Abstract
Background
In recent years encouraging progress has been made in developing vaccine treatments for cancer, particularly with melanoma. However, the overall rate of clinically significant results has remained low. The present research used microarray datasets from previous investigations to examine gene expression patterns in cancer cell lines with the goal of better understanding the tumor microenvironment.
Methods
Principal Components Analyses with Promax rotational transformations were carried out with 90 cancer cell lines from 3 microarray datasets, which had been made available on the internet as supplementary information from prior publications.
Results
In each of the analyses a well defined melanoma component was identified that contained a gene coding for the enzyme, glutaminyl cyclase, which was as highly expressed as genes from a variety of well established biomarkers for melanoma, such as MAGE-3 and MART-1, which have frequently been used in clinical trials of melanoma vaccines.
Conclusion
Since glutaminyl cyclase converts glutamine and glutamic acid into a pyroglutamic form, it may interfere with the tumor destructive process of vaccines using peptides having glutamine or glutamic acid at their N-terminals. Finding ways of inhibiting the activity of glutaminyl cyclase in the tumor microenvironment may help to increase the effectiveness of some melanoma vaccines.
Background
In recent decades considerable progress has been made in developing methods which use the immune system to treat cancer. One particularly noteworthy series of events was the discovery of the first genes encoding human tumor antigens recognized by cytolytic T lymphocytes (CTLs) [1-3]. Since then, dozens of other antigen genes have been identified [4], many of which contain several short sequences of DNA, coding for peptides called "epitopes." When epitopes are bound by members of the human leukocyte antigen (HLA) family, presented on the tumor cell surface, and recognized by CTLs; destruction of the tumor cell tends to occur. Scientists have become so adept at using such methods for reproducibly destroying tumor cells in the laboratory that many small scale clinical vaccine trials have been carried out [5]. Unfortunately, although there have been reports of some dramatic remissions, the overall degree of successful therapy has remained low [6].
Explanations for the difficulty in translating findings from the laboratory to the clinic in the field of tumor antigen (TA) specific immunotherapy have focussed upon a variety of factors, one of which has been the importance of better understanding the tumor microenvironment [7]. In this regard, it has been suggested that breakthroughs in the development of microarray technology may be able to help overcome tumor resistance to effective vaccine responses [5,8].
The present report is of a retrospective microarray datamining study looking for information relevant to the tumor microenvironment in TA specific immunotherapy.
Methods
The first dataset analysed in the current research was published in a study by Ross et al [9] using the National Cancer Institute (NCI) group of 60 cancer cell lines from 9 different tissues of origin: melanoma, colon, renal, breast, CNS, leukemia, prostate, lung, and ovarian. For each of the NCI60 cell lines, expression levels of approximately 8000 genes were obtained from a microarray upon which 9706 cDNAs had been physically deposited. This dataset is available at the Genomics and Bioinformatics Group Microarray Datasets website [10].
The second dataset was collected by Staunton et al [11] using the NCI60 cell lines and an Affymetrix microarray, upon which short (25-mer) oglionucleoties had been directly synthesized. Using this form of microarray platform 7130 gene expression levels were measured. The raw data has been deposited at the Genomics and Bioinformatics Group Microarray Datasets website [12].
The third dataset used here was published in conjunction with a paper by Györffy et al [13] using an Affymetrix HGU133 microarray chip containing 42297 estimates of gene expression levels. This study utilized 30 cancer cell lines from 9 different types of human tissue: breast, liver, lung, melanoma, ovarian, pancreas, colon, gastric and prostate. The microarray raw dataset was made available in supplementary Table 1 published with the article.
The primary method used in the present research was the multivariate statistical technique of principal components analysis (PCA), previously utilized with microarray data in a study by Crescenzi and Giuliani [14]. However, unlike the Crescenzi and Giuliani research, in which cDNAs with values for all 60 cell lines were used, the present PCA was carried out after replacing any missing values with the mean for each cell line. This meant that, instead of using 1416 cDNAs, there were 9706 values included in the PCA. Another important difference was that, following the main principal components extraction, Promax rotational transformations were performed [15]. These alternative, complementary procedures were undertaken to see if PCA could produce results in addition to the interesting findings obtained in earlier research [14].
Additional analyses were carried out using: (a) Pearson product-moment correlation coefficients, (b) linear discriminant analysis, and (c) unpaired two-tailed Student t tests for the difference between means from independent samples. Prior to each t test, F ratios were calculated to ascertain whether to assume equality of variances.
All analyses were performed with SPSS for Windows XP (Version 12.0) using default values for each procedure unless otherwise indicated. Prior to the PCA, the NCI60 Affymetrix microarray expression estimates were converted to standardized z-scores.
Results
Ross et al cDNA microaarray data
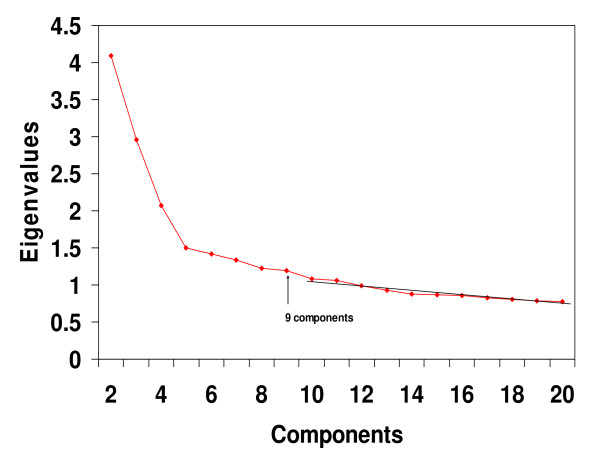
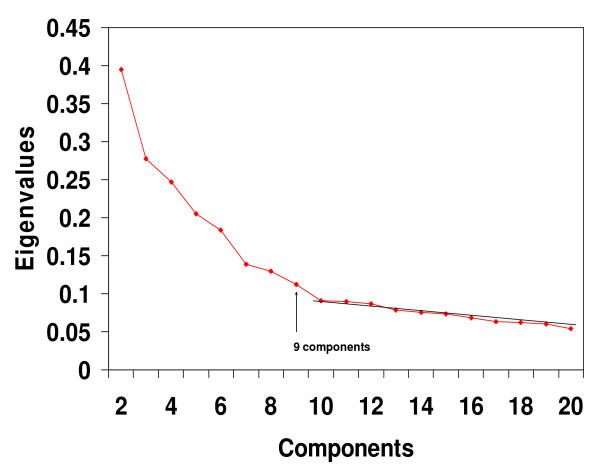
Pearson product-moment correlation coefficients were calculated between the cell lines, and then eigenvalues for the 60 × 60 correlation matrix were calculated. A scree test [14,16,17], presented in Figure 1, was performed using the eigenvalues plot. The scree results indicated that it would be appropriate to extract 9 components. Following extraction of 9 principal components, Promax rotations were carried out. As may be seen in the Promax component pattern loading values presented in Table 1 (see Additional file 1), there were 68% of the loadings within the +/-.10 hyperplane, which indicated that a high degree of simple structure had been achieved [17-19]. In addition, it may be seen in Table 1 (see Additional file 1) that Component #1 consisted of the melanoma cell lines: MALME3M, SLMEL2, SKMEL5, SKMEL28, M14, UACC62 and UACC257 together with MDAMB435 and MDAN. This group of cell lines is identical with the Melanoma Cluster found previously by hierarchical clustering in two separate research reports (see Figure 1 of Ross et al [9] and cluster 3 in Figure 3 of Crescenzi and Giuliani [14]).
Figure 1.
Scree test for the Ross et al dataset. Eigenvalues from the 60 × 60 cell line correlation matrix showed that, beginning with the break in the plot after component 9, a straight scree line could be fitted to the remaining values. Such a finding suggests that the eigenvalues after component 9 represent only error variance. The term "scree" was borrowed from geology where it refers to debris (i.e. error) that has fallen to the base of a mountain. Note that for clarity of presentation, the first eigenvalue of 13.787 was not plotted.
It should be noted that there is evidence the cell lines, MDAMB435 and MDAN (derived from the same patient), originally believed to be of breast cancer in origin, may have resulted from melanoma metastasis [9]. At the same time, the allegedly melanoma cell line, LOXIMVI, which had previously fallen outside of the Melanoma Cluster [9,14], also did not appear in the present Melanoma Component. Because of these questions about the cancer type of these three cell lines, as a conservative measure, their scores were not included in further statistical analyses.
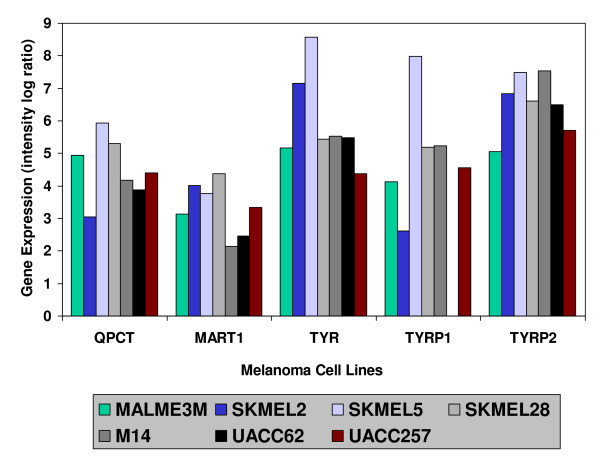
In order to identify the most highly expressed genes in the cell lines comprising the Melanoma Component, regression scores were calculated for each of the 9706 cDNAs. The 26 highest scoring genes of the Melanoma Component (together with the component regression score for each gene) are listed in Table 2 (see Additional file 2), followed by the 26 genes found by Ross et al within their Melanoma Cluster. In Table 2 (see Additional file 2) it may be seen that 14 of the genes identified by Ross et al were among the highest scoring in the present Melanoma Component. From the 14 genes falling in both the Ross et al and the present melanoma group, several genes were chosen for presentation in Figure 2, these genes were: Melanoma Antigen recognized by T-cells (MART1), Tyrosinase (TYR), Tyrosinase Related Protein 1 (TYRP1) and Tyrosinase Related Protein 2 (TYRP2). These four genes were selected because they are well known biomarkers for melanoma and have been utilized as epitopes in tumor immunotherapy [5]. Also presented in Figure 2 was the gene, Glutaminyl Cyclase (QPCT). The reason for selecting QPCT was that, as may be seen in later subsections, it was found to be highly expressed in two other microarray datasets.
Figure 2.
Highly expressed genes within the Ross et al Melanoma Component. Gene expression levels of 5 genes within each of the 7 cell lines in the Ross et al cDNA Melanoma Component. Measurements were log ratios between the gene expression level of each cell line and a reference sample of 12 of the 60 tumor cell lines. Further details may be found in the original Ross et al publication. The means for each gene for the non-melanoma cell lines (followed by the Student t test significances for the mean difference between melanoma and non-melanoma cell lines) were as follows: QPCT -.514 (p < .001), MART1 -.055 (p = .001), TYR -.031 (P < .001), TYRP1 -.449 (P < .001), and TYRP2 -.041 (p < .001). It may be seen that the means of the non-melanoma cell lines were all very close to zero, and therefore were not plotted simply for greater clarity of presentation.
Next, to determine how a combination (and not only a gene-by-gene analysis) of the selected genes perform on the melanoma identification, a linear discriminant analysis [20] was performed using MART1, TYR, TYRP1, TYRP2 and QPCT as independent variables and melanoma vs non-melanoma PCA scores as the grouping variable. It was found that the overall gene combination was related to the melanoma distinction (Wilks Lambda = .057, p < .001) with the standardized canonical discriminant function coefficients being MART1 = .267, TYR = .420, TYRP1 = .361, TYRP2 = .595, and QPCT = .808.
Staunton et al Affymetrix microaarray data
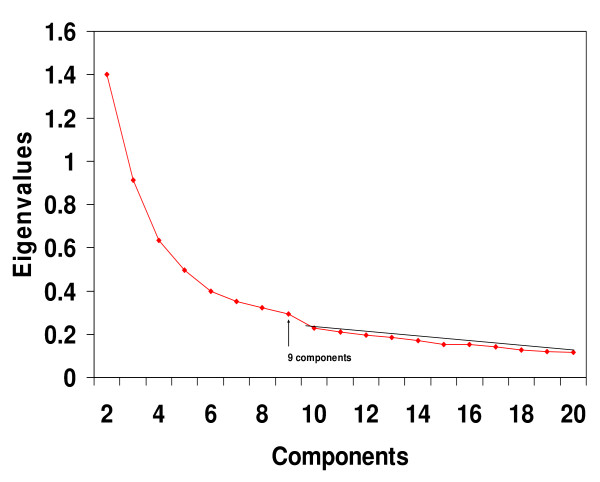
As with the cDNA analysis, a scree test with the eigenvalues from the 60 × 60 NCI Affymetrix correlation matrix indicated that 9 components should be extracted (Figure 3). PCA was performed on the NCI60 Affymetrix gene expression estimates. The PCA produced results that were highly similar to those obtained with the cDNA values analysed in the original Ross et al study [9]. It was found that PROMAX rotations (with the maximum iterations for convergence parameter set at 50 instead of the default value of 25) resulted in a readily identifiable Melanoma Component (component pattern loading values are presented in Table 3 – see Additional file 3) made up of the cell lines, MALME3M, SLMEL2, SKMEL5, SKMEL28, M14, UACC62 and UACC257.
Figure 3.
Scree test for the Staunton et al dataset. Eigenvalues from the 60 × 60 cell line correlation matrix showed that, beginning with the break in the plot after component 9, a straight scree line could be fitted to the remaining values. Such a finding suggests that the eigenvalues after component 9 represent only error variance. Note that for clarity of presentation, the first eigenvalue of 51.392 was not plotted.
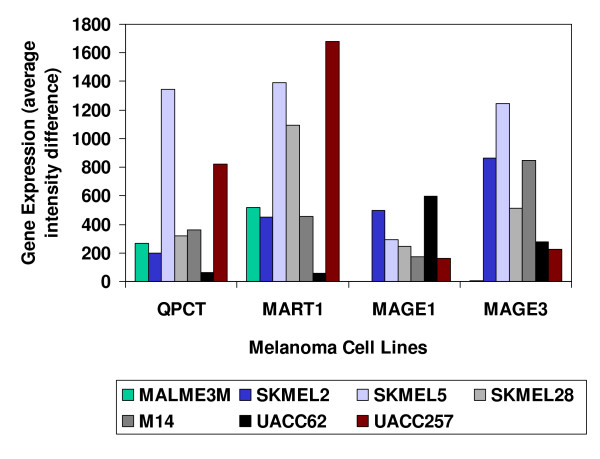
When component scores were calculated for each of the genes on the microarray, the enzyme QPCT again was found to be as highly expressed (see Figure 4) as some well known proteins used in melanoma immunotherapy [5]: MART1, Melanoma Antigen 1 (MAGE1) and Melanoma Antigen 3 (MAGE3).
Figure 4.
Highly expressed genes within the Staunton et al Melanoma Component. Gene expression levels of 4 genes within each of the 7 cell lines in the Staunton et al Affymetrix Melanoma Component together with the mean expression levels for the combined non-melanoma cell lines. The expression values were average intensity difference units determined with Affymetrix GENECHIP software that assigned a value of 100 to all expression measurements of less than 100 units. Further details may be found in the Staunton et al publication. The Student t test significance levels for each gene, calculated between the melanoma and the non-melanoma cell lines, were as follows: QPCT (p = .033), MART1 (p = .012), MAGE1 (p = .019) and MAGE3 (p = .024). Note that for clarity of presentation, this figure used expression average intensity difference units, as they were prior to conversion to the standardized z-scores that were used in the PCA and t test calculations.
Combined Ross et al and Staunton et al data analysis
In order to show the effective consistence of the Staunton et al Melanoma Component (shown in Table 3 – see Additional file 3) with the previously identified Melanoma Component from the Ross et al data (depicted in Table 1 – see Additional file 1), a Pearson Correlation Coefficient was calculated between the loading values. The obtained value of .905 (p < .001) supported a hypothesis of identity between these component patterns.
Györffy et al Affymetrix microarray data
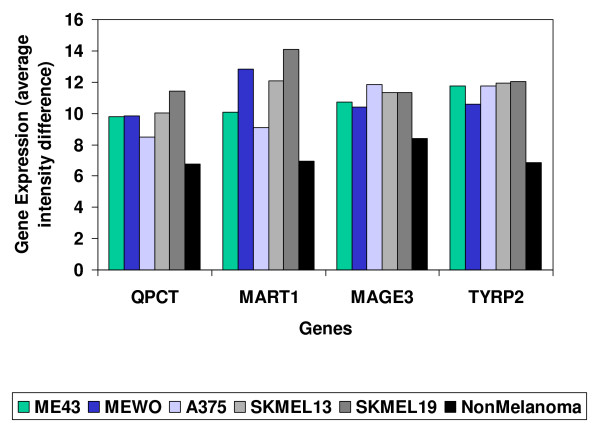
As with the other 2 analyses, PCA results with the Affymetrix HGU133 microarray chip produced results that matched well with previous findings. A scree test indicated that 9 components should be extracted (Figure 5). Following components extraction and Promax rotations, one of the 9 components clearly corresponded to the Melanoma Cluster found in the original Györffy et al study [13]. The Melanoma Component (component pattern loading values are presented in Table 4 – see Additional file 4) was composed primarily of the melanoma cell lines ME43, MEWO, A375, SKMEL13 and SKMEL19. One lung cell line, COLO699, loaded significantly on the Melanoma Component, and one melanoma cell line, C8161, did not load. However, both of these findings also occurred in the hierarchical cluster analysis of the Györffy et al study (see their Figure 5) [13].
Figure 5.
Scree test for the Györffy et al Affymetrix dataset. Eigenvalues from the 30 × 30 cell line correlation matrix showed that, beginning with the break in the plot after component 9, a straight scree line could be fitted to the remaining values. Such a finding suggests that the eigenvalues after component 9 represent only error variance. Note that for clarity of presentation, the first eigenvalue of 27.020 was not plotted.
Interestingly, the QPCT enzyme again was expressed at a level similar to well established antigens used in melanoma vaccines [5]: MART1, MAGE3 and TYRP2 (Figure 6).
Figure 6.
Highly expressed genes within the Györffy et al Melanoma Component. Gene expression levels of 4 genes within each of the 5 cell lines in the Györffy et al Melanoma Component together with the mean expression levels for the combined non-melanoma cell lines. The expression values were average intensity difference units determined with Affymetrix MAS 5.0 software. Further details may be found in the Györffy et al publication. The Student t test significance levels for each gene, calculated between the melanoma and the non-melanoma cell lines, were as follows: QPCT (p < .001), MART1 (p = .010), MAGE3 (p < .001) and TYRP2 (p < .001).
Discussion
Prior to the present findings there had been several indications that the gene coding for glutaminyl cyclase tends to be highly expressed in melanoma. In an exploratory microarray study aimed at identifying potential immunological targets in tumor microenvirnoments, Wang et al [21] published an additional file listing 2044 clones which appeared to be upregulated in metastatic cutaneous melanoma. Included in this list of cDNA clones was glutaminyl cyclase.
In another study, employing the high throughput method of Serial Analysis of Gene Expression to survey the gene expression patterns of melanoma tumor samples, Weeraratna et al [22] identified glutaminyl cyclase as being among those most abundantly expressed [see their Figure 1 (c)].
Consequently, when combined with the present findings, there is now evidence from a variety of sources supporting the notion that glutaminyl cyclase tends to be highly expressed in melanoma.
Evidence about the potential significance of this finding for immunological treatment of cancer comes from several sources. Firstly, it is interesting to note that several highly researched epitopes, such as MAGE3 (EVDPIGHLY) and the synthetic MART1 (ELAGIGILTV), which are among those having produced the most clinically interesting results [23-26], have an E at their N-terminal. Secondly, it has been shown that the main function of glutaminyl cyclase is to accelerate the transformation of glutamine (Q) or glutamic acid (E) into pyroglutamic acid (pE) [27,28]. Thirdly, when either Q or E occur at the N terminal of an epitope, their cyclic transformation to pE has been shown to decrease the affinity with which the peptide will bind with HLA molecules [29,30]. Hence, high levels of glutaminyl cyclase within melanoma cells may interfere with the avidity with which T cells recognize and destroy tumors.
The plausibility of such a scenario is increased by knowledge that the generation of antigenic peptides is a multistep process. Part of this process takes place in the endoplasmic reticulum (ER), and the glutaminyl cyclase gene codes for an ER targeting signal [31].
Should laboratory research confirm the present microarray-derived indication of QPCT activity within the melanoma tumor microenvironment, attention may be focused upon finding ways to overcome QPCT interference. In this regard it is interesting to note that potent new inhibitors have been discovered recently for QPCT [32]. However, application of such inhibitors in a clinical setting is likely to present difficulties, because QPCT plays a significant role in many normal physiological processes, such as influencing the bioactivity of hormones like gonadotropin-releasing hormone and thyrotropin-releasing hormone [33]. It is also noteworthy that QPCT may be essential for the activity of monocyte chemotactic protein 2 (MCF-2), since MCF-2 is practically inactive without a pE at its N-terminal [34]. Perhaps the development of tissue specific inhibitors could help to overcome problems due to the widespread activity of glutaminyl cyclase [35].
Another possible difficulty is that there is some evidence that glutaminyl cyclase, like MART-1, TYR, and TYRP2, may be highly expressed in normal melanocytes as well as in melanoma (see Table 1 of Dooley et al [36]). Further research is needed to find ways to overcome such potential problems in translating results from the present basic research into clinical immunotherapy.
Competing interests
The author(s) declare that they have no competing interests.
Supplementary Material
Table 1: Promax component pattern loading values for the first 4 components of the Ross et al dataset.
Table 2: Biomarker genes in (a) the present melanoma component and (b) the original Ross et al cluster.
Table 3: Promax melanoma component pattern loadings for the Staunton et al dataset.
Table 4: Promax melanoma component pattern loading values for the Györffy et al dataset.
Acknowledgments
Acknowledgements
This research was facilitated by a grant to St. Thomas University from the Canadian Foundation for Innovation. Thanks are also extended to the authors of the Ross et al, Staunton et al, and Györffy et al studies for making their data publicly available on the internet.
References
- van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- Gaugler B, Van den Eynde B, van der Bruggen P, Romero P, Gaforio JJ, De Plaen E, Lethe B, Brasseur F, Boon T. Human gene MAGE-3 codes for an antigen recognized on a melanoma by autologous cytolytic T lymphocytes. J Exp Med. 1994;179:921–930. doi: 10.1084/jem.179.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Sakaguchi K, Appella E, Yannelli JR, Adema GJ, Miki T, Rosenberg SA. Identification of a human melanoma antigen recognized by tumor-infiltrating lymphocytes associated with in vivo tumor rejection. Proc Natl Acad Sci U S A. 1994;91:6458–6462. doi: 10.1073/pnas.91.14.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novellino L, Castelli C, Parmiani G. A listing of human tumor antigens recognized by T cells: March 2004 update. Cancer Immunol Immunother. 2005;54:187–207. doi: 10.1007/s00262-004-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon T, Coulie PG, Eynde BJ, Bruggen PV. Human T Cell Responses against Melanoma. Annu Rev Immunol. 2006;24:175–208. doi: 10.1146/annurev.immunol.24.021605.090733. [DOI] [PubMed] [Google Scholar]
- Slingluff CLJ, Speiser DE. Progress and controversies in developing cancer vaccines. J Transl Med. 2005;3:18. doi: 10.1186/1479-5876-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Panelli MC, Marincola FM. Understanding the response to immunotherapy in humans. Springer Semin Immunopathol. 2005;27:105–117. doi: 10.1007/s00281-004-0198-7. [DOI] [PubMed] [Google Scholar]
- Gabriele L, Moretti F, Pierotti MA, Marincola FM, Foa R, Belardelli F. The use of microarray technologies in clinical oncology. J Transl Med. 2006;4:8. doi: 10.1186/1479-5876-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- cDNA Microarray Data of the NCI 60 Cancer Cell Lines http://discover.nci.nih.gov/datasetsNature2000.jsp
- Staunton JE, Slonim DK, Coller HA, Tamayo P, Angelo MJ, Park J, Scherf U, Lee JK, Reinhold WO, Weinstein JN, Mesirov JP, Lander ES, Golub TR. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci U S A. 2001;98:10787–10792. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix Data of the NCI 60 Cancer Cell Lines http://discover.nci.nih.gov/datasetsPnas2001.jsp
- Gyorffy B, Surowiak P, Kiesslich O, Denkert C, Schafer R, Dietel M, Lage H. Gene expression profiling of 30 cancer cell lines predicts resistance towards 11 anticancer drugs at clinically achieved concentrations. Int J Cancer. 2006;118:1699–1712. doi: 10.1002/ijc.21570. [DOI] [PubMed] [Google Scholar]
- Crescenzi M, Giuliani A. The main biological determinants of tumor line taxonomy elucidated by a principal component analysis of microarray data. FEBS Lett. 2001;507:114–118. doi: 10.1016/S0014-5793(01)02973-8. [DOI] [PubMed] [Google Scholar]
- Dien J. Progressing towards a consensus on PCA of ERPs. Clin Neurophysiol. 2006 doi: 10.1016/j.clinph.2005.09.029. [DOI] [PubMed] [Google Scholar]
- Cattell RB. The scree test for the number of factors. Multivariate Behavioral Research. 1966;1:245–276. doi: 10.1207/s15327906mbr0102_10. [DOI] [PubMed] [Google Scholar]
- Cattell RB. The scientific use of factor analysis in behavioral and life sciences. New York, Plenum; 1978. [Google Scholar]
- Cattell RB. Factor analysis: An introduction to essentials, I The purpose and underlying models. Biometrics. 1965;21:190–215. doi: 10.2307/2528364. [DOI] [PubMed] [Google Scholar]
- Cattell RB. Factor analysis: An introduction to essentials, II The role of factor analysis in research. Biometrics. 1965;21:405–435. doi: 10.2307/2528100. [DOI] [PubMed] [Google Scholar]
- Mendez MA, Hodar C, Vulpe C, Gonzalez M, Cambiazo V. Discriminant analysis to evaluate clustering of gene expression data. FEBS Lett. 2002;522:24–28. doi: 10.1016/S0014-5793(02)02873-9. [DOI] [PubMed] [Google Scholar]
- Wang E, Panelli MC, Zavaglia K, Mandruzzato S, Hu N, Taylor PR, Seliger B, Zanovello P, Freedman RS, Marincola FM. Melanoma-restricted genes. J Transl Med. 2004;2:34. doi: 10.1186/1479-5876-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeraratna AT. Serial analysis of gene expression (SAGE): advances, analysis and applications to pigment cell research. Pigment Cell Res. 2003;16:183–189. doi: 10.1034/j.1600-0749.2003.00042.x. [DOI] [PubMed] [Google Scholar]
- Lurquin C, Lethe B, De Plaen E, Corbiere V, Theate I, van Baren N, Coulie PG, Boon T. Contrasting frequencies of antitumor and anti-vaccine T cells in metastases of a melanoma patient vaccinated with a MAGE tumor antigen. J Exp Med. 2005;201:249–257. doi: 10.1084/jem.20041378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germeau C, Ma W, Schiavetti F, Lurquin C, Henry E, Vigneron N, Brasseur F, Lethe B, De Plaen E, Velu T, Boon T, Coulie PG. High frequency of antitumor T cells in the blood of melanoma patients before and after vaccination with tumor antigens. J Exp Med. 2005;201:241–248. doi: 10.1084/jem.20041379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanagiri T, van Baren N, Neyns B, Boon T, Coulie PG. Analysis of a rare melanoma patient with a spontaneous CTL response to a MAGE-A3 peptide presented by HLA-A1. Cancer Immunol Immunother. 2006;55:178–184. doi: 10.1007/s00262-005-0063-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Baren N, Bonnet MC, Dreno B, Khammari A, Dorval T, Piperno-Neumann S, Lienard D, Speiser D, Marchand M, Brichard VG, Escudier B, Negrier S, Dietrich PY, Maraninchi D, Osanto S, Meyer RG, Ritter G, Moingeon P, Tartaglia J, van der Bruggen P, Coulie PG, Boon T. Tumoral and immunologic response after vaccination of melanoma patients with an ALVAC virus encoding MAGE antigens recognized by T cells. J Clin Oncol. 2005;23:9008–9021. doi: 10.1200/JCO.2005.08.375. [DOI] [PubMed] [Google Scholar]
- Bateman RCJ, Temple JS, Misquitta SA, Booth RE. Evidence for essential histidines in human pituitary glutaminyl cyclase. Biochemistry. 2001;40:11246–11250. doi: 10.1021/bi011177o. [DOI] [PubMed] [Google Scholar]
- Schilling S, Hoffmann T, Manhart S, Hoffmann M, Demuth HU. Glutaminyl cyclases unfold glutamyl cyclase activity under mild acid conditions. FEBS Lett. 2004;563:191–196. doi: 10.1016/S0014-5793(04)00300-X. [DOI] [PubMed] [Google Scholar]
- Beck A, Bussat MC, Klinguer-Hamour C, Goetsch L, Aubry JP, Champion T, Julien E, Haeuw JF, Bonnefoy JY, Corvaia N. Stability and CTL activity of N-terminal glutamic acid containing peptides. J Pept Res. 2001;57:528–538. doi: 10.1034/j.1399-3011.2001.00895.x. [DOI] [PubMed] [Google Scholar]
- Thompson LW, Hogan KT, Caldwell JA, Pierce RA, Hendrickson RC, Deacon DH, Settlage RE, Brinckerhoff LH, Engelhard VH, Shabanowitz J, Hunt DF, Slingluff CLJ. Preventing the spontaneous modification of an HLA-A2-restricted peptide at an N-terminal glutamine or an internal cysteine residue enhances peptide antigenicity. J Immunother. 2004;27:177–183. doi: 10.1097/00002371-200405000-00001. [DOI] [PubMed] [Google Scholar]
- Pohl T, Zimmer M, Mugele K, Spiess J. Primary structure and functional expression of a glutaminyl cyclase. Proc Natl Acad Sci U S A. 1991;88:10059–10063. doi: 10.1073/pnas.88.22.10059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz M, Heiser U, Schilling S, Niestroj AJ, Zunkel K, Demuth HU. The first potent inhibitors for human glutaminyl cyclase: synthesis and structure-activity relationship. J Med Chem. 2006;49:664–677. doi: 10.1021/jm050756e. [DOI] [PubMed] [Google Scholar]
- Huang KF, Liu YL, Wang AH. Cloning, expression, characterization, and crystallization of a glutaminyl cyclase from human bone marrow: a single zinc metalloenzyme. Protein Expr Purif. 2005;43:65–72. doi: 10.1016/j.pep.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Blaszczyk J, Coillie EV, Proost P, Damme JV, Opdenakker G, Bujacz GD, Wang JM, Ji X. Complete crystal structure of monocyte chemotactic protein-2, a CC chemokine that interacts with multiple receptors. Biochemistry. 2000;39:14075–14081. doi: 10.1021/bi0009340. [DOI] [PubMed] [Google Scholar]
- Sykes PA, Watson SJ, Temple JS, Bateman RCJ. Evidence for tissue-specific forms of glutaminyl cyclase. FEBS Lett. 1999;455:159–161. doi: 10.1016/S0014-5793(99)00872-8. [DOI] [PubMed] [Google Scholar]
- Dooley TP, Curto EV, Davis RL, Grammatico P, Robinson ES, Wilborn TW. DNA microarrays and likelihood ratio bioinformatic methods: discovery of human melanocyte biomarkers. Pigment Cell Res. 2003;16:245–253. doi: 10.1034/j.1600-0749.2003.00036.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1: Promax component pattern loading values for the first 4 components of the Ross et al dataset.
Table 2: Biomarker genes in (a) the present melanoma component and (b) the original Ross et al cluster.
Table 3: Promax melanoma component pattern loadings for the Staunton et al dataset.
Table 4: Promax melanoma component pattern loading values for the Györffy et al dataset.