Abstract
Local chromatin compaction undergoes dynamic perturbations to regulate genetic processes. To address this, the direct measurement of the fluidity of chromatin structure is carried out in single live cells using steady-state anisotropy imaging and polarization modulation microscopy. Fluorescently tagged core and linker histones are used to probe different structural aspects of chromatin compaction. A graded spatial heterogeneity in compaction is observed for the chromatin besides the distinct positional ordering of core and linker histones. These spatio-temporal features are maintained by active processes and perturbed during death. With cell cycle, the distribution in compaction heterogeneity continually changes maximizing during M-G1 transition where it displays bimodal behavior. Such measurements of spatio-temporal chromatin fluidity could have broader implications in understanding chromatin remodeling within living cells.
INTRODUCTION
The DNA in eukaryotic cells is present in the nucleus as chromatin, a structure formed by the wrapping of DNA around highly basic octameric proteins called histones and the further organization of this basic repeating unit (1) to various levels of folded structures. In vivo the chromatin has been shown to be a dynamic structure both in the context of its local packing and nuclear positioning (2). An interphase nucleus has its chromatin present as the silent heterochromatin and the active euchromatin. Though it is fairly established that heterochromatin has tighter folding, it is not clear how distinctly different they are from each other in terms of packing and what are the spatial gradients in compaction that separate them. This is an important issue to resolve because it is now clear that the spatio-temporal regulation of genetic processes is strongly linked to the dynamic architectural reorganization of the chromatin (3–5). The spatial regulation demands heterogeneity in compaction whereby regions of the chromatin are appropriately either fluidic enough to relocate within the nucleus, or rigid enough to assert structural hindrance. For temporal regulation of DNA accessibility, the folding needs to be modulated in real time to provide the appropriate kinetic barrier. In this article the chromatin compaction has been mapped within a single live cell and the changes brought about by cellular perturbation and growth phase have been probed with a view to understand how the functional state of the cell links to the alterations in chromatin's compaction profile.
MATERIALS AND METHODS
Cell culture
The model systems used are Drosophila larvae salivary gland cells and HeLa cells. Salivary gland cells (with polytene chromosomes) were derived from the Drosophila larvae (transgenic flies bearing histone H2B-EGFP). The C-terminal sequence of histone protein (H2B) is genetically fused with a reporter gene coding for enhanced green fluorescent protein (EGFP). The salivary glands, from the third instar larvae, are dissected in Ringer's medium (or medium containing 600 mM NaCl for control experiments) using standard protocols. In brief, under an inspection microscope, the middle portion of the larvae is held using one microneedle. Another pair of fine-tipped forceps is used to pull out the mouth region of the larvae. The salivary glands are dissected from this open preparation and are then transferred onto a clean microscope No. 1 coverslip in the Ringer's medium and sealed using another coverslip. Care is taken to ensure that the glands are intact during the sample preparation procedure and the samples are stable for microscopic observations.
HeLa cells were transfected with an expression vector encoding histone H2B-EGFP, H1.1-EGFP, or plain EGFP. The promoters driving the expression were EF1α for H2B-EGFP and CMV for H1.1 and EGFP vectors. Stables of HeLa-H2B-EGFP and HeLa-EGFP were prepared by antibiotic selection using blasticidin and G418, respectively. HeLa-H1.1-EGFP was transiently transfected 12–16 h before the experiment. Cells were grown in DMEM (Gibco, Life Technologies, Paisley, UK) supplemented with 10% FBS (Gibco), glutamine and penicillin/streptavidin at 37°C, and 5% CO2 in glass-bottom petri dishes. Before imaging, the medium was replaced by M1 medium (150 mM NaCl, 20 mM HEPES, 1 mM MgCl2) supplemented with 1% glucose.
Cell preparation
For depleting adenosine triphosphate (ATP), dishes at midlog phase were taken and first washed with phosphate buffered saline (PBS) after which they were treated with 10 mM NaN3 (Sigma, St. Louis, MO) and 6 mM 2-deoxy-D-glucose (Sigma) in M1 without glucose and were left in the incubator at 37°C for 1 h after which their medium was replaced by the imaging medium. For inducing apoptosis cells were treated with 10 μM of staurosporine (Sigma) and used for imaging after 4 h of incubation. Cells were again washed and M1 medium was used for imaging. For cell phase experiments, dishes were used with cells at midlog phase of their growth and were further grown on the microscope using an incubator (Olympus, Tokyo, Japan) whose temperature was maintained at 37°C and humidity and CO2 (∼5%) maintained optimal for cell growth. Cells were grown in nonphenol red DMEM supplemented with 5% FBS.
Imaging
Anisotropy images were captured on a Nikon/Olympus (Tokyo, Japan) microscope with 100×/1.4 NA objective and images captured with ICCD cameras (Roper Scientific, Duluth, GA). Mercury arc source was used for the excitation light, which was then selected for vertical polarization using a sheet polarizer (Melles Griot, Carlsbad, CA). The light collected was split into its parallel and perpendicular polarization components using a polarizing beam splitter (Melles Griot) or by swapping between two polarizers (Melles Griot) parallel and perpendicular to the excitation. Images were captured using V++ Digital Optics (Auckland, New Zealand) software and analyzed using LabView. The cell population imaged is asynchronous but excludes mitosis altogether except for the growth phase tracking experiment. For polarization modulation experiments, linearly polarized light from an Argon-ion laser (50 mW, 488 nm, Spectra-Physics, Mountain View, CA) is rotated through 180° using an electro-optic modulator (EOM, NewFocus) followed by a quarter wave plate (Melles Griot) and directed to the 100×/1.4 NA objective in a microscope (IX70, Olympus) for confocal excitation. The fluorescence collected from the sample using a 50-μm pinhole is selected for parallel polarization using a polarizer (Melles Griot) and detected using an avalanche photodiode (EG&G, Wellesey, MA). For polarization modulation, the voltage input to the EOM is modulated in steps with an angular resolution of 2.8° or 11.25° and moved 180° each scan (see Supplementary Material). At each angle the fluorescence is measured using NIDAQ and LabView and the process is repeated for all angles. The raw fluorescence data are analyzed by first subtracting a baseline to account for the polarization-dependent response of the detection system and then the base-subtracted data are checked for peaks. It is observed that the results are the same for both angular resolutions (2.8° and 11.25°). However, to save time per scan so as not to have artifacts due to chromatin movements, all experiments reported were done at 11.25° angular resolution. Controls were done on rhodamine crystal and EGFP solution to establish the method of order parameterization by peak heights (see Supplementary Material, Fig. S3).
RESULTS AND DISCUSSIONS
Chromatin is heterogeneously packed in the nucleus
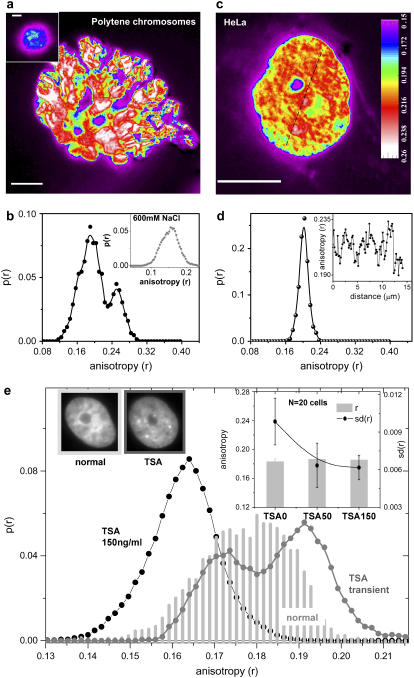
To understand the levels of compaction present in the chromatin, fluorescence anisotropy (6,7) imaging was performed for HeLa and polytene cells expressing enhanced green fluorescent protein tagged to core histone protein—H2B-EGFP (8). Fluorescence anisotropy (r) measures the rotational mobility of the fluorophore, which in this case is EGFP tagged to H2B. Here, anisotropy reflects the rotational mobility of H2B and therefore also of the local chromatin structure to which H2B is bound. The anisotropy image is calculated from individual parallel and perpendicular polarized images  and plotted with color coding to differentiate various levels of fluidic regions. Highly fluidic regions (lower anisotropy) are violet to blue in color and compacted regions are white (higher anisotropy). For all the above experiments it was ensured that the changes in anisotropy values were not an artifact of variations in fluorescence intensity values (Supplementary Material, Fig. S1). It is demonstrated using polytene chromosomes from Drosophila larvae salivary glands (Fig. 1 a; Fig. S1 a) that anisotropy can distinguish between the two states of chromatin assembly showing distinct band and interband regions (9). The corresponding distribution of anisotropy of an internal region (Fig. 1 b) exhibits a clear bimodal distribution confirming the existence of spatially separated euchromatin (first peak) and heterochromatin (second peak). In a high-salt environment (600 mM NaCl) due to screening of electrostatic interactions, the chromatin higher order structure is known to break down (10). Under such conditions the disappearance of bands (Fig. 1 a, inset) is observed besides an overall reduction of the mean anisotropy (Fig. 1 b, inset) indicating global decondensation of the chromatin and thereby further confirming compaction measurements by fluorescence anisotropy (Supplementary Material, Fig. S2). HeLa cells at interphase, unlike polytene chromosomes, do not have regular spatial patterning of the chromatin but are known to show appreciable spatial segregation by highly regulated mechanisms. Fig. 1 c is a snapshot of anisotropy of a H2B-EGFP in an interphase HeLa cell and the corresponding anisotropy line scan shown in Fig. 1 d (inset) indicates that the packing is indeed nonuniform at micrometer length scales and visible as heterogeneous structures in Fig. 1 c. However, the conspicuous absence of a bimodal distribution in Fig. 1 d, suggests that the chromatin takes a repertoire of intermediate compaction states perhaps in dynamic equilibrium rather than distinctly open and tight states as in the case of polytene cells.
and plotted with color coding to differentiate various levels of fluidic regions. Highly fluidic regions (lower anisotropy) are violet to blue in color and compacted regions are white (higher anisotropy). For all the above experiments it was ensured that the changes in anisotropy values were not an artifact of variations in fluorescence intensity values (Supplementary Material, Fig. S1). It is demonstrated using polytene chromosomes from Drosophila larvae salivary glands (Fig. 1 a; Fig. S1 a) that anisotropy can distinguish between the two states of chromatin assembly showing distinct band and interband regions (9). The corresponding distribution of anisotropy of an internal region (Fig. 1 b) exhibits a clear bimodal distribution confirming the existence of spatially separated euchromatin (first peak) and heterochromatin (second peak). In a high-salt environment (600 mM NaCl) due to screening of electrostatic interactions, the chromatin higher order structure is known to break down (10). Under such conditions the disappearance of bands (Fig. 1 a, inset) is observed besides an overall reduction of the mean anisotropy (Fig. 1 b, inset) indicating global decondensation of the chromatin and thereby further confirming compaction measurements by fluorescence anisotropy (Supplementary Material, Fig. S2). HeLa cells at interphase, unlike polytene chromosomes, do not have regular spatial patterning of the chromatin but are known to show appreciable spatial segregation by highly regulated mechanisms. Fig. 1 c is a snapshot of anisotropy of a H2B-EGFP in an interphase HeLa cell and the corresponding anisotropy line scan shown in Fig. 1 d (inset) indicates that the packing is indeed nonuniform at micrometer length scales and visible as heterogeneous structures in Fig. 1 c. However, the conspicuous absence of a bimodal distribution in Fig. 1 d, suggests that the chromatin takes a repertoire of intermediate compaction states perhaps in dynamic equilibrium rather than distinctly open and tight states as in the case of polytene cells.
FIGURE 1.
(a) Color-coded anisotropy map of polytene chromosomes extracted from salivary glands of Drosophila larvae; (inset) polytene chromosomes in 600 mM NaCl; bar, 10 μm. (b) Anisotropy distribution for polytene chromosomes; (inset) anisotropy distribution of polytene chromosomes treated with 600 mM NaCl. (c) Color-coded anisotropy map of interphase HeLa cell nucleus; bar, 10 μm. (d) Anisotropy distribution of interphase HeLa cell nucleus; (inset) line scan of anisotropy values across the HeLa cell nucleus shown as the dashed line in Fig. 1 c. (e) Anisotropy distributions of normal cells as well as cells treated with TSA. (Inset) Bar graph for mean anisotropy (r) and standard deviation (sd(r)) of anisotropy across single nucleus averaged over 20 cells for normal and TSA-treated cells: TSA50 (50 ng/ml TSA, 4 h) and TSA150 (150 ng/ml TSA, 4 h) cells; representative intensity images of normal and TSA-treated (150 ng/ml, 4 h) cells.
Chromatin compaction dynamics is brought about by the constant impact of various remodeling enzymes—acetylation being one such example causing overall decondensation (11). To test this, a small molecule inhibitor of histone deacetylase (HDAC) is used, which blocks deacetylation reaction resulting in a buildup of acetylation and decondensation of chromatin (12,13). Within 4 h of trichostatin-A (TSA) (150 ng/ml) treatment there is a lowering of heterogeneity as well as a transient exposure of heterochromatin perhaps because their acetylation is much slower compared to the intermediate structures that decondense and spread out at relatively faster timescales. Fig. 1 e shows anisotropy distributions for various cases where the chromatin is either decondensed and less heterogeneous or shows a second peak emerging at a higher anisotropy. However, heterochromatin exposure is accompanied by the global unfolding of intermediate structures leading to a lowering of overall heterogeneity (Fig. 1 e, inset). It was also observed that the response of the cells to TSA is highly stochastic and therefore responsible for the higher cell-to-cell variability. To scrutinize the biological significance of the observed gradation or heterogeneity in compaction, the next investigation addresses the effect of perturbations to cell state either by altering the ATP levels or inducing cell death.
Local chromatin fluidity is sensitive to ATP depletion or cell death
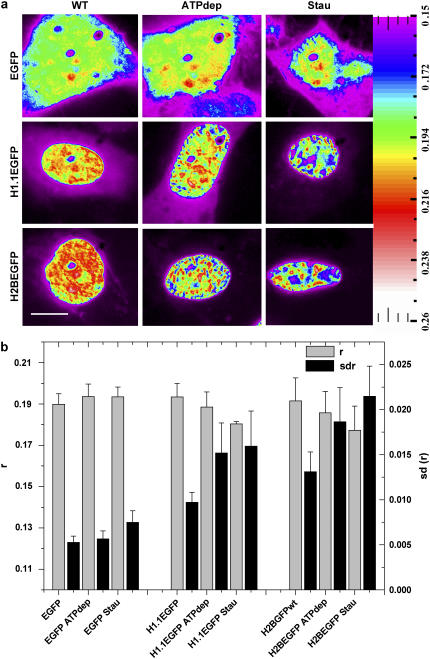
The dependence of packing heterogeneity on the active state of the cell was studied by using both core histones (H2B-EGFP) and linker histones (H1.1-EGFP) as probes for the chromatin structure. Although H2B is an integral part of chromatin for all levels of folding, H1.1 and other linker subtypes are known to stabilize higher order structures of the chromatin. Therefore, comparing heterogeneity maps observed with H2B and H1.1 should contrast the contribution of chromatin structural perturbations as opposed to the individual responses of the probes. Spatial heterogeneity was quantified by plotting anisotropy line scans and distributions (Supplementary Material, Fig. S1) for the nucleus of the cells imaged. The mean (r) and standard deviation (sd(r)) of the anisotropy values were obtained and compared between various systems. Because different cells of the same population have variability in their compaction status originating from the dissimilar growth phases as well the intrinsic causes, trends in r and sd(r) reported here are single cell features observed for the whole population. The heterogeneity maps observed with H2B-EGFP, H1.1-EGFP, and EGFP molecules evidenced that the histones are packed into micron-scale substructures in the nucleus, as shown in Fig. 2 a whereas the noninteracting EGFP is found diffused in the nucleus. H2B-EGFP shows various rotational mobilities in the nucleus with a relatively narrow standard deviation of anisotropy (H2B-EGFPsdWT = 0 .013 (6.8% deviation)). In contrast, the map of the linker histone, H1.1-EGFP, showed a less heterogeneous environment (H1.1-EGFPsdWT = 0.010 (5% deviation)). The lower sd(r) for linker proteins is consistent with the dynamic interaction model of the linker histones (14,15); whereas the core histones are mostly immobile, a large fraction of linker histones are under continuous transit from one binding site to another. To find out the contribution to the heterogeneity arising due to the nuclear architecture itself, histone heterogeneity maps were contrasted with free EGFP, a noninteracting molecule in the cell nucleus. Free EGFP shows negligible heterogeneity as compared to the histones (EGFPsdWT = 0.005 (2.7% deviation)). The spatial compaction for all three cases is compared by plotting both the mean anisotropy value (r) over the nucleus as well as the standard deviation (sd(r)) (Fig. 2 b). The values reported are averaged over 45 cells each and the error bars correspond to the standard deviation corresponding to the cell-to-cell variations in the quantities plotted. Fig. 2 b shows increasing sd(r) from EGFP to H1.1-EGFP to H2B-EGFP cells.
FIGURE 2.
(a) Color-coded anisotropy map of HeLa-EGFP, HeLa-H1.1-EGFP, and HeLa-H2B-EGFP cells when untreated (WT), ATP depleted (ATP dep), and treated with staurosporine (Stau). (b) Bar graph representation of mean anisotropy (r) and standard deviation of anisotropy (sd(r)) of all pixels inside a single nucleus averaged over 45 different cells in each case. Bar, 10 μm.
A number of chromatin remodeling proteins use ATP to bring about conformational changes to the chromatin (16). The differences in chromatin compaction heterogeneity were investigated in the presence and absence of ATP to understand if the heterogeneity also is actively configured. Upon depleting the cells of ATP (17), the micron-scale substructures as observed previously form larger clumps (Fig. 2), that show a broader standard deviation (H2B-EGFPsdATPdep = 0.019 (10% deviation)) indicating an increase in compaction heterogeneity with an overall loosening of the chromatin (Supplementary Material, Fig. S1). The heterogeneity map observed by labeling the linker histone, H1.1-EGFP, also shows an increase in the heterogeneity (H1.1-EGFPsdATPdep = 0.012 (8% deviation)) accompanied by the redistribution of the proteins into bigger domains. However for plain EGFP, the heterogeneity still remains at a much lower level (EGFPsdATPdep = 0.006 (2.9% deviation)) with no specific structures emerging—indicating that the chromatin gets reorganized causing the changes in both the length scales involved and the heterogeneity of the labeled histones.
Apoptosis is yet another process where the state of chromatin is drastically altered and the chromatin is known to both fragment as well as aggregate (18). When apoptosis is induced (10 μM staurosporine), the heterogeneity is altered to a state where regions on the chromatin show anisotropy values that are distinctly either low or high (Fig. 2) (H2B-EGFPsdStau = 0.022 (12% deviation)). H1.1-EGFP showed less increase (H1.1-EGFPsdStau = 0.016 (8.4% deviation)) and for plain EGFP (EGFPsdStau = 0.008 (3.8% deviation)) the values remain low though higher than the untreated population. Once again the disruption of the gradation in compaction is observed in consequence to the activation of the programmed death machinery of the cell. It is therefore inferred that the maintenance of the normal chromatin heterogeneity requires active cellular processes that keep a balance between opened up and closed structures by creating a large number of intermediate structures. Fluorescence anisotropy gives here the range of the chromatin compaction levels present in the cell. Core histones due to their greater interaction with DNA and higher packing in the chromatin show maximal heterogeneity in case of structural perturbations. The higher mobility of linker histones keeps them less heterogeneously distributed in the nucleus than core histones but their binding to the chromatin fiber at specific linker DNA coordinates results in the spatial distribution still partially resembling that of chromatin. However, while anisotropy gives the compaction states it is essential to exploit different approaches to probe the positional order. For this, a fluorescence polarization-based method has been used next to address the long-range positional organization of the compacted chromatin state.
Positional order of local chromatin assembly and its remodeling
Various imaging techniques suggest that the packing of chromatin within the cell nucleus may be associated with a degree of long-range nucleosome ordering. To address this, a fluorescence polarization-based method has been devised to quantify the extent of positional ordering of chromatin. Long-range ordering of the chromatin should in principle result in a spatially periodic arrangement of the associated fluorescently labeled histones, H2B-EGFP and H1.1-EGFP. Thus, the dipole of the EGFP tag should ideally reflect the positional ordering by having a different dipole angle distribution from a randomly oriented sample. The logic of the technique used here is to measure signatures of nonrandom angle distribution from a small volume by varying the excitation polarization angles in steps from 0° to 180° (Fig. S3 a) and measuring the fluorescence response at each angle (Fig. 3 a). Experiments on model systems as well as numerical calculations as outlined in the Supplementary Material demonstrate that whereas a random sample or a sample with a unique dipole orientation responds monotonically to the polarization modulation (19), a sample with semiordered dipole orientations shows nonmonotonic behavior where certain dipole orientations are more favored, resulting in enhanced fluorescence for specific excitation polarization angles. The amplitude of these fluorescence peaks, therefore, are a measure of the number of ordered dipoles. Normalizing with the net intensity yields an approximate ratio of ordered dipoles to random ones. The normalized peak heights averaged over ∼100 scans are plotted for each cell type for any comparison. Measuring similar responses from live cell nuclei involved repetitions over different regions of a given cell nucleus as well as on nuclei of different cells. Fig. 3 b shows the mean height of fluorescence peaks for H2B-EGFP, H1.1-EGFP, and free EGFP in normal cells, ATP-depleted cells and apoptotic cells plotted as a bar graph where the error bars correspond to the standard deviation showing cell-to-cell variability of the mean peak height over various trials. It is observed that in normal cells, the core histones show the highest mean peak height (Fig. 3 b) reflective of maximal long-range ordering of these particles. The linker histones, however, show less ordering again in line with their dynamic association with chromatin. In ATP-depleted cells, H2B-EGFP shows a large change in peak height confirming the role of active processes in the maintenance of the orientational ordering. On the other hand, H1.1-EGFP shows a lesser change in peak height in ATP-depleted cells indicating that it is less sensitive to changes in long-range ordering than H2B-EGFP. Upon induction of apoptosis, H2B-EGFP cells show both regions of high and low order. However, the mean peak height is relatively reduced (Fig. 3 b), while increasing the statistically significant standard deviation. H1.1-EGFP (Fig. 3 b), in contrast, shows negligible alteration in the positional ordering whereas free EGFP still showed low long-range ordering. The EGFP data show that measurements are sensitive to the interaction of the chromatin with the probes (H1.1-EGFP, H2B-EGFP). Core histones interact maximally with the DNA in chromatin and hence reflect the chromatin positional ordering with the greatest fidelity, thereby being sensitive to reagents that perturb chromatin structure. Given that the chromatin exhibits spatial heterogeneity in its packing as evidenced by steady-state anisotropy measurements next, the temporal aspect of chromatin reorganization was explored. In particular the compaction states achieved by the nucleus have been mapped at different time points of its cell cycle.
FIGURE 3.
(a) Schematic of polarization modulation experiment. (b) Mean peak height and standard deviation (error bars) for untreated (WT) HeLa-H2B-EGFP, HeLa-H1.1-EGFP, HeLa-EGFP, and on ATP depletion (ATP dep) as well as staurosporine (Stau) treatment.
Transitions in local chromatin fluidity during cell cycle
Cell-cycle progression and cell-cycle checkpoints are guided by dynamic changes in gene expression that require the concerted actions of chromatin remodeling (20) and transcription machinery. This involves large-scale chromatin reorganization during various stages in the cell cycle (21). The aforementioned methods were applied to measure chromatin compaction in a single cell as it proceeds from prometaphase or late anaphase into G1-interphase. Steady-state time-lapse anisotropy images of a single HeLa cell expressing H2B-EGFP have been captured during different growth phases. Fig. 4 b show anisotropy snapshots of a single cell undergoing mitosis and reaching the interphase. The distributions of observed anisotropy values (Fig. 4 a) show the transitions from a bimodal distribution with two distinct high-compaction states to a single state at late interphase. After the completion of mitosis both the cells were individually followed in time and the data for the left cell have been reported in Fig. 4 a where normalized distribution of pixel anisotropy values over the whole nucleus has been plotted. Though the compaction levels are much higher initially, so is the heterogeneity in the compaction, which increases during the transition to the interphase and decreases as interphase is attained. The high heterogeneity observed is evidence of large conformational freedom possible for the mitotic chromatin and the increase in heterogeneity during late M phase is consistent with the increased exchange rates observed for core and linker histones (22). The two distinct compaction states at early G1 represent an unfolding intermediate (second peak) and the partially decondensed chromatin (first peak) consistent with the chromonema model for interphase chromatin (23). Distributions for the entire range have been shown in the Supplementary Material (Fig. S4). Measuring compaction details of the nucleus provides the handle to be able to directly study differential chromatin dynamics either across the nucleus or in time.
FIGURE 4.
(a) Anisotropy distributions of H2B-EGFP from metaphase to interphase in a single HeLa cell nucleus. Anaphase (t = 0:17 min) onwards the anisotropy distributions of the left daughter cell has been plotted. (b) Panel shows color-coded anisotropy snapshots at corresponding time points. Bar, 10 μm.
CONCLUSIONS
Chromatin in the interphase cell nucleus goes through cycles of packing transitions to accommodate a number of DNA-related processes (24). A close relationship exists between such large-scale chromatin folding and genetic activity (25–27). To be able to understand the basic rules that govern chromatin assembly mediated regulation it is important to map globally the relative activity and chromatin compaction state dynamics (28–30). This article reveals fluidity maps of chromatin assembly within the nucleus and it then becomes visually evident from these maps that such a spatial distribution is not only maintained actively by the cell, but also altered dynamically with histone deacetylase inhibitors and biological cues. Using a new polarization modulation measurement, signatures of the positional ordering of histones are found in the nucleus; the measurements on large-scale chromatin remodeling induced by ATP depletion and apoptosis indicate that one can extend the technique to follow the dynamics in a live cell undergoing various chromatin-related processes. Measurements of positional ordering of chromatin to date have been done only on in vitro samples under harsh treatments. Here it is shown that fluidity as well as the order of the assembly are crucial for the chromatin-mediated regulation and indeed can be probed spatio-temporally in a live cell nucleus. Such compaction states of the chromatin are under continuous evolution during its cell cycle and perhaps key to the maintenance of the right cell-cycle dependent gene expression patterns. The development of decondensation of the chromatin has been mapped following it through mitosis and interphase and it is observed that the transitions are noisy and get stabilized at interphase. We suggest that such live cell measurements of compaction/order in the genome are crucial to further investigate in detail the basic physical mechanisms of chromatin-related processes. Experiments in the lab are in progress to dissect the mechanistic principles that underlie chromatin remodeling and transcription control within living cells.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
References
- 1.Luger, K., A. W. Maeder, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 389:251–260. [DOI] [PubMed] [Google Scholar]
- 2.Marshall, W. F. 2002. Order and disorder in the nucleus. Curr. Biol. 12:R185–R192. [DOI] [PubMed] [Google Scholar]
- 3.Peterson, C. L. 2002. Chromatin remodeling enzymes: taming the machines. EMBO Rep. 3:319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spector, D. L. 2003. The dynamics of chromosome organization and gene regulation. Annu. Rev. Biochem. 72:573–608. [DOI] [PubMed] [Google Scholar]
- 5.Huebert, D. J., and B. E. Bernstein. 2005. Genomic views of chromatin. Curr. Opin. Genet. Dev. 15:476–481. [DOI] [PubMed] [Google Scholar]
- 6.Lakowicz, J. R. 1983. Principles of Fluorescence Spectroscopy. Plenum Press, New York.
- 7.Sinha, D. K., D. Bhattacharya, B. Banerjee, F. M. Hameed, and G. V. Shivashankar. 2004. Development of single-molecule tracking confocal microscope combined with force spectroscopy for gene-expression analysis. Curr. Sci. 87:239–244. [Google Scholar]
- 8.Kanda, T., K. F. Sullivan, and G. F. Wahl. 1998. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8:377–385. [DOI] [PubMed] [Google Scholar]
- 9.Pardue, M. L. 1994. Looking at polytene chromosomes. Methods Cell Biol. 44:333–351. [DOI] [PubMed] [Google Scholar]
- 10.Hansen, J. C. 2002. Conformation dynamics of the chromatin fiber in solution: determinants, mechanisms and functions. Annu. Rev. Biophys. Biomol. Struct. 31:361–392. [DOI] [PubMed] [Google Scholar]
- 11.Eberharter, A., and P. B. Becker. 2002. Histone acetylation: a switch between repressive and permissive chromatin. EMBO Rep. 3:224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toth, K. F., T. A. Knoch, M. Wachsmuth, M. Frank-Stohr, M. Stohr, C. P. Bacher, G. Muller, and K. Rippe. 2004. Trichostatin A-induced histone acetylation causes decondensation of interphase chromatin. J. Cell Sci. 117:4277–4287. [DOI] [PubMed] [Google Scholar]
- 13.Taddei, A., D. Roche, W. A. Bickmore, and G. Almouzni. 2005. The effects of histone deacetylase inhibitors on heterochromatin: implications for anticancer therapy? EMBO Rep. 6:520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Misteli, T., A. Gunjan, R. Hock, M. Bustin, and D. T. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature. 408:877–881. [DOI] [PubMed] [Google Scholar]
- 15.Lever, M. A., J. P. Th'ng, X. Sun, and M. J. Hendzel. 2000. Rapid exchange of histone H1.1 on chromatin in the living human cells. Nature. 408:873–876. [DOI] [PubMed] [Google Scholar]
- 16.Becker, P. B., and W. Horz. 2002. ATP-dependent nuclear remodeling. Annu. Rev. Biochem. 71:247–273. [DOI] [PubMed] [Google Scholar]
- 17.Richard, J. P., K. Melikov, E. Vives, C. Ramos, B. Verbeure, M. J. Gait, L. V. Chernomordik, and B. Lebleu. 2003. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J. Biol. Chem. 278:585–590. [DOI] [PubMed] [Google Scholar]
- 18.Robertson, J. D., S. Orrenius, and B. Zhivotovsky. 2000. Review: nuclear events in apoptosis. J. Struct. Biol. 129:346–358. [DOI] [PubMed] [Google Scholar]
- 19.Asenjo, A. B., N. Krohn, and H. Sosa. 2003. Configuration of the two kinesin motor domains during ATP hydrolysis. Nat. Struct. Biol. 10:836–842. [DOI] [PubMed] [Google Scholar]
- 20.Bradbury, E. M. 1992. Reversible histone modifications and the chromosome cell cycle. Bioessays. 14:9–16. [DOI] [PubMed] [Google Scholar]
- 21.Manders, E. M., A. E. Visser, A. Koppen, W. C. de Leeuw, R. van Liere, G. J. Brakenhoff, and R. van Driel. 2003. Four-dimensional imaging of chromatin dynamics during the assembly of the interphase nucleus. Chromosome Res. 11:537–547. [DOI] [PubMed] [Google Scholar]
- 22.Chen, D., M. Dundr, C. Wang, A. Leung, A. Lamond, T. Misteli, and S. Huang. 2005. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J. Cell Biol. 168:41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belmont, A. S., and K. Bruce. 1994. Visualization of G1 Chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. J. Cell Biol. 127:287–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cremer, T., and C. Cremer. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2:292–301. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert, N., S. Boyle, H. Fiegler, K. Woodfine, N. P. Carter, and W. A. Bickmore. 2004. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 118:555–566. [DOI] [PubMed] [Google Scholar]
- 26.Chambeyron, S., N. R. Da Silva, K. A. Lawson, and W. A. Bickmore. 2005. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development. 132:2215–2223. [DOI] [PubMed] [Google Scholar]
- 27.Szutorisz, H., C. Canzonetta, A. Georgiou, C. M. Chow, L. Tora, and N. Dillon. 2005. Formation of an active tissue-specific chromatin domain initiated by epigenetic marking at the embryonic stem cell stage. Mol. Cell. Biol. 25:1804–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerlich, D., J. Beaudouin, B. Kalbfuss, N. Daigle, R. Eils, and J. Ellenberg. 2003. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 112:751–764. [DOI] [PubMed] [Google Scholar]
- 29.Levi, V., Q. Ruan, M. Plutz, A. S. Belmont, and E. Gratton. 2005. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys. J. 89:4275–4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tramier, M., K. Kemnitz, C. Durieux, J. Coppey, P. Denjean, R. B. Pansu, and M. Coppey-Moisan. 2000. Restrained torsional dynamics of nuclear DNA in living proliferative mammalian cells. Biophys. J. 78:2614–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]