Abstract
The LuxS protein is required for the biosynthesis of the type 2 autoinducer (AI-2), which is involved in quorum sensing in a wide range of bacterial species. We have determined the effects of a defined luxS mutation on the virulence of Streptococcus pneumoniae. Although the luxS mutant displayed reduced virulence relative to its wild-type parent, the type 2 strain D39, it was by no means avirulent in a mouse model. After intranasal administration, the luxS mutant was able to colonize the nasopharynx of the mouse as efficiently as the wild type. However, it was less able to spread from the nasopharynx to the lungs or the blood. Intraperitoneal coadministration studies indicated that the luxS mutant was less fit and was readily outcompeted by wild-type D39. However, when administered on its own by this route, the mutant was able to proliferate and cause fatal systemic disease, albeit at a lower rate than the wild type. Western blot analysis of whole-cell lysates of the mutant and its parent did not reveal any differences in the levels of several well-characterized virulence proteins. However, analysis of Coomassie blue-stained protein profiles after separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed that mutation of luxS had pleiotropic effects on protein expression in all cellular compartments. This is consistent with the product of luxS having a regulatory role in S. pneumoniae. This is the first report of a direct role for luxS (and by inference, AI-2) in the virulence of a gram-positive pathogen. However, the fact that mutagenesis of luxS does not completely attenuate S. pneumoniae has implications for the possible use of AI-2 antagonists for treatment of pneumococcal infections.
Streptococcus pneumoniae is a major cause of invasive diseases such as pneumonia, meningitis, and bacteremia, as well as less severe but highly prevalent infections such as otitis media (23, 42). The elderly and the very young are most at risk from pneumococcal disease, which causes the deaths of over 1 million children under 5 years of age each year (13, 47). Treatment of patients with pneumococcal disease is being complicated by increasing rates of resistance to penicillin and other antimicrobial drugs and the rapid global spread of multiply resistant S. pneumoniae clones. Disease prevention programs have also been frustrated by the low clinical efficacy of pneumococcal polysaccharide vaccines, which are poorly immunogenic in young children and other high-risk groups (9, 31). Recently licensed polysaccharide-protein conjugate vaccines are more immunogenic but are expensive and provide protection against only a limited range of S. pneumoniae serotypes (18).
The ongoing threat to human health posed by the pneumococcus has prompted extensive research aimed at understanding its mechanism of pathogenesis, with a view to identifying alternative targets for vaccines and novel drugs. One potential new class of antimicrobial agents includes compounds that interfere with the phenomenon of quorum sensing (21, 32). This is a mechanism whereby bacteria coordinate their gene expression in response to population density. Small autoinducer molecules are secreted by the bacterium, and as the population density increases, cell surface receptors sense the increasing extracellular concentration of autoinducer, triggering alterations in gene expression (2, 16, 46). The functions influenced by quorum sensing in various species are wide ranging and include the production of virulence factors, motility, biofilm formation, and differentiation, as well as bioluminescence (2, 22, 26, 43). The number of organisms which have been shown to regulate some of their key virulence genes by quorum sensing is steadily growing, and the list currently includes the genes for the enterohemorrhagic Escherichia coli type III secretion apparatus (15, 34), toxin expression in Erwinia carotovora and Pseudomonas aeruginosa (17, 28), the hemagglutinin of Porphyromonas gingivalis (6), and toxin production in Clostridium perfringens (27). The signaling molecule in these organisms is autoinducer 2 (AI-2), which has been studied extensively in Vibrio harveyi. The luxS gene is required for synthesis of AI-2, which is released from the cell and builds up in the environment. It is then captured on a specific receptor/sensor complex (LuxP/Q), and subsequent phosphorelay via either LuxU or a LuxU homologue leads to modification of the transcriptional activator LuxO. This, in turn, relieves repression of the Vibrio lux operon (10, 11).
Interestingly, the LuxS protein appears to play a dual role in many bacterial species; in addition to its involvement in AI-2 synthesis, it is important in the methyl cycle (44). LuxS, together with the methylthioadenosine/S-adenosylhomocysteine nuclease (MTA/SAHase), converts S-adenosylhomocysteine to homocysteine and adenosine via a two-step process. The exact role LuxS plays in the production of AI-2 is still unclear, but AI-2 may be a spontaneous by-product of the conversion of 4,5-dihydroxy-2,3-pentanedione to 4-hydroxy-5-methyl-3(2H)furanone (44, 45). The AI-2 molecule produced by V. harveyi has been reported to be a furanosyl borate diester (8), but whether this holds for AI-2 molecules produced by other organisms is still unclear. What is known is that the periplasmic AI-2 binding protein proposed to be involved in quorum sensing in Salmonella enterica serovar Typhimurium (LsrB) shows little homology to the AI-2 binding protein of V. harveyi (35).
S. pneumoniae has a well characterized quorum-sensing system involved in regulation of competence for genetic transformation. Indeed, this was the first such system to be described in bacteria (37). The signaling molecule is a peptide which is encoded by comC, exported from the cell by ComAB and then sensed by the two-component signal transduction system ComDE (reviewed by Morrison and Lee [22]). The pneumococcus also has a luxS homologue, but whether this is involved in a second quorum-sensing circuit has not previously been investigated. Mutation of luxS in Streptococcus pyogenes was recently shown to have marked effects on the expression of several virulence-related genes, resulting in increased hemolytic activity and reduced secretion of SpeB; the in vitro growth rate in certain media was also affected (20). In the present study, we have examined the role of LuxS in the pathogenesis of S. pneumoniae infection with a view to assessing the potential of AI-2 antagonists for treatment of infections caused by this organism.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S. pneumoniae strain used in this study was the virulent type 2 strain D39 (1). Bacteria were grown overnight on blood agar and were inoculated into either serum broth (meat extract broth supplemented with 10% horse serum) or Todd-Hewitt broth with 0.5% yeast extract (THY) for growth prior to inoculation or challenge. S. pneumoniae D39 was transformed as described previously (4, 49), and transformants were selected on blood agar containing 0.2 μg of erythromycin/ml. V. harveyi strains BB152 and BB170 (3) were obtained from S. Kjelleberg, University of New South Wales, Australia. They were grown at 30°C either on Luria-Bertani agar containing 0.16 M NaCl or in autoinducer bioassay (AB) medium (0.3 M NaCl, 0.05 M MgSO4, and 0.2% vitamin-free Casamino Acids [Difco], adjusted to pH 7.5 with KOH, sterilized, and supplemented with 10 ml of 1 M potassium phosphate [pH 7.0], 10 ml of 0.1 M l-arginine, 20 ml of glycerol, 1 ml of 10-μg/ml riboflavin and 1 ml of 1-mg/ml thiamine per liter).
DNA manipulations.
S. pneumoniae chromosomal DNA used in Southern hybridization experiments was extracted and purified with the Wizard genomic DNA purification kit (Promega Corporation, Madison, Wis.). Chromosomal DNA was purified according to the manufacturer's instructions, except that cell lysis was induced by the addition of 0.1% (wt/vol) deoxycholate followed by incubation at 37°C for 10 min. PCR amplification was performed in a Hybaid Touchdown thermal cycler, and the 50-μl reaction volume contained PCR buffer (1.5 mM MgCl2, 10 mM Tris [pH 8.4], 50 mM KCl, and 100 μg of gelatin/ml), 1.5 U of Taq polymerase, 1 μM (each) primer, 100 ng of DNA template, and 200 μM (each) four deoxynucleoside triphosphates. The program comprised 25 cycles consisting of denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and elongation at 72°C for 1 to 4 min (1 min for each kilobase of expected product). A 5-μl sample of the PCR product was analyzed by agarose gel electrophoresis.
The oligonucleotides used for PCR amplification of luxS were designed with reference to the luxS sequence in the genome of S. pneumoniae R6 (a derivative of D39) (accession number NC_003098). The primers were LuxSI (GCCCTGCAGGGACCAAAAGGAGACATC) and LuxSII (GGCGGATCCAACAGCTGCGATTTTAGCACT), which contain PstI and BamHI sites, respectively (underlined). The resultant 220-bp PCR product was cloned into the PstI and BamHI sites in the suicide vector pACH74 (14). All restriction enzymes and Taq DNA polymerase were purchased from Roche Molecular Biochemicals (Rotkreuz, Switzerland) and used according to the manufacturer's recommendations.
Southern hybridization analysis.
Chromosomal DNA (2.5 μg) was digested with appropriate restriction enzymes, and the digests were electrophoresed on agarose gels in Tris-borate-EDTA buffer. DNA was then transferred to a positively charged nylon membrane (Hybond N+; Amersham, Little Chalfont, England) as described by Southern (33), hybridized to digoxigenin (DIG)-labeled probe DNA, washed, and then developed with anti-DIG-alkaline phosphatase conjugate (Roche) and 4-nitro blue tetrazolium-X-phosphate substrate according to the manufacturer's instructions. DIG-labeled lambda DNA, restricted with HindIII, was used as a DNA molecular size marker.
Cell fractionation and SDS-PAGE analysis.
Cells were grown in THY to an A600 of approximately 0.5 and harvested by centrifugation at 5,000 × g for 10 min. The culture supernatant was collected, and proteins were precipitated in 12% trichloroacetic acid. After centrifugation, precipitated proteins were redissolved in sodium dodecyl sulfate (SDS) loading buffer and stored at −20°C. Meanwhile, the bacterial cells were resuspended in phosphate-buffered saline (PBS) and lysed with a French pressure cell (SLM Aminco Instruments) at 12,000 lb/in2. Unbroken cells were removed by centrifugation at 5,000 × g for 10 min, and the supernatant was centrifuged at 100,000 × g for 1 h at 4°C. The high-speed supernatant (soluble fraction) and the pellet (insoluble fraction) were stored at −20°C prior to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. Protein concentrations in the various fractions were determined by using the method of Bradford (5). Samples were dissolved in SDS loading buffer and subjected to SDS-PAGE as described by Laemmli (19). Loadings were adjusted so that fractions from D39 and D39luxS contained the same amount of total protein. Gels were stained with Coomassie brilliant blue R250.
Western immunoblotting.
Bacterial cell lysates from 1 ml of log-phase culture (A600, 0.25; 5 × 108 CFU/ml) were separated on 12% SDS-PAGE gels (17) prior to transfer onto nitrocellulose (41). Filters were probed with mouse antibodies specific for various pneumococcal virulence proteins, used at a dilution of 1:3,000, followed by a 1:5,000 dilution of goat anti-mouse immunoglobulin G conjugated to alkaline phosphatase (Bio-Rad Laboratories, Hercules, Calif.). Enzyme-labeled bands were visualized with the 4-nitro blue tetrazolium-X-phosphate substrate system (Roche). A benchmark prestained protein ladder (Life Technologies, Gaithersburg, Md.) was used as molecular size markers.
Determination of AI-2 activity.
The method used is essentially that described previously (3). Cell-free supernatants were prepared as follows. V. harveyi strain BB152 was grown in AB medium overnight and then subcultured into fresh AB medium and grown to an A600 of approximately 0.9; bacterial cells were removed by centrifugation, and the supernatant was subsequently filter sterilized. S. pneumoniae was grown overnight on blood agar plates and subcultured into THY and then grown to an A600 of approximately 0.45; the culture was subsequently treated as described above. The indicator strain B172 was grown overnight in AB medium and diluted 1:5,000 in fresh AB medium. The cell-free supernatants were added to the diluted BB172 cells at a 1:10 ratio, and the luminescence was monitored over a period of 5 h using a TD-20/20 illuminometer (Turner Designs, Sunnyvale Calif.).
Intranasal colonization model.
The method used was a modification of that described by Wu et al. (48). Bacterial strains used for the intranasal challenge were grown in THY broth to an A600 of approximately 0.25. Ten mice were anaesthetized prior to challenge by intraperitoneal injection with Nembutal (pentobarbitone sodium; Rhone-Merieux) at a dose of 66 μg per gram of body weight. Subsequently, 20 μl of S. pneumoniae D39 or its luxS-negative derivative, at concentrations of approximately 1 × 108 CFU/ml, was pipetted into the nares and involuntarily inhaled. Five mice were sacrificed by carbon dioxide asphyxiation at both 24 and 48 h postchallenge. Blood (75 μl) was collected from each mouse by retro-orbital bleeding with heparinized capillary tubing prior to sacrifice, of which 45 μl was diluted in 160 μl of sterile PBS. After exposure of the trachea, the nasopharynx was washed with 1 ml of buffer (0.5% trypsin-0.02% EDTA in sterile PBS) by insertion of a 26-gauge needle sheathed in tubing into the tracheal end of the upper respiratory tract. Buffer was allowed to drip into the nasopharynx slowly and was collected from the nose, with each wash taking approximately 40 seconds. Both lungs were entirely removed, rinsed twice in PBS to remove excess blood, weighed, and placed in 2 ml of sterile PBS. The lungs were then homogenized with a CAT X120 homogenizer (CAT, M, Zipperer GmbH, Staufen, Germany) at 30,000 rpm for approximately 10 s. All blood, lung, and nasal washout samples were then serially diluted and plated on blood agar (supplemented with 0.2 μg of erythromycin/ml in the case of the luxS mutant) to determine the number of viable pneumococci. Differences in the levels of colonization were analyzed using Student's unpaired t test (two-tailed).
Intraperitoneal challenge.
Bacterial strains were grown in serum broth to an A600 of 0.08. Groups of mice were then injected intraperitoneally with 0.1 ml of diluted cultures containing approximately 1 × 105 CFU of S. pneumoniae D39 or its luxS-negative derivative (confirmed by viable count). Mice were monitored at approximately 4-h intervals, and the survival time of each mouse was recorded. The statistical significance of differences in median survival times between groups was analyzed using the Mann-Whitney U test (two-tailed).
Intraperitoneal competition model.
Ten mice were injected intraperitoneally with 0.1 ml of serum broth containing 1 × 104 CFU each of S. pneumoniae D39 and its luxS-negative derivative. Five mice were sacrificed by carbon dioxide asphyxiation at each of 24 and 48 h postchallenge. Blood (75 μl) was removed from each mouse by retro-orbital bleeding by heparinized capillary tubing prior to sacrifice, of which 45 μl was diluted in 160 μl of sterile PBS. The peritoneal cavity was also washed by injection of 4 ml of sterile PBS, and the washout was collected to determine the number of CFU in the peritoneum. The peritoneal cavity was then opened, and the spleen was removed and placed in 2 ml of sterile PBS. The spleens were then homogenized at 30,000 rpm for approximately 10 s. All blood, spleen, and peritoneal washout samples were serially diluted and plated onto blood agar or blood agar supplemented with erythromycin to determine the numbers of wild-type and luxS mutant S. pneumoniae.
Adherence assay.
Adherence of pneumococci to A549 cells (human type II pneumocytes) and HEp-2 cells (human laryngeal carcinoma) was determined essentially as described previously (36). Briefly, pneumococci were suspended at a density of 1 × 106 CFU/ml (confirmed by viable count) in Dulbecco's modified Eagle's medium buffered with 20 mM HEPES and supplemented with 2 mM l-glutamine. Washed A549 or HEp-2 monolayers in 24-well tissue culture plates were infected with 1-ml aliquots of bacterial suspension. After incubation at 37°C for 2 h, the culture medium was removed, and the monolayers were washed four times with PBS to remove nonadherent bacteria. The cell monolayers were then detached from the plate by treatment with 100 μl of 0.25% trypsin-0.02% EDTA. Cells were then lysed by addition of 400 μl of 0.025% Triton X-100, and 50-μl aliquots (and serial tenfold dilutions thereof) were plated on blood agar to determine the total number of adherent bacteria per well. Assays were carried out in quadruplicate, and results were expressed as the mean CFU/well ± standard error. The significance of differences in adherence was analyzed using Student's unpaired t test (two-tailed).
RESULTS
Construction of S. pneumoniae with an insertion-duplication mutation in luxS.
A 220-bp fragment of the luxS gene was PCR amplified from S. pneumoniae D39 chromosomal DNA by using oligonucleotides LuxSI and LuxSII, digested with PstI and BamHI, and cloned into the suicide vector pACH74. The resulting plasmid pLuxS01 was then transformed into D39. Interruption of the luxS open reading frame by insertion-duplication in a selected erythromycin-resistant transformant (designated D39luxS) was confirmed by PCR amplification using a combination of oligonucleotides LuxSI, LuxSII, and the forward and reverse M13 sequencing primers that anneal to pACH74 (result not shown). The mutation in D39luxS was also confirmed by Southern hybridization analysis of genomic DNA using the 220-bp DIG-labeled LuxSI/LuxSII PCR product as probe. This labeled a single 12-kb fragment in BamHI digests of D39 chromosomal DNA but labeled 10- and 8-kb fragments in BamHI digests of D39luxS DNA (result not shown). Examination of the DNA sequence flanking luxS in the genome sequence of S. pneumoniae R6 (a derivative of D39) (accession number NC_003098) indicated that clpL (which encodes a heat shock protease) is located 295 nucleotides downstream of luxS, but it is on the opposite DNA strand. This intergenic region also contains a weak stem-loop structure (ΔG = −10.3 kcal/mol). Thus, insertion-duplication mutagenesis of luxS is unlikely to have polar effects.
In vitro growth.
LuxS appears to be involved in the activated methyl cycle within the cell (45), and hence mutations in luxS might impact upon growth rate. Furthermore, if LuxS plays a role in quorum sensing, it may influence the growth of S. pneumoniae at higher culture densities. The growth rate of D39luxS was compared with that of wild-type D39 using various starting densities of cells resuspended in either serum broth or THY medium after growth overnight on blood agar. Experiments were carried out at least twice and showed no significant difference in the growth rates between the two strains, regardless of the culture medium or initial inoculum (data not shown).
Determination of AI-2 activity in D39luxS.
AI-2 produced by other bacterial species has been shown to induce bioluminescence in V. harveyi (3). We therefore examined the ability of both D39 and its isogenic luxS mutant to induce bioluminescence in the V. harveyi BB170 indicator strain, as described in Materials and Methods. Although the level of bioluminescence induced by the cell-free culture supernatant of D39 was markedly lower than that of the BB152 V. harveyi positive control strain, there was a significant difference in AI-2 activity between D39 and its luxS mutant. The D39luxS induced only 8 to 14% of the luminescence induced by D39.
Intranasal challenge studies.
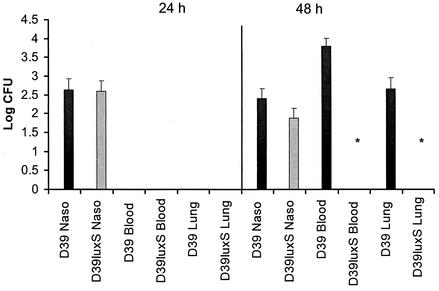
The abilities of D39luxS and wild-type D39 to colonize the mouse nasopharynx and to invade the lungs and blood were compared. Ten Swiss mice were inoculated intranasally with a 1:1 mixture of D39luxS and D39 (total inoculum, 2 × 106 CFU). Five mice were sacrificed after 24 and 48 h, and the relative numbers of D39 and D39luxS organisms in the nasopharynx, lung, and blood were determined as described in Materials and Methods (Fig. 1). There was no significant difference in the numbers of D39 and D39luxS organisms in the nasopharynx at either time point, and after 48 h, the output ratio for the wild-type and mutant strains remained similar to the input ratio. After 24 h, no bacteria were found in either the blood or the lungs. However, after 48 h, three out of the five mice sacrificed had significant levels of wild-type D39 in both the lungs and the blood (Fig. 1). Interestingly, D39luxS was not detected in either of these tissues, indicating that the mutant had a reduced capacity either to translocate from the nasopharynx and survive in the lung or to enter the bloodstream. The experiment was repeated with a higher dose of D39luxS relative to D39 (the input ratio was approximately 8:1). The ratios of D39luxS:D39 recovered from the nasopharynx after 24 and 48 h were again essentially unchanged from the input ratios (7.0:1 and 7.2:1, respectively). As in the previous experiment, neither strain was detected in lungs or blood after 24 h. However, after 48 h, two out of the five mice examined had significant levels of wild-type D39, but not D39luxS, in these tissues (result not presented). An additional experiment involving intranasal inoculation with D39 and D39luxS separately indicated that both strains were capable of colonizing the nasopharynx to a similar extent, suggesting that cross-complementation of D39luxS with AI-2 released from D39 was not essential for survival of the mutant in this niche (data not shown).
FIG. 1.
Numbers of D39 and D39luxS organisms in the nasopharynx, lungs, and blood after intranasal challenge. Ten mice were challenged with approximately 2 × 106 CFU of a 1:1 mixture of D39 and D39luxS organisms. Five mice were sacrificed after 24 or 48 h, and the number of bacteria in the various tissues was determined as described in Materials and Methods. Data are mean log10 CFU for the five mice, and the bars indicate the standard error. The asterisk denotes numbers significantly different from that of the wild type (P < 0.001) (unpaired Student's t test, two-tailed).
Intraperitoneal challenge studies.
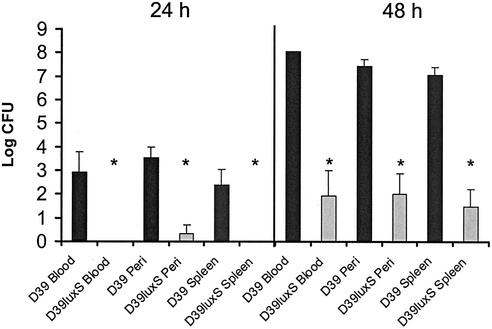
Ten BALB/c mice were injected intraperitoneally with approximately 1 × 104 CFU of a mixture of D39 and D39luxS at an input ratio of approximately 1:3. Five mice were sacrificed 24 and 48 h after challenge, and the relative numbers of organisms of the two strains were determined in peritoneal washout fluid as well as in the blood and in spleen homogenates (Fig. 2). After 24 h, no D39luxS bacteria were recovered from either the blood or spleen (the lower limit of detection was approximately 102 CFU/ml), and only one of the five mice had detectable levels of D39luxS in the peritoneal washout. However, high numbers of wild-type D39 organisms were present in the peritoneal washout of all five mice and in the blood and spleens of four. Although the D39:D39luxS input ratio was approximately 1:3, the output ratio was of the order of 1,000:1 for all sites examined. After 48 h, all five mice had high numbers of D39 organisms in all tissues examined. Interestingly, D39luxS was now detected in the peritoneal fluid, blood, and spleens of three of the five mice, albeit at levels approximately 105-fold lower than D39. Clearly D39luxS is less able than D39 to proliferate in vivo, but it is nevertheless capable of increasing its numbers between 24 and 48 h.
FIG. 2.
Numbers of D39 and D39luxS organisms in peritoneal washout, blood, and spleen after intraperitoneal challenge. Ten mice were challenged with approximately 1 × 104 CFU of a mixture of D39 and D39luxS organisms at an input ratio of approximately 1:3. Five mice were sacrificed after 24 or 48 h, and the numbers of bacteria in the various tissues were determined as described in Materials and Methods. Data are mean log10 CFU for the five mice, and the bars indicate the standard error. The asterisks denote numbers significantly different from that of the wild type (P < 0.001) (unpaired Student's t test, two-tailed).
Comparative virulence of D39 and D39luxS.
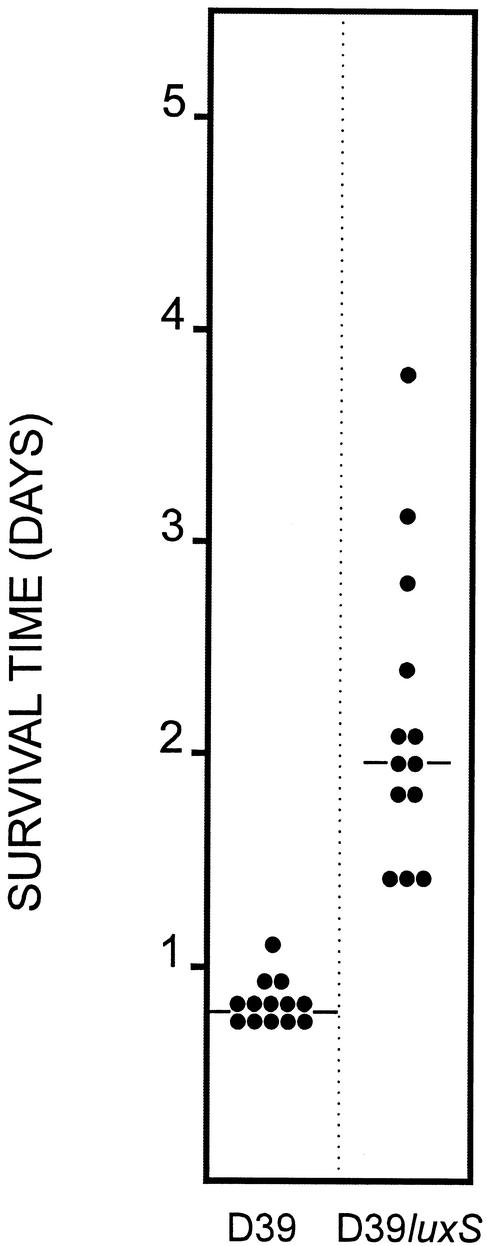
In view of the fact that D39luxS was able to increase in numbers between 24 and 48 h after intraperitoneal challenge, its capacity to kill mice was compared with that of D39. Two groups of 13 BALB/c mice were challenged by intraperitoneal inoculation with approximately 6.5 × 104 CFU of either D39luxS or D39, and their survival was followed (Fig. 3). All challenged mice succumbed, although the median survival time for the D39luxS group (44 h) was significantly longer than for the D39 group (19 h) (P < 0.001, Mann-Whitney U test). The appropriate S. pneumoniae strain was isolated from heart blood collected post mortem from each of the mice.
FIG. 3.
Survival times of mice after intraperitoneal challenge. Groups of 13 BALB/c mice were challenged with approximately 6.5 × 104 CFU of either D39 or D39luxS. Survival time for each mouse was recorded. The two horizontal lines denote the median survival times for each group.
Adherence to A549 and HEp-2 cells.
The nasopharyngeal challenge data suggesting that D39luxS and D39 could colonize the nasopharynx to similar extents but that only D39 could translocate to the lungs suggested the possibility of differences in their capacities to adhere to epithelial cells from the upper and lower respiratory tract. Accordingly, the abilities of these strains to adhere in vitro to cell lines derived from either the lung (A549) or the larynx (HEp-2) were compared. At an initial inoculum of 1.0 × 107 CFU per well, total adherence after 2 h to A549 cells was (2.95 ± 0.71) × 104 and (2.94 ± 0.53) × 104 CFU per well for D39 and D39luxS, respectively. Under the same conditions, total adherence to HEp-2 cells was (8.88 ± 0.97) × 104 and (1.05 ± 0.13) × 105 CFU per well for D39 and D39luxS, respectively. Interestingly, although there was no significant difference between the adherences of the two strains to either cell line, both D39 and D39luxS adhered more efficiently to HEp-2 cells than to A549 cells (P < 0.05).
Comparative protein expression in D39 and D39luxS.
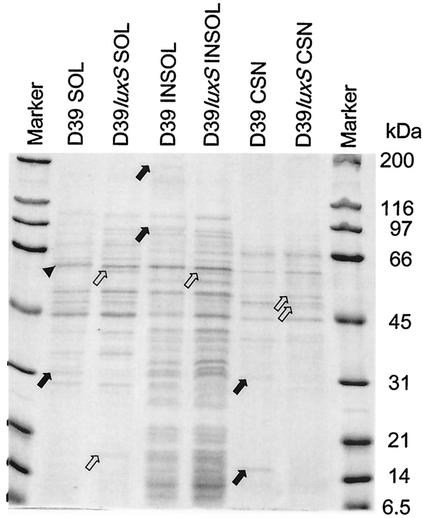
The protein expression profiles of D39 and D39luxS were then compared. The strains were grown in THY broth to an A600 of approximately 0.5, and the cells were fractionated as described in Materials and Methods. Fractions of the soluble (cytoplasmic) and insoluble (membrane and cell wall) extracts, as well as the culture supernatant, were analyzed by SDS-PAGE stained with Coomassie blue (Fig. 4). At least 10 changes were detected in the protein expression profiles for the various cell fractions between D39 and D39luxS, indicating that the mutation has pleiotropic effects. Both strains were in the opaque phase, and so these changes in expression pattern were not due to phase variation. Furthermore, both increases and decreases in staining intensity for particular protein bands, as well as one example of an apparent shift in mobility, were observed between fractions from the mutant and its parent. The apparent molecular mass of the proteins affected ranged from approximately 180 to 12 kDa. Similar differences in protein expression profile between D39 and D39luxS fractions were also seen when lower-cell-density cultures were tested (A600 of 0.1 rather than 0.5) (result not shown).
FIG. 4.
Protein profiles of subcellular fractions of D39 and D39luxS. Supernatants (100,000 × g) (SOL) and pellets (INSOL) of French pressure cell lysates of S. pneumoniae cells, as well as trichloroacetic acid-precipitated culture supernatant from the original THY cultures (CSN), were separated by SDS-PAGE (12% gel) and stained with Coomassie blue, as described in Materials and Methods. Solid arrows indicate protein species present in D39 but absent in D39luxS. Open arrows indicate protein species present in D39luxS but absent in D39. The arrowhead indicates a protein band with altered mobility between D39 and D39luxS. Molecular masses of size markers are indicated on the right of the figure.
Levels of expression of previously characterized virulence-related proteins (29, 30) were also compared by Western immunoblot analysis. D39 and D39luxS were grown in THY broth, and whole-cell extracts were separated by SDS-PAGE. Proteins were then transferred to nitrocellulose, and filters were probed with polyclonal antisera specific for pneumolysin (Ply), choline binding protein A (CbpA), neuraminidase (NanB), autolysin (LytA), pneumococcal surface protein A (PspA), and pneumococcal surface antigen A (PsaA). No difference in labeling intensity was observed between the mutant and parent strains for any of the virulence proteins tested (result not shown).
DISCUSSION
At least three major classes of quorum-sensing molecules involved in cell density-dependent gene regulation have been described in bacteria. The first to be studied were the small peptide pheromones associated with development of genetic competence in gram-positive bacteria, most notably S. pneumoniae and Bacillus subtilis (37, 38, 39, 40). In the late 1970s, various acylhomoserinelactones were shown to be involved in quorum-sensing circuits in gram-negative bacteria, and studies by Bassler and colleagues elucidated the mechanism whereby these so-called class I autoinducers (AI-1) regulated luminescence in certain Vibrio spp. (3, 7, 16, 24, 25). Both the peptide pheromones and acylhomoserinelactones exhibit species specificity as far as cognate receptor interactions are concerned, and they are viewed as mechanisms whereby bacteria sense and respond to the presence of their own species in an environmental niche (12). More recently, an additional broader-spectrum quorum-sensing molecule, AI-2, has been described. AI-2 has been implicated in regulation of virulence gene expression in several pathogens on the basis of studies involving mutagenesis of luxS, a gene required for AI-2 biosynthesis. The presence of luxS homologues in both gram-positive and gram-negative bacteria implied that AI-2-mediated quorum-sensing circuits might play regulatory roles in many species, including virulence gene regulation in important pathogens. Thus, AI-2 antagonists capable of interfering with virulence gene expression might represent a novel class of antimicrobial agents. The significantly reduced ability of D39luxS culture supernatant relative to that of wild-type D39 to elicit bioluminescence in a V. harveyi AI-2 reporter strain clearly shows that luxS is important for the production of AI-2-like activity in S. pneumoniae.
S. pneumoniae is one of the foremost human pathogens, responsible for millions of deaths each year, and increasing rates of resistance to conventional antibiotics are a major concern. The S. pneumoniae genome contains a luxS homologue, and in the present study, we have examined the involvement of this gene in pathogenesis of disease in a mouse model with a view to assessing the potential of AI-2 antagonists for treatment of pneumococcal infections. The genetic organization of the luxS region of the pneumococcal chromosome is such that insertion-duplication mutagenesis of this gene is unlikely to result in polar effects on neighboring genes. Comparison of in vitro growth rates at differing starting densities and in either serum broth or THY also did not reveal any differences between wild-type S. pneumoniae D39 and D39luxS, unlike the situation in S. pyogenes (20). Moreover, when administered intranasally, both the wild type and the mutant were capable of establishing colonization of the nasopharynx, and neither strain outcompeted the other in this niche. These findings indicate that mutation of luxS does not appear to have any detrimental effect on basic cellular metabolic processes required for growth in vitro or in the nasopharynx. However, unlike D39, D39luxS was not found in either the lungs or the blood of mice after intranasal challenge with the mixture of strains. This may be a consequence of inability to translocate from the nasopharyngeal niche or an inability to resist host defenses (e.g., polymorphonuclear leukocytes and alveolar macrophages) in the deeper tissues. Whether this defect is a direct consequence of inability to adjust gene expression in response to AI-2 is uncertain. Given that the wild type and mutant strains cocolonized the nasopharynx at similar densities, D39luxS may have been able to respond to exogenous AI-2 released from D39. However, when the strains were administered individually, the numbers of D39luxS and D39 organisms recovered from the nasopharynx were not significantly different.
The higher fitness of D39 relative to D39luxS was also evident when the two strains were coadministered intraperitoneally. The wild-type strain readily outcompeted the mutant, and after 24 h, only D39 could be found in the blood or spleen. Nevertheless, the numbers of D39luxS organisms in peritoneal washout, blood, and spleen increased by 48 h, although the numbers of D39 organisms were at least 105-fold higher at all three sites. The capacity of D39luxS to survive and proliferate after intraperitoneal challenge was not dependent on coadministration of D39, because all mice challenged with D39luxS alone died within 4 days. However, all mice challenged with the same dose of D39 succumbed within 28 h, indicating that the wild-type strain is significantly more virulent.
This is the first report that mutagenesis of luxS directly affects virulence of a gram-positive pathogen. A recent study by Lyon et al. (20) reported both positive and negative effects on expression of virulence-related genes in a luxS mutant of S. pyogenes, but whether these have a net impact on virulence in animal models is not known. In the present study, we did not observe differences in expression of any of six previously characterized virulence-related proteins, but both increases and decreases in levels of expression of at least 10 unidentified proteins in various cell fractions were evident from SDS-PAGE analysis of D39 and D39luxS. Some of these changes in protein expression may account for the altered virulence phenotype of D39luxS. Additional changes may also be detectable using higher-resolution separation protocols (e.g., two-dimensional gels) and more sensitive detection techniques such as silver staining. It also seems likely that proteomic analysis of the two strains will identify additional pneumococcal proteins directly involved in pathogenesis of disease. Notwithstanding the clear contribution of luxS to pneumococcal virulence, it is evident from this study that disruption of the gene does not completely attenuate S. pneumoniae. This unequivocal finding argues against the likely efficacy of AI-2 antagonists for treatment of pneumococcal disease.
Acknowledgments
This work was supported by grants from the National Health and Medical Research Council of Australia and the Meningitis Research Foundation (UK).
Editor: A. D. O'Brien
REFERENCES
- 1.Avery, O. T., C. M. MacLeod, and M. McCarty. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a deoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79:137-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signaling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 4.Berry, A. M., J. Yother, D. E. Briles, D. Hansman, and J. C. Paton. 1989. Reduced virulence of a defined pneumolysin-negative mutant of Streptococcus pneumoniae. Infect. Immun. 57:2037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Burgess, N. A., D. F. Kirke, P. Williams, K. Winzer, K. R. Hardie, N. L. Meyers, J. Aduse-Opoku, M. A. Curtis, and M. Camara. 2002. LuxS-dependent quorum sensing in Porphyromonas gingivalis modulates protease and haemagglutinin activities but is not essential for virulence. Microbiology 148:763-772. [DOI] [PubMed] [Google Scholar]
- 7.Cao, J. G., and E. A. Meighen. 1989. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J. Biol. Chem. 264:21670-21676. [PubMed] [Google Scholar]
- 8.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 9.Douglas, R. M., J. C. Paton, S. J. Duncan, and D. Hansman. 1983. Antibody response to pneumococcal vaccination in children younger than five years of age. J. Infect. Dis. 148:131-137. [DOI] [PubMed] [Google Scholar]
- 10.Freeman, J. A., and B. L. Bassler. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31:665-677. [DOI] [PubMed] [Google Scholar]
- 11.Freeman, J. A., and B. L. Bassler. 1999. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J. Bacteriol. 181:899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuqua, C., S. C. Winans, and E. P. Goldberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garenne, M., C. Ronsmans, and H. Campbell. 1995. The magnitude of mortality from acute respiratory infections in children under 5 years in developing countries. World Health Stat. Q. 46:180-191. [PubMed] [Google Scholar]
- 14.Giammarinaro, P., and J. C. Paton. 2002. Role of RegM, a homologue of the catabolite repressor protein CcpA in virulence of Streptococcus pneumoniae. Infect. Immun. 70:5454-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giron, J. A., A. G. Torres, E. Freer, and J. B. Kaper. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361-379. [DOI] [PubMed] [Google Scholar]
- 16.Hastings, J. W., C. J. Potrikas, S. C. Gupta, M. Kurfurst, and J. C. Makemson. 1985. Biochemistry and physiology of bioluminescent bacteria. Adv. Microb. Physiol. 26:235-291. [DOI] [PubMed] [Google Scholar]
- 17.Jones, S., B. Yu, N. J. Bainton, M. Birdsall, B. W. Bycroft, S. R. Chhabra, A. J. Cox, P. Golby, P. J. Reeves, S. Stephens, et al. 1993. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 12:2477-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein, D. L. 2000. Pneumococcal disease and the role of conjugate vaccines, p. 467-477. In A. Tomasz (ed.), Streptococcus pneumoniae molecular biology and mechanisms of disease. Mary Ann Liebert Inc., New York, N.Y.
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 21.Manefield, M., L. Harris, S. A. Rice, R. de Nys, and S. Kjelleberg. 2000. Inhibition of luminescence and virulence in the black tiger prawn (Penaeus monodon) pathogen Vibrio harveyi by intercellular signal antagonists. Appl. Environ. Microbiol. 66:2079-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrison, D. A., and M. S. Lee. 2000. Regulation of competence for genetic transformation in Streptococcus pneumoniae: a link between quorum sensing and DNA processing genes. Res. Microbiol. 151:445-451. [DOI] [PubMed] [Google Scholar]
- 23.Musher, D. M. 1992. Infections caused by Streptococcus pneumoniae: clinical spectrum, pathogenesis, immunity and treatment. Clin. Infect. Dis. 14:801-809. [DOI] [PubMed] [Google Scholar]
- 24.Nealson, H. K., and J. W. Hastings. 1979. Bacterial bioluminescence: its control and ecological significance. Microbiol. Rev. 43:496-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nealson, H. K., T. Platt, and J. W. Hastings. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novick, R. P., and T. W. Muir. 1999. Virulence gene regulation by peptides in staphylococci and other Gram-positive bacteria. Curr. Opin. Microbiol. 2:40-45. [DOI] [PubMed] [Google Scholar]
- 27.Ohtani, K., H. Hayashi, and T. Shimizu. 2002. The luxS gene is involved in cell-cell signaling for toxin production in Clostridium perfringens. Mol. Microbiol. 44:171-179. [DOI] [PubMed] [Google Scholar]
- 28.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 29.Paton, J. C., P. W. Andrew, G. J. Boulnois, and T. J. Mitchell. 1993. Molecular analysis of the pathogenicity of Streptococcus pneumoniae: the role of pneumococcal proteins. Annu. Rev. Microbiol. 47:89-115. [DOI] [PubMed] [Google Scholar]
- 30.Paton, J. C., A. M. Berry, and R. A. Lock. 2000. Molecular analysis of putative pneumococcal virulence proteins, p. 261-270. In A. Tomasz (ed.), Streptococcus pneumoniae molecular biology and mechanisms of disease: update for the 1990s. Mary Ann Liebert, Inc., New York, N.Y.
- 31.Paton, J. C., I. R. Toogood, R. Cockington, and D. Hansman. 1986. Antibody response to pneumococcal vaccine in children aged 5 to 15 years. Am. J. Dis. Child. 140:135-138. [DOI] [PubMed] [Google Scholar]
- 32.Ren, D., J. J. Sims, and T. K. Wood. 2001. Inhibition of biofilm formation and swarming of Escherichia coli by (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone. Environ. Microbiol. 3:731-736. [DOI] [PubMed] [Google Scholar]
- 33.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 34.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 36.Talbot, U., A. W. Paton, and J. C. Paton. 1996. Uptake of Streptococcus pneumoniae by respiratory epithelial cells. Infect. Immun. 64:3772-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomasz, A. 1965. Control of the competent state in Pneumococcus by a hormone-like cell product: an example for a new type of regulatory mechanism in bacteria. Nature 208:155-159. [DOI] [PubMed] [Google Scholar]
- 38.Tomasz, A. 1966. Model for the mechanism controlling the expression of competence state in pneumococcus cultures. J. Bacteriol. 29:1050-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomasz, A., and R. D. Hotchkiss. 1964. Regulation of the transformability of pneumococcal cultures by macromolecular cell products. Proc. Natl. Acad. Sci. USA 29:480-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tortosa, P., and D. Dubanau. 1999. Competence for transformation: a matter of taste. Curr. Opin. Microbiol. 2:588-592. [DOI] [PubMed] [Google Scholar]
- 41.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuomanen, E. I., R. Austrian, and H. R. Masure. 1995. The pathogenesis of pneumococcal infections: correlation of clinical events with molecular mechanisms. N. Engl. J. Med. 332:1280-1284. [DOI] [PubMed] [Google Scholar]
- 43.Williams, P., M. Camara, A. Hardman, S. Swift, D. Milton, V. J. Hope, K. Winzer, B. Middleton, D. I. Pritchard, and B. W. Bycroft. 2000. Quorum sensing and the population-dependent control of virulence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:667-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winzer, K., K. R. Hardie, and P. Williams. 2002. Bacterial cell-to-cell communication: sorry, can't talk now—gone to lunch! Curr. Opin. Microbiol. 5:216-222. [DOI] [PubMed] [Google Scholar]
- 45.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 46.Withers, H., S. Swift, and P. Williams. 2001. Quorum sensing as an integral component of gene regulatory networks in Gram-negative bacteria. Curr. Opin. Microbiol. 4:186-193. [DOI] [PubMed] [Google Scholar]
- 47.World Health Organization. 1997. Global program for vaccines and immunization (Vaccine Research and Development). Report of the technical review group meeting. July 1997-June 1998: achievements and plans of activities: meningococcal and pneumococcal disease vaccines, p. 26-30. World Health Organization, Geneva, Switzerland.
- 48.Wu, H. Y., A. Virolainen, B. Mathews, J. King, M. W. Russell, and D. E. Briles. 1997. Establishment of a Streptococcus pneumoniae nasopharyngeal colonization model in adult mice. Microb. Pathog. 23:127-137. [DOI] [PubMed] [Google Scholar]
- 49.Yother, J., L. S. McDaniel, and D. E. Briles. 1986. Transformation of encapsulated Streptococcus pneumoniae. J. Bacteriol. 168:1463-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]