Abstract
The pathogenicity of Shiga-like toxin (stx)-producing Escherichia coli (STEC), notably serotype O157, the causative agent of hemorrhagic colitis, hemolytic-uremic syndrome, and thrombotic thrombocytopenic purpura, is based partly on the presence of genes (stx1 and/or stx2) that are known to be carried on temperate lambdoid bacteriophages. Stx phages were isolated from different STEC strains and found to have genome sizes in the range of 48 to 62 kb and to carry either stx1 or stx2 genes. Restriction fragment length polymorphism patterns and sodium dodecyl sulfate-polyacrylamide gel electrophoresis protein profiles were relatively uninformative, but the phages could be differentiated according to their immunity profiles. Furthermore, these were sufficiently sensitive to enable the identification and differentiation of two different phages, both carrying the genes for Stx2 and originating from the same STEC host strain. The immunity profiles of the different Stx phages did not conform to the model established for bacteriophage lambda, in that the pattern of individual Stx phage infection of various lysogens was neither expected nor predicted. Unexpected differences were also observed among Stx phages in their relative lytic productivity within a single host. Two antibiotic resistance markers were used to tag a recombinant phage in which the stx genes were inactivated, enabling the first reported observation of the simultaneous infection of a single host with two genetically identical Stx phages. The data demonstrate that, although Stx phages are members of the lambdoid family, their replication and infection control strategies are not necessarily identical to the archetypical bacteriophage λ, and this could be responsible for the widespread occurrence of stx genes across a diverse range of E. coli serotypes.
Strains of Shiga-like toxin (Stx)-producing Escherichia coli (STEC), especially serotype O157, are increasingly implicated as causative agents of human disease, including hemorrhagic colitis, hemolytic-uremic syndrome, and thrombotic thrombocytopenic purpura (21). There are many virulence factors that contribute to the pathogenic personalities of STEC strains, including the locus of enterocyte effacement (17), which provides genes for promoting both attachment and effacement lesions and a type III secretion system (10), two different hemolysins (29, 35), attachment fimbriae (11), a heat-stable enterotoxin (28), an iron transport system (14, 19), and Stx (22). Genes encoding Stx are most typically associated with Shigella dystenteriae and E. coli O157:H7, but they have been found in E. coli strains encompassing >100 serotypes, as well as other species of gram-negative bacteria, including Citrobacter freundii (30, 37), Vibrio cholerae (20), Enterobacter cloacae (23), and Aeromonas spp. (7).
There are two forms of Stx: Stx1 is almost identical to the toxin from Shigella dysenteriae type 1; Stx2 shares only 50 to 60% amino acid homology with Stx1, and there is a greater degree of antigenic heterogeneity among members of this toxin class (18). Although both toxins have the same mode of action, their effects in vitro and in vivo differ considerably. Stx2 production alone is reported to be associated with more serious disease than strains that produce either Stx1 or a combination of Stx1 and Stx2 (12). Stx1 binds more efficiently to the mammalian globotriaosylceramide receptor and is more potent against Vero cells (36). Stx2, when injected intravenously, is 400 times more lethal for mice (36) and 1,000 times more active on human renal microvascular endothelial cells (16), the putative target for the development of hemolytic-uremic syndrome (15). These observations should, however, be interpreted with caution, since there is also a report that Stx1 is as potent an inhibitor of protein synthesis as Stx2 when challenged against highly purified human glomerular microvascular endothelial cells (39).
Both Stx1 and Stx2 have been shown to be encoded on bacteriophages (34, 38), and such phages have been isolated from a variety of E. coli strains of human, bovine, and porcine origin. The phages appear to belong to the lambdoid family of bacteriophages (27). E. coli strains that produce both Stx1 and Stx2 have been shown to carry two separate phages, each of which encodes only one of the toxins although possessing similarities in gross morphology and genome size and a high degree of DNA homology (27). The potential phage-mediated transfer of stx genes between E. coli strains and other bacterial species is cause for concern. Stx expression has been described in species representing several genera of gram-negative bacteria, not all of which are members of the Enterobacteriaceae. Thus, the presence of stx genes in combination with other virulence factors could lead to the emergence of new pathogens. Phages involved in stx dissemination must be relatively nonspecific, belong to a heterogeneous group, and/or interact with a highly conserved ligand or receptor to account for this etiology.
Recent studies characterizing Stx phages have been limited because of the assumption that immunity to subsequent infection was conferred to a lysogen by related or closely related phages (27, 33, 41). The host range of the recombinant Stx phage described in the present study has been reported (9). Here we characterize the group of Stx phages from which this was isolated, with emphasis on the investigation of Stx phage immunity profiles because of the observance of multiple phage infection events in a single host cell. The bacteriophage λ immunity mechanism is well studied, and multiple infections of genetically identical phage are obviated (26). An important objective of the present study reported here was to determine whether Stx phage deviated from this immunity model to account for the aetiology of multiple Stx phage and stx genes within a single host cell.
MATERIALS AND METHODS
Bacterial strains and phage isolates.
Thirty-one wild-type STEC strains were obtained from the Central Public Health Laboratory (CPHL; Colindale, London, United Kingdom), one strain was from the Department of Medical Microbiology (University of Liverpool, Liverpool, United Kingdom), and six were isolated on sorbitol-MacConkey agar plates from samples of cattle feces. Phages obtained from all of these STEC strains were named according to their bacterial strain origin and the type of toxin produced. The E. coli K-12 derivative strain, MC1061 (42), was used as the nontoxigenic host for Stx phage infections and propagation.
Isolation of phage particles from STEC lysogens.
Putative STEC lysogens were cultured overnight in 10 ml of nutrient broth (NB) at 37°C, and the cultures were membrane filtered (Millipore; 0.45-μm pore diameter). These potential phage-containing suspensions were used to infect E. coli strain MC1061, and individual plaques were picked and suspended in LB medium plus 0.01 M CaCl2 to isolate a single phage type. Where indicated, UV induction of lysis was used to enhance the recovery of phage (20).
Phage infections.
Nontoxigenic strains of E. coli were grown to an optical density at 600 nm (OD600) of 0.5 in either NB or Luria-Bertani (LB) medium containing 0.01 M CaCl2 at 37°C with shaking. Aliquots (200 μl) of these cultures were incubated at 37°C with 200 μl of serially diluted phage suspensions. After 30 min, 6 ml of Stx top agar (1% tryptone, 0.5% yeast extract, 1% NaCl, 0.4% [wt/vol] Difco agar, 0.01 M CaCl2) was added to the infection mix and poured onto the LB agar. The agar plates were vented and incubated overnight at 37°C before the plaques were identified, counted, and picked.
Isolation of colicin-resistant MC1061 mutants.
Briefly, 200 ml of MC1061 culture at OD600 of 0.5 was mixed with 1 ml of culture filtrate from an overnight culture of a putative colicin-producing STEC strain. This mixture was plated on LB agar, and any colonies formed were examined for their ability to inhibit naive cultures of E. coli MC1061 in addition to producing phage. Colonies that were resistant to the initial colicin and incapable of producing colicin or phage were designated colicin-resistant mutants.
Production of high-titer phage suspensions.
E. coli MC1061 was cultured in NZCYM medium (1% [wt/vol] NZ amine (Difco), 0.5% [wt/vol] NaCl, 0.5% [wt/vol] Bacto yeast extract, 0.1% [wt/vol] Casamino Acids, 0.2% [wt/vol] MgSO4 · 7H2O) with shaking (100 rpm) at 37°C to an OD600 of 0.5. Bacteriophages were added at a multiplicity of infection of 0.1, and the infected culture was further incubated overnight at 37°C with shaking. Phages were then recovered after the removal of whole cells and cellular debris by centrifugation at 12,000 × g for 10 min at 4°C, followed by membrane filtration. All phage stocks were stored at 4°C with a few drops of chloroform. With the exception of phage φPS14, which decreased in titer overnight from 109 to 102 PFU ml−1, phage stocks were stable under these conditions for at least 6 months.
Stx oligonucleotide probes.
Oligonucleotides that correspond to the published sequences of the stx1 and stx2 structural genes (8) were purchased from Genset SA (Paris, France). The oligonucleotide sequences were 5′-CTG ATG ATT GAT AGT GGC TCA-3′ and 5′-TCT GTT ATT AAC CAC ACC CAC-3′, respectively. These oligonucleotides were labeled by using polynucleotide kinase according to the manufacturer's instructions (Boehringer Mannheim) with 4,500 Ci of 32P (ICN)/mmol.
Colony hybridization.
Stx top agar overlay plates were incubated at 4°C for 1 h, and nylon membrane disks (Hybond N+; Amersham Pharmacia Biotech) were then laid on the top agar overlay and removed after 1 min. The membranes were then placed sequentially on Whatman 3MM paper soaked in denaturing solution (1.5 M NaCl, 0.5 M NaOH) for 7 min, in neutralization solution (1.5 M NaCl, 0.5 M Tris-HCl [pH 7.2], 1 mM EDTA) for 3 min (two times), and finally in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 1 min. The DNA was UV cross-linked to the membrane. Cell debris was removed by a wash in 5× SSC. The membranes were placed in prehybridization solution (50 mM Tris-HCl [pH 7.5], 1% [wt/vol] sodium dodecyl sulfate [SDS], 10% [wt/vol] dextran sulfate, 1 M NaCl, 10% [vol/vol] deionized formamide) for 2 to 3 h at 37°C. Radiolabeled probes were then applied and allowed to hybridize overnight at 37°C. Membranes were rinsed several times in 2× SSC containing 0.1% (wt/vol) SDS for 40 min at 42°C until the background radiation was no longer detected. The positive clones were detected by autoradiography.
Electron microscopy.
Phage particles were prepared for electron microscopy by dialysis of 1010 PFU ml−1 against double-distilled H2O overnight. Samples were then negatively stained with 2% (wt/vol) uranyl acetate and then examined by transmission electron microscopy.
Phage DNA preparation.
Phage DNA was isolated from 10 ml of the high-titer phage suspensions by using the Promega Wizard DNA purification system according to the manufacturer's instructions.
Southern hybridization.
Restriction enzyme-digested phage genomic DNA samples were subjected to 40 mM Tris-acetate-1 mM EDTA-agarose gel electrophoresis. Upon completion, the DNA was depurinated in 0.25 M HCl, denatured in 0.4 M NaOH for 10 min, and neutralized in 1.5 M NaCl-0.5 M Tris-HCl (pH 7.2)-1 mM EDTA. DNA was transferred to a positively charged nylon membrane (Hybond N+) by capillary transfer and fixed to the membrane by UV cross-linking. The prehybridization and hybridization parameters used were identical to those described above for colony hybridization analysis.
SDS-PAGE analysis.
Phage particles were precipitated by using 2/5 volume of PEG solution (33% [wt/vol] polyethylene glycol 8000, 3.3 M NaCl), followed by incubation on ice for 1 h prior to centrifugation at 17,500 × g for 20 min at 4°C. The pellet was suspended in 6 ml of SM buffer (0.1 M NaCl, 0.01 M MgSO4 · 7H2O, 0.05 M Tris [pH 7.5], 0.01% [wt/vol] gelatin). PEG and cell debris were removed by mixing the sample with 1 volume of chloroform, and the phages were recovered by centrifugation at 1,000 × g for 10 min at 4°C. The phage-containing aqueous layer was harvested, and 0.57 g of CsCl ml−1 was added. After ultracentrifugation at 90,000 × g for 24 h at 4°C, the phage particles were harvested and dialyzed against SM buffer, and the titer was determined. SDS-polyacrylamide gel electrophoresis (PAGE) gels were prepared by the method of Laemmli (13) with a 12% separating gel and a 5% stacking gel. The proteins were separated overnight at 0.5 V cm−1 and then subjected to silver staining (Bio-Rad).
Lysogen production.
E. coli MC1061 lysogens were created by infecting 0.2 ml of culture in NB (OD600 = 0.5) with 5 × 109 PFU of Stx phage. This infection mix was added to 6 ml of top agar and poured onto LB agar base plates. After overnight incubation, any colonies were regarded as possible lysogens and subcultured on to NB agar plates. Any colonies that could produce phages and were immune to subsequent phage infection were considered lysogens.
Characterization of phages produced by STEC lysogens.
E. coli MC1061 lysogens were created by using phage preparations obtained from a single plaque obtained from an STEC culture. The culture filtrates from the original STEC strain were then plated against the confirmed MC1061 lysogen. Where plaques were evident, the presence of more than one phage was considered likely, since bacterial lysogens should be immune to subsequent infection by their parent phage.
Recombinant mutant phage construction.
Plasmid NTP707 (43), which was constructed in pACYC184 and contains a 5-kb EcoRI-fragment possessing the Stx operon from φ933W, served as a mutagenic template for the construction of a selectable Stx-deficient phage. This plasmid was digested with PstI, which interrupts the a subunit gene (stxA2) of the stx2 operon. A kanamycin resistance gene, aph3, was obtained from pUC4K (Amersham Pharmacia Biotech) after digestion with PstI. These two DNA fragments were ligated together by using T4 ligase (Boehringer Mannheim). The resultant plasmid, NTP707-Kan, was transformed into E. coli strain MC1061, which was subsequently infected with φE86654-Stx2. The culture filtrates from this infection were then used to subsequently infect naive MC1061 cultures. The infection mix was plated onto NB agar (with 50 μg of kanamycin ml−1) and then replica plated onto NB agar (with 50 μg of kanamycin and 5 μg of tetracycline ml−1). Kanamycin-resistant, tetracycline-sensitive colonies were harvested as putative lysogens of the recombinant phage and further examined for their ability to produce plaques.
A second recombinant phage was created by using a similar strategy. Briefly, the plasmid NTP707x, the mutagenic vector, was created from NTP707 by the removal of a previously added green fluorescent protein (Clontech) reporter gene construct (32) that had been cloned into the region between the SmaI and PstI sites within stxA2 of the toxin operon. Digestion of this reporter gene construct with PstI and religation of the plasmid resulted in the removal of 0.9-kb fragment of the stxA2 gene, as well as the conservation of the PstI site. The chloramphenicol resistance gene from pLysS (Novagen) was used as a selectable marker. This cat gene was amplified from the vector by using the oligonucleotides 5′ChlorAmp (5′-AACTGCAGAAATGAGACGTTGATCGG-3′) and 3′ChlorAmp (5′-AACTGCAGCCTTAAAAAAATTACGCC-3′ [Sigma GenoSys]) by using Platinum Pfx (Invitrogen Life Technologies) according to the following amplification parameters: denaturation at 94°C for 2 min and 30 cycles of denaturation at 94°C for 1 min, primer annealing at 55°C for 1 min, and extension at 68°C for 1 min. A restriction endonuclease site, PstI, was present at the 5′ end of both oligonucleotides so that it could be cloned into the PstI site of NTP707x to create NTP707x-Cat. The strategy for isolation of the recombinant phage was then performed as indicated above with the exception that chloramphenicol-resistant (50 μg ml−1), tetracycline-sensitive (5 μg ml−1) colonies were picked for further study.
Confirmation of lysogen production by using the recombinant phage.
A strategy with Taq DNA polymerase (MBI) was used to amplify the stx2 operon containing either the aph3 or cat genes from suspected lysogens with the oligonucleotide primers Stx2A3′ (5′-TCTGTTCAGAAACGCTGC-3′) and Stx2A5′ (5′-TACTGTGCCTGTTACTGG-3′) that were designed from the published sequence of the 933W Stx2-phage (GenBank accession no. NC_000924) (25) to amplify the stxA2 subunit gene (9). Southern blot analysis was also used to confirm the identity of a double lysogen. Chromosomal DNA was treated as described above for Southern blot analysis. Prehybridization, hybridization, and detection methods for the analysis of the double lysogen were performed according to the manufacturer's guidelines by using the DIG nucleic acid detection kit (Roche). Probes for this Southern blot were labeled by using the PCR DIG probe synthesis kit (Roche) with the primer pairs 5′-AAT GTC GGG CAA TCA GG-3′ and 5′-GAA TCC GGT GAG AAT GG-3′, 5′-AAC TGC AGA AAT GAG ACG TTG ATC GG-3′ and 5′-AAC TGC AGC CTT AAA AAA ATT ACG CC-3′, 5-CAC TGG CGA TAA AGA AGG-3′ and 5′-CAC TGG CGA TAA AGA AGG-3′, and 5′-GCA TAT ACC AAG GCC TCT-3′ and 5′-GCG TCA CTG TAT GTC CC-3′ that amplify the genes for the kanamycin resistance cassette, the chloramphenicol resistance cassette, Q, and substxB2, respectively.
RESULTS
Screening E. coli strains for the presence of Stx phages.
The presence of Stx phages was detected by infecting strain MC1061 with overnight culture filtrates of 48 STEC strains and examining overlaid plates for plaques. This screening procedure was initially hampered by the apparent production of colicins by 13 STEC strains, which killed the indicator strain MC1061. Consequently, two spontaneous colicin-resistant mutants of E. coli (MC1061) were isolated to enable examination of phage susceptibility. One of these strains, MC1061-CR1, was isolated by using the colicin-producing O157 STEC strain, PS14, and the other resistant mutant, MC1061-CR2, was isolated by using the colicin-producing O91 STEC strain, E45037. Neither colicin-resistant mutant produced phages or colicins, but both were still susceptible to phage infection. This suggested that their resistance was not mediated by the acquisition of a colicin-producing plasmid or lysogenization by phage. All three MC1061 hosts were used to unequivocally screen all 48 STEC strains for phage production. Culture filtrates from five serotype O157 strains (E83642, E86654, E85539, E83819, and PS14), one O111:H− strain (E45040), and two of the bovine STEC isolates that were not serotyped (CF16 and D155) gave rise to plaques on the MC1061 indicator strains. Of the 48 STEC strains screened, evidence for phage production was obtained in 8 strains.
Confirmation of the presence of multiple phages within a single host strain.
STEC strains produce one or more types of Stx. To determine whether these different toxins are encoded by more than one phage, experiments were designed to differentiate the phages. Culture filtrates from strains that produced more than one Stx were used to infect MC1061. The resultant plaques were transferred to nylon membranes and sequentially probed with stx1 and then stx2 gene-specific oligonucleotide probes (Table 1). Only plaques produced by CF16 culture filtrates failed to hybridize with either probe, possibly indicating that the plaques formed on the indicator strain by CF16 culture filtrates were not due to Stx phages. Although the CPHL had reported that this strain produced Stx1, we were unable to confirm this through either Southern blot analysis or hybridization to plaques. The phage titer produced by CF16 (ca. 103 PFU ml−1) was many orders of magnitude lower than all of those produced by the other STEC strains (ca. 105 to 109 PFU ml−1) (Table 1). Except for strain E86654, phages encoding only one toxin type were detected.
TABLE 1.
Oligonucleotide probing of plaques on an MC1061 bacterial lawn infected with culture filtrates from various STEC strainsa
| STEC strain | Serotype | Phage type | Phage titer/ml of culture filtrate | No. of plaques probed | No. of plaques hybridizing to: |
|
|---|---|---|---|---|---|---|
| Stx1 probe | Stx2 probe | |||||
| CF16 | NDc | ND | 8.3 × 102 | 332 | 0 | 0 |
| E45040 | 1.2 × 106 | 121 | 0 | 121 | ||
| E45040 (UV)b | O111:H− | ND | 6.7 × 108 | 670 | 0 | 670 |
| E83819 | 1.7 × 106 | 171 | 171 | 0 | ||
| E83819 (UV) | O157:H7 | 14 | 1.6 × 108 | 160 | 160 | 0 |
| E85539 | O157:H7 | 4 | 1.4 × 106 | 175 | 0 | 175 |
| E86654 | O157:H7 | 4 | 7.1 × 104 | 126 | 121 | 5 |
| PS14 | 1.5 × 106 | 510 | 0 | 510 | ||
| PS14 (UV) | O157H7 | ND | 2.4 × 107 | 240 | 0 | 240 |
Plaques were probed with oligonucleotides specific to Stx1 and Stx2 toxin genes. Data for serotypes and phage types were obtained from the CPHL.
(UV), culture was subjected to UV irradiation to induce phage production.
ND, not determined.
In order to isolate as many different phages as possible from the STEC strains, an alternative procedure was used. It was based on infecting an MC1061 lysogen with phages such that any virion in the filtrate able to plaque on that lysogen was assumed to be different from the original bacteriophage. Again, the presence of both Stx1 and Stx2 phages in strain E86654 was revealed, but two other strains (E83819 and E85539) were now shown to produce more than one Stx phage (Table 2). This approach also allowed the identification and isolation of two different Stx2 phages from the same strain (E85539), which would not have been possible by using the simple plaque hybridization assay (Table 1). Although strains E45040 and PS14 (both from the CPHL) were originally characterized as possessing stx1 and stx2 genes, we could only isolate a single Stx2 phage from each, even when UV induction was also applied.
TABLE 2.
Isolation and differentiation of phages from STEC strains after MC1061 lysogen infection
| Strain | Culture filtrate PFUb | Primary phage isolated | PFU of secondary phagec | Secondary phage isolated | Ratio of primary to secondary phage isolate |
|---|---|---|---|---|---|
| E45040 | 1.23 × 105 | φE45040-Stx2 | 0 | None | NAd |
| E45040 (UV)a | 8.34 × 108 | φE45040-Stx2 | 0 | None | NA |
| E83819 | 2.90 × 105 | φE83819-Stx1 | 5.60 × 103 | φE83819-Stx2 | 50:1 |
| E85539 | 1.41 × 106 | φE85539-Stx21 | 1.01 × 106 | φE85539-Stx22 | 2:5 |
| E86654 | 6.90 × 104 | φE86654-Stx1 | 6.60 × 103 | φE86654-Stx2 | 10:1 |
| PS14 | 1.47 × 106 | φPS14-Stx2 | 0 | None | NA |
| PS14 (UV) | 1.90 × 109 | φPS14-Stx2 | 0 | None | NA |
(UV), culture was subjected to UV irradiation to induce phage production.
That is the total amount of PFU found in the culture filtrates.
Calculated as the titer against MC1061 − the titer against the primary phage lysogen.
NA, not applicable.
Characterization of Stx phages.
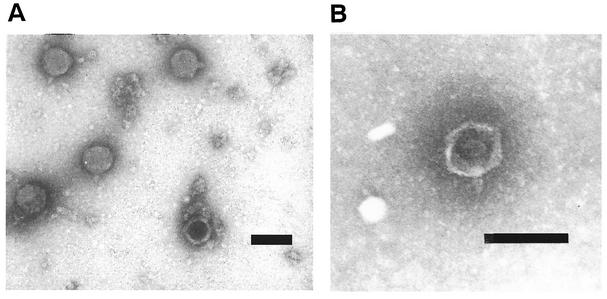
Two of the Stx phages were examined by transmission electron microscopy. Both φE86654-Stx2 and φE86654-Stx1 particles had regular hexagonal heads and short stubby tails (Fig. 1) that were similar in morphology to Stx phages described previously (27).
FIG. 1.
Transmission electron micrographs of φE86654-Stx1 and φE86654-Stx2. (A) φE86654-Stx1; (B) φE86654-Stx2. Both phages appear to be lambda-like in that they possess icosahedral heads and short stubby tails. Bar, 100 nm.
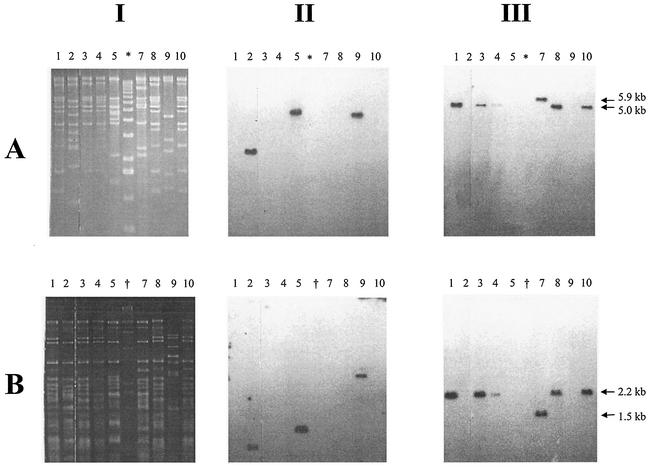
Restriction patterns of phage genomic DNA digested with EcoRI and HincII (Fig. 2) indicated that the Stx2 phages had broadly similar restriction fragment length polymorphism (RFLP) patterns (Fig. 2, lanes 1, 3, 4, 7, and 8 to 10); the Stx1 phages were more heterogeneous (Fig. 2, lanes 2, 5, and 9). After TAE-gel electrophoresis, the restriction endonuclease-digested phage genomic DNA samples were transferred to a nylon membrane and probed with oligonucleotides specific for either stx1 or stx2 (8). The stx2 oligonucleotide probe usually identified a 5.0-kb EcoRI or 2.2-kb HincII fragment (Fig. 2, column III). However, the stx2 gene was present on a 5.9-kb EcoRI fragment and 1.5 HincII fragment in phage φE83819. The size of the EcoRI and HincII fragments harboring the stx1 gene varied in all three Stx1-phages. The phage genome sizes were estimated from the molecular weights of DNA fragments produced by the RFLP analysis (Fig. 2). Four of the five Stx2-phages possessed 61.7-kb genomes, the remaining Stx2-phage had a 49.2-kb genome, and the three Stx1-phages had genomes ranging from 48.2 to 59.7 kb.
FIG. 2.
RFLP and Southern blot analysis of phage genomic DNA. (A) EcoRI-digested DNA; (B) HincII-digested DNA. Columns: I, RFLP patterns on TAE-agarose gel; II, Southern blot analysis of I with a Stx1-specific probe; III, Southern blot analysis of I with an Stx2-specific probe. Phage DNA were from the following sources: lane 1, φE86654-Stx2; lane 2, φE86654-Stx1; lane 3, φE85539-Stx2; lane 4, φE85539-Stx2′; lane 5, φE83819-Stx1; lane 7, φE83819-Stx2; lane 8, φE45040-Stx2; lane 9, φD155-Stx1; lane 10, φPS14-Stx2; lane ✽, molecular mass marker X (Roche); lane †, molecular weight marker II (Roche). Sizes: ✽, 12,216, 1,1198, 10,180, 9,162, 8,144, 7,126, 6,108, 5,090, 4,072, 3,054, 2,036, 1,636, 1,018, 517, 506, 396, 433, 298, 220, 201, 154, 134, and 75 bp; †, 23,130, 9,416, 6,557, 4,361, 2,322, 2,027, 564, and 125 bp.
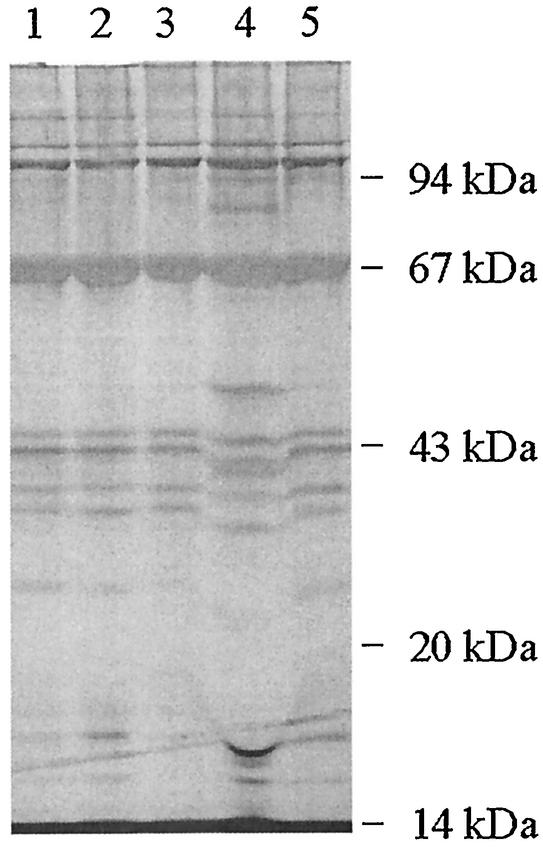
Total phage proteins were analyzed by SDS-PAGE for some of the Stx phages, and most had very similar gross protein profiles (Fig. 3). In some cases, Stx phages that could be readily distinguished by the RFLP analysis described above exhibited identical gross protein profiles. However, φE85539-Stx22 had an unusually distinct protein profile but had an RFLP pattern identical to that for φE85539-Stx21.
FIG. 3.
Silver-stained SDS-PAGE analysis of protein profiles of phages isolated from various STEC strains. The locations of the molecular mass markers are indicated. Lane numbers represent phage proteins from φE86654-Stx1 (lane 1), φE86654-Stx2 (lane 2), φE85539-Stx2 (lane 3), φE85539-Stx2′ (lane 4), and φE45040-Stx2 (lane 5).
Immunity profiles of Stx phages and their lysogens.
Phage immunity patterns were derived by plating each phage against every lysogen, and all phages failed to infect their own lysogens. The immunity profiles varied considerably; some phages infected all lysogens, while others, e.g., φPS14-Stx2, infected few lysogens (Table 3). Three of the STEC strains each yielded two different Stx phages. Their immunity patterns were not uniform in that one phage could infect the MC1061 lysogen of the other, but the reciprocal infection was either not obtained or was at a much reduced level (Table 3). Both phages obtained from STEC strain E86654 were unusual because they were able to infect many lysogens but often with much reduced efficiency. There are a number of examples of an absence of infection reciprocity between two phages and their lysogens (i.e., φE83819 Stx1 and φE45040 Stx2 [see Table 3]), and this finding was unexpected because it does not fit the established lambda bacteriophage immunity model (4), wherein the order of infection would have no effect on the immunity of the lysogen to the second phage.
TABLE 3.
Immunity patterns of MC1061 lysogens and phages isolated from STEC strains
| Lysogen | Immunity patterna with phage: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| φD155- Stx1 | φE45040- Stx2 | φE83819- Stx1 | φE83819- Stx2 | φE85539- Stx21 | φE85539- Stx22 | φE86654- Stx1 | φE86654- Stx2 | φPS14- Stx2 | |
| MC1061(φD155-Stx1) | (−) | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| MC1061(φE45040-Stx2) | ++ | (−) | ++ | ++ | ++ | ++ | ++ | ++ | − |
| MC1061(φE83819-Stx1) | ++ | − | (−) | − | ++ | ++ | ++ | + | − |
| MC1061(φE83819-Stx2) | ++ | ++ | ++ | (−) | ++ | ++ | ++ | ++ | ++ |
| MC1061(φE85539-Stx21) | ++ | − | ++ | ++ | (−) | ++ | ++ | ++ | − |
| MC1061(φE85539-Stx22) | ++ | − | ++ | ++ | − | (−) | + | − | − |
| MC1061(φE86654-Stx1) | ++ | ++ | ++ | ++ | ++ | ++ | (−) | ++ | ++ |
| MC1061(φE86654-Stx2) | ++ | ++ | ++ | ++ | ++ | ++ | + | (−) | − |
| MC1061(φPS14-Stx2) | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | (−) |
Pattern: (−), phage plated against its own lysogen; ++, lytic infection (PFU count was identical to that obtained from infection of naive MC1061); +, lytic infection (PFU count was reduced several orders of magnitude compared to infection of naive MC1061); −, no plaques found.
Construction of an Stx phage for lysogen selection.
A recombinant phage, φ24B::Kan, was created from phage φE86654-Stx2. The laboratory E. coli strain MC1061 was very susceptible to lytic infection by this wild-type phage, so a derived recombinant phage could provide an experimental system that does not require high-level laboratory containment. Plasmid NTP707, which contains the stx2 operon from φ933W and a tetracycline resistance gene in its vector backbone (43), was used to create a mutagenic template. The stxA2 gene from the toxin operon was interrupted with a kanamycin resistance cassette (see Materials and Methods). This recombinant plasmid was introduced into E. coli MC1061, and the resulting transformant was infected with phage φE86654-Stx2. Double recombinational events at both the 5′ and the 3′ ends of the Stx operon were selected for on LB agar containing 50 μg of kanamycin ml−1, and loss of the plasmid backbone was confirmed by screening for loss of the tetracycline resistance phenotype. Kanamycin-resistant, tetracycline-sensitive clones were further examined for the production of phage particles with or without UV induction. Phages obtained in this manner were also shown to produce kanamycin-resistant lysogens in strain MC1061. Plaques obtained with the recombinant phage were phenotypically identical to those produced by the wild-type phage φE86654-Stx2 (data not shown). No differences in phage morphology could be detected by electron microscopy. The MC1061 lysogens produced with this recombinant phage could easily be detected and enumerated by culturing infection mixtures on LB agar containing 50 μg kanamycin ml−1.
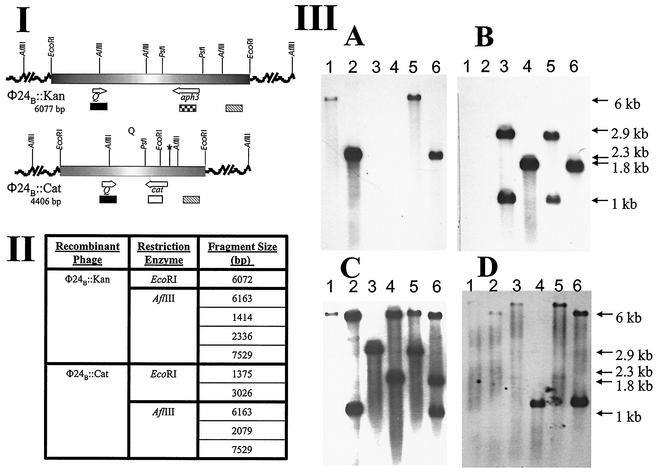
An alternative selective marker, the gene for chloramphenicol resistance, was introduced into the stxA2 gene of the Stx operon in φE86654-Stx2 to create φ24B::Cat. This enabled experiments in which both φ24B::Kan and φ24B::Cat could be used to doubly infect E. coli, and the markers allowed for strong selection to aid in the identification of lysogens and/or double lysogens. The latter would be unexpected if the underlying mechanism for immunity in these lambdoid Stx phages conformed to the lambda bacteriophage model (4). Regardless of the order of infection, both phages were able to produce lytic infection on the lysogen of the other, and double lysogens were detected at a very low frequency (data not shown). A single colony of a PCR-confirmed double lysogen was subjected to Southern analysis. The chromosomal DNAs of naive strain MC1061, a PCR-confirmed φ24B::Kan lysogen, a PCR-confirmed φ24B::Cat lysogen, and the double lysogen were digested with either EcoRI or AflIII (Fig. 4). Labeled probes specific to aph3, cat, and Q and to a region, substxB2, located downstream of the stxB2 genes were synthesized. After Southern hybridization analysis it was definitively demonstrated that two intact toxin operons existed within the chromosome of the double lysogen. As shown in Fig. 4, the aph3 gene probe hybridized to the predicted fragments in the φ24B::Kan lysogen and in the φ24B::Kan φ24B::Cat double lysogen, only (Fig. 4IIIA, lanes 1, 2, 5, and 6). When the cat gene probe was used, it hybridized to DNA only from the lysogens containing φ24B::Cat (Fig. 4IIIB, lanes 3, 4, 5, and 6). The Q- and substxB2-specific probes hybridized to DNA fragments of the predicted sizes in both single lysogens (Fig. 4IIIC and D, lanes 1, 2, 3, and 4), whereas the double lysogens possessed all DNA fragments that were unique to each single lysogen (Fig. 4IIIC and D, lanes 5 and 6). Since all of the DNA fragments detected were of the expected sizes and both AflIII and EcoRI sites were found outside the stx2 toxin operon, it is clear that both toxin operons must be intact. No probes recognized DNA fragments in uninfected E. coli MC1061 (data not shown). It was also demonstrated that both φ24B::Cat and φ24B::Kan were released as infective phage particles from the double-lysogen host (Table 4). The lysogens that were formed after infection of naive host with phage released from the double lysogens were characterized by their growth on selective media containing kanamycin and/or chloramphenicol and by PCR as described previously (9). Recombinant phages containing both selectable markers were never produced by a double lysogen.
FIG. 4.
Confirmation of the residence of two complete toxin operons within the double lysogen genome. (I) Genetic maps showing the location of relevant restriction endonuclease sites and areas recognized by the relevant probes: aph3 ( ), cat (□), Q (▪), and substxB2 (▧). The asterisk (✽) indicates the position of a PstI site that was lost after a single nucleotide substitution, as confirmed by DNA sequencing. (II) Tabulated data of the RFLP lengths of the relevant AflIII and EcoRI fragments from each isogenic recombinant phage. Fragment sizes are listed in consecutive order beginning at the 5′ end of the maps in part I. (III) Southern blots with probes specific to aph3 (A), cat (B), Q (C), and substxB2 (D). Lanes 1, 3, and 5 on blots A to D contain EcoRI-digested DNA, and lanes 2, 4, and 6 contain AflIII-digested DNA. The sources of the DNA are E. coli MC1061 lysogens containing the following prophages: lanes 1 and 2, φ24B::Kan; lanes 3 and 4, φ24B::Cat; lanes 5 and 6, φ24B::Kan and φ24B::Cat.
TABLE 4.
Characterization of phage released from the double lysogens
| Double lysogena | Total PFU released (SEM) | LFU (SEM)b |
||
|---|---|---|---|---|
| φ24B::Kan | φ24B::Cat | φ24B::Kan φ24B::Cat | ||
| φ24B::Kan φ24B::Cat | ||||
| Uninduced | 8.0 × 105 (5.3 × 104) | 355 (45) | 0 (0) | 0 (0) |
| Induced | 2.1 × 108 (1.3 × 107) | 1,030 (141) | 673 (64) | 0 (0) |
| φ24B::Cat φ24B::Kan | ||||
| Uninduced | 7.6 × 105 (1.6 × 104) | 3 (3) | 0 (0) | 0 (0) |
| Induced | 2.6 × 108 (4.0 × 106) | 1,430 (57) | 837 (22) | 0 (0) |
Uninduced, phage spontaneously released after overnight culture of the double lysogen in LB medium plus Ca; induced phage released from the double lysogen after norfloxacin treatment (1 μg/ml, 1 h).
LFU, lysogen-forming units. Lysogens were detected after infection of naive MC1061 cells with culture filtrate from double-lysogen culture.
DISCUSSION
There are many reports of bacteria that can produce Stx, although not all species are normally associated with toxin production (7, 20, 23, 30, 37). The production of Stx-like toxins in strains of C. freundii and Aeromonas spp., for example, may be due to bacteriophage-mediated transfer of vt genes from the STEC reservoir. This could have serious implications for the increase and spread of STEC-like diseases, and the presence of stx genes in “unusual” hosts may further complicate the detection of new Stx-producing pathogens in outbreaks. Consequently, characterization of Stx phages, and particularly their infection and immunity profiles, is important but rarely reported.
Here, Stx phages were isolated from a variety of STEC serotypes that had been isolated previously from various animal or human hosts. Stx phages were isolated by infecting laboratory strains of E. coli with culture filtrates from each of the STEC cultures. In a few cases, where inhibitory products (probably colicins) were produced by STEC strains, resistant host strains were generated to enable phage detection. Only 3 of 17 non-O157 STEC strains and 5 of 31 O157 STEC strains spontaneously released phages that produced plaques on E. coli MC1061. All of these plaques hybridized to either the stx1 or the stx2 gene probe. Approximately 10% of these STEC strains spontaneously released Stx phages, a frequency similar to those reported previously (27, 33). If phages are present in the other strains, they may be defective, unable to be induced, not induced into the lytic cycle within the parameters of the present study, or incapable of infecting strain MC1061.
The phages that were isolated in the present study displayed similar, though not identical, genomic DNA restriction patterns. Two phages were examined by electron microscopy and exhibited typical lambdoid phage morphology (26). The calculated genome sizes for nine of the Stx phages were in the range of 48 to 62 kb, with a pronounced cluster of phages with genomes of 61.7 kb in size. These general features are of limited significance, but they did reveal that Stx1- and Stx2-phages from the same strain (φE83819-Stx1 and φE83819-Stx2) are more similar to one another than to Stx1- and Stx2-phages from different E. coli strains. This finding supports previous work in which stx1 and stx2 genes were always located on separate phages within a single bacterial host, and the restriction patterns of both phage genomes displayed strong similarity to one another (27). Strains E83819, E85694, and E86654 possessed more than one Stx phage and produced both types of phage progeny simultaneously. Interestingly, the ratio of the yield of the two phages was highly variable, and in strain E83819, the Stx1/Stx2 ratio was 50:1. It is not known whether these variations are due to differences in the controls of the lytic cycles of the relevant bacteriophages, to the assembly of virion particles, to the location and hence the activity in the host genome, or to the overall fitness of the phages. It has been reported that genotypic variation in Stx phages does affect the expression of late genes (40). This indicates that it is possible for infectious particles to remain undetected, especially if the host is multiply infected, and one phage is a more efficient pathogen of the host. If a strain was infected with a phage that was biased toward the lysogenic cycle due to stringent regulatory controls, it would be difficult, if not impossible, to detect a lytic cycle and subsequent phage release. The full genome sequence analysis of a strain of O157:H7 has confirmed the presence of at least one complete Stx phage and multiple areas in the genome that appear to represent sequences of otherwise-uncharacterized cryptic, or presumed cryptic, Stx-prophages (24). The results presented here indicate that at least some of these cryptic phages could be competent viruses whose activity has yet to be detected.
Most of the properties of the Stx phages isolated here are similar to those described previously (27, 33). However, one of the interesting and distinguishing features of the Stx phages studied here was their immunity profile pattern. Unexpectedly, two phages with identical RFLP patterns, φE85539-Stx21 and φE85539-Stx22, that would normally have been indistinguishable were isolated from the same strain. Identification of different phages by superinfection of lysogens had the limitation of only separating phages with different immunity profiles. Rietra et al. (27) showed that Stx1- and Stx2 phages from the same strain had identical immunity patterns. The results reported here indicate that this is not always the case, although the RFLP patterns of Stx1- and Stx2 phages isolated from the same host exhibited a level of similarity that was not evident in phages isolated from different hosts. Recombination between prophages within a single host could result in the minimization of heterogeneity. The implication of the data presented here is that the immunity conferred on an E. coli Stx phage lysogen is not absolute. Multiple, complete Stx phages can be carried by a single E. coli host strain, and this has clear implications for the pathogenicity and epidemiology of STEC.
All of the genes necessary to propagate phage lambda under laboratory conditions are present on 30 kb of DNA (1). The remainder of the genome, sometimes greater than 30 kb as found in the Stx phages reported here, presumably comprises genes that are not essential for propagation, at least in E. coli K-12. However, these genes are clearly maintained, suggesting that they offer some selective advantage, either to the Stx phage itself or to a lysogen once the phage genome has become integrated. This is the case with lambda, which encodes additional virulence determinants lom and bor that are expressed by lysogens, and may aid fitness in its natural environment and increase host growth rate in laboratory conditions (1). Sequences homologous to these two genes have been found on 933W and other stx2-carrying Stx phages (25). Thus, in nature, the carrier phages and possibly the Stx genes might confer some selective advantage assisting in their maintenance within bacterial populations. From an evolutionary standpoint, a phage with a different immunity grouping will have some selective advantage within a population simply because it is able to escape immunity control and grow on lysogens of otherwise closely related phages. This could enable phage infection of gut commensals or other bacteria not typically associated with Stx production, and result in the creation of new pathogens or add to the refractory nature of the disease process.
The mechanisms underlying the immunity patterns observed here need to be examined further because lysogens of Stx phages are not necessarily immune to superinfection. The experiment in which we created a double lysogen by using the same phage with different selectable markers, i.e., φ24B::Kan and φ24B::Cat, is unique. The data in Fig. 4 provide unequivocal evidence that genetically identical phages can coexist in the same E. coli host despite similar genetic arrangement of the Stx phage loci homologous to the lambda phage immunity region (5). In previous studies on lambda phage, double lysogens resulted from the integration of a second phage into the first prophage genome by homologous recombination (2, 3), which resulted in a population of mosaic phage particles upon entry into the lytic cycle. In later unrelated studies, double lysogens were obtained when immunity mutants were used (6, 31). We used powerful antibiotic resistance selection criteria and never detected phage particles that possessed both resistance markers. These selection criteria would assure the detection of every instance of phage incorporation into the host cell chromosome, regardless of the mechanism. We are therefore confident that the double lysogens of this Stx phage have been formed by single phage-directed integration into the host chromosome, in contrast to the observations of Calef et al. (2, 3) in bacteriophage lambda. We also used a phage background that was only recently acquired from a pathogenic E. coli strain and, other than the replacement of the stx2 operon, is unlikely to have undergone mutations that make it markedly different from Stx phages found in the environment. It is therefore unlikely that we are studying a laboratory-induced immunity mutant, but even if the double lysogen is derived from a naturally occurring immunity mutant of the Stx phage, then the data presented here are still very relevant to phage-mediated dissemination of vt genes. The numbers of double lysogens obtained by using the isogenic recombinant phage were found at a frequency sufficiently high to reinforce the notion that this mechanism could play a significant role in stx gene dissemination. Double lysogens could also have a role in creating a genetic pool within a host cell for the production of novel mosaic phages after recombination events in situ. Future studies with our recombinant Stx phage may help explain the unusual immunity data reported here, as well as enable general Stx phage carriage investigations, both in vitro and in vivo.
Acknowledgments
We thank Brian Getty of the Department of Medical Microbiology at the University of Liverpool for electron microscopy and Susan Mattinson for technical assistance.
This research was supported by a Natural Environment Research Council (United Kingdom) studentship and a research grant from the Biotechnology and Biological Sciences Research Council (United Kingdom).
Editor: D. L. Burns
REFERENCES
- 1.Barondess, J. J., and J. Beckwith. 1990. A bacterial virulence determinant encoded by the lysogenic coliphage λ. Nature 346:871-874. [DOI] [PubMed] [Google Scholar]
- 2.Calef, E. 1967. Mapping of integration and excision crossovers in superinfection double lysogens for phage lambda in Escherichia coli. Genetics 55:547-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calef, E., C. Marchelli, and F. Guerrini. 1965. The formation of superinfection-double lysogens of phage lambda in Escherichia coli K-12. Virology 27:1-10. [DOI] [PubMed] [Google Scholar]
- 4.Eisen, H., P. Brachet, L. P. da Silva, and F. Jacob. 1970. Regulation of repressor expression in lambda. Proc. Natl. Acad. Sci. USA 66:855-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fattah, K. R., S. Mizutani, F. J. Fattah, A. Matsushiro, and Y. Sugino. 2000. A comparative study of the immunity region of lambdoid phages including Shiga-toxin-converting phages: molecular basis for cross immunity. Gene Genet. Sys. 75:223-232. [DOI] [PubMed] [Google Scholar]
- 6.Freifelder, D., and I. Kirschner. 1971. The formation of homoimmune double lysogens of phage lambda and the segregation of single lysogens from them. Virology 44:633-637. [DOI] [PubMed] [Google Scholar]
- 7.Haque, Q. M., A. Sugiyama, Y. Iwade, Y. Midorikawa, and T. Tamauchi. 1997. Diarrheal and environmental isolates of Aeromonas spp. produce a toxin similar to Shiga-like toxin I. Curr. Microbiol. 32:239-245. [DOI] [PubMed] [Google Scholar]
- 8.Jackson, P. J., R. J. Neill, A. D. O'Brien, R. K. Holmes, and J. W. Newland. 1987. Nucleotide sequence and comparison of the structural genes for Shiga-like toxin I and Shiga-like toxin II encoded by bacteriophages from Escherichia coli 933. FEMS Microbiol. Lett. 44:109-114. [Google Scholar]
- 9.James, C. E., K. N. Stanley, H. E. Allison, H. J. Flint, C. S. Stewart, R. J. Sharp, J. R. Saunders, and A. J. McCarthy. 2001. Lytic and lysogenic infection of diverse Escherichia coli and Shigella strains with a verocytotoxigenic bacteriophage. Appl. Environ. Microbiol. 67:4335-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarvis, K. G., and J. B. Kaper. 1996. Secretion of extracellular proteins by enterhaemorrhagic Escherichia coli via a putative type III secretion system. Infect. Immun. 64:4826-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karch, H., J. Heeseman, R. Laufs, A. D. O'Brien, C. O. Tacket, and M. M. Levine. 1987. A plasmid of enterohaemorrhagic Escherichia coli O157:H7 is required for expression of a new fimbrial antigen for adhesion to epithelial cells. Infect. Immun. 55:455-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleanthous, H., H. R. Smith, S. M. Scotland, R. J. Gross, B. Rowe, C. M. Taylor, and D. V. Milford. 1990. Haemolytic uraemic syndromes in the British Isles, 1985-88: association with verocytotoxin producing Escherichia coli. Part 2. Microbiological aspects. Arch. Dis. Child. 65:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.Law, D. 2000. Virulence factors of Escherichia coli O157 and other shiga toxin-producing E. coli. J. Appl. Microbiol. 88:729-745. [DOI] [PubMed] [Google Scholar]
- 15.Louise, C. B., and T. G. Obrieg. 1995. Specific interactions of Escherichia coli O157:H7-derived Shiga-like toxin II with human renal endothelial cells. J. Infect. Dis. 172:1397-1401. [DOI] [PubMed] [Google Scholar]
- 16.Louise, C. B., and T. G. Obrig. 1991. Shiga toxin-associated hemolytic-uremic syndrome: combined cytotoxic effect of Shiga toxin, interleukin-1, and tumor necrosis factor alpha on human vascular endothelial cells in vitro. Infect. Immun. 59:4173-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melton-Celso, A. R., and A. D. O'Brien. 1998. Structure, biology, and relative toxicity of Shiga toxin family members for cells and animals, p. 121-128. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology, Washington, D.C.
- 19.Mills, M., and S. M. Payne. 1995. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coli O157:H7. J. Bacteriol. 177:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Brien, A. D., M. E. Chen, R. K. Holmes, J. Kaper, and M. M. Levine. 1984. Environmental and human isolates of Vibrio cholerae and Vibrio parahaemolyticus produce a Shigella dysenteriae 1 (Shiga)-like cytotoxin. Lancet i:77-78. [DOI] [PubMed]
- 21.O'Brien, A. D., and J. B. Kaper. 1998. Shiga toxin-producing Escherichia coli: yesterday, today, and tomorrow, p. 1-11. In J. B. Kaper and A. D. O'Brien (ed.), Escherchia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Soceity for Microbiology, Washington, D.C.
- 22.O'Brien, A. D., and G. D. LaVeck. 1983. Purification and characterization of a Shigella dysenteriae 1-like toxin produced by Escherichia coli. Infect. Immun. 40:657-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paton, A. W., and J. C. Paton. 1996. Enterobacter cloacae producing a Shiga-like toxin II-related cytotoxin associated with a case of hemolytic-uremic syndrome. J. Clin. Microbiol. 34:463-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perna, N. T., G. I. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 25.Plunkett, G., D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157: H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ptashne, M. 1992. A genetic switch: phage λ and higher organisms, 2nd ed. Blackwell Scientific Publications, Oxford, England.
- 27.Rietra, P. J. G. M., G. A. Willshaw, H. R. Smith, A. M. Field, S. M. Scotland, and B. Rowe. 1989. Comparison of verocytotoxin-encoding phages from Escherichia coli of human, and bovine origin. J. Gen. Microbiol. 135:2307-2318. [DOI] [PubMed] [Google Scholar]
- 28.Savarino, S. J., A. McVeigh, J. Watson, J. Molina, A. Cravioto, P. Echeverria, M. K. Bhan, M. M. Levine, and A. Fasano. 1992. Enteroaggregative Escherichia coli heat-stable enterotoxin is not restricted to enteroaggregative Escherichia coli. J. Infect. Dis. 173:1019-1022. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt, H., H. Karch, and L. Beutin. 1994. The large-sized plasmids of enterohaemorrhagic Escherichia coli O157 strains encode haemolysins which are presumably members of the E. coli alpha-haemolysin family. FEMS Microbiol. Lett. 117:189-196. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt, H., M. Montag, J. Bockemuhl, J. Heeseman, and H. Karch. 1993. Shiga-like toxin II related cytotoxins in Citrobacter freundii strains from human and beef samples. Infect. Immun. 61:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider, J., and H. B. Strack. 1977. Analysis of the phase variation in lambda reduced immunity lysogens. Mol. Gen. Genet. 151:181-187. [DOI] [PubMed] [Google Scholar]
- 32.Sergeant, M. J. 1998. Molecular biological characterisation of verotoxigenic bacteriophage in E. coli. Ph.D. thesis. University of Liverpool, Liverpool, United Kingdom.
- 33.Smith, H. W., P. Green, and Z. Parsell. 1983. Vero cell toxins in Escherichia coli and related bacteria-transfer by phage and conjugation and toxic action in laboratory animals, chickens and pigs. J. Gen. Microbiol. 129:3121-3137. [DOI] [PubMed] [Google Scholar]
- 34.Stockbine, N. A., L. R. M. Marques, J. W. Newland, H. W. Smith, R. K. Holmes, and A. D. O'Brien. 1986. Two toxin-converting phages from Escherichia coli O157-H7 strain 933 encode antigenically distinct toxins with similar biological activities. Infect. Immun. 53:135-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stroeher, U. H., L. Bode, L. Beutin, and P. A. Manning. 1993. Characterization and sequence of a 33 kDa enterohaemolysin (Ehly 1) associated protein in Escherichia coli. Gene 132:89-94. [DOI] [PubMed] [Google Scholar]
- 36.Tesh, V. L., J. A. Burris, J. E. Owens, V. M. Gordon, E. A. Waldolkowski, A. D. O'Brien, and J. E. Samuel. 1993. Comparison of relative toxicities of Shiga-like toxins type I and type II for mice. Infect. Immun. 62:3392-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tschape, H., R. Prager, W. Streckel, A. Fruth, H. Tietre, and A. Bohme. 1995. Verotoxigenic Citrobacter freundii associated with severe gastroenteritis and cases of haemolytic uraemic syndrome in a nursery school: green butter as the infection source. Epidemiol. Infect. 114:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unkmeir, A., and H. Schmidt. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Setten, P. A., V. W. van Hinsbergh, T. J. van der Velden, N. C. van de Kar, M. Vermeer, J. D. Mahan, K. J. Assmann, L. P. van den Heuvel, and L. A. Monnens. 1997. Effects of TNF alpha on verocytotoxin cytotoxicity in purified human glomerular microvascular endothelial cells. Kidney Int. 51:1245-1256. [DOI] [PubMed] [Google Scholar]
- 40.Wagner, P. L., D. W. Acheson, and M. K. Waldor. 1999. Isogenic lysogens of diverse Shiga toxin 2-encoding bacteriophages produce markedly different amounts of Shiga toxin. Infect. Immun. 67:6710-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watarai, M., T. Sato, M. Kobayashi, T. Shimizu, S. Yamasaki, T. Tobe, C. Sasakawa, and Y. Takeda. 1998. Identification and characterization of a newly isolated Shiga toxin 2-converting phage from Shiga-toxin-producing Escherichia coli. Infect. Immun. 66:4100-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wertman, K. F., A. R. Wyman, and D. Botstein. 1986. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene 49:253-262. [DOI] [PubMed] [Google Scholar]
- 43.Willshaw, G. A., H. R. Smith, S. M. Scotland, A. M. Field, and B. Rowe. 1987. Heterogeneity of Escherichia coli phages encoding Vero cytotoxins: comparison of cloned sequences determining Stx1 and Stx2 and development of specific gene probes. J. Gen. Microbiol. 133:1309-1317. [DOI] [PubMed] [Google Scholar]