Abstract
Germin-like proteins (GLPs) have been shown to be encoded by multigene families in several plant species and a role of some subfamily members in defense against pathogen attack has been proposed based on gene regulation studies and transgenic approaches. We studied the function of six GLP subfamilies of barley (Hordeum vulgare) by selecting single mRNAs for gene expression studies as well as overexpression and gene-silencing experiments in barley and Arabidopsis (Arabidopsis thaliana). Expression of all six subfamilies was high in very young seedlings, including roots. The expression pattern gradually changed from developmental to conditional with increasing plant age, whereby pathogen attack and exogenous hydrogen peroxide application were found to be the strongest signals for induction of several GLP subfamilies. Transcripts of four of five GLP subfamilies that are expressed in shoots were predominantly accumulating in the leaf epidermis. Transient overexpression of HvGER4 or HvGER5 as well as transient silencing by RNA interference of HvGER3 or HvGER5 protected barley epidermal cells from attack by the appropriate powdery mildew fungus Blumeria graminis f. sp. hordei. Silencing of HvGER4 induced hypersusceptibility. Transient and stable expression of subfamily members revealed HvGER5 as a new extracellular superoxide dismutase, and protection by overexpression could be demonstrated to be dependent on superoxide dismutase activity of the encoded protein. Data suggest a complex interplay of HvGER proteins in fine regulation of basal resistance against B. graminis.
Germin, long known as a marker protein in germinating wheat (Triticum aestivum) seed, is an apoplastic and glycosylated protein with oxalate-oxidase (OXOX) activity and with resistance to heat, proteolysis, and denaturing agents like SDS or low pH (Lane et al., 1993, 1994). Wheat germin was found to possess sequence similarity to stress-related spherulin of the slime mold Physarum polycephalum and represents the founding member of a functionally diverse protein superfamily, designated as cupins, based on a common, highly conserved β-barrel (Lane et al., 1991; Dunwell et al., 2004). A basic cupin domain has been duplicated during plant evolution to form bicupins, including, for instance, a spore-specific fern protein as well as the storage globulins legumin and vicilin of higher plants (Baumlein et al., 1995; Shutov et al., 1998, 1999). Other cupins include various isomerases, cyclases, dioxygenases, as well as sugar- and auxin-binding proteins thought to be involved in abiotic-stress responses, including exposure to heat, salt, submergence, and aluminum (Dunwell et al., 2000). Plant genomes encode a subgroup of cupins called germin-like proteins (GLPs) with mostly unknown function. Like germin (Woo et al., 2000), GLPs have been described to assemble into homohexameric complexes in vivo with an apparent molecular mass of approximately 100 kD in seminative SDS-PAGE (Zhang et al., 1995; Vallelian et al., 1998; Christensen et al., 2004), and they are also often robust against denaturation and degradation by heat, detergents, or proteinases (e.g. see Vallelian et al., 1998). It is likely that the three-dimensional structural similarity among cupins is related to this remarkable protein stability.
So far, germin and all analyzed GLPs possess N-terminal secretory signals, suggesting a role in cell wall function or defense against invading pathogens. The latter assumption is supported by OXOX or superoxide dismutase (SOD) activities of several GLPs leading to hydrogen peroxide (H2O2) production, which has been proposed as a signaling molecule for a range of defense reactions, including cell death, and as a cofactor for cell wall reenforcement by cross-linking (Yamahara et al., 1999; Christensen et al., 2004; Laloi et al., 2004). Moreover, several GLP-encoding genes have been found to be induced by pathogen attack. More direct evidence for a defensive role of GLPs comes from transgenic plants ectopically expressing OXOX (Donaldson et al., 2001; Liang et al., 2001; Ramputh et al., 2002; Schneider et al., 2002; Cober et al., 2003; Livingstone et al., 2005) from virus-mediated gene-silencing experiments (Lou and Baldwin, 2006) and from transient expression studies in single epidermal cells. Resistance of single epidermal cells transiently overexpressing GLP-encoding cDNAs was observed in wheat and barley (Hordeum vulgare) leaf segments that had been bombarded with DNA-coated gold particles, followed by challenge inoculation with either the wheat powdery mildew Blumeria graminis f. sp. tritici or the barley powdery mildew Blumeria graminis f. sp. hordei (Bgh; Schweizer et al., 1999a; Christensen et al., 2004). In a complementary approach, TaGLP4 and HvGLP4 (previous nomenclature) were suppressed in their expression by transient-induced gene silencing (TIGS), a recently established test system based on RNA interference (RNAi; Douchkov et al., 2005). Data obtained in these experiments indicated an important role of subfamily 4 of GLPs in defense against B. graminis (Christensen et al., 2004).
Because of extensive sequence information from genome or expressed sequence tag (EST) sequencing programs, it became clear that germin and GLPs belong to a multigene family in plants, including Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and barley subsp. vulgare. Initial family-wide descriptive studies of GLPs have been carried out in Arabidopsis and barley, leading to a conceptual framework for GLP nomenclature within each species (Carter et al., 1998; Druka et al., 2002). In barley, five GLP subfamilies have been described and named HvGER1 to 5.
Here, we provide an updated and extended overview of the GLP multigene family of barley based on analysis of 370,000 ESTs, plus functional data of individual subfamily members with respect to defense against Bgh.
RESULTS
The HvGER Multigene Family of Barley
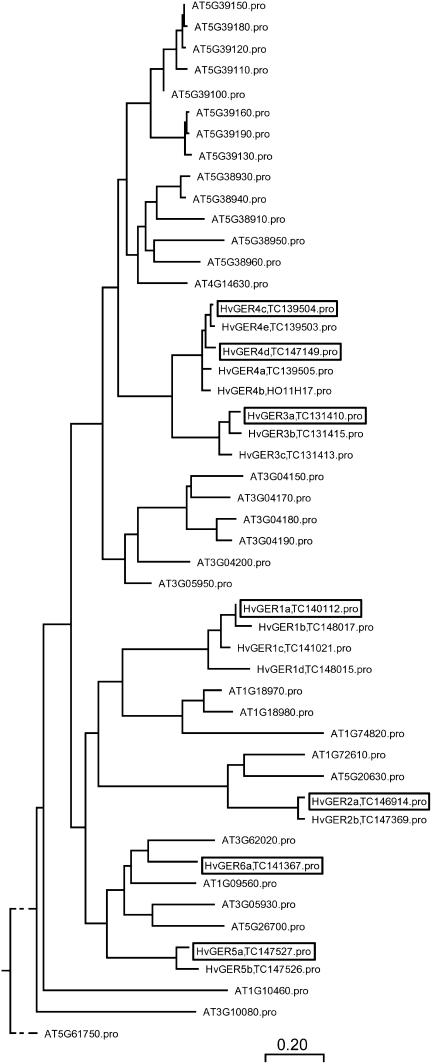
The Crop-EST database of the Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK; http://pgrc.ipk-gatersleben.de/est/index.php) was searched by using the keyword combination germin OR GLP. Matching EST sequences were compared to subfamily prototypes introduced by Druka et al. (2002) and, for each subfamily, one full coding sequence (CDS) cDNA clone was selected from the clone collection of the IPK based on graphic display of EST clustering results, BLASTN searches against annotated HvGER complete mRNA sequences, and prediction of an N-terminal signal peptide by using the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP). One EST (ID HK04D11; accession no. AL450783) did not match previously described subfamilies and was therefore considered as a founder of a new subfamily, designated HvGER6. In the case of HvGER3 and HvGER6 subfamilies, no full CDS cDNA clones were available and, therefore, corresponding PCR fragments were obtained from genomic barley DNA by using primers based on unigene tentative consensi (TC) in The Institute of Genomic Research (TIGR) Gene Index of barley (http://www.tigr.org/tdb/tgi/plant.shtml; release 9.0). Using sequences from these selected subfamily representatives, all TCs with a DNA sequence identity of at least 81%, which was set as a limit for the identification of paralogous subfamily members, were obtained from TIGR database. In addition, all putative GLPs encoded by the Arabidopsis genome were extracted from The Arabidopsis Information Resource database (TAIR). Multiple sequence alignment of the encoded proteins in both species revealed GLP multigene families of intermediate complexity, with 32 and 21 predicted genes in Arabidopsis and barley, respectively (Fig. 1). In both species, GLP families are subdivided into several subfamilies that often contain two or more duplicated genes of high sequence similarity. Only members of subfamilies 2 and 6 of barley were found to possess gene orthologs in Arabidopsis.
Figure 1.
Phylogenetic tree of GLP proteins from barley and Arabidopsis. Multiple sequence alignment was carried out by using ClustalW with default parameter settings, including the Gonnet series of protein weight matrix (MegAlign software package; DNASTAR). Scale bar indicates amino acid substitutions. Boxed HvGER proteins correspond to genes selected for this study. TC numbers of Barley Gene Index at TIGR (release 9.0) corresponding to HvGER proteins are indicated, too. Only HvGER proteins represented by full CDS sequences are included in the multiple sequence alignment. Three partial cDNAs encoding additional putative HvGER4 paralogs were excluded from this analysis (TIGR TC139506, TC139507, and TC139508).
The choice of experimental design for our study was based on the high degree of sequence similarity within barley GLP subfamilies. Therefore, one subfamily member (cDNA clone) was chosen as a representative for the corresponding subfamily and its entire CDS was used for 32P-labeled probe synthesis, overexpression, or TIGS (Table I). Due to expected cross hybridization and cross silencing, gene expression and TIGS data will be discussed at the level of GLP subfamilies only. Overexpression data can be discussed at the single-gene level, although functional redundancy of the encoded proteins per subfamily appears likely. Although we cannot exclude haplotype-specific effects in the overexpression system depending on barley genotypes used for cDNA cloning, this scenario is unlikely due to the very narrow genetic basis of modern barley cultivars, resulting in an estimated 5% chance for one nonsilent mutation per allele (Caldwell et al., 2006).
Table I.
The multigene family encoding GERs of barley
| Subfamilya | Previous Names | Complexityb | Representative Gene | Full CDS cDNA or Genomic Clone | Cultivar | Accession No. | TIGR TCc | Length DNAd | Size Protein (Amino Acid) |
|---|---|---|---|---|---|---|---|---|---|
| bp | |||||||||
| HvGER1 | Germin, OXOXe | 5 | HvGER1a | HW09M16 | Barke | DQ647619 | 140112 | 990 | 225 |
| HvGER2 | HvGLP1f | 2 | HvGER2a | HK03D12 | Barke | DQ647620 | 146914 | 810 | 213 |
| HvGER3 | GerminFg | 3 | HvGER3a | Genomic PCR fragment | Steffi | DQ647621 | 131410 | 1,025h | 227 |
| – | HvGER4c | HO01J19 | Ingrid | DQ647622 | 139504 | 914 | |||
| HvGER4 | OxOLPi, HvGLP4j | 8 | HvGER4d | HO08H12 | Ingrid | DQ647623 | 147149 | 881 | 230 |
| HvGER5 | – | 2 | HvGER5a | HW05P16 | Barke | DQ647624 | 147527 | 835 | 217 |
| HvGER6 | – | 1 | HvGER6a | Genomic PCR fragment | Steffi | DQ647625 | 141367 | 1,581h | 219 |
Number of TCs of the TIGR Gene Index of barley that represent likely subfamily members, based on DNA sequence identity of at least 81% in the CDS.
TC identifier of TIGR gene index barley (release 9.0) corresponding to representative DNA clone used in this study.
Size refers to representative cDNA or genomic clone.
Unspliced genomic DNA.
HvGER Gene Expression during Barley Development
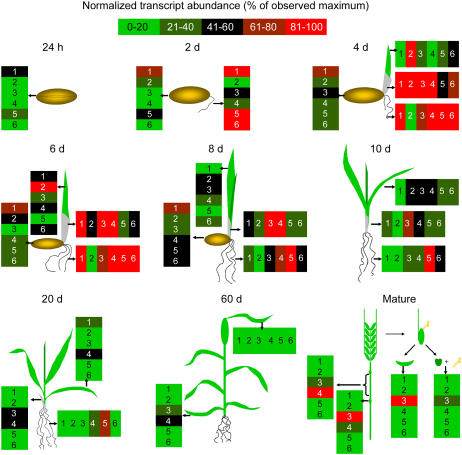
Figure 2 shows the results obtained in RNA-blot experiments from seed germination to grain filling in mature plants. No significant cross hybridization between subfamilies was observed, except for the HvGER6a probe that also weakly detected HvGER3 and HvGER5 transcripts (Supplemental Fig. 1). As expected, transcripts of HvGER1 encoding OXOX, which has been described as a marker for seed germination, accumulated first after imbibition followed by HvGER5. A transient wave of high GLP expression was observed that started in germinating seeds and ended in 10-d-old seedlings. With the exception of HvGER2, which turned out to be a marker for young elongating leaves, all GLP transcripts accumulated to high levels in roots. In older seedlings and mature plants, GLP mRNA was barely detectable, except for HvGER3 in spikelets 1 to 2 weeks after flowering and HvGER3 plus HvGER4 in nodes and the uppermost internode of mature plants. However, node- and internode-specific expression of these two subfamilies could not be confirmed in later experiments. Therefore, expression might have been triggered by stress conditions, like insect attack, for example, which could not be excluded because no spray treatments were performed on these experimental plants in the greenhouse. In contrast, HvGER3 expression in vegetative and generative spikelet tissues was reproducibly found and also previously reported (Wu et al., 2000). In general, expression data presented here are in good agreement with an expression summary based on EST abundance in different barley cDNA libraries (Supplemental Table I). Only in the case of HvGER5, EST abundance in coleoptiles and shoots was not reflected by mRNA abundance of the RNA blot.
Figure 2.
Developmental regulation of the HvGER multigene family of barley. Transcript abundance detected on RNA blots was normalized to 26S rRNA signals after rehybridization of blots. Data are expressed as percentage of maximum signal intensity observed per cDNA probe over all RNA samples (for primary data, see Supplemental Fig. 2). RNA from mature plants was extracted three times during a period of 3 weeks (starting approximately 1 week after pollination and covering the yellow-to-white pollen stage). Mean values of all three time points are shown. Numbers (1–6) inside boxes correspond to HvGER1 to HvGER6.
Stress-Induced Regulation of HvGER Gene Expression
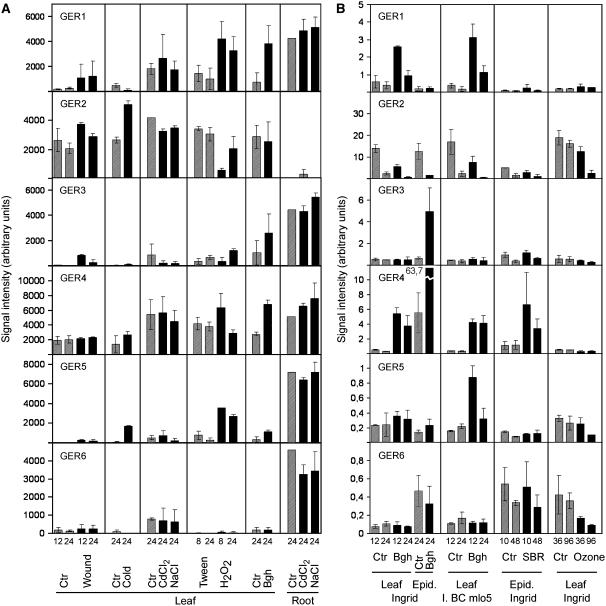
Expression of GLP subfamilies was studied in 7-d-old barley seedlings subjected to a range of biotic and abiotic stress conditions by performing RNA-blot analysis (Fig. 3A) as well as reverse RNA-blot analysis (Fig. 3B; see “Materials and Methods” for details). In general, abiotic stress treatments were poor inducers of HvGER genes and, only in the case of cold (4°C) and wound treatments, a reproducible induction of HvGER5 and HvGER3 was found. The apparent induction of HvGER2 transcript accumulation by cold treatment was difficult to interpret because these genes are under circadian control superimposed onto possible stress regulation (Vallelian et al., 1998). Moreover, HvGER2 transcript abundance was found to oscillate in inner leaf tissues only resulting in variable transcript ratios between the epidermis and the rest of the leaf (data not shown). An important exception to the weak induction of HvGER genes by abiotic stress was exogenous treatment with H2O2. This treatment strongly induced HvGER1 and HvGER5 and also caused accumulation of HvGER4 mRNA despite a considerable basal level of mRNA abundance of this subfamily in mock-treated plants (Fig. 3A). Interestingly, ozone that was applied at concentrations leading to visible stress symptoms after 5 d (data not shown) did not induce any HvGER mRNA accumulation in leaves (Fig. 3B). Ozone treatment of barley was shown to lead primarily to accumulation of a superoxide radical, suggesting that HvGER genes were not induced by that form of reactive oxygen (Wu and von Tiedemann, 2002). Pathogen attack by Bgh provided strong signals for induction of HvGER1, HvGER3, HvGER4, and HvGER5, as reflected by results from both RNA-blot and reverse RNA-blot analysis. Attack by the nonhost pathogen soybean rust fungus (Phakopsora pachyrhizi), which directly penetrates the host epidermis, caused induction of HvGER4.
Figure 3.
Stress-regulated expression of the HvGER multigene family of barley. A, HvGER transcript abundance detected by RNA-blot analysis was normalized to 26S rRNA signals after rehybridization of blots. Mean signal intensities ± range of two biological replicates are shown. Data of both biological replicates were normalized to each other by using control values of root samples, except for HvGER2 (leaf sample, controls for salt treatments). Black bars, Treatment; striped bars, corresponding control. B, HvGER transcript abundance detected by reverse RNA-blot analysis was normalized by median centering as described in “Materials and Methods.” Mean signal intensities ± range of two biological replicates are shown. Black bars, Treatment; striped bars, corresponding control. A and B, Units of signal intensities were defined by the different software packages used for RNA-blot or reverse RNA-blot analysis and are therefore not directly comparable. Numbers below bars correspond to duration of treatments (h). Ctr, Control; Epid, epidermis; SBR, soybean rust fungus.
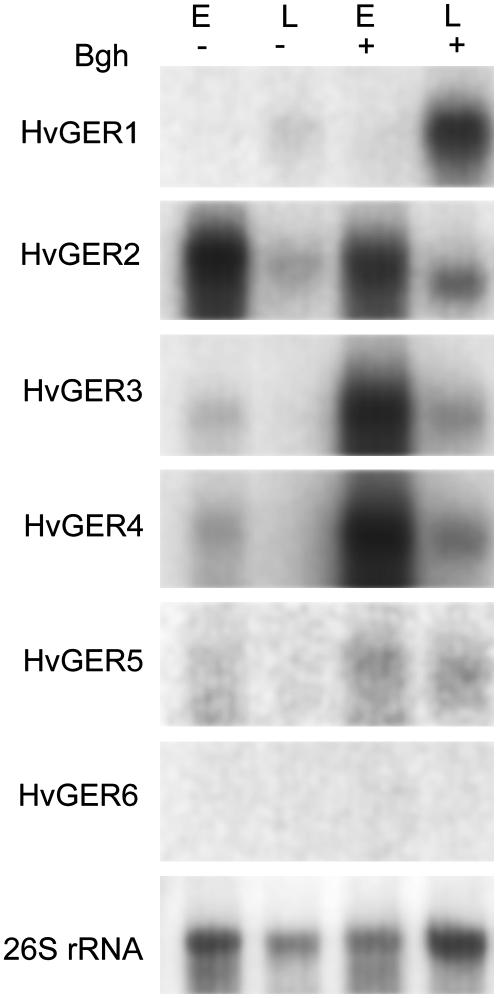
Bgh-induced or background transcript accumulation was localized in the epidermis versus inner leaf tissues (Fig. 4). Four of five GLP subfamilies that are expressed in leaves showed epidermal localization of their transcripts, suggesting an important role in either protection or general function of this specialized monocell layer. The weak signals observed in remaining leaf tissue after stripping off the abaxial epidermis probably represent diluted transcripts from the adaxial epidermis. In the case of HvGER2, distribution of mRNA abundance was dependent on the circadian time point chosen for RNA extraction (see above).
Figure 4.
Localization of HvGER transcripts in leaf epidermis versus inner leaf tissues. Equal amounts of RNA extracted from stripped abaxial epidermis (E) or from the remaining leaf (L) were loaded per lane. RNA was extracted simultaneously 24 h postinoculation with Bgh (+) or from noninoculated control plants (−).
Functional Analysis of the HvGER Gene Family
A transient assay based on overexpression or silencing by RNAi (TIGS) of candidate genes in single epidermal cells was used to study the effect of misexpression of HvGER-encoding genes on basal resistance against Bgh. In a previous study, we found that the effect of TIGS or overexpression of HvGER4 was dependent on the barley genotype used for bombardment and that different cultivars responded differently to either silencing or overexpression (Christensen et al., 2004). Therefore, transient overexpression was carried out in cv Pallas, whereas TIGS was carried out in cv Golden Promise. Overexpression of HvGER2a, HvGER4c, HvGER4d, and HvGER5a significantly enhanced resistance against Bgh (Table II). TIGS of HvGER3 and HvGER5 also enhanced resistance against the same pathogen, whereas TIGS of HvGER4 induced hypersusceptibility. The effects obtained by overexpression or TIGS of HvGER4 are in agreement with previous results from wheat and barley (Schweizer et al., 1999a; Christensen et al., 2004). Remarkably, transient overexpression of either of two selected paralogous genes of subfamily 4 induced protection, supporting the initial speculation about functional redundancy of closely related gene paralogs within GLP subfamilies. The data for HvGER1 differ from those obtained in a transient assay system of wheat, where protection by overexpressing the wheat germin gf-2.8 gene against B. graminis f. sp. tritici was found (Schweizer et al., 1999a). Both HvGER1a and GF-2.8 proteins have been shown to possess OXOX enzymatic activity (Schweizer et al., 1999b; Table IV). One possible explanation of these different results may be limiting or absent oxalate availability in barley in contrast to wheat.
Table II.
Transient overexpression and silencing of HvGER genes influences basal resistance of barley
NS, Not significant (p > 0.05); n.a., not analyzed.
| Gene | Overexpression
|
TIGS
|
||
|---|---|---|---|---|
| Relative Haustorial Indexa | p (t Test)b | Relative Haustorial Indexa | p (t Test)b | |
| % | % | |||
| HvGER1a | 78.5 ± 12.5 | NS | n.a. | n.a. |
| HvGER2a | 58.4 ± 9.9 | 0.0138 | 106.7 ± 33.3 | NS |
| HvGER3a | 69.9 ± 16.1 | NS | 56.0 ± 11.1 | 0.017 |
| HvGER4c | 54.8 ± 9.8 | 0.0098 | n.a. | n.a. |
| HvGER4d | 48.3 ± 4.2 | 0.0002 | 145.1 ± 11.2 | 0.016 |
| HvGER5a | 61.8 ± 9.2 | 0.0090 | 70.4 ± 7.1 | 0.025 |
| HvGER6a | 70.9 ± 13.3 | NS | n.a. | n.a. |
| HvSNAP34 | n.a. | n.a. | 257.0 ± 34.5 | 0.020 |
Barley leaf segments were cobombarded with pUbiGUS and overexpression constructs encoding HvGER proteins or with RNAi (TIGS) constructs targeting HvGER genes, followed by challenge with Bgh 4 h (overexpression) or 48 h (TIGS) postbombardment. Overexpression and TIGS experiments were performed by using cv Pallas and cv Golden Promise, respectively. Mean value ± sem of five independent bombardments, relative to the empty-vector controls.
Significant difference from empty-vector control (one-sample t test against hypothetical value 100).
Table IV.
Summary of functional characterization of HvGER genes with respect to the interaction of barley with B. graminis
n.a., Not analyzed.
| Subfamily | Tested Genea | Experiment in Leaf | Induced by Bgh | Effect of Overepression | Effect of TIGS | OXOX Activityb | SOD Activityc | Enzymatic Activity Required for Resistance |
|---|---|---|---|---|---|---|---|---|
| HvGER1 | HvGER1a | Inner tissues | Yes | None | n.a. | Yes | Nod | n.a. |
| HvGER2 | HvGER2a | Epidermis and inner | No | Uncleare | None | No | No | Unclearf |
| HvGER3 | HvGER3a | Epidermis | Yes | None | Resistance | No | n.a. | n.a. |
| HvGER4 | HvGER4d | Epidermis | Yes | Resistance | Hypersusceptibility | No | Yes | Unclearg |
| HvGER5 | HvGER5a | Epidermis | Yes | Resistance | Resistance | No | Yes | Yes |
| HvGER6 | HvGER6a | None | No | None | n.a. | No | n.a. | n.a. |
For ID and accession number of cDNA or genomic clone corresponding to this gene, see Table I.
Determined by transient expression in barley leaf epidermal cells.
Determined by stable expression in transgenic Arabidopsis.
Determined for wheat ortholog Gf-2.8 (germin gf-2.8 possessing OXOX activity; Schweizer et al., 1999b).
Resistance-enhancing effect could not be reproduced in a second experimental series.
Resistance-enhancing effect could not be reproduced in a second experimental series.
Not conclusive due to enhanced instability of recombinant mutant proteins in transgenic Arabidopsis.
We next addressed the question of whether the protection observed by transient overexpression of HvGER2a, HvGER4d, and HvGER5a was due to known or proposed enzymatic activities of the encoded proteins. For this purpose, site-directed mutagenesis of one of three conserved His residues of the catalytic center that has been proposed to be involved in Mn2+ binding was carried out by replacing it with Ser. This nonconservative amino acid replacement was expected to eliminate Mn2+ binding without drastically changing the polarity of the side chain or the pI of the amino acid. As shown in Table III, the mutant HvGER proteins had lost their ability to protect epidermal cells against Bgh. However, in the case of HvGER2a, wild-type protein was inefficient in protecting cells against Bgh in contrast to the first set of experiments (Table II). Although we currently do not understand this lack of reproducibility, it might reflect a complex situation of partial redundancy of HvGER function or dependence on endogenous factors like substrate availability, which might vary between experiments or experimental series. Interestingly, overexpression of HvGER5a (H109S) caused hypersusceptibility of epidermal cells compared to control cells that had been transformed with the empty vector plus the β-glucuronidase-encoding reporter plasmid pUbiGUS. This dominant negative effect of the presumably nonfunctional protein might be explained by competition with related, functional GLP proteins for binding sites or interaction partners in the epidermal cell wall.
Table III.
Site-directed mutagenesis eliminates the resistance-mediating effect of overexpressed HvGER proteins
| Gene | Relative Haustorial Indexa | p (t Test)b |
|---|---|---|
| % | ||
| HvGER4d wild type | 76.0 ± 4.7 | |
| HvGER4d (H110S) | 106.1 ± 12.5 | 0.033 |
| HvGER2 wild type | 98.4 ± 6.2 | |
| HvGER2 (H103S) | 122.4 ± 18.6 | 0.134 |
| HvGER5 wild type | 76.1 ± 11.0 | |
| HvGER5 (H109S) | 125.4 ± 2.3 | 0.002 |
Barley leaf segments were cobombarded with pUbiGUS and overexpression constructs encoding wild-type or mutated HvGER proteins, followed by challenge with Bgh 4 h later. Mean value ± sem of four independent bombardments relative to the empty-vector control pIPKTA9.
Significance of difference between wild-type and mutant forms (unpaired t test).
Characterization of Recombinant HvGER Proteins
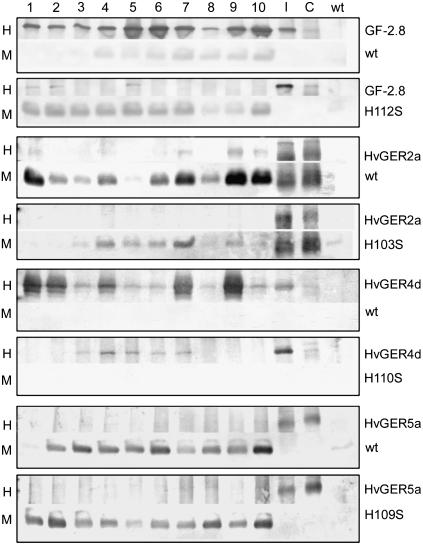
Genes encoding wild-type or mutant forms of HvGER2a, HvGER4d, and HvGER5a, which had been found in the first experimental series to protect epidermal cells against Bgh, were stably expressed in Arabidopsis transgenic plants. This experiment aimed to test whether the point mutation affected the accumulation of stable proteins in the transient assay system of barley. Moreover, it allowed testing of putative SOD activity of wild-type and mutant forms of the selected HvGERs. Gene gf-2.8 encoding wheat germin that has been successfully expressed in dicotyledonous transgenic plants (Berna and Bernier, 1997; Donaldson et al., 2001) was included as a positive control in this experiment. The polyclonal antibodies used to detect recombinant GF-2.8 and HvGER proteins were found not to cross-react with proteins from Arabidopsis, except for the antibodies used for GF-2.8 (Supplemental Fig. 5, band CR). However, this cross-reaction was weak and did not interfere with specific signals from transgene products. As shown in Figure 5, recombinant wild-type GF-2.8 and HvGER4d accumulated predominantly as homohexameric complexes with the same migration behavior in semidenaturing SDS-PAGE as endogenous barley GLPs. On the other hand, HvGER2a and HvGER5a predominantly migrated as monomers, indicating reduced stability at least as a recombinant protein expressed in Arabidopsis.
Figure 5.
Accumulation of recombinant HvGER proteins in transgenic Arabidopsis plants. Acid-soluble proteins were extracted from leaves of 10 T0 plants (nos. 1–10) and used for western blotting. I, Extract from Bgh-inoculated wheat (Gf-2.8 blot) or barley (all other blots) leaves; C, extract from noninoculated wheat (Gf-2.8 blot) or barley (all other blots) leaves; wt, extract from Arabidopsis wild-type plants; H, high molecular mass complex (approximately 100-kD apparent molecular mass); M, monomeric protein band (approximately 22-kD apparent molecular mass). HvGER5a was detected by using a polyclonal serum directed against HvGER1a. Five micrograms of protein were loaded per lane.
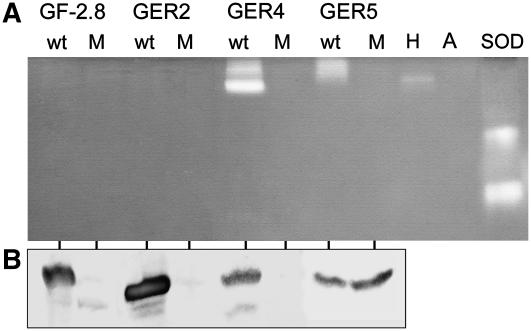
Exchange of one His by a Ser caused a destabilization of the recombinant GF-2.8, HvGER2a, and HvGER4d proteins, resulting in a shift from the homohexameric to the monomeric form or in a general decrease in protein abundance probably resulting from elimination of misfolded proteins by the cellular quality control system. This indicates that an intact Mn2+-binding center has a major impact on protein three-dimensional structure, which, by itself, affects the stability of quaternary complexes. It is currently unknown, however, to what extent the acidic extraction procedures used here (pH 2.8) negatively influence the stability of mutated proteins. By contrast, the three-dimensional structure of HvGER5a appeared to be less affected by the His-to-Ser exchange because abundance of this mutated protein was not decreased. Figure 6 shows that HvGER5a was identified as a new extracellular SOD migrating as a homohexameric complex in native PAGE, similar to HvGER4d. Mutation of one His to Ser eliminated SOD activity of both HvGER4d and HvGER5a. Therefore, the absence of protection of barley epidermal cells by transient overexpression of HvGER4d (H110S) can either be explained by decreased accumulation of accurately folded homohexameric protein, which was speculated also to have a structural role in cell wall reinforcement (Schweizer et al., 1999a) or by abolished SOD activity. On the other hand, we can conclude from data shown in Figures 5 and 6 that enhanced basal resistance caused by transient overexpression of HvGER5a was due to SOD activity of the encoded protein. As an additional result from testing recombinant cereal GLPs expressed in Arabidopsis, we could not confirm previous findings about SOD activity of purified GF-2.8 and HvGER2a proteins (Woo et al., 2000; Segarra et al., 2003). Similarly, OXOX but no SOD activity of recombinant HvGER1a expressed in yeast (Saccharomyces cerevisiae) was reported (Whittaker and Whittaker, 2002).
Figure 6.
HvGER4 and HvGER5 possess SOD activity that is destroyed by mutation of one conserved His residue. A, SOD in-gel assay with protein extracts from one selected Arabidopsis T0 line expressing either wild-type (wt) or mutant (M) GLP. H, Protein extract from Bgh-attacked barley leaves; A, protein extract from Arabidopsis wild-type leaves; SOD, commercial SOD from horseradish (Sigma-Aldrich). B, Same protein extracts as shown in A were subjected to western blotting.
OXOX activity of HvGER1a to 6a was tested in the transient assay system of barley by using a method described before (Schweizer et al., 1999b). GF-2.8 and HvGER1a showed strong OXOX activity, but none of the other HvGER proteins did so (data not shown).
DISCUSSION
Plant GLPs are encoded by multigene families. Despite a considerable amount of work that has been invested in this group of proteins, their biological function has largely remained a matter of speculation. Here, we present an extended overview of the HvGER multigene family in barley, including functional data related to basal defense against Bgh.
Multisequence alignments of GLPs in Arabidopsis and barley have revealed some overlap of multigene family structure. However, four subfamilies per species appeared to be species specific and contained no putative gene orthologs. Due to the fact that GLPs often appear to be noncovalently bound to cell walls (Vallelian et al., 1998; Lane, 2002) it may be speculated that these subfamilies reflect family- or class-specific roles of GLPs linked, for example, to specific structural features of corresponding cell walls. Another striking feature is the frequent occurrence of gene paralogs with very high sequence similarity and with low ka/ks ratios (Hurst, 2002) within the open reading frames, indicating purifying selection (Supplemental Table II). Functional redundancy of two proteins of the HvGER4 subfamily has been shown here because both protected barley from Bgh attack upon transient overexpression. Together, this suggests that promoters rather than CDS might have diversified after local or large-scale genome duplication events resulting in neofunctionalization. To address the question of divergence of expression patterns, we carried out metadata analysis of HvGER4 mRNA abundance in BarleyBase (http://www.barleybase.org), based on publicly available data from several experiments using the Barley1 chip from Affymetrix (Caldo et al., 2004; Boddu et al., 2006; Druka et al., 2006). Supplemental Figure 4 shows that, indeed, HvGER4 paralogs are differentially regulated, although global patterns still resemble each other, suggesting fine tuning of HvGER4 expression. On the other hand, subfunctionalization has recently been suggested as a driving force for maintenance of two active paralogs of HvGER3 (Federico et al., 2006). HvGER proteins might play an important role in young germinating seedlings, especially in their nongreen tissues, which are roots and coleoptiles, because mRNA abundance there was very high. The only exception was HvGER2 encoding an extracellular ADP-Glc pyrophosphatase/phosphodiesterase of unknown biological function that was highly expressed in very young primary leaves without detectable transcripts in roots (Rodriguez-Lopez et al., 2001). Due to a demonstrated, protective role of a few GLPs in transgenic plants or in transiently expressing cells, it is tempting to speculate about a role of HvGER proteins in preformed protection against opportunistic or true soil-borne pathogens. In leaves of 7-d-old seedlings, HvGER1, HvGER3, HvGER4, and HvGER5 expression became pathogen and H2O2 inducible and, in most cases, mRNA abundance was highest in the epidermal cell layer. Therefore, HvGERs might be important for defense against directly penetrating fungal pathogens in older plants. Regulation of HvGER2 was antagonistic to the remaining four HvGERs that are expressed in leaves because its transcripts declined upon pathogen attack, H2O2, or ozone treatment, as reported before (Vallelian et al., 1998).
A possible role of HvGERs in inducible host defense was tested in a transient assay system based on single transformed epidermal cells, which were subsequently challenge inoculated with Bgh (data summarized in Table IV). Interestingly, several HvGER genes not only enhanced basal resistance upon overexpression, but also upon TIGS. HvGER4d and HvGER5a were found to encode active enzymes with SOD activity that protected barley epidermal cells from Bgh attack upon overexpression. At least in the case of HvGER5a, SOD activity could be demonstrated to be required for resistance-enhancing activity. In several barley-B. graminis interactions, local accumulation of H2O2, the product of SOD, has been found to be correlated with mlo-mediated host or nonhost resistance (Piffanelli et al., 2002). This means that H2O2 production by HvGER4 and HvGER5 might be generally important for race-nonspecific resistance, including basal resistance. H2O2 is the product of HvGER1, HvGER4, and HvGER5, and exogenous H2O2 application was found to lead to transcript accumulation of several HvGERs. Therefore, a positive feedback loop of defense- or disease-related signaling involving HvGERs may be operating in barley.
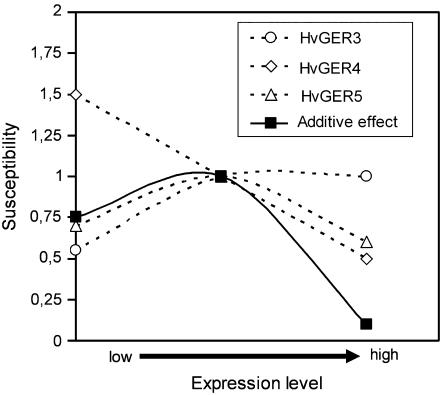
The picture is complicated by the fact that TIGS of HvGER4 and HvGER5 induced host hypersusceptibility and partial resistance, respectively. Therefore, unlike HvGER4, which behaved as a simple factor of basal defense with a linear correlation between protein accumulation and resistance, HvGER5 may have dual function: defense related due to its SOD activity and participation in a susceptibility-mediating protein-protein or protein-cell wall complex. The defense-related, SOD-dependent function would be predicted to be more important at higher levels of HvGER5 expression, whereas the susceptibility-mediating function might play a role at moderate levels of expression. Finally, HvGER3 behaved as a simple susceptibility-mediating factor.
In summary, a complex picture of fine regulation of basal host resistance or, depending on the point of view, basal susceptibility by a number of HvGER genes that encode proteins with partially overlapping functions emerges from data presented here (Fig. 7). Tunable mechanisms of quantitative resistance of hosts to their pathogens are likely to play an important role in the coevolution of plants with obligate biotrophic pathogens like Bgh and have the potential to stabilize host and pathogen populations over long periods of time due to their durable nature. It remains to be examined to what extent the strong constitutive expression of HvGERs in nongreen tissues of germinating seedlings provides protection against soil-borne (opportunistic) pathogens. In particular, the role of HvGER5 that was shown to encode an extracellular SOD with antifungal activity against Bgh and transcripts of which accumulate to very high levels in, for instance, young roots will have to be addressed with respect to preformed protection.
Figure 7.
Model of fine regulation of basal resistance by HvGER protein expression in barley. The model is based on TIGS (x axis low) and overexpression (x axis high) data of this study. The y axis shows the relative susceptibility compared to the empty-vector controls (set to 1), which are assumed to correspond to median expression levels. For calculation of the hypothetical additive effect of all three HvGER proteins, the differences between relative susceptibility values and control value 1 were added to value 1. Please note that all three HvGER subfamilies in the model are predominantly expressed in leaf epidermis (Fig. 4).
MATERIALS AND METHODS
Plants and Bgh
Barley (Hordeum vulgare) plants cv Ingrid, cv Golden Promise, or cv Pallas were grown in pots of compost soil (from IPK nursery) in a growth chamber (16-h light from metal halogen lamps; 8-h dark, 70% relative humidity, 20°C constant temperature). Blumeria graminis DC Speer f. sp. hordei (Bgh; isolate 4.8 carrying AvrMla9) was maintained at 22°C and 16-h light by weekly transfer to fresh barley cv Golden Promise. For RNA extraction from germinating seedlings, seeds were surface sterilized (5 min 70% [v/v] ethanol, 15 min 0.8% [v/v] sodium hypochlorite, four times for 10 min with water) followed by incubation on sterile, wet filter paper in a closed glass container. Water was replaced daily. For RNA extraction from mature plants, plants were grown in a greenhouse with automatic shading and supplementary light (sodium-halogen lamps), resulting in a light period of 16 h. Temperature ranged from 18°C (night) to 21°C (day).
Arabidopsis Transformation
Transformation of 6-week-old Arabidopsis (Arabidopsis thaliana ecotype Columbia-0) was performed by the vacuum infiltration method as described by Bechtold et al. (1993).
Stress Treatments
For all stress treatments, 7-d-old seedlings cv Ingrid were used, except for ozone treatments, where 21-d-old plants were used. For the duration of stress treatments, see Figure 3 (bottom). Cold: Plants were incubated at 4°C under a normal 16-h d/8-h night rhythm. Wound: Primary leaves were squeezed with a pair of tweezers at an interval of 7 mm. H2O2: Primary leaves were sprayed four times with a solution of 100 mm H2O2 in 0.05% (v/v) Tween 20 at an interval of 2 h. Control plants were sprayed with Tween 20 solution alone. No macroscopically visible tissue damage was induced by this H2O2 treatment. Ozone: Plants were acclimatized in the experimental growth chamber during 4 d (14-h light/10-h dark, 18°C/16°C during the light/dark period) and then treated daily with 190 ppb ozone for 9 h starting 2 to 3 h after onset of illumination. Sodium chloride: Plants grown on filter paper were watered with a solution of 150 mm NaCl. Cadmium chloride: Plants grown on filter paper were watered with a solution of 100 μm CdCl2. B. graminis: Plants were heavily inoculated by dusting Bgh spores from inoculated donor plants over the test plants. Phakopsora pachyrhizi: Plants were inoculated with a field isolate from Brazil by using standard procedures, except for dusting spores from inoculated soybean (Glycine max) plants directly onto test plants prewetted with 0.05% Tween 20 (Feng et al., 2005).
Clones and Constructs
For transient overexpression in barley, the selected cDNA or genomic clones (see Table I) were subcloned into pIPKTA9 (Supplemental Fig. 3) by using restriction sites BamHI and ApaI (HvGER1a, HvGER3a, HvGER4c, and HvGER6a), NotI and ApaI (HvGER2a), or SpeI and ApaI (HvGER5a). This allowed directional cloning into pIPKTA9, except for HvGER2a, where the ends of the excised cDNA insert were polished by T4 DNA polymerase followed by ligation into the SmaI cloning site of pIPKTA9. The final constructs were verified by sequencing. For HvGER3a, full CDS cDNA clones were not available within the IPK clone collection and, therefore, nested PCR from genomic barley DNA was performed by using primers 5′-GCTCACATGCAAGTTCATGCATATCA (HvGER3a sense outer), 5′-GCTCTGTCAATCTACGGCTAT (HvGER3a sense nested), 5′-GCTCACATGCAAGTTCATGC (HvGER3a antisense outer), and 5′-CTAGCATTACATATGAACTTTCCA (HvGER3a antisense nested). For HvGER6a, full CDS cDNA clones were not available either and, therefore, nested PCR from genomic barley DNA was performed by using primers 5′-TTGTAGGACACCATGATGATGG (HvGER6a sense outer), 5′-GATGATGGCACGTGTTTCC (HvGER6a sense nested), 5′-CGACTGCATTAGCATGACATG (HvGER6a antisense outer), and 5′-CGAACCACTGAGAATGGTACT (HvGER6a antisense nested). PCR fragments were subcloned into pCRII-TOPO (Invitrogen) and verified by sequencing.
For TIGS in barley, RNAi constructs were produced in vector pIPKTA30N as described (Douchkov et al., 2005), except for the use of GATEWAY entry vector pIPKTA33 instead of pIPKTA38. The two vectors differ in their multiple cloning sites (details available from authors upon request). Initial PCR fragments were produced by using primers 5′-GCCAACGCAATGTTGCTC (HvGER2a sense) and 5′-CGCGTAGTCAGTGATCTGGA (HvGER2a antisense), 5′-GCGAGTCAATGGATTTGCTT (HvGER3a sense) and 5′-GCCGTGGCTAGAACATCATC (HvGER3a antisense), 5′-TAGCAAGCAAGCATTGACCA (HvGER5a sense), and 5′-CCCCTGTTTTGCTGGAAGT (HvGER5a antisense). For TIGS of HvGER4, a previously described construct was used (Christensen et al., 2004).
For stable expression in Arabidopsis, expression cassettes containing the cauliflower mosaic virus 35S promoter, wild-type or mutagenized HvGER2a, HvGER4d, or HvGER5a, followed by the 35S terminator were excised from pIPKTA9 by EcoRI and subcloned into pBINPLUS (Vanengelen et al., 1995), cut with the same enzyme, and dephosphorylated. Wild-type or mutagenized Gf-2.8 (wheat [Triticum aestivum] OXOX gene gf-2.8) was subcloned from the expression cassette pOXOX described previously by using EcoRI (Schweizer et al., 1999a). Recombinant clones with inserts in tandem orientation to the kanamycin resistance cassette were selected for transformation. Correct integration of effector genes was confirmed by sequencing using primer pBIN_reverse, 5′-TGACCATGATTACGCCAAGC.
Site-Directed Mutagenesis
Site-directed mutagenesis was performed using the QuickChange kit (Stratagene) and overexpression constructs in pIPKTA9 as templates. Primers used for Gf-2.8 (H111S) were 5′-GGCACCAACCCACCAAGCATCCACCCGCGTGCC and 5′-GGCACGCGGGTGGATGCTTGGTGGGTTGGTGCC; for HvGER2a (H103S), 5′-GGCGTCGTGCCGATGAGCACCCACCCGGCCGCC and 5′-GGCGGCCGGGTGGGTGCTCATCGGCACGACGCC; for HvGER4d (H110S), 5′-TTGGGTCAGAACCCGCCAAGCACGCACCCGCGGGCCACTGAGATCCTCA and 5′-TGAGGATCTCAGTGGCCCGCGGGTGCGTGCTTGGCGGGTTCTGACCCAA; and for HvGER5a (H109S), 5′-GGCCAGAACCCGCCGAGCACCCACCCGCGCGCC and 5′-GGCGCGCGGGTGGGTGCTCGGCGGGTTCTGGCC. Mutations were verified by sequencing.
Transcript Analysis
RNA was isolated from different tissues and developmental stages of barley and blotted onto nylon membranes (Hybond N) as described (Vallelian et al., 1998), except for stripped leaf epidermis from which RNA was extracted by using the NucleoSpin RNA L kit (Machery-Nagel).
For RNA-blot analysis, 10 μg of total RNA were loaded per lane of formaldehyde-containing agarose gels. 32P-labeled cDNA probes covering the entire open reading frame of HvGERs were produced by using random-prime labeling and hybridized to RNA blots overnight at 65°C in CHURCH buffer (0.5 m sodium phosphate, pH 7.2, 1% [w/v] bovine serum albumin, 7% [w/v] SDS, 1 mm sodium EDTA) followed by washing three times for 20 min in 0.1× SSC, 0.1% (w/v) SDS at 65°C. Phosphor imager screens (Amersham) were exposed to blots and scanned by using a BAS 3000 instrument (Fuji). All RNA blots were rehybridized with a diluted, 32P-labeled 26S rRNA probe of Phaseolus vulgaris as an internal control of gel-loading and blotting efficiency. Hybridization signals (bands) were quantified by using Tina 2.08 software (Raytest) and normalized to quantified signals from 26S rRNA hybridizations.
For reverse RNA-blot analysis, cDNA clones HW09M16 (for HvGER1a; accession no. AL505876), HK03B24 (for HvGER2a; accession no. AL450548), HK04J17 (for HvGER3a; accession no. AL450860), HO03H17 (for HvGER4d; accession no. CD053788), HW05P16 (for HvGER5a; accession no. AL504660), and HK04D13 (for HvGER6a; accession no. AL450784) were PCR amplified (30 cycles 95°C, 65°C, 72°C) in four 50-μL reactions by using in-house-produced Taq polymerase and primers M13-21PE (5′-ACGACGTTGTAAAACGACGGCCAG) and MVR26 (5′-CTCACTAAAGGGAACAAAAGCTGG) together with an additional 10,274 unigenes from barley. Pooled PCR fragments from the four parallel reactions were quality controlled by agarose gel electrophoresis, purified, and 2-fold concentrated by ultrafiltration (MinElute; Qiagen), resulting in an estimated DNA concentration of 50 ng/μL. DNA was spotted in duplicate onto nylon membranes (Biodyne B; PALL Company) by using a BioGrid II Robot (Genomics Solutions) equipped with a 0.2-mm pin tool as described (Zierold et al., 2005). This resulted in a cDNA macroarray, which can be used for general transcript-profiling experiments as well as for targeted reverse-northern approaches of selected mRNAs as described here. Membrane hybridization with 33P-labeled cDNA probes, spot detection, and data processing were done as described (Zierold et al., 2005). Signal intensities of HvGER transcripts were normalized by median centering of spot intensities of all 10,280 unigenes of the cDNA macroarray.
Protein Analysis
HvGERs were described as acid-soluble, extracellular proteins (Vallelian et al., 1998). Therefore, to enrich protein extracts from transgenic Arabidopsis plants for recombinant TaGLP or HvGER, protein leaves were homogenized in phosphate/citrate buffer at pH 2.8 as described (Schweizer et al., 1997). Five micrograms of protein per lane were separated on 12% SDS-PAGE gels followed by blotting onto nitrocellulose (Bio-Rad). Seminative SDS-PAGE was carried out by separating proteins dissolved in SDS sample buffer without β-mercaptoethanol and without boiling (Zhang et al., 1995). Detection of Gf-2.8 and HvGER5a was performed by a polyclonal antiserum directed against HvGER1 at a dilution of 1:2,000 (Zhou et al., 1998). Detection of HvGER2a was performed by using a polyclonal antiserum directed against HvGER2 at a dilution of 1:2,000 (Vallelian et al., 1998). Detection of HvGER4d was performed by using a polyclonal antiserum directed against HvGER4d at a dilution of 1:1,000 (Wei et al., 1998). Cross-reactive bands were stained by using a secondary anti-rabbit antibody coupled to alkaline phosphatase at a dilution of 1:2,000.
Enzymatic Assays
OXOX in situ activity after bombardment of barley leaf segments was performed as described (Schweizer et al., 1999b). SOD in gel assay of recombinant HvGERs was carried out as described (Mock et al., 1998). As a positive control, horseradish SOD (catalog no. S4636; Sigma-Aldrich) was used.
Transient Overexpression and TIGS
HvGER genes were transiently overexpressed in bombarded barley epidermal cells of cv Pallas by using a PDS-1000 System (Bio-Rad) as described (Christensen et al., 2004). Leaf segments were challenge inoculated with Bgh 4 h postbombardment at a density of 150 to 200 conidia/mm2 and stained for GUS reporter-protein activity 40 h postinoculation. TIGS was performed in barley cv Golden Promise as described (Douchkov et al., 2005). Bombarded leaf segments were challenge inoculated with Bgh 48 h postbombardment. The selection of cultivars for overexpression or TIGS was based on previous observations about genotype-dependent effects of HvGERs (Christensen et al., 2004).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ647619 to DQ647625.
Supplementary Material
Acknowledgments
The technical assistance of Elke Liemann for the production of transgenic Arabidopsis and of Annegret Wolf and Petra Linow for SOD in-gel assay is acknowledged. The authors thank Dr. A. Schützendübel for ozone treatments, Dr. H. Schultheiss for inoculation with the soybean rust fungus, Dr. H. Thordal-Christensen for the generous gift of anti-HvGER1 and anti-HvGER4 antisera, Maria L. Federico for making available data in press, and both anonymous reviewers for very valuable comments.
This work was supported by the Deutsche Forschungsgemeinschaft (project no. SCHW–848/1–1 and /1–2) and by the Leibniz Institute of Plant Genetics and Crop Plant Research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Patrick Schweizer (schweiz@ipk-gatersleben.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Baumlein H, Braun H, Kakhovskaya IA, Shutov AD (1995) Seed storage proteins of spermatophytes share a common ancestor with desiccation proteins of fungi. J Mol Evol 41: 1070–1075 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In-planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Ser III Sci Vie 316: 1194–1199 [Google Scholar]
- Berna A, Bernier F (1997) Regulated expression of a wheat germin gene in tobacco: oxalate oxidase activity and apoplastic localization of the heterologous protein. Plant Mol Biol 33: 417–429 [DOI] [PubMed] [Google Scholar]
- Boddu J, Cho S, Kruger WM, Muehlbauer GJ (2006) Transcriptome analysis of the barley-Fusarium graminearum interaction. Mol Plant Microbe Interact 19: 407–417 [DOI] [PubMed] [Google Scholar]
- Caldo RA, Nettleton D, Wise RP (2004) Interaction-dependent gene expression in Mla-specified response to barley powdery mildew. Plant Cell 16: 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell KS, Russell J, Langridge P, Powell W (2006) Extreme population-dependent linkage disequilibrium detected in an inbreeding plant species, Hordeum vulgare. Genetics 172: 557–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Graham RA, Thornburg RW (1998) Arabidopsis thaliana contains a large family of germin-like proteins: characterization of cDNA and genomic sequences encoding 12 unique family members. Plant Mol Biol 38: 929–943 [DOI] [PubMed] [Google Scholar]
- Christensen AB, Thordal-Christensen H, Zimmermann G, Gjetting T, Lyngkjaer MF, Dudler R, Schweizer P (2004) The germinlike protein GLP4 exhibits superoxide dismutase activity and is an important component of quantitative resistance in wheat and barley. Mol Plant Microbe Interact 17: 109–117 [DOI] [PubMed] [Google Scholar]
- Cober ER, Rioux S, Rajcan I, Donaldson PA, Simmonds DH (2003) Partial resistance to white mold in a transgenic soybean line. Crop Sci 43: 92–95 [Google Scholar]
- Donaldson PA, Anderson T, Lane BG, Davidson AL, Simmonds DH (2001) Soybean plants expressing an active oligomeric oxalate oxidase from the wheat gf-2.8 (germin) gene are resistant to the oxalate-secreting pathogen Sclerotina sclerotiorum. Physiol Mol Plant Pathol 59: 297–307 [Google Scholar]
- Douchkov D, Nowara D, Zierold U, Schweizer P (2005) A high-throughput gene-silencing system for the functional assessment of defense-related genes in barley epidermal cells. Mol Plant Microbe Interact 18: 755–761 [DOI] [PubMed] [Google Scholar]
- Druka A, Kudrna D, Kannangara CG, von Wettstein D, Kleinhofs A (2002) Physical and genetic mapping of barley (Hordeum vulgare) germin-like cDNAs. Proc Natl Acad Sci USA 99: 850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druka A, Muehlbauer G, Druka I, Caldo R, Baumann U, Rostoks N, Schreiber A, Wise R, Close T, Kleinhofs A, et al (2006) An atlas of gene expression from seed to seed through barley development. Funct Integr Genomics 6: 202–211 [DOI] [PubMed] [Google Scholar]
- Dunwell JM, Khuri S, Gane PJ (2000) Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol Mol Biol Rev 64: 153–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell JM, Purvis A, Khuri S (2004) Cupins: the most functionally diverse protein superfamily? Phytochemistry 65: 7–17 [DOI] [PubMed] [Google Scholar]
- Federico ML, Iniguez-Lui FL, Skadsen RW, Kaeppler HF (2006) Spatial and temporal divergence of expression in duplicated barley germin-like protein-encoding genes. Genetics (in press) [DOI] [PMC free article] [PubMed]
- Feng PCC, Baley GJ, Clinton WP, Bunkers GJ, Alibhai MF, Paulitz TC, Kidwell KK (2005) Glyphosate inhibits rust diseases in glyphosate-resistant wheat and soybean. Proc Natl Acad Sci USA 102: 17290–17295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD (2002) The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet 18: 486–487 [DOI] [PubMed] [Google Scholar]
- Laloi C, Apel K, Danon A (2004) Reactive oxygen signalling: the latest news. Curr Opin Plant Biol 7: 323–328 [DOI] [PubMed] [Google Scholar]
- Lane BG (1994) Oxalate, germin, and the extracellular matrix of higher plants. FASEB J 8: 294–301 [DOI] [PubMed] [Google Scholar]
- Lane BG (2002) Oxalate, germins, and higher-plant pathogens. IUBMB Life 53: 67–75 [DOI] [PubMed] [Google Scholar]
- Lane BG, Bernier F, Dratewkakos E, Shafai R, Kennedy TD, Pyne C, Munro JR, Vaughan T, Walters D, Altomare F (1991) Homologies between members of the germin gene family in hexaploid wheat and similarities between these wheat germins and certain Physarum spherulins. J Biol Chem 266: 10461–10469 [PubMed] [Google Scholar]
- Lane BG, Dunwell JM, Ray JA, Schmitt MR, Cuming AC (1993) Germin: a protein marker of early plant development is an oxalate oxidase. J Biol Chem 268: 12239–12242 [PubMed] [Google Scholar]
- Liang H, Maynard CA, Allen RD, Powell WA (2001) Increased Septoria musiva resistance in transgenic hybrid poplar leaves expressing a wheat oxalate oxidase gene. Plant Mol Biol 45: 619–629 [DOI] [PubMed] [Google Scholar]
- Livingstone DM, Hampton JL, Phipps PM, Grabau EA (2005) Enhancing resistance to Sclerotinia minor in peanut by expressing a barley oxalate oxidase gene. Plant Physiol 137: 1354–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou YG, Baldwin IT (2006) Silencing of a germin-like gene in Nicotiana attenuata improves performance of native herbivores. Plant Physiol 140: 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock HP, Keetman U, Kruse E, Rank B, Grimm B (1998) Defense responses to tetrapyrrole-induced oxidative stress in transgenic plants with reduced uroporphyrinogen decarboxylase or coproporphyrinogen oxidase activity. Plant Physiol 116: 107–116 [Google Scholar]
- Piffanelli P, Zhou FS, Casais C, Orme J, Jarosch B, Schaffrath U, Collins NC, Panstruga R, Schulze-Lefert P (2002) The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol 129: 1076–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramputh AI, Arnason JT, Cass L, Simmonds JA (2002) Reduced herbivory of the European corn borer (Ostrinia nubilalis) on corn transformed with germin, a wheat oxalate oxidase gene. Plant Sci 162: 431–440 [Google Scholar]
- Rodriguez-Lopez M, Baroja-Fernandez E, Zandueta-Criado A, Moreno-Bruna B, Munoz FJ, Akazawa T, Pozueta-Romero J (2001) Two isoforms of a nucleotide-sugar pyrophosphatase/phosphodiesterase from barley leaves (Hordeum vulgare L.) are distinct oligomers of HvGER1, a germin-like protein. FEBS Lett 490: 44–48 [DOI] [PubMed] [Google Scholar]
- Schneider M, Droz E, Malnoe P, Chatot C, Bonnel E, Metraux JP (2002) Transgenic potato plants expressing oxalate oxidase have increased resistance to oomycete and bacterial pathogens. Potato Res 45: 177–185 [Google Scholar]
- Schweizer P, Buchala A, Silverman P, Seskar M, Raskin I, Metraux JP (1997) Jasmonate-inducible genes are activated in rice by pathogen attack without a concomitant increase in endogenous jasmonic acid levels. Plant Physiol 114: 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer P, Christoffel A, Dudler R (1999. a) Transient expression of members of the germin-like gene family in epidermal cells of wheat confers disease resistance. Plant J 20: 541–552 [DOI] [PubMed] [Google Scholar]
- Schweizer P, Pokorny J, Abderhalden O, Dudler R (1999. b) A transient assay system for the functional assessment of defense-related genes in wheat. Mol Plant Microbe Interact 12: 647–654 [Google Scholar]
- Segarra CI, Casalongue CA, Pinedo ML, Ronchi VP, Conde RD (2003) A germin-like protein of wheat leaf apoplast inhibits serine proteases. J Exp Bot 54: 1335–1341 [DOI] [PubMed] [Google Scholar]
- Shutov AD, Blattner FR, Baumlein H (1999) Evolution of a conserved protein module from Archaea to plants. Trends Genet 15: 348–349 [DOI] [PubMed] [Google Scholar]
- Shutov AD, Braun H, Chesnokov YV, Baumlein H (1998) A gene encoding a vicilin-like protein is specifically expressed in fern spores—evolutionary pathway of seed storage globulins. Eur J Biochem 252: 79–89 [DOI] [PubMed] [Google Scholar]
- Vallelian BL, Mosinger E, Metraux JP, Schweizer P (1998) Structure, expression and localization of a germin-like protein in barley (Hordeum vulgare L.) that is insolubilized in stressed leaves. Plant Mol Biol 37: 297–308 [DOI] [PubMed] [Google Scholar]
- Vanengelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ (1995) Pbinplus—an improved plant transformation vector based on Pbin19. Transgenic Res 4: 288–290 [DOI] [PubMed] [Google Scholar]
- Wei YD, Zhang ZG, Andersen CH, Schmelzer E, Gregersen PL, Collinge DB, Smedegaard-Petersen V, Thordal-Christensen H (1998) An epidermis/papilla-specific oxalate oxidase-like protein in the defence response of barley attacked by the powdery mildew fungus. Plant Mol Biol 36: 101–112 [DOI] [PubMed] [Google Scholar]
- Whittaker MM, Whittaker JW (2002) Characterization of recombinant barley oxalate oxidase expressed by Pichia pastoris. J Biol Inorg Chem 7: 136–145 [DOI] [PubMed] [Google Scholar]
- Woo EJ, Dunwell JM, Goodenough PW, Marvier AC, Pickersgill RW (2000) Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat Struct Biol 7: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Wu SP, Druka A, Horvath H, Kleinhofs A, Kannangara CG, von Wettstein D (2000) Functional characterization of seed coat-specific members of the barley germin gene family. Plant Physiol Biochem 38: 685–698 [Google Scholar]
- Wu YX, von Tiedemann A (2002) Evidence for oxidative stress involved in physiological leaf spot formation in winter and spring barley. Phytopathology 92: 145–155 [DOI] [PubMed] [Google Scholar]
- Yamahara T, Shiono T, Suzuki T, Tanaka K, Takio S, Sato K, Yamazaki S, Satoh T (1999) Isolation of a germin-like protein with manganese superoxide dismutase activity from cells of a moss, Barbula unguiculata. J Biol Chem 274: 33274–33278 [DOI] [PubMed] [Google Scholar]
- Zhang ZG, Collinge DB, Thordal-Christensen H (1995) Germin-like oxalate oxidase, a H2O2-producing enzyme, accumulates in barley attacked by the powdery mildew fungus. Plant J 8: 139–145 [Google Scholar]
- Zhou FS, Zhang ZG, Gregersen PL, Mikkelsen JD, de Neergaard E, Collinge DB, Thordal-Christensen H (1998) Molecular characterization of the oxalate oxidase involved in the response of barley to the powdery mildew fungus. Plant Physiol 117: 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierold U, Scholz U, Schweizer P (2005) Transcriptome analysis of mlo-mediated resistance in the epidermis of barley. Mol Plant Pathol 6: 139–151 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.