Abstract
Two acidified nitrite-inducible genes of Salmonella enterica serovar Typhimurium were identified with a green fluorescent protein-based promoter-trap screen. The nitrite-inducible promoters were located upstream of loci that we designated nipAB and nipC, which correspond to hcp-hcr (hybrid cluster protein) of Escherichia coli and norA of Alcaligenes eutrophus, respectively. Maximal induction of the promoters by nitrite was dependent on pH. The nipAB promoter was regulated by oxygen in an Fnr-dependent manner. The nipC promoter was also regulated by oxygen but in an Fnr-independent manner. The promoters were upregulated in activated RAW264.7 macrophage-like cells, which produce NO via the inducible nitric oxide synthase (iNOS), and the induction was inhibited by aminoguanidine, an inhibitor of iNOS. Although the nipAB and nipC mutants displayed no defects under a variety of in vitro conditions or in tissue culture infections, they exhibited lower oral 50% lethal doses (LD50s) than did the wild type in C57BL/6J mouse infections. The lower LD50s reflected an unexpected increased ability of small inoculating doses of the mutant bacteria to cause lethal infection 2 to 3 weeks after challenge, compared to a similar challenge dose of wild-type bacteria. We conclude that these genes are regulated by physiological nitrogen oxides and that the absence of these bacterial genes in some way diminishes the ability of mice to clear a low dose infection.
A hallmark of Salmonella enterica serovar Typhimurium (hereafter referred to as serovar Typhimurium) infection of the mouse is the presence of intracellular bacteria, primarily in macrophages, during all systemic phases of the disease (29, 33). Macrophages ordinarily phagocytose particles and bacteria, which are then degraded in phagolysosomes by various means, including acid, cationic peptides, and oxidative radicals (18). In contrast, serovar Typhimurium is able to survive these challenges and exploit the macrophage as a site of intracellular replication and persistence. In order to circumvent or survive this hostile environment, serovar Typhimurium has evolved properties that allow it to avoid and persist in the presence of the innate and adaptive host immune systems.
Reactive nitrogen intermediates (RNI) are also part of the arsenal of antibacterial measures produced by macrophages (13, 18, 35). Macrophages produce the inducible nitric oxide synthase (iNOS), which catalyzes the generation of nitric oxide (NO), upon stimulation with cytokines and/or mitogens. iNOS has been demonstrated to play a role in host defense from infection by a number of microbial agents, including serovar Typhimurium (25, 34, 45). While reactive oxygen intermediates seem to play a bactericidal role early during the infection process, NO displays a bacteriostatic effect at later time points (45), when most of the bacteria are intracellular. Although the localization of iNOS is not clear, recent work suggests that iNOS is present in multiple subcellular locations within macrophages (47). Furthermore, NO is abundant in both the intracellular and extracellular environments due to its membrane permeability (2, 19). As serovar Typhimurium cells do not normally inhibit production of NO (11), it is likely that intracellular bacteria experience high concentrations of NO. However, serovar Typhimurium cells do not display the high sensitivity to NO that Mycobacterium tuberculosis cells do (35), suggesting that serovar Typhimurium could possess mechanisms of resistance to RNI.
Multiple modes of antimicrobial activity have been observed in association with NO exposure. NO possesses a free electron that is reactive with many other intracellular molecules, which can result in the generation of even more destructive RNI such as peroxynitrite (13, 23). These RNI are capable of causing widespread damage to proteins and lipids (13). In addition, RNI can cause damage to DNA by a variety of chemical mechanisms and are thereby mutagenic (3). Moreover, NO also possesses signaling roles for host processes, playing roles in inflammatory responses including vasodilation and cytokine regulation (24, 35). It is not known if the effects of NO are largely due to direct action on the bacteria or if they are mediated by other antimicrobial compounds that are activated through the signaling capacity of NO. It therefore remains unclear which of the multiple physiological activities of NO are the major factors contributing to its antimicrobial activity during infection.
An in vitro model system used to mimic RNI is acidified nitrite (14). Nitrogen oxides, including NO, are generated when nitrite is subjected to acidic conditions. In order to further our understanding of how Salmonella responds to NO, we utilized differential fluorescence induction (42, 43) to identify loci of serovar Typhimurium that respond to RNI generated by acidified nitrite.
MATERIALS AND METHODS
Bacterial cultures and promoter identification.
Strains used in this study are described in Table 1. Bacteria were grown in Luria-Bertani (LB) broth or M9 minimal medium as indicated. Agitated cultures were grown in a shaking floor incubator rotating at 200 to 300 rpm, unagitated cultures were grown standing (without shaking), and anaerobic cultures were grown in 2.5-liter GasPak jars containing Oxoid AnaeroGen packets. Differential fluorescence induction, a technique based on promoter fusions to green fluorescent protein (GFP), was performed as previously described (42, 43). In brief, standing LB cultures (pH 5) of promoter-GFP fusion pools (two pools containing 3,000 to 50,00 clones each) were grown for 4 h, followed by addition of 1 mM NaNO2 for 4 h and fluorescence-activated cell sorter (FACS)-based recovery of fluorescent clones. Three total rounds of sorting were performed, selecting highly GFP-fluorescent bacteria, followed by a sort for nonfluorescent bacteria under noninducing conditions, with a final positive fluorescence sort. Individual clones were identified by screening as described in the next paragraph. The isolated plasmids were transferred to a clean SL1344 background by electroporation.
TABLE 1.
Strains, constructs, and gene nomenclature
| Strain, plasmid, or gene nomenclature | Genotype or alternate name | References or source |
|---|---|---|
| Strains | ||
| SL1344 | xyl hisG rpsL | 37 |
| ANI1 | SL1344/pANI1 | This work |
| ANI2 | SL1344/pANI2 | This work |
| CK1 | SL1344 ΔnipAB::Kan | This work |
| CK2 | SL1344 ΔnipC::Kan | This work |
| CK60 | SL1344 ΔnipC::pCE36 | 10; this work |
| SL1344 fnr | SL1344 fnr2::Tn10 | Charles Miller |
| SL1344 arcA | SL1344 arcA2-1::Tn10d-tet | John Roth |
| Plasmids | ||
| pFPV25 | colE1 mob bla promoterless gfpmut3 | 42 |
| pANI1 | pFPV25 with nipAB promoter | This work |
| pANI2 | pFPV25 with nipC promoter | This work |
| pRS415 | colE1 bla promoterless lacZYA | 36 |
| pCK32 | pRS415 with nipAB promoter | This work |
| Gene nomenclature | ||
| nipA | hcp, ybjW | 1, 5, 44 |
| nipB | hcr, b0872 | 1, 5, 44 |
| nipC | norA, orf1, ytfE | 6, 31 |
Broth inductions for FACS.
Two milliliters of LB broth (pH 5) was inoculated from a single colony and incubated standing for 4 h at 37°C. The culture was split into two 1-ml cultures, and the inducing agent was added to one tube. The cultures were incubated for 4 h at 37°C and analyzed by flow cytometry on a Becton Dickinson FACSCalibur by using CellQuest software with gates set to forward and side scatters characteristic of the bacteria.
LacZ assays.
Overnight cultures were diluted 1:1,000 into 100 μl of LB broth (pH 7) in 96-well plates. After 4 h of growth at 37°C, the bacteria were harvested and resuspended in LB broth with differing pH values. Sodium nitrite was added to 1 mM where appropriate. Bacteria were permeabilized after a 20-min incubation with 1 drop of chloroform plus 1 drop of 1% sodium dodecyl sulfate. Fifty microliters of luminescent lacZ reagent (Gal-Screen; Applied Biosystems) was added to 50 μl of bacterial lysate in a white 96-well plate. Assays were incubated at room temperature for 1 h and were analyzed on a Tropix TR717 microplate luminometer by using WinGlow software. Light units were normalized for bacterial levels by dividing the measured arbitrary light units by the optical density at 630 nm at the time of harvesting.
Intracellular inductions.
RAW264.7 murine macrophage-like cells were maintained in Dulbecco’s modified Eagle minimal essential medium (DMEM) containing 10% fetal bovine serum. Cells were seeded in 24-well dishes at 2 × 106 to 3 × 106 cells/well in 1 ml of medium. Cells were activated with 10 to 100 U of gamma interferon (IFN-γ) (no. 1276 905; Boehringer Mannheim)/ml and 10 to 50 ng of serovar Typhimurium lipopolysaccharide (LPS) (L6143; Sigma)/ml for 24 h. Aminoguanidine (A7009; Sigma) was added at 200 μg/ml where indicated, and nitrite production was monitored by the Griess assay. Supernatants from unstimulated and aminoguanidine-treated activated cells showed background levels of 1 μM nitrite, while cells activated with 100 U of IFN-γ/ml and 50 ng of LPS/ml produced 20 to 30 μM nitrite. Macrophages were infected with standing overnight cultures of bacteria at a multiplicity of infection of 30:1 in the presence of fresh DMEM containing 10% fetal bovine serum with 100 μg of ampicillin/ml (for plasmid maintenance). The media were replaced every hour (maintaining aminoguanidine where appropriate) to limit replication of extracellular bacteria. After 4 h, supernatants were collected and saved for FACS analysis. The cells were washed four times with 1 ml of phosphate-buffered saline and were lysed in 1% Triton X-100 for 15 min, followed by vigorous pipetting to release intracellular bacteria. The extracellular (supernatants) and intracellular bacteria were analyzed for GFP induction by flow cytometry by gating on forward and side scatters.
Strain construction.
All regulatory mutations were moved into the SL1344 background by using P22 transductions. The nipAB operon was PCR amplified and was cloned into pCR-XL TOPO (Invitrogen) with the primers GGCTTTATCCTCAGCCTGCTG and TTATGCGAGAACAAGATCGCC. The nipC gene was PCR amplified and was cloned into the same vector with the primers CGCCAGCCAAAAATTGCCAAC and TAACCGGGCTTACAGCGTAAC. Deletions were created in the two vectors by inverse PCR with the primers AAGGCCTGTCGACAACCTTGCAAAGCCGCAATC and AAGGCCTGTCGACAAAGAGAAGTTCTTCACGCC for nipAB and AAGGCCTGTCGACTTTTCCGCCGCAGCAGTAATC and AAGGCCTGTCGACCACCAAAAATGTAACGCCGCC for nipC. The nipAB and nipC genes were cloned into pCVD442 by XbaI and SacI digest. A kanamycin resistance cassette from pUC4K was cloned into the deleted region at the SalI site for nipAB and nipC. The suicide plasmids were electroporated into SM10λpir and were conjugated to SL1344. Double recombinants were selected on plates containing 30 μg of streptomycin/ml, 50 μg of kanamycin/ml, and 10% sucrose. PCR and Southern blot analyses confirmed the mutations.
The nipAB-lacZ reporter was constructed in pRS415 (36). A DraI digest of the TA-cloned nipAB region (described above) yielded a fragment that included the nipAB promoter and the Kan cassette, which was cloned into the SmaI site of pRS415 to create pCK32.
CK60, a chromosomally integrated nipC-lacZ reporter strain, was constructed by using a PCR-based knockout system (8) combined with an FLP-mediated recombination of a promoterless lacZYA construct (pCE36) into the Flp recombination target scar left in the chromosome (10). Primer sequences for the deletion construct are available on our website (http://falkow.stanford.edu).
Mouse infections.
Seven- to 9-week-old female C57BL/6 mice were used for all mouse experiments. Food was withheld overnight before intragastric infection. For the 50% lethal dose (LD50) experiments, five C57BL/6 mice per dose were inoculated intragastrically with 10-fold serial dilutions. Seven dilutions (103 to 109 CFU) were used in the first experiment, and six dilutions (102 to 107 CFU) were used for the second experiment. Survival was monitored for 30 days. LD50s were calculated by the method of Reed and Muench (32). Calculations were automated in Perl scripts available at http://falkow.stanford.edu or from C. Kim as source code or Windows executables.
RESULTS
Isolation of acidified nitrite-inducible genes.
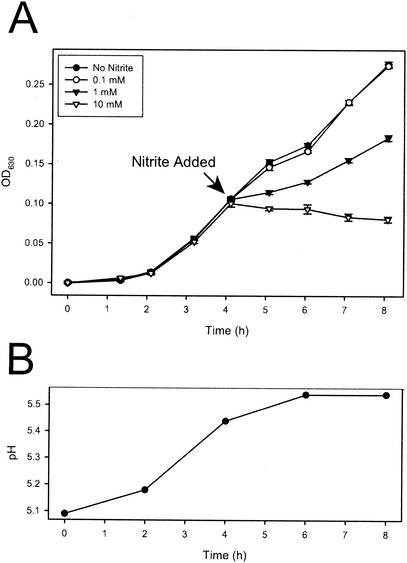
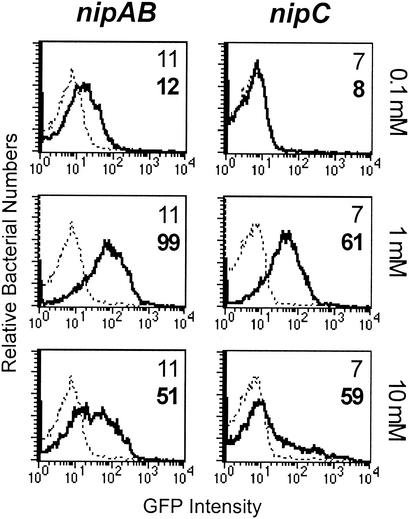
We performed a nonsaturating screen for promoters induced by acidified nitrite, a system for generating NO (14). Previously described promoter-GFP fusion libraries of serovar Typhimurium SL1344 (42) were screened with differential fluorescence induction. Monitoring culture turbidity (Fig. 1A) and the number of CFU (data not shown) showed that 10 mM nitrite caused slight killing of SL1344 cells in LB broth (pH 5), while 1 mM nitrite caused moderate growth inhibition. We chose to use 10 mM nitrite for our inducing conditions. We isolated two distinct clones displaying inducible promoters fused to GFP. The first clone, designated ANI1 (SL1344/pANI1), was identified in a 4-h exposure to 10 mM nitrite in an unagitated culture of LB broth (pH 5). Further analysis of the induction properties of this clone indicated that optimal induction occurred with 1 mM nitrite (Fig. 2, nipAB). Subsequently, we used LB broth (pH 5) with 1 mM nitrite to isolate a second clone, designated ANI2 (SL1344/pANI2), which exhibited induction properties similar to ANI1 (Fig. 2, nipC).
FIG. 1.
Inhibition of Salmonella growth by acidified nitrite. (A) Typical growth curve. One millimolar nitrite significantly slows growth of Salmonella, and 10 mM nitrite is slightly bactericidal. Error bars represent standard error (n = 5). OD630, optical density at 630 nm. (B) Typical pH curve starting in LB broth, pH 5. The unbuffered media change pH over the time of the experiment but not dramatically. Typical pH values ranged from 5.3 to 5.5 at the time of nitrite addition and from 5.4 to 5.6 after 8 h of standing incubation.
FIG. 2.
nipAB and nipC promoter expression is maximally induced by 1 mM nitrite at pH 5. Four-milliliter LB cultures (pH 5) were grown standing for 4 h at 37°C. Each culture was split into four 1-ml cultures, to which 0 (dashed lines), 0.1, 1, or 10 mM nitrite (solid lines) was added. Cultures were incubated standing for another 4 h. FACS analysis was performed to monitor GFP expression from the nipAB promoter or the nipC promoter. Numbers indicate the mean GFP intensity values for the uninduced (normal font) and induced (boldface) conditions.
BLAST analysis of the cloned promoter region in pANI1 revealed homology to a region located upstream of a two-gene operon in serovar Typhimurium LT2 (26) (STM0936-STM0937). We designated these genes nipA and nipB (nitrite-inducible promoter), but a recent publication designated them hcp and hcr, respectively (44). The first gene, hcp (hybrid cluster protein), encodes a redox protein containing one normal and one atypical [4Fe-4S] cluster. The second gene, hcr, encodes an oxidoreductase that is thought to participate in electron transfer to hcp. Interestingly, the genes are regulated by nitrite in Escherichia coli (44). Although the crystal structure of hcp has been solved (1, 44), its substrates and reactions remain unidentified.
The promoter region of pANI2 possessed homology to a region upstream of a hypothetical gene in E. coli, ytfE (89% amino acid identity with serovar Typhimurium, STM4399 in the LT2 sequence [26]). We designated this gene nipC. The nipC gene of serovar Typhimurium possesses homology to dnrN (50% amino acid identity) of Pseudomonas stutzeri and to norA (46% amino acid identity) of Alcaligenes eutrophus H16. The functions of DnrN and NorA are unknown, but they are regulated by the NO-responsive regulators DnrD and NorR, respectively (6, 31, 46).
Nitrite-induced activation of the nipAB and nipC promoters exhibits pH dependence.
A range of nitrite concentrations was tested for optimal upregulation of the GFP-inducing promoters in ANI1 and ANI2. After 4 h of induction in LB broth (pH 5), 1 mM nitrite caused higher levels of GFP induction of both ANI1 and ANI2 than did 0.1 mM or 10 mM nitrite (Fig. 2). Lower levels of induction at pH 5 plus 10 mM nitrite were probably due to toxicity, as confirmed by measurement of culture turbidity, enumeration of CFU, and monitoring of a constitutive rpsM-gfp fusion, which exhibited decreased expression at the high nitrite concentration but constitutive levels under all other conditions (data not shown).
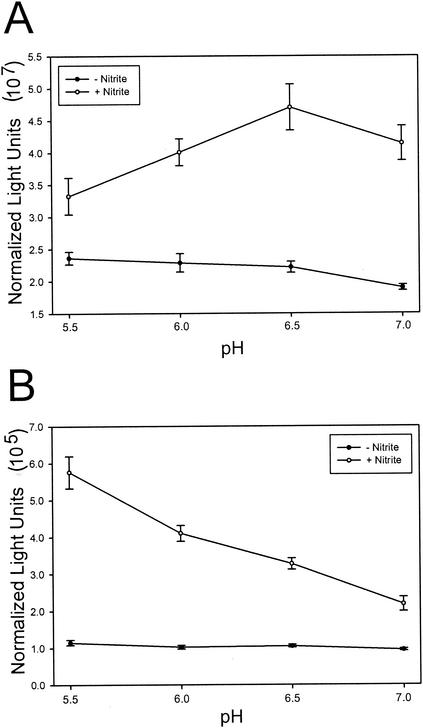
We also wished to assess the effects of pH on induction by nitrite, since different nitrogen oxide species are expected to be formed at different pH values. We observed that the pH of the medium increased during the 4-h incubation (Fig. 1B). Because maturation of GFP is relatively slow (40), it is not possible to assess promoter activity on short time scales. In order to examine the pH dependence of induction on a time scale during which the pH varied minimally, we constructed lacZ fusions to each of these promoters, which allowed us to conduct assays after a 20-min incubation. The nipAB fusion displayed maximal induction near pH 6.5, while nipC displayed maximal induction at pH 5.5 (Fig. 3). Induction was abolished in both reporters at pH 5.0, which we presume to be due to toxicity. The observation that the nipAB and nipC promoters display maximal expression at different pH levels suggests that the nipAB and nipC promoters may respond to different nitrogen oxide species.
FIG. 3.
pH dependence of inductions. LB cultures (pH 5) were grown standing for 4 h. Bacteria were harvested and resuspended in LB broth (pH 5 to 7) with or without 1 mM nitrite. LacZ activity was measured after 20 min for the nipAB (A) and nipC (B) promoters. Standard error is reported for triplicate wells.
nipAB is a member of the Fnr regulon.
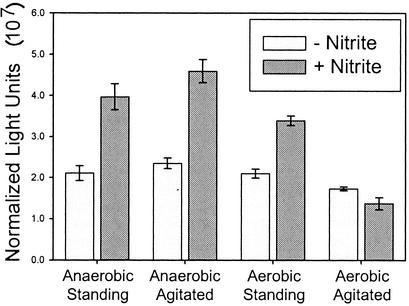
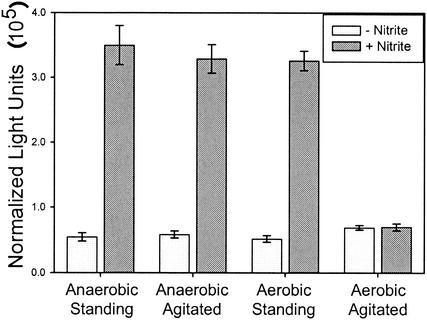
ANI1 exhibits bright green fluorescence by FACS and microscopy (data not shown) upon static (unagitated) incubation with 1 mM nitrite. However, GFP expression is absent in cultures agitated during the induction period (data not shown). There are at least two possible explanations for the agitation-dependent regulation. One possibility is that oxygen tension of the media plays a role in the regulation of the promoters. A second possibility is that optimally inducing concentrations of NO or other gaseous RNI are not achieved due to rapid dissipation during agitation. In order to distinguish between these mechanisms, cultures were subjected to induction under agitated and unagitated growth in aerobic and anaerobic conditions and assayed for LacZ activity. The promoters were induced under low-oxygen conditions, even in the presence of agitation (Fig. 4). These data are in accord with the nipAB promoter being regulated by oxygen tension.
FIG. 4.
nipAB promoter expression is repressed by oxygen. Four-milliliter LB cultures (pH 7) were grown standing for 4 h at 37°C. Bacteria were harvested and resuspended in LB broth (pH 6.5) with or without 1 mM nitrite and were subjected to every permutation of agitated/standing and aerobic/anaerobic. LacZ activity was assayed after 20 min. Standard error is reported for triplicate cultures.
Sequence analysis of the region upstream of the nipA open reading frame in serovar Typhimurium with Mfold, a program designed to detect secondary structure in nucleic acids (49), revealed inverted repeats. The region contained an 8/10 match to the consensus sequence for the Fnr-binding site (39), separated by one nucleotide from another upstream inverted repeat sequence (TTTGGCGGTAGGCATCTGCCGCCAAAATTGCGCTAAATCAA [underline indicates inverted repeat; boldface indicates predicted Fnr-binding site]). This arrangement is reminiscent of the promoters of other members of the Fnr regulon, such as nirB and narG, in which a nitrogen oxide-responsive element acts in conjunction with the oxygen-responsive Fnr (41). A similar arrangement of the region upstream of E. coli hcp was previously noted (44).
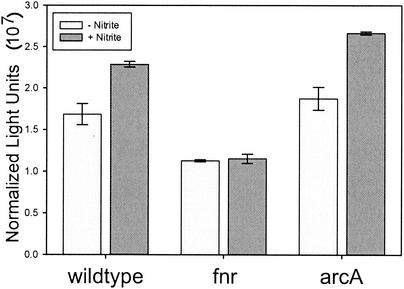
The Fnr regulon, in a complex interplay with the ArcAB regulon, controls expression of many anaerobically expressed genes (22). The nipAB promoter completely failed to induce in the SL1344 fnr background, while ArcA did not appear to have an effect on regulation as measured by LacZ activity (Fig. 5). These results, considered together with regulatory sequence homologies and the agitation experiments, indicate that nipAB is likely to be positively regulated by Fnr.
FIG. 5.
The nipAB promoter requires Fnr for activation by nitrite. Two-milliliter LB cultures (pH 7) were grown agitated for 4 h at 37°C. Bacteria were resuspended in LB broth (pH 6.5) and were split into two 1-ml cultures, to one of which 1 mM nitrite was added. LacZ activity was assayed after 20 min. Standard error is reported for triplicate cultures.
Many members of the Fnr regulon exhibit complex expression patterns involving multiple regulatory proteins. In addition, the inverted repeat upstream of the putative Fnr -binding site suggests the involvement of another regulator. We tested SL1344 mutants in soxS, oxyR, narL, slyA, phoP, ssrB, orgA, ompR, rpoS, and rpoE for regulation of nipAB. None of these backgrounds displayed dramatic effects on regulation of nipAB (data not shown). In addition, we constructed deletion mutants in glnG (STM4005) and ygaA (STM2839) (26), which display the highest similarity in the LT2 genome sequence to the NO-responsive regulator norR of A. eutrophus. These mutations did not affect GFP expression from the nipAB promoter.
nipC expression is affected by oxygen.
Induction of the nipC promoter by nitrite was repressed by oxygen in a pattern similar to that for the nipAB promoter (Fig. 6). Sequence analysis of the promoter region revealed no obvious regulatory sequences, although large regions of secondary structure are predicted by MFold (data not shown). Two major classes of alternative structures are predicted, reminiscent of promoter regions that are regulated by transcriptional attenuation mechanisms. However, none of the mutant backgrounds tested for regulation of nipAB displayed any significant effects on expression of nipC, including the norR homologues. We conclude that, although the nipC promoter is regulated by oxygen tension, it is not dependent on the regulatory pathways that control the nipAB promoter.
FIG. 6.
nipC promoter expression is repressed by oxygen. LacZ assays were conducted identically to those described in the Fig. 4 legend, except that the bacteria were resuspended in LB broth, pH 5.5.
nipAB and nipC are induced by NO-producing macrophages.
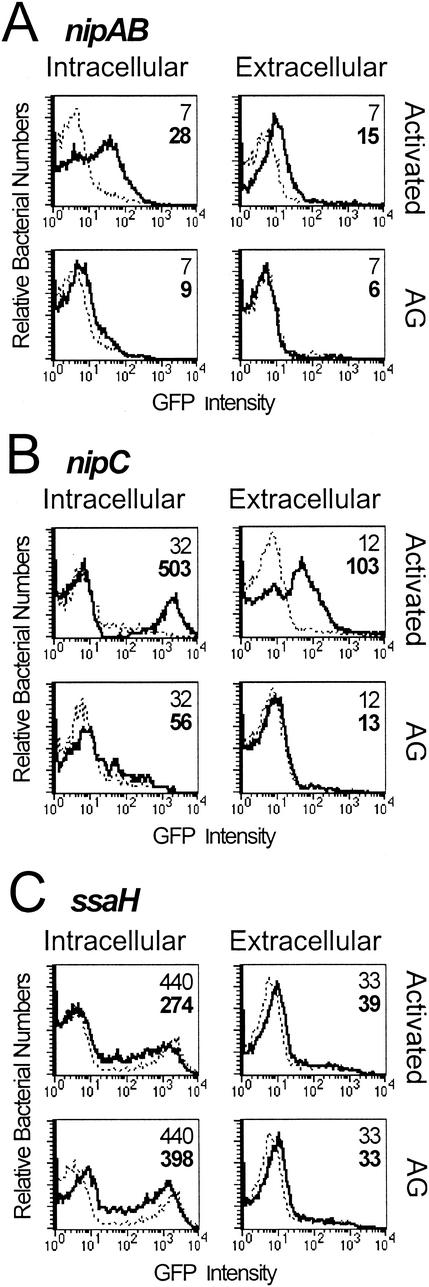
Macrophages can be stimulated by mitogens and cytokines to produce large quantities of NO (23). RAW264.7 mouse macrophage-like cells were activated for 24 h with 50 ng of S. enterica serovar Typhimurium LPS/ml plus 100 U of IFN-γ/ml, followed by infection with SL1344 carrying nipAB-gfp and nipC-gfp. The Griess assay (7) was used to monitor nitrite concentration in culture supernatants, which is used to infer levels of NO production. Intracellular bacteria carrying nipAB-gfp and nipC-gfp harvested at 4 h displayed induction of GFP in activated macrophages but did not express GFP in unstimulated macrophages (Fig. 7A and B). Aminoguanidine, an inhibitor of iNOS (38), reduced nitrite accumulation by greater than 96% when macrophages were treated during activation (data not shown). Treatment of activated macrophages with aminoguanidine also prevented upregulation of the nipAB and nipC promoters, suggesting that a product of iNOS is responsible for the induction. Aminoguanidine had no effect on the induction of nipAB and nipC by nitrite in the absence of cells, further indicating that the inhibition of promoter induction is a result of iNOS inhibition (data not shown). In contrast, an SPI-2 reporter (ssaH-gfp) that is specifically expressed in the intracellular environment (4, 43) is induced under all intracellular conditions regardless of NO production (Fig. 7C). This indicates that nipAB and nipC are specifically upregulated in macrophages producing RNI. Interestingly, nipAB and nipC also appear to display extracellular induction, although to lower levels than intracellular bacteria (Fig. 7A and B). As expected, the SPI-2 reporter displays no extracellular induction (Fig. 7C). Conditioned media from activated macrophages did not cause induction of the promoters in the absence of macrophages, and the promoters displayed the same responsiveness to nitrite in DMEM containing FBS that they did to nitrite in LB broth (data not shown), suggesting that proximity to the host cells may be important for GFP induction in extracellular bacteria. This observation is in accord with the membrane-diffusible nature of NO and the extracellular presence of NO near activated macrophages (2, 19).
FIG. 7.
nipAB and nipC are induced inside activated macrophages. RAW264.7 mouse macrophage-like cells were activated with LPS and IFN-γ prior to a 4-h infection with ANI1 or ANI2 (multiplicity of infection, 30:1). For aminoguanidine (AG) treatment, macrophages received treatment both during activation and infection. Inductions in activated or aminoguanidine-treated macrophages (solid lines) are compared with values for unactivated macrophages (dashed lines) in each plot. Extracellular bacteria were collected from the supernatant, while intracellular bacteria were recovered by Triton X-100 lysis of host cells. Inductions are shown for nipAB-gfp (A), nipC-gfp (B), and ssaH-gfp (SPI-2) (C). Numbers indicate the mean GFP intensities for the unstimulated (normal font) and activated and/or aminoguanidine (boldface) infections. The number of CFU did not change significantly during the course of the 4-h infection.
Mutants in nipAB and nipC do not display growth defects under inducing conditions.
In order to begin to assess the functions of nipAB and nipC, we constructed mutations in these genes in an SL1344 background. CK1 contains a deletion spanning the nipAB operon marked with a kanamycin resistance marker. CK2 contains a deletion spanning nipC and also contains a kanamycin cassette insertion. These mutants did not display any discernible difference from wild-type SL1344 in a variety of assays where we tested for growth or survival with the following variables: media (LB and M9), pHs (5 and 7), electron acceptors (oxygen, nitrite, and nitrate), and carbon sources (glucose, glycerol, and lactate) (data not shown). This was in contrast to an fnr mutant, which displayed growth defects under anaerobic growth with nonfermentable carbon sources (data not shown). As NO has been reported to have mutagenic activity (3), we also tested CK1 and CK2 for increased susceptibility to mutagenesis by acidified nitrite. While we did observe that acidified nitrite causes significant mutagenesis in SL1344, CK1, and CK2 harboring pKM101 (a plasmid used in the Ames tester strains to increase sensitivity of the carrying strain to mutagenesis [27]), we were unable to demonstrate in vitro that CK1/pKM101 or CK2/pKM101 displays any more sensitivity to mutagenesis by nitrite than SL1344/pKM101 (data not shown).
In addition, we tested CK1 and CK2 for survival and replication in RAW264.7 cells that were unstimulated, activated with 10 ng of IFN-γ/ml, activated with 100 ng of IFN-γ/ml, or activated with 100 ng of IFN-γ/ml and 50 ng of LPS/ml. While the IFN-γ + LPS-stimulated cells produced NO as measured by NO2− levels in the cell supernatants, neither CK1 nor CK2 was compromised for intracellular survival or replication under any of the tested conditions when compared to the wild-type strain SL1344 as assessed by CFU counts over a 24-h period (data not shown). In addition, CK1- and CK2-infected RAW264.7 cells produced NO at levels similar to SL1344-infected cells (data not shown).
Low doses of mutants in nipAB and nipC cause late death in C57BL/6 mice.
CK1 and CK2 were tested for virulence in mice by oral challenge. The ability of these mutants to colonize various mouse tissues and organs was assessed in mixed infections with wild-type SL1344 upon oral challenge with 107 to 109 CFU of each strain. Cecum, Peyer's patches, mesenteric lymph nodes, and spleen did not contain significant differences in bacterial load of the wild-type and mutant strains over a 5-day infection (matched t tests, 95% confidence level) (data not shown).
Although we did not observe any differences in the competition experiments over a short-term high-dose infection, we determined the LD50s for the strains. Five C57BL/6J mice per dose at seven different doses were inoculated intragastrically. Reed-Muench analysis (32) on day 7 yielded LD50s that were similar for SL1344, CK1, and CK2 (Table 2). LD50s on day 14 were also largely similar between mutants and wild type. On day 30, however, the LD50s for the mutants were substantially lower than for the wild type. These unexpected results suggested that, compared to the wild type, smaller doses of CK1 and CK2 are able to cause lethal infection in mice.
TABLE 2.
Reed-Muench LD50s
| Background or mutant | Results
|
|||||
|---|---|---|---|---|---|---|
| 1st expt on:
|
2nd expt on:
|
|||||
| Day 7 | Day 14 | Day 30 | Day 7 | Day 14 | Day 30 | |
| SL1344 | 4.49 × 108 | 8.96 × 105 | 5.65 × 105 | NCa | 2.81 × 106 | 6.66 × 105 |
| CK1 | 2.27 × 108 | 4.60 × 105 | 6.73 × 104 | NCa | 2.67 × 106 | 1.19 × 105 |
| CK2 | 1.73 × 108 | 1.73 × 106 | 3.51 × 103 | 1.42 × 107 | 8.61 × 105 | 8.96 × 103 |
NC, not calculable due to insufficient deaths.
A second repetition of the LD50 experiment was performed but with experimenters being blind to the strain identities. As in the first LD50 progression, the initial LD50 progression was unrevealing (Table 2). However, on day 19, we were able to correctly predict the strain identities (data not shown). On day 30, the LD50s exhibited the same pattern as that seen in the first LD50 experiment. The Pearson correlation coefficient between the day 30 LD50s for the two experimental repetitions is 0.91.
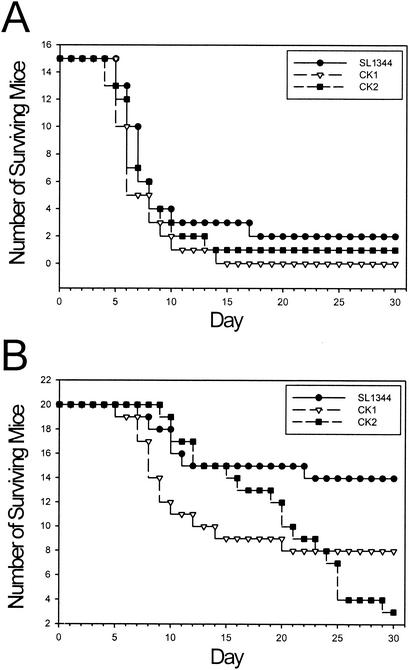
Closer analysis of the survival progression of the mice in the first LD50 experiment revealed that the observed differences were related to dose. We grouped the doses into high (107 to 109 CFU) and low (103 to 106 CFU) dose categories and tallied survival of the mice over time. All strains killed mice at similar rates when inoculated at high doses (Fig. 8A). In contrast, infecting with small inoculating doses of bacteria revealed strain-related differences during the course of the infection: while wild-type-infected mice generally survive low-dose infections, the mutant-infected mice exhibited an increased tendency to succumb to the infection at 2 or 3 weeks postinoculation (Fig. 8B). A similar pattern was observed for both of the LD50 experiments. We conclude that, while infections with small inoculum doses of SL1344 are normally cleared by the mice, identical doses of CK1 and CK2 cause late death by some unknown mechanism.
FIG. 8.
Survival of C57BL/6J mice infected with serovar Typhimurium. Raw survival numbers for the first LD50 experiment were classified into two groups representing high (107 to 109 CFU) (A) and low (103 to 106 CFU) (B) doses. The surviving number of mice represents the total tally from all doses in the high- and low-dose classes. The same analysis on the second LD50 experiment yielded similar results.
DISCUSSION
While NO is required for the control of serovar Typhimurium infection (25, 34, 45), little is known regarding its mechanism of action. Due to the wide range of biological activities exhibited by NO, it remains unclear if NO is acting directly on the bacteria as an antimicrobial, if it is acting in a signaling capacity in other host processes, or if a combination of both is acting. In order to better to understand the effects of NO on serovar Typhimurium, we identified two acidified nitrite-inducible promoters using a promoter-GFP fusion screen. These promoters regulate loci that we designated nipAB and nipC.
The first promoter identified in our screen regulated expression of the nipAB operon. These genes are homologous to the hybrid cluster protein redox couple, Hcp and Hcr. It is believed that Hcr transfers electrons from an unknown source to Hcp, which subsequently transfers the electrons to a substrate. The electron-accepting substrate of Hcp is unknown, although evidence from the crystal structure suggests that it is a diatomic molecule or smaller (1). Furthermore, the substrate is believed to be a gas, given the presence of hydrophobic solvent channels in the structure (5). Taken together with the observation that the operon is regulated by nitrite and nitrate under anaerobic conditions in E. coli and Morganella morganii (44), the data suggest that NO is a good candidate for the substrate of Hcp. However, assuming that the promoter is induced by the substrate of the gene products of this operon, our observation that maximal induction occurs around pH 6.5 in Salmonella suggests that the substrate may be a different nitrogen oxide, since more NO would be expected under more acidic conditions. A recent report that Hcp displays hydroxylamine reductase activity suggests that this may be the case, although the in vivo function of Hcp has not yet been demonstrated (48).
Our results also provide evidence that nipAB expression is regulated by Fnr, as induction is completely abolished in an fnr mutant background. A second repeat motif is located directly upstream of the putative Fnr-binding site. This site is likely to be the binding site for the regulatory protein which responds to the nitrogen oxide inducer, as a similar arrangement is observed in the promoter regions of other members of the Fnr regulon, such as nirB (41). Interestingly, ArcA was recently reported to play a role in acidified nitrite resistance in serovar Enteritidis (20). However, the ArcA-binding site consensus (WGTTAATTAW [21]) is quite disparate in sequence from the second inverted repeat located upstream of nipAB, and our induction studies utilizing the lacZ reporter indicate that ArcA does not regulate nipAB. Nonetheless, the data strongly suggest that the nipAB operon is involved in anaerobic metabolism of a nitrogen oxide.
The second isolated promoter was located upstream of nipC, the function of which is even less well understood. A close homologue of nipC (50% amino acid identity), dnrN, is found in the denitrifying bacterium P. stutzeri (46). During the process of denitrification, nitrate is reduced to ammonia through a succession of electron transfers, with NO being an intermediary product. The reduction of NO to N2O is regulated by DnrD, a member of the FNR-CRP family of transcriptional regulators. DnrN is the first gene of the operon containing DnrD, and this operon is regulated by NO. However, the function of DnrN remains unknown. Based on the expectation that stronger acid generates more NO, our observation that the nipC promoter displayed a correlation between induction levels and acidity is consistent with the possibility that expression of nipC is controlled by NO. As was the case for nipAB, oxygen represses expression of nipC. However, we were unable to identify any regulators of nipC.
Another close homologue of nipC (46% amino acid identity) is the norA gene of A. eutrophus, which is also a denitrifying bacterium. The norA open reading frame is located just upstream of an NO reductase encoded by norB (6). The promoter that drives expression of the NO reductase is located upstream of norA, implying that the genes are coregulated. Expression of this operon is dependent on a regulatory protein designated NorR (31). Interestingly, while E. coli K-12 and serovar Typhimurium possess norA homologues, neither species contains an obvious norB or norR homologue. Nevertheless, the fact that the nipC promoter has maintained regulation by a variety of NO donors (see below) even in the absence of an obvious reductase or response regulator suggests a role in NO metabolism. The presence of this gene in Salmonella is especially interesting in light of the fact that enterobacteria do not perform denitrification (15).
We constructed mutations in nipAB and nipC and tested the mutants for defects in growth under various nonfermentative and stress conditions. We did not detect any differences in the mutants compared to wild-type serovar Typhimurium. van den Berg et al. also reported that they could not identify a growth phenotype in their E. coli hcp-hcr mutants (44). It is possible that the mutants may exhibit subtle differences from the wild type or that the proper conditions have not yet been tested. In any case, it is interesting that these genes are dispensable under inducing conditions. Further work is needed to clarify the role of these genes in nitrogen oxide metabolism.
The physiological relevance of acidified nitrite has previously been questioned (14), so we consequently tested our fusions under more physiologically relevant conditions. In addition to being induced by a variety of NO donors (S-nitrosoglutathione, S-nitroso-N-acetylpenicillamine, and Spermine-NO adduct [data not shown]), the promoters are induced in activated, NO-producing macrophages in an iNOS inhibitor-sensitive manner. Although it is not possible to dissect the exact nature of the inducing agent due to the rapid formation of a complex equilibrium of nitrogen oxides both in acidified nitrite and in macrophages, it is clear that nipAB and nipC respond to nitrogen oxides in both environments.
Interestingly, bacteria that are extracellular to activated macrophages also display nipAB and nipC promoter induction, though to a lesser degree than intracellular bacteria are. Supernatants from activated macrophages did not cause induction of the promoters, nor did DMEM that contained nitrite levels comparable to the levels measured in the supernatants. This indicates that (i) the bacteria experience high local concentrations of the inducing agent within a certain distance from an NO-producing cell, (ii) the bacteria are not responding to nitrite but to a different inducing agent, or (iii) both of the above apply. NO, which can diffuse across membranes, is detected both in the intracellular and extracellular environments with high concentrations close to the cell (2, 19). While this does not predict the precise nature of the inducing agent, since other RNI would presumably be generated in a gradient proportional to the NO gradient, these data provide an explanation for our observation that extracellular bacteria also appear to detect a host-derived nitrogen oxide.
Unexpectedly, the mutant strains CK1 and CK2 both exhibited considerably lower LD50s than the wild-type parent strain at later time points (day 30 versus days 7 and 14). These results prompted us to perform a second repetition in which the experimenters did not know the identity of the strains, which yielded results very similar to those from the first trial. The apparent decrease in LD50s could be attributed to an increased ability of low doses of the mutant bacteria to cause lethal infections in mice. The lethality caused by low-dose infections was primarily observed at later time points (2 to 4 weeks), during the time when adaptive immune responses are normally able to control infection by wild-type serovar Typhimurium. There are therefore two facets to the phenotype that we observed: first, that low doses of our mutants were able to kill mice while similar doses of wild-type bacteria do not, and second, that the observed death occurred at relatively late time points.
Exacerbation of disease as a result of deletion of a gene is uncommon but not unprecedented. One recent publication describes grvA, which is present on the Gifsy-2 phage of serovar Typhimurium (16). Both overexpression and deletion of this locus caused increased replication in BALB/c mice as measured in competition with wild-type serovar Typhimurium. Also of particular interest are the GAM mutants generated in a screen for mutants with increased growth yield in tissue culture macrophage-like cells (J774.1) (11). One of the mutants, GAM4, outcompeted the wild-type parent strain in mice. Furthermore, five of the isolated mutants were able to inhibit the production of host-derived NO. However, CK1- and CK2-infected macrophages did not display any reduction in NO production compared to wild type-infected cells. Additional examples of mutants that appear to have increased fitness in host organisms are described by Ho and Slauch (16).
Most of the effects of NO in wild-type serovar Typhimurium-infected mice manifest themselves later in infection (2 to 3 weeks postinoculation [25]). In fact, the kinetics of survival of iNOS knockout mice are strikingly similar to those of the survival of our C57BL/6J mice infected with CK1 and CK2 (Fig. 1 in reference 25 compared to Fig. 8B here). Moreover, DBA/2 mice, which are deficient in mounting adaptive immune responses to serovar Typhimurium, initially control rampant replication of the bacteria by means of the NrampR locus but eventually succumb to infection with kinetics that are similar to those of the iNOS knockouts (17, 28, 30). Interleukin 4 knockouts also exhibit delayed death kinetics compared to wild-type mice, although overall survival of the knockouts is similar to that of wild-type mice (12). Given the late kinetics of death observed in the mice infected with our mutants and in the iNOS knockout mice, it seems reasonable to hypothesize that NO may play a role in the development of adaptive immune responses during serovar Typhimurium infection of the mouse. Perhaps CK1 and CK2 fail to metabolize NO to the same degree as does the wild type, leading to excessive concentrations of NO and subsequent host damage and/or immunosuppression (9). Alternatively, we speculate that the mutant bacteria may somehow be able to escape detection by adaptive immune responses and are able to progress to full-blown systemic disease at late time points because of this early avoidance.
Regardless of the mechanism that underlies these results, it is clear that Salmonella responds to both chemically generated NO and host-derived NO in a complex manner. Our data suggest that wild-type serovar Typhimurium may be responding to NO in a manner that either modulates virulence traits or host responses.
Acknowledgments
We thank Michael Shiloh and Carl Nathan for initiating our interest in bacterial responses to NO and for helpful ideas and discussions. Ferric Fang, Hotcherl Jeong and Renée Dawson (John Roth lab) and Tina Knox (Charles Miller lab) and Francois Norel kindly provided the regulatory mutant strains.
C. C. Kim is supported by a Howard Hughes Medical Institute Predoctoral Fellowship and a Stanford Graduate Fellowship. This work was funded by NIH grant AI26195.
Editor: V. J. DiRita
REFERENCES
- 1.Arendsen, A. F., J. Hadden, G. Card, A. S. McAlpine, S. Bailey, V. Zaitsev, E. H. M. Duke, P. F. Lindley, M. Kroeckel, A. X. Trautwein, M. C. Feiters, J. M. Charnock, C. D. Garner, S. J. Marritt, A. J. Thomson, I. M. Kooter, M. K. Johnson, W. A. M. van den Berg, W. M. A. M. van Dongen, and W. R. Hagen. 1998. The “prismane” protein resolved: X-ray structure at 1.7 angstroms and multiple spectroscopy of two novel 4Fe clusters. JBIC 3:81-95. [Google Scholar]
- 2.Barker, S. L., H. A. Clark, S. F. Swallen, R. Kopelman, A. W. Tsang, and J. A. Swanson. 1999. Ratiometric and fluorescence-lifetime-based biosensors incorporating cytochrome c′ and the detection of extra- and intracellular macrophage nitric oxide. Anal. Chem. 71:1767-1772. [DOI] [PubMed] [Google Scholar]
- 3.Christen, S., P. Gee, and B. N. Ames. 1996. Mutagenicity of nitric oxide in base pair-specific Salmonella tester strains: TA7000 series. Methods Enzymol. 269:267-278. [DOI] [PubMed] [Google Scholar]
- 4.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 5.Cooper, S. J., C. D. Garner, W. R. Hagen, P. F. Lindley, and S. Bailey. 2000. Hybrid-cluster protein (HCP) from Desulfovibrio vulgaris (Hildenborough) at 1.6 angstrom resolution. Biochemistry 39:15044-15054. [DOI] [PubMed] [Google Scholar]
- 6.Cramm, R., R. A. Siddiqui, and B. Friedrich. 1997. Two isofunctional nitric oxide reductases in Alcaligenes eutrophus H16. J. Bacteriol. 179:6769-6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniels, L., R. S. Hanson, and J. A. Phillips. 1994. Chemical analysis, p. 512-554. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. ASM Press, Washington, D.C.
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenstein, T. K. 2001. Implications of Salmonella-induced nitric oxide (NO) for host defense and vaccines: NO, an antimicrobial, antitumor, immunosuppressive and immunoregulatory molecule. Microbes Infect. 3:1223-1231. [DOI] [PubMed] [Google Scholar]
- 10.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 11.Eriksson, S., J. Bjorkman, S. Borg, A. Syk, S. Pettersson, D. I. Andersson, and M. Rhen. 2000. Salmonella typhimurium mutants that downregulate phagocyte nitric oxide production. Cell. Microbiol. 2:239-250. [DOI] [PubMed] [Google Scholar]
- 12.Everest, P., J. Allen, A. Papakonstantinopoulou, P. Mastroeni, M. Roberts, and G. Dougan. 1997. Salmonella typhimurium infections in mice deficient in interleukin-4 production: role of IL-4 in infection-associated pathology. J. Immunol. 159:1820-1827. [PubMed] [Google Scholar]
- 13.Fang, F. C. 1997. Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J. Clin. Investig. 99:2818-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feelisch, M., and J. S. Stamler. 1996. Donors of nitrogen oxides, p. 71-115. In M. Feelisch and J. S. Stamler (ed.), Methods in nitric oxide research. John Wiley & Sons, New York, N.Y.
- 15.Gennis, R. B., and V. Stewart. 1996. Respiration, p. 217-261. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 16.Ho, T. D., and J. M. Slauch. 2001. Characterization of grvA, an antivirulence gene on the Gifsy-2 phage in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:611-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hormaeche, C. E. 1979. Genetics of natural resistance to salmonellae in mice. Immunology 37:319-327. [PMC free article] [PubMed] [Google Scholar]
- 18.Langermans, J. A., W. L. Hazenbos, and R. van Furth. 1994. Antimicrobial functions of mononuclear phagocytes. J. Immunol. Methods 174:185-194. [DOI] [PubMed] [Google Scholar]
- 19.Leone, A. M., V. W. Furst, N. A. Foxwell, S. Cellek, and S. Moncada. 1996. Visualisation of nitric oxide generated by activated murine macrophages. Biochem. Biophys. Res. Commun. 221:37-41. [DOI] [PubMed] [Google Scholar]
- 20.Lu, S., P. B. Killoran, F. C. Fang, and L. W. Riley. 2002. The global regulator ArcA controls resistance to reactive nitrogen and oxygen intermediates in Salmonella enterica serovar Enteritidis. Infect. Immun. 70:451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch, A. S., and E. C. Lin. 1996. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J. Bacteriol. 178:6238-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynch, A. S., and E. C. C. Lin. 1996. Responses to molecular oxygen, p. 1526-1538. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 23.MacMicking, J., Q. W. Xie, and C. Nathan. 1997. Nitric oxide and macrophage function. Annu. Rev. Immunol. 15:323-350. [DOI] [PubMed] [Google Scholar]
- 24.Mashimo, H., and R. K. Goyal. 1999. Lessons from genetically engineered animal models. IV. Nitric oxide synthase gene knockout mice. Am. J. Physiol. 277:G745-G750. [DOI] [PubMed] [Google Scholar]
- 25.Mastroeni, P., A. Vazquez-Torres, F. C. Fang, Y. Xu, S. Khan, C. E. Hormaeche, and G. Dougan. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J. Exp. Med. 192:237-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 27.Mortelmans, K. E., and B. A. Stocker. 1979. Segregation of the mutator property of plasmid R46 from its ultraviolet-protecting property. Mol. Gen. Genet. 167:317-327. [DOI] [PubMed] [Google Scholar]
- 28.Nickol, A. D., and P. F. Bonventre. 1981. Acquired immunity against mouse typhoid: genetic restriction and comparative efficacy of ribosomal and conventional vaccines. J. Med. Microbiol. 14:419-433. [DOI] [PubMed] [Google Scholar]
- 29.Nnalue, N. A., A. Shnyra, K. Hultenby, and A. A. Lindberg. 1992. Salmonella choleraesuis and Salmonella typhimurium associated with liver cells after intravenous inoculation of rats are localized mainly in Kupffer cells and multiply intracellularly. Infect. Immun. 60:2758-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Brien, A. D., B. A. Taylor, and D. L. Rosenstreich. 1984. Genetic control of natural resistance to Salmonella typhimurium in mice during the late phase of infection. J. Immunol. 133:3313-3318. [PubMed] [Google Scholar]
- 31.Pohlmann, A., R. Cramm, K. Schmelz, and B. Friedrich. 2000. A novel NO-responding regulator controls the reduction of nitric oxide in Ralstonia eutropha. Mol. Microbiol. 38:626-638. [DOI] [PubMed] [Google Scholar]
- 32.Reed, L. J., and H. Muench. 1938. A simple method of Estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 33.Richter-Dahlfors, A., A. M. J. Buchan, and B. B. Finlay. 1997. Murine salmonellosis studied by confocal microscopy: Salmonella typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J. Exp. Med. 186:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiloh, M. U., J. D. MacMicking, S. Nicholson, J. E. Brause, S. Potter, M. Marino, F. Fang, M. Dinauer, and C. Nathan. 1999. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity 10:29-38. [DOI] [PubMed] [Google Scholar]
- 35.Shiloh, M. U., and C. F. Nathan. 2000. Reactive nitrogen intermediates and the pathogenesis of Salmonella and mycobacteria. Curr. Opin. Microbiol. 3:35-42. [DOI] [PubMed] [Google Scholar]
- 36.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 37.Smith, B. P., M. Reinaguerra, S. K. Hoiseth, B. A. D. Stocker, F. Habasha, E. Johnson, and F. Merritt. 1984. Aromatic-dependent Salmonella-typhimurium as modified live vaccines for calves. Am. J. Vet. Res. 45:59-66. [PubMed] [Google Scholar]
- 38.Southan, G. J., and C. Szabo. 1996. Selective pharmacological inhibition of distinct nitric oxide synthase isoforms. Biochem. Pharmacol. 51:383-394. [DOI] [PubMed] [Google Scholar]
- 39.Spiro, S., and J. R. Guest. 1990. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol. Rev. 6:399-428. [DOI] [PubMed] [Google Scholar]
- 40.Tsien, R. Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509-544. [DOI] [PubMed] [Google Scholar]
- 41.Tyson, K. L., A. I. Bell, J. A. Cole, and S. J. Busby. 1993. Definition of nitrite and nitrate response elements at the anaerobically inducible Escherichia coli nirB promoter: interactions between FNR and NarL. Mol. Microbiol. 7:151-157. [DOI] [PubMed] [Google Scholar]
- 42.Valdivia, R. H., and S. Falkow. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22:367-378. [DOI] [PubMed] [Google Scholar]
- 43.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bac-terial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 44.van den Berg, W. A., W. R. Hagen, and W. M. van Dongen. 2000. The hybrid-cluster protein (′prismane protein') from Escherichia coli. Characterization of the hybrid-cluster protein, redox properties of the [2Fe-2S] and [4Fe-2S-2O] clusters and identification of an associated NADH oxidoreductase containing FAD and [2Fe-2S]. Eur. J. Biochem. 267:666-676. [DOI] [PubMed] [Google Scholar]
- 45.Vazquez-Torres, A., J. Jones-Carson, P. Mastroeni, H. Ischiropoulos, and F. C. Fang. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J. Exp. Med. 192:227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vollack, K. U., and W. G. Zumft. 2001. Nitric oxide signaling and transcriptional control of denitrification genes in Pseudomonas stutzeri. J. Bacteriol. 183:2516-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webb, J. L., M. W. Harvey, D. W. Holden, and T. J. Evans. 2001. Macrophage nitric oxide synthase associates with cortical actin but is not recruited to phagosomes. Infect. Immun. 69:6391-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolfe, M. T., J. Heo, J. S. Garavelli, and P. W. Ludden. 2002. Hydroxylamine reductase activity of the hybrid cluster protein from Escherichia coli. J. Bacteriol. 184:5898-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244:48-52. [DOI] [PubMed] [Google Scholar]