38% lower in brain extracts from PTPN22 knockout compared with wild-type mice (Fig. 4b). These findings indicate that PTPN22 does contribute to the dephosphorylation of pAEA in vivo, but is not the only phosphatase that can do so.
LPS-Induced Generation of AEA from NAPE Is Mediated via the PLC/Phosphatase Pathway.
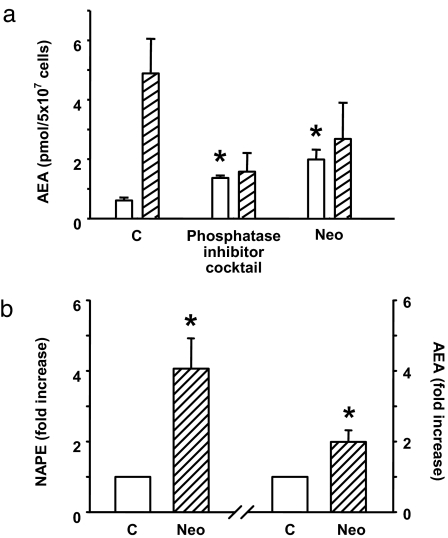
The above findings strongly suggest that NAPE is converted to AEA in vivo via a two-step process involving its cleavage by a PLC to yield pAEA, which is then dephosphorylated by phosphatases including PTPN22. To further test the possible role of this pathway in LPS-induced AEA synthesis, RAW264.7 cells were preincubated with the PLC inhibitor neomycin or a phosphatase inhibitor mixture before their exposure to vehicle or 10 ng/ml LPS for 90 min, after which cellular AEA levels were quantified by LC/MS. As shown in Fig. 6a, LPS treatment failed to increase AEA levels under these conditions, which indicates that the LPS-induced increase in AEA synthesis proceeds through the PLC/phosphatase pathway. Blocking this pathway by neomycin resulted in a parallel increase in the basal levels of AEA and NAPE (Fig. 6b).
Fig. 6.
Blocking PLC or tyrosine phosphatases prevents LPS-induced, but not basal, AEA synthesis. (a) RAW264.7 cells were incubated for 2 h with 3 mM neomycin or with a mixture of tyrosine phosphatase inhibitors containing 2 mM imidazole, 1 mM NaF, 1.15 mM sodium molybdate, 1 mM NaVO3, and 4 mM sodium tartrate dihydrate. AEA was then quantified in lipid extracts of vehicle (open columns) or LPS-treated (hatched columns) cells as described in Materials and Methods. Values represent means ± SE from two to four experiments. Asterisk indicates significant difference (P < 0.05) from values in control cells. (b) Incubation of RAW264.7 cells with neomycin (NEO) increases both NAPE and AEA levels.
Discussion
It has been generally accepted that the endogenous cannabinoid AEA is produced through a single-step, phosphodiesterase-mediated cleavage of its membrane precursor NAPE (10). The present findings document the existence in both brain and macrophages of a parallel pathway through which AEA is generated from NAPE by a two-step process involving the PLC-catalyzed cleavage of NAPE to yield pAEA, which is subsequently dephosphorylated by phosphatases, including PTPN22, originally described as a protein tyrosine phosphatase (13, 14). The present findings also indicate that the regulated (i.e., LPS-induced) synthesis of AEA in macrophages proceeds through the PLC/phosphatase pathway, and NAPE–PLD, which is considered to be a constitutively active rather than a regulated enzyme (17), may function as a salvage pathway of AEA synthesis when the PLC/phosphatase pathway is compromised. The nonexclusive role of NAPE–PLD in the conversion of NAPE to AEA is clearly indicated by the unchanged brain levels of AEA in NAPE–PLD knockout mice, as documented in a recent paper that appeared after the present manuscript had been submitted for publication (18).
The existence of a regulated PLC/phosphatase pathway is strongly suggested by several lines of evidence. First, a pAEA intermediate has been identified in both brain tissue and macrophages, and its quantity is markedly increased when its degradation is blocked by nonselective inhibition of phosphatase activity; this may explain why in the absence of phosphatase inhibition it escaped detection with earlier, less sensitive methods (14). Conclusive identification of pAEA in both brain tissue and macrophages was made possible through the application of the highly selective SRM monitoring mode, using LC/ES-MS/MS, which enables molecule-selective detection with high sensitivity for quantification (19). Second, macrophage or brain tissue extracts rapidly convert synthetic pAEA to AEA in a heat- and phosphatase inhibitor-sensitive manner. Third, incubation of macrophages with either the PLC inhibitor neomycin or a mixture of phosphatase inhibitors prevents the LPS-induced increase in cellular AEA levels. These treatments also lead to the cellular accumulation of NAPE, suggesting that the parallel modest increase in cellular AEA, which is likely mediated by constitutively active NAPE–PLD, is due to increased substrate availability, although a modest increase in enzyme expression is also possible.
Several lines of evidence suggest the involvement of PTPN22 in the generation of AEA. LPS induces the expression of PTPN22 in RAW264.7 cells, and overexpression of PTPN22 results in increased conversion of pAEA to AEA by extracts of such cells. More importantly, recombinant PTPN22 expressed in a wheat germ system can dephosphorylate pAEA. The finding that this reaction was reduced but not eliminated in the absence of PTPN22 indicates that additional as yet unidentified phosphatases are also involved. Given that the levels of PTPN22 are much lower in brain than in immune cells (see http://symatlas.gnf.org/SymAtlas/), other phosphatases may play a more dominant role in anandamide synthesis in the nervous system. However, the relative contribution of such phosphatases to AEA synthesis may be overestimated by the present findings in PTPN22 knockout mice, in which such phosphatases may be overexpressed to compensate for the life-long loss of PTPN22.
PTPN22 and its mouse homolog, PEST domain-enriched tyrosine phosphatase (PEP) are predominantly expressed in lymphoid and hematopoietic tissues (13, 14, 20), and have been implicated in susceptibility to various autoimmune disorders (14, 21). For example, mice deficient in PEP have enhanced effector/memory T cell functions, which can lead to the development of autoimmunity (14). We now find that PTPN22 is also present in the brain, and is involved in the inducible generation of the lipid messenger AEA. Thus, PTPN22 may act as a lipid phosphatase, even though it had earlier been characterized as a protein tyrosine phosphatase (13, 14); this is not unprecedented, because the tumor suppressor PTEN phosphatase can act on both polypeptide and phosphoinositide substrates (22).
There is evidence that an LPS-induced increase in AEA production in circulating macrophages mediates the early hypotensive phase of septic shock (3, 4), and also plays a key role in the vasodilated state of advanced liver cirrhosis (5, 6), a condition that contributes to potentially fatal complications such as ascites and variceal rupture. The observed exclusive role of the PLC/phosphatase pathway in LPS-induced AEA synthesis may offer therapeutic targets for the treatment of these conditions. Furthermore, cannabinoids have immunosuppressive effects in autoimmune models of multiple sclerosis (23) and diabetes (24), and mice deficient in CB1 receptors show increased susceptibility to neuronal damage found in autoimmune encephalitis (25). Also, AEA limits immune responses after primary CNS damage in multiple sclerosis in humans (9). It is tempting to speculate that the robust link between PTPN22 and autoimmunity may be related, at least in part, to the role of PTPN22 in the regulated synthesis of the endocannabinoid AEA.
Materials and Methods
Gene Screening and Plasmid Transfection.
A custom LPS-stimulated RAW264.7 cell ZAP express EcoRI/XhoI cDNA library was obtained from Stratagene (La Jolla, CA). The pBK-CMV phagemid vectors from the ZAP Express vector were released by in vivo excision following the manufacturer’s protocol. Positive clones were screened with [α-32P]dCTP-labeled cDNAs differentially expressed in response to LPS, as generated by using a PCR-Select cDNA subtraction kit (Clontech, Palo Alto, CA). Individual positive clones in pBK-CMV phagemid vectors were transiently transfected into RAW264.7 cells. Using an initial seeding density of 104 cells per ml, RAW264.7 cells were ready for transfection at 18–24 h after seeding. For each 75-cm2 flask of cells to be transfected, 60 μl of Lipofactamine 2000 reagent and 5 μg of DNA were diluted separately in 1,500 μl of Opti-MEM I-reduced serum medium for <5 min. Solutions were combined, gently mixed, and incubated for 20 min at room temperature. The DNA–lipid complexes were added subsequently to each flask containing 6 ml of normal medium and mixed gently. Transfected cells were harvested for AEA measurement or real-time PCR after 24 h. Stable transformants were selected by using growth medium containing 1 mg/ml G418.
siRNA Knockdown and Real-Time PCR.
Transfection of RAW264.7 cells with 300 pmol of siRNA for NAPE–PLD was as described above. The degree of reduction of NAPE–PLD mRNA at 48 h after transfection was verified by real-time PCR, using the NAPE–PLD probe 6FAM-GGTTCCAAAGAGGAACT-MGB, forward primer ATGGCTGATAATGGAGAAGAATCAC, and reverse primer CGTCTTCAGGGTCACTGACAAA. Probe and primer mix for real-time PCR of PTPN22 were from Applied Biosystems (Foster City, CA). For siRNA knockdown of PTPN22, predesigned siRNAs 1–7 were purchased from Qiagen (Valencia, CA), and the degree of knockdown was established by real-time PCR for each probe set.
TLC.
To detect pAEA, 1 mg of RAW264.7 cell homogenate was incubated with 5 × 105 dpm [14C]NAPE at 37°C for 1 h. [14C]NAPE was synthesized as described (26). Lipids were extracted and separated by TLC with chloroform/methanol/NH4OH (80:20:1) as the mobile phase. Radioactivity of the lipid spots was quantified by PhosphorImaging (Typhoon 8600). The synthetic pAEA used as standard was prepared as described (15).
LC/MS.
To prepare samples for LC/MS analyses, RAW264.7 cells or brain homogenate were extracted three times by using CHCl3/MeOH/H2O (0.5:0.5:1). Organic layers were collected, dried under nitrogen flow, and reconstituted with MeOH after precipitating proteins with ice-cold acetone. For measuring AEA without simultaneously measuring pAEA, LC/MS atmospheric pressure chemical ionization was used with [2H4]AEA as internal standard, as described (27). For simultaneous measurement of AEA and pAEA, the reconstituted samples were subjected to HPLC/electrospray ionization-MS/MS analysis using a Finnigan TSQ Quantum Ultra triple stage quadrupole mass spectrometer (San Jose, CA) equipped with an Agilent 1100 series microflow HPLC system. Separation was performed on a BDS Hypersil C18 Pioneer column (2.1 × 50 mm; Thermo Electron, Waltham, MA). The flow rate was set at 400 μl/min. Elution solvent A consisted of 0.1% formic acid in water, and solvent B was methanol. As solvent C, 100 mM ammonium acetate was used to assist in removing the carryover. The solvent composition was changed from 70% A/30% B to 0% A/100% B in 1 min and held at 100%B for 2 min. At 3.1 min, the solvent composition returned to 70% A/30% B and maintained until 5 min. Subsequently, the column was washed with 100% C for 1 min, and the solvent composition returned to the initial composition of 70% A/30% B. The TSQ Quantum Ultra was operated with electrospray voltage set at 4,500 V and ion transfer tube temperature at 350°C. The sheath and auxiliary gas pressures were 39 and 5 psi, respectively. Collision-induced dissociation (CID) was performed by using argon as the collision gas at 1.5 mTorr. Quantitation was performed by SRM in the positive ion mode using [2H4]AEA as the internal standard. Recorded reactions were as follows: m/z 428 → 330 for pAEA, m/z 348 → 287 for AEA, and m/z 352 → 287 for the internal standard, [2H4]AEA.
Radioligand Binding.
The binding of pAEA and AEA to mouse brain cannabinoid receptors was assessed as described (28), except that the membranes were treated with PMSF (28). The membranes were subsequently treated with or without 1 mM NaVO3 for 1 h on ice before the initiation of the assay. Binding of 0.5 nM [3H]CP55,940 was tested in competition assays by using eight concentrations (10 nM to 30 μM) of the displacing ligands. IC50 values were obtained by nonlinear regression of log concentration percent displacement data and then converted to KD values by using Prism 3.03 software.
Phosphatase Activity.
RAW264.7 cell or mouse brain homogenates (10 μg of protein unless indicated otherwise) were incubated with 5 pmol to 5 nmol of synthetic pAEA in 50 mM Tris buffer with no added calcium or magnesium, pH 8.0, containing 1 μM URB597 with or without 1 mM NaVO3 for the indicated time. Enzymatic reactions were stopped by adding two volumes of chloroform/methanol (2:1) containing 5 nmol of [2H4]AEA as internal standard, and the AEA generated was measured as above. The reaction was linear between 20 s and 5 min. Endogenous AEA in the samples was <1% of the AEA generated from 5 nmol of synthetic pAEA.
Wheat Germ Cell-Free Protein Translation.
Full-length mouse PTPN22 cDNA was obtained by RT-PCR using LPS stimulated RAW264.7 cell mRNA as template. An NcoI and an SmaI restriction site were generated at the two ends of the amplicon, which was then inserted into a wheat germ his6-tag vector pIVEX1.4WG. Mouse PTPN22 protein was translated from the mPTPN22-pIVEX1.4WG construct by using RTS 100 wheat germ CECF kit, following the manufacturer’s instructions (Roche Diagnostics, Indianapolis, IN).
Northern Blot Hybridization.
Northern blot hybridization was performed as described (29). A 342-bp PTPN22 cDNA probe was generated with plasmid mPTPN22-pIVEX1.4WG as a template for amplification by PCR with forward primer ATAGCAACCCACACGACTCC and reverse primer GTGGAGGAGAACCATCCTGA. The cDNA probe was labeled with alkaline phosphatase by using the AlkPhos Direct kit (Amersham Pharmacia, Piscataway, NJ) according to the manufacturer’s instructions.
Western Blotting and Immunohistochemistry.
Native PTPN22 in mouse brain homogenate was detected by Western immunoblotting using a rabbit polyclonal antibody against PTPN22 (Novus Biologicals, Littleton, CO), as described (30). The same antibody was used in immunohistochemical localization of PTPN22 in mouse brain sections. Double staining using the PTPN22 antibody and a rabbit anti GFAP antibody (Chemicon, Temecula, CA) or the neuron-specific mouse anti-MAP-2 antibody (Chemicon) was done as described (31). For blocking/competition, PTPN22 antibody was combined with a 5-fold excess of blocking peptide in 500 μl of 1× PBS and incubated overnight at 4°C before being diluted into blocking buffer.
NAPE Levels.
NAPE in lipid extracts of cells was isolated by TLC and quantified by the amount of AEA released through its digestion with Streptomyces chromofuscus PLD, as described (4, 26).
PTPN22 Knockout Mice.
PTPN22−/− mice and their wild-type controls were developed and maintained as described (14). The brain was removed by decapitation and frozen on dry ice until used. Procedures have been approved by the Institutional Animal Use and Care Committee.
Acknowledgments
We thank R. L. Veech for critically reading the manuscript. This work was supported by intramural funds of the National Institutes of Health.
Abbreviations
- AEA
anandamide
- pAEA
phosphoanandamide
- NAPE
N-arachidonoyl phosphatidylethanolamine
- PLD
phospholipase D
- SRM
selected reaction monitoring
- LC
liquid chromatography.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Giuffrida A., Parsons L. H., Kerr T. M., Rodriguez de Fonseca F., Navarro M., Piomelli D. Nat. Neurosci. 1999;2:358–363. doi: 10.1038/7268. [DOI] [PubMed] [Google Scholar]
- 2.Di Marzo V., De Petrocellis L., Sepe N., Buono A. Biochem. J., 1996;316:977–984. doi: 10.1042/bj3160977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varga K., Wagner J. A., Bridgen D. T., Kunos G. FASEB J. 1998;12:1035–1044. doi: 10.1096/fasebj.12.11.1035. [DOI] [PubMed] [Google Scholar]
- 4.Liu J., Bátkai S., Pacher P., Harvey-White J., Wagner J. A., Cravatt B. F., Kunos G. J. Biol. Chem. 2003;278:45034–45039. doi: 10.1074/jbc.M306062200. [DOI] [PubMed] [Google Scholar]
- 5.Bátkai S., Járai Z., Wagner J. A., Goparaju S. K., Varga K., Liu J., Wang L., Mirshahi S., Khanolkar A. D., Makriyannis A., et al. Nat. Med. 2001;7:827–832. doi: 10.1038/89953. [DOI] [PubMed] [Google Scholar]
- 6.Ros J., Claria J., To-Figueras J., Planaguma A., Cejudo-Martin P., Fernandez-Varo G., Martin-Ruiz R., Arroyo V., Rivera F., Rodes J., et al. Gastroenterology. 2002;122:85–93. doi: 10.1053/gast.2002.30305. [DOI] [PubMed] [Google Scholar]
- 7.Cabral G. A., Toney D. M., Fischer-Stenger K., Harrison M. P., Marciano-Cabral F. Life Sci. 1995;56:2065–2072. doi: 10.1016/0024-3205(95)00190-h. [DOI] [PubMed] [Google Scholar]
- 8.Molina-Holgado F., Molina-Holgado E., Guaza C., Rothwell N. J. J. Neurosci. Res. 2002;67:829–836. doi: 10.1002/jnr.10165. [DOI] [PubMed] [Google Scholar]
- 9.Eljaschewitsch E., Witting A., Mawrin C., Lee T., Schmidt P. M., Wolf S., Hoertnagl H., Raine C. S., Schneider-Stock R., Nitsch R., et al. Neuron. 2006;49:67–79. doi: 10.1016/j.neuron.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 10.Di Marzo V., Fontana A., Cadas H., Schinelli S., Cimino G., Schwartz J. C., Piomelli D. Nature. 1994;372:686–691. doi: 10.1038/372686a0. [DOI] [PubMed] [Google Scholar]
- 11.Okamoto Y., Morishita J., Tsuboi K., Tonai T., Ueda N. J. Biol. Chem. 2004;279:5298–5305. doi: 10.1074/jbc.M306642200. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto Y., Morishita J., Wang J., Schmid P. C., Krebsbach R. J., Schmid H. H., Ueda N. Biochem. J. 2005;389:241–247. doi: 10.1042/BJ20041790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen S., Dadi H., Shaoul E., Sharfe N., Roifman C. M. Blood. 1999;93:2013–2024. [PubMed] [Google Scholar]
- 14.Hasegawa K., Martin F., Huang G., Tumas D., Diehl L., Chan A. C. Science. 2004;303:685–689. doi: 10.1126/science.1092138. [DOI] [PubMed] [Google Scholar]
- 15.Sheskin T., Hanus L., Slager J., Vogel Z., Mechoulam R. J. Med. Chem. 1997;40:659–667. doi: 10.1021/jm960752x. [DOI] [PubMed] [Google Scholar]
- 16.Kathuria S., Gaetani S., Fegley D., Valino F., Duranti A., Tontini A., Mor M., Tarzia G., La Rana G., Calignano A., et al. Nat. Med. 2003;9:76–81. doi: 10.1038/nm803. [DOI] [PubMed] [Google Scholar]
- 17.Hansen H. S., Moesgaard B., Hansen H. H., Petersen G. Chem. Phys. Lipids. 2000;108:135–150. doi: 10.1016/s0009-3084(00)00192-4. [DOI] [PubMed] [Google Scholar]
- 18.Leung D., Saghatelian A., Simon G. M., Cravatt B. F. Biochemistry. 2006;45:4720–4726. doi: 10.1021/bi060163l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hopfgartner G., Bourgogne E. Mass. Spectr. Rev. 2003;22:195–214. doi: 10.1002/mas.10050. [DOI] [PubMed] [Google Scholar]
- 20.Chien W., Tidow N., Williamson E. A., Shih L. Y., Krug U., Kettenbach A., Fermin A. C., Roifman C. M., Koeffler H. P. J. Biol. Chem. 2003;278:27413–27420. doi: 10.1074/jbc.M304575200. [DOI] [PubMed] [Google Scholar]
- 21.Gregersen P. K. Nat. Genet. 2005;37:1300–1302. doi: 10.1038/ng1205-1300. [DOI] [PubMed] [Google Scholar]
- 22.Lee J. O., Yang H., Georgescu M. M., Di Cristofano A., Maehama T., Shi Y., Dixon J. E., Pandolfi P., Pavletich N. P. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 23.Pryce G., Ahmed Z., Hankey D. J., Jackson S. J., Croxford J. L., Pocock J. M., Ledent C., Petzold A., Thompson A. J., Giovannoni G., et al. Brain. 2003;126:2191–2202. doi: 10.1093/brain/awg224. [DOI] [PubMed] [Google Scholar]
- 24.Li X., Kaminski N. E., Fischer L. J. Int. Immunopharmacol. 2001;1:699–712. doi: 10.1016/s1567-5769(01)00003-0. [DOI] [PubMed] [Google Scholar]
- 25.Jackson S. J., Pryce G., Diemel L. T., Cuzner M. L., Baker D. Neuroscience. 2005;134:261–268. doi: 10.1016/j.neuroscience.2005.02.045. [DOI] [PubMed] [Google Scholar]
- 26.Sugiura T., Kondo S., Sukagawa A., Tonegawa T., Nakane S., Yamashita A., Ishima Y., Waku K. Eur. J. Biochem. 1996;240:53–62. doi: 10.1111/j.1432-1033.1996.0053h.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang L., Liu J., Harvey-White J., Zimmer A., Kunos G. Proc. Natl. Acad. Sci. USA. 2003;100:1393–1398. doi: 10.1073/pnas.0336351100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin S., Khanolkar A. D., Fan P., Goutopoulos A., Qin C., Papahadjis D., Makriyannis A. J. Med. Chem. 1998;41:5353–5361. doi: 10.1021/jm970257g. [DOI] [PubMed] [Google Scholar]
- 29.Osei-Hyiaman D., DePetrillo M., Pacher P., Liu J., Radaeva S., Batkai S., Harvey-White J., Mackie K., Offertaler L., Wang L., Kunos G. J. Clin. Invest. 2005;115:1298–1305. doi: 10.1172/JCI23057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., Gao B., Mirshahi F., Sanyal A. J., Khanolkar A. D., Makriyannis A., Kunos G. Biochem. J. 2000;346:835–840. [PMC free article] [PubMed] [Google Scholar]
- 31.Osei-Hyiaman D., Depetrillo M., Harvey-White J., Bannon A. W., Cravatt B. F., Kuhar M. J., Mackie K., Palkovits M., Kunos G. Neuroendocrinology. 2005;81:273–282. doi: 10.1159/000087925. [DOI] [PubMed] [Google Scholar]