Abstract
The APETALA2 (AP2) domain defines a large family of DNA-binding proteins that play important roles in plant morphology, development, and stress response. We describe isolation and characterization of a gene (CAP2) from chickpea (Cicer arietinum) encoding a novel AP2-family transcription factor. Recombinant CAP2 protein bound specifically to C-repeat/dehydration-responsive element in gel-shift assay and transactivated reporter genes in yeast (Saccharomyces cerevisiae) one-hybrid assay. CAP2 appeared to be a single/low copy intronless gene, and the protein product localized in the nucleus. Transcript level of CAP2 increased by dehydration and by treatment with sodium chloride, abscisic acid, and auxin, but not by treatment with low temperature, salicylic acid, and jasmonic acid. The 35S promoter-driven expression of CAP2 in tobacco (Nicotiana tabacum) caused drastic increase in the leaf cell size, and, thereby, in leaf surface area and number of lateral roots. Transgenic plants demonstrated more tolerance to dehydration and salt stress than the wild-type plants. Transgenic plants expressed higher steady-state transcript levels of abiotic stress-response genes NtERD10B and NtERD10C and auxin-response genes IAA4.2 and IAA2.5. Taken together, our results indicated a mutual interrelation between plant growth-development and abiotic stress-response pathways and a probable involvement of CAP2 in both the signaling pathways.
The APETALA2 (AP2)/ethylene-responsive factor (ERF) family of proteins regulates diverse processes of plant development and metabolism, such as vegetative and reproductive development, cell proliferation, secondary metabolism, biotic and abiotic stress responses, and responses to different plant hormones. These transcription regulators are characterized by the presence of approximately 60-amino acid-long AP2/ERF DNA-binding domains that directly interact with GC-rich cis-acting elements (GCC box/C-repeat) in the promoter of their target genes. Proteins belonging to the AP2 subgroup contain two copies of the DNA-binding domain (BD) separated by a spacer region (Meyerowitz, 1994; Okamuro et al., 1997). In Arabidopsis (Arabidopsis thaliana), the homeotic gene AP2 is best known for its central role in establishment of floral meristem, determination of sepal and petal identity, and expression of other floral homeotic genes (Irish and Sussex, 1990; Bowman et al., 1993). Loss-of-function AP2 mutation causes a relative increase in the seed mass (Ohto et al., 2005). AP2 gene also expresses at RNA level in the nonfloral organs, suggesting its involvement in the development processes of other organs as well; however, AP2-mutant plants do not show apparent defects in vegetative development (Jofuku et al., 1994). Another homeotic gene of this subgroup, AINTEGUMENTA (ANT), is critical in regulation of ovule and female gametophyte development (Klucher et al., 1996). BABY BOOM (BBM) expresses preferentially in developing embryo and seeds in Arabidopsis and Brassica napus. Its ectopic expression induces pleiotropic phenotypes such as spontaneous formation of somatic embryo, neoplastic growth, and hormone-free regeneration of explants (Boutilier et al., 2002). PLETHORA1 and PLETHORA2 (PLT1 and PLT2) genes encoding repeated AP2 domain proteins are transcribed in response to auxin in the basal embryo region, embryonic root primordium, and later in root meristem stem cells, and are essential in maintaining the position of the stem cell niche in the Arabidopsis root (Aida et al., 2004). ERF/EREBPs, referred to as the ethylene-responsive element binding proteins, were first identified in Nicotiana and possess a single copy of the AP2 domain (Ohme-Takagi and Shinshi, 1995). ESR1 gene (enhancer of shoot regeneration), encoding a single AP2-domain protein, when overexpressed promotes cytokinin-independent shoot regeneration from Arabidopsis root explants (Banno et al., 2001). Semidominant expression of TINY, a similar protein, suppresses cell proliferation during vegetative and floral organogenesis (Wilson et al., 1996). Genetic and molecular studies with several other proteins related to AP2 (RAP2) also indicated that the AP2/ERF family of proteins functions in several tissues and organs and plays important roles in general plant development (Okamuro et al., 1997).
Transcriptional activation of the osmotic stress-response genes is regulated by abscisic acid (ABA)-dependent and -independent signal transduction pathways and by their mutual cooperation. Promoter analysis of these genes has identified ABA-dependent and ABA-independent cis-acting elements (Ingram and Bartels, 1996; Yamaguchi-Shinozaki and Shinozaki, 1997). A 9-bp conserved sequence (TACCGAC), referred to as dehydration-responsive element (DRE)/C-repeat, which functions independent of ABA, was found essential and sufficient for the high expression of an Arabidopsis gene, rd29A, under drought, low temperature, and high salinity (Yamaguchi-Shinozaki and Shinozaki, 1994). Subsequently, DRE-related sequences were identified in the promoters of several other abiotic stress-inducible genes. Transcription factor(s) (DREB) that specifically interacts with DRE sequences has been cloned from a number of plant systems (Shinozaki et al., 2003). The DREB group of proteins contains a single AP2 domain-like ERF. Transcription of DREB1A gene is induced quickly at low temperature, while the DREB2A is induced by dehydration and high salinity. Ectopic expression of DREB1A alone, but not DREB2A, induced expression of many of the stress-response genes under normal growth condition and improved tolerance of the transgenic plants to drought, high salinity, and cold. However, expression of DREB1A under strong constitutive promoter affected the general development of the transgenic plants and resulted in severe growth retardation (Liu et al., 1998; Kasuga et al., 1999).
The plant hormone auxin regulates a number of cellular and developmental processes depending on the context. It plays critical roles in patterning different tissues and organs, for example, promotion or inhibition of organ development according to tropic responses, and maintenance of the positions of the root and shoot stem cells during embryogenesis, initiation, and emergence of lateral roots. It also influences cell division, cell growth, and differentiation. At the molecular level, auxin exerts its effect by regulating expression of numerous auxin-responsive genes (e.g. auxin/indole-3-acetic acid [AUX/IAA]). A family of transcription factors (auxin-responsive factor [ARF]) binds to the auxin-response element in the promoters of auxin-inducible genes to promote auxin-mediated gene induction response (Leyser, 2001, 2002; Reed, 2001; Ljung et al., 2002).
Cross interactions of signal transduction pathways keep balance between the growth and development of the plants and their responses to different external stimuli. Mechanical wounding induces genes related to abiotic stress and hormonal responses (Cheong et al., 2002). Ectopic expression of a pathogenesis-related transcription factor (OPBP1) induces tolerance against salinity stress (Guo et al., 2004). Exposure to reactive oxygen species enhances auxin-driven cell division and promotes formation of lateral roots (Pasternak et al., 2005). Loss-of-function mutation of a positive regulator of ABA signaling, AtMYC2, results in photomorphological growth but compromised lateral root formation (Yadav et al., 2005).
In this study, we report full-length cloning and characterization of a chickpea (Cicer arietinum) expressed sequence tag (EST), earlier reported by us, that is induced by dehydration (Boominathan et al., 2004). The gene CAP2 (for C. arietinum AP2) encodes a 202-amino acid protein with an AP2/ERF domain. CAP2 protein binds specifically to DRE in vitro. Transgenic tobacco (Nicotiana tabacum) lines expressing CAP2 showed higher constitutive expression of abiotic stress-responsive genes, improved tolerance to high salinity and osmotic stresses, and, in addition, had morphological features characteristic of auxin response, demonstrating interaction of two signal transduction pathways and a potential involvement of a transcription factor in both the pathways.
RESULTS
CAP2 Encodes an AP2/ERF Family Protein
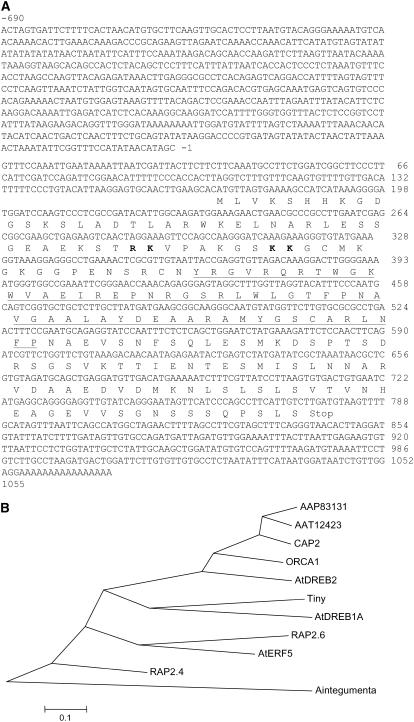
The 2 × 105 plaque-forming unit of a cDNA library constructed with mRNA isolated from 6-d-old chickpea seedlings dehydrated for 5 h was screened with a chickpea EST (GenBank accession no. CD051361), which was induced by 5 h of dehydration treatment (Boominathan et al., 2004). A clone of 1,055-bp size was obtained that possesses a conserved AP2/ERF domain in the deduced amino acid sequence and is therefore named C. arietinum AP2 (CAP2; GenBank accession no. DQ321719). CAP2 possesses a 609-bp open reading frame (ORF) encoding a predicted protein of 202 amino acids (Fig. 1A). Transcription start site was mapped at a G residue 169 bp upstream of the translation initiation site determined by primer extension analysis with a primer encompassing the translational start site (data not shown). A fragment of 690 bp, 5′ to the transcription start site, was also cloned by genome walking (Fig. 1A). The CAP2 protein possesses a 57-amino acid ERF-type AP2 domain with a conserved YRG element and an 18-amino acid core region (RLWLG) predicted to form an amphipathic α-helix. In silico analyses of the CAP2 protein sequence indicated a number of posttranslational modifications, as it contains five potential casein kinase 2 phosphorylation sites (at amino acids 14, 31, 129, 133, and 148); four potential protein kinase C phosphorylation sites (at amino acids 38, 47, 133, and 145); two potential N-glycosylation sites (at amino acids 127 and 174); and six potential myristoylation sites (at amino acids 46, 56, 92, 99, 144, and 193). There are other amino acids (Lys-4, Trp-22, Lys-23, Asn-26) also in the N terminus found to be invariant with most other AP2 family proteins. The basic amino acid-rich region at the N terminus may function for nuclear localization. We compared the amino acid sequence of CAP2 with other known AP2 family proteins. Only two proteins of similar size (GenBank accession nos. AAP83131 and AAT12423) from another legume, soybean (Glycine max), showed extensive homology throughout the length of the protein. No other proteins shared any significant stretch of homology outside the AP2/ERF domain. Phylogenetic analysis showed that CAP2 and the two soybean proteins are of the same lineage in a cluster having ORCA1 of Catharanthus roseus and DREB2 of Arabidopsis (Fig. 1B).
Figure 1.
A, cDNA, deduced amino acid, and genomic DNA (5′ to the transcription start site −1 to −690) sequence of CAP2. The AP2 domain is indicated by underline. Bold fonts indicate the basic amino acids that may act as nuclear localization signal. B, Phylogenetic tree showing relationship between CAP2 and other well-studied AP2/ERF family proteins. The tree was generated using the neighbor-joining algorithm of MEGA 2.0 software, version 2.1. The bar indicates the scale for branch length.
Stress-Induced Expression of CAP2
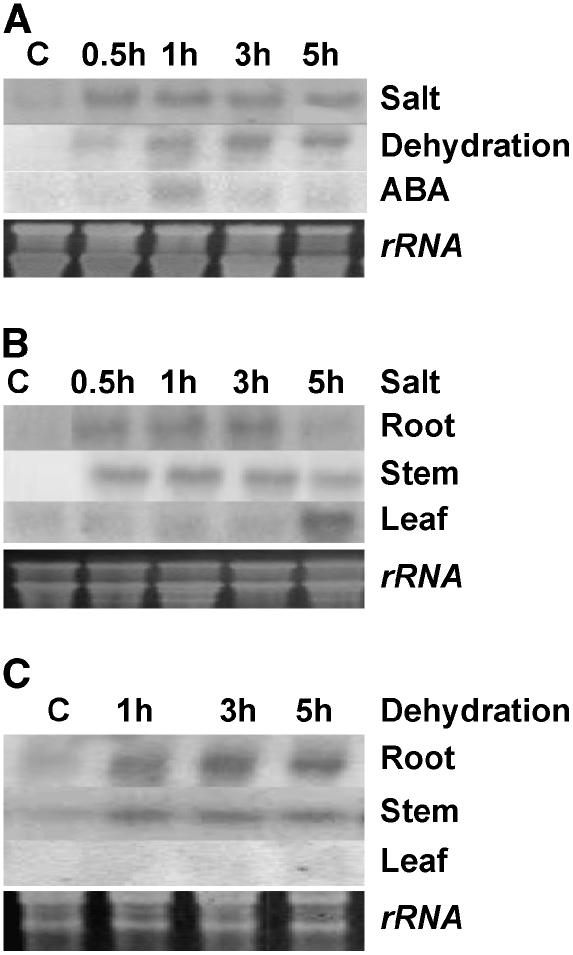
Expression pattern of CAP2 mRNA was analyzed using RNA-gel blot (Fig. 2A). A CAP2 cDNA fragment representing the C terminus of the protein was used as a probe. In the whole seedling, very low levels of CAP2 mRNA expression can be detected in normal growth conditions, indicating its requirement in normal developmental process. The steady-state transcript of CAP2 quickly reached the maximum level within one-half hour of treatment with 150 mm NaCl and maintained the level up to 5 h. Under dehydration, CAP2 mRNA began to accumulate within 30 min and reached the highest level within 3 h and remained the same at 5 h. Accumulation of CAP2 transcript in root reached its maximum level within 30 min under salt stress and remained the same up to 3 h, but drastically reduced to almost a basal level at 5 h. In stem, the CAP2 transcript accumulation followed the same course as in the root, but reduction at 5 h was not so drastic. Accumulation of CAP2 transcript in leaf, under salt stress, was totally different and interesting. It appeared that down-regulation of CAP2 transcription in root and stem at 5 h under salt stress was accompanied by up-regulation in leaf (Fig. 2B), which indicated that either the mechanism of CAP2 induction in leaf in salt stress is different from that in root and stem, or a high threshold amount of salt accumulation in leaf is required to induce CAP2 expression. Expression of CAP2 in root and stem is similar in response to dehydration. There was no detectable expression in leaf in response to the same treatment (Fig. 2C). Treatment with 100 μm ABA transiently induced CAP2 mRNA accumulation in the whole seedling after 1 h, then declined and maintained at a very low level up to 5 h. Cold (4°C), wound, salicylic acid (5 mm), and jasmonic acid (100 μm) treatments could not induce detectable CAP2 mRNA expression (data not shown).
Figure 2.
Expression pattern of CAP2 after different stresses to chickpea seedlings. A, Salt (150 mm), dehydration (withdrawal from soil), and ABA (100 μm). B and C, Expression of CAP2 in different organs under salt stress (B) and under dehydration (C). Northern transfer of 20 μg of total RNA was hybridized with a cDNA fragment representing the C-terminal part of CAP2 (“Materials and Methods”) used as a radiolabeled probe. Ethidium bromide-stained rRNA was used for equivalent loading control.
CAP2 Protein Binds in Vitro to C-Repeat/DRE and Activates Transcription in Yeast
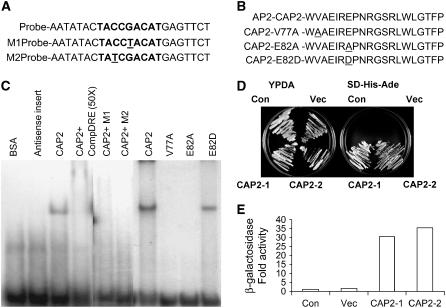
A gel-shift experiment was conducted to demonstrate that the CAP2 protein binds specifically to C-repeat/DRE. The dimer of a 23-bp stretch from the promoter of Arabidopsis RD29A gene containing C-repeat/DRE (TACCGACAT) was used as the probe (Fig. 3A). The full-length ORF of CAP2 cDNA was cloned in pGEX4T-2 vector (Amersham) and expressed in Escherichia coli DH5α to produce CAP2 protein fused to glutathione S-transferase (GST) at its N terminus. Bacterially expressed GST-CAP2 produced gel shift when wild-type probe was used, while a GST-fused antisense clone of CAP2 could not. The binding was competed out by excess cold probe. None of the mutated probes produced by substituting either the cytidine at the third or the guanosine at the fifth position of the C-repeat could produce gel shift, demonstrating specificity of the binding to this element (Fig. 3C). Val-13 and Glu-18 in the AP2 domain have been shown to be essential for the DNA binding (Sakuma et al., 2002). Therefore, we individually substituted the corresponding amino acids (Val-77 and Glu-82) in CAP2 protein with Ala (Fig. 3B). Both the mutations caused failure to bind the probe. But the mutant where Glu-82 was replaced with another acidic amino acid (Asp) weakly bound to the probe, showing that acidic moiety is essential at that position (Fig. 3C).
Figure 3.
A, The C-repeat/DRE sequences tested for gel-shift assay are either wild-type version found in the promoter of Arabidopsis RD29A (bold fonts represent 9-bp element) or mutant versions M1 and M2. Monomers are shown, and dimers were used in the experiments. Modified bases are underlined. B, Part of AP2 domains of the wild-type CAP2 and the mutated proteins V77A, E82A, and E82D used in the gel-shift assay. Modified amino acids are underlined. C, Gel-shift assays demonstrating that CAP2 binds to the C-repeat/DRE probe. The probes (1 ng) used in all reactions were 32P-labeled dimers of the oligonucleotides shown in A. Mutated and wild-type recombinant CAP2 proteins, described in B, were expressed in E. coli DH5α as GST-fused proteins and purified by GST-agarose columns. CAP2 cDNA was cloned in antisense orientation and purified similarly to make antisense protein. D, Activation of two reporter genes in yeast by CAP2. CAP2 cDNA cloned in yeast expression vector to produce CAP2 protein fused with GAL4 DNA BD was used for transformation into a yeast strain carrying three reporter genes, HIS3, ADE2, and LacZ, under the control of the GAL4 promoter. Yeast colonies carrying no vector (Con), vector only (Vec), and two transformants (CAP2-1 and CAP2-2) carrying CAP2 cDNA cloned in vector were grown on YPDA and on synthetic medium lacking His and Ade. E, Activation of the third reporter gene as in D is shown by β-galactosidase assay of the transformants presented as fold increase in activity.
Transactivation property of CAP2 was demonstrated by yeast (Saccharomyces cerevisiae) one-hybrid assay. CAP2 ORF was cloned at NdeI-EcoRI sites of the yeast expression vector pGBKT7 (CLONTECH) to express the protein as a fusion to GAL4 DNA-BD. Yeast reporter strain PJ694-A carrying HIS3, ADE2, and LacZ reporter genes under GAL4 promoter was transformed with the BD-CAP2 construct. Colonies transformed with BD-CAP2 construct grew on selective medium lacking His and Ade unlike the wild type and vector-transformed colonies (Fig. 3D). BD-CAP2-transformed cells also showed higher β-galactosidase activity (Fig. 3E).
CAP2 Is a Single/Low Copy Gene, and CAP2 Protein Is Localized in the Nucleus
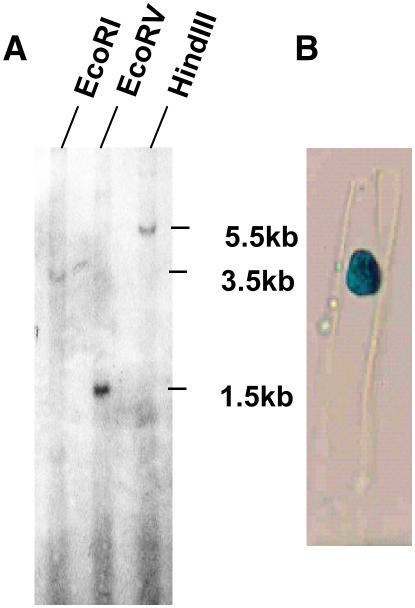
Southern transfer of chickpea genomic DNA digested separately with various restriction enzymes was hybridized at high stringency with 3′ terminus of CAP2 cDNA as a probe. A small number of bands were observed in each of the digests, indicating the CAP2 gene is either single or low copy number (Fig. 4A). A PCR reaction was performed with a pair of primers designed from the 5′ and 3′ ends of CAP2 cDNA and chickpea genomic DNA as template. The sequence of the PCR product entirely matched the CAP2 cDNA sequence, indicating CAP2 gene does not have introns. To determine the intracellular localization of CAP2 protein, CAP2 ORF was cloned at the XbaI site of the vector pBI121 (CLONTECH) to express CAP2 protein fused with β-glucuronidase (GUS) at the C terminus. Nicotiana leaf explants were transformed with the construct using Agrobacterium. Samples from stem epidermal layer showed GUS activity in the nucleus, demonstrating CAP2 is a nuclear protein (Fig. 4B).
Figure 4.
A, CAP2 is a unique or low copy number gene. Chickpea DNA (approximately 10 μg) was digested with indicated restriction endonucleases, and Southern transfer was hybridized with 32P-labeled probe used for northern hybridization. B, CAP2 protein localizes in nucleus. CAP2 ORF was cloned in pBI121 (CLONTECH) to make CAP2 protein fused with GUS at its C terminus and introduced in tobacco leaf explant through Agrobacterium. Stem peel of the transgenic plant was stained for GUS activity. The sample was restained with orcein to confirm the position of nucleus (not shown).
Morphological Features of CAP2 Expressing Transgenic Tobacco Seedlings
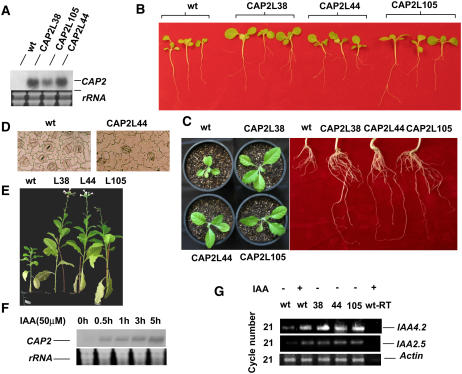
To establish the functional significance of the CAP2 gene, the complete ORF was cloned in the plant expression vector pBI121 by replacing the GUS gene and mobilized into tobacco plants using Agrobacterium-mediated transformation. Preliminary screening of the kanamycin-resistant transgenic lines was performed by PCR amplification of genomic DNA with CAP2-specific primers. We have grown 12 independently transformed lines to maturity; however, three randomly selected lines were taken for subsequent analyses. All three transgenic lines (CAP2L38, CAP2L44, and CAP2L105) carried a single copy of the transgene, as confirmed by Southern analyses and constitutively expressed CAP2 as determined by northern analyses; however, expression in CAP2L105 was more than 3-fold lower than that of other two lines (Fig. 5A).
Figure 5.
A, Expression of CAP2 in leaves of untransformed (wt) and transformed (CAP2L38, CAP2L44, and CAP2L105) T1 tobacco plants. Bottom segment shows rRNA stained with ethidium bromide. B, Morphology of the 16-d-old transgenic T1 seedlings showing lateral roots and expanded leaves in comparison to the wild-type (wt) seedlings. C, Comparison of 50-d-old soil-grown representative wild-type (wt) and transgenic plants and the roots of the same plants. D, Epidermal peels from the ventral surface of the middle lamina of the wild-type (wt) and transgenic (CAP2L44) leaves showing cell size. E, Comparison of 90-d-old soil-grown representative wild-type (wt) and transgenic plants. F, Expression of CAP2 in root of chickpea seedlings in response to 50 μm IAA. Northern transfer was hybridized as mentioned before. G, Semiquantitative RT-PCR of IAA4.2, IAA2.5, and Actin. RNA was isolated from IAA untreated (−) and treated (+; for 5 h) wild-type (wt) and transgenic seedlings (L38, L44, and L105). No PCR product was detected from the treated wild-type sample without RT (wt-RT).
T1 seedlings of the transgenic lines were morphologically different from the wild-type plants (Fig. 5B). No difference was observed between the transgenic lines and the wild-type seeds in the rate and period taken for germination. Average surface area of the third and fourth leaves of 10 seedlings (16 d after germination in soil) from each transgenic line was measured (25.92 ± 2.14 mm2) and found to be more than 2-fold of that of the wild type (11.11 ± 2.36 mm2). Similarly, average fresh weight of the transgenic seedlings of the same age was almost 3-fold that of wild-type seedlings (35.73 ± 2.90 mg and 12.30 ± 2.15 mg, respectively). The leaves of the transgenic lines remain larger up to senescence stage (Supplemental Fig. 1; Fig. 5E). We further explored the reason behind the increase in the leaf area in the transgenic plants. Ventral epidermal peels from both sides of the midrib of the leaf base, middle lamina, and leaf tip of the wild-type and transgenic lines were compared for cell size. Six samples from each leaf and leaves from four plants of each line were taken. Calculated average cell size (from 24 samples of each line) of the ventral epidermal surface of the fourth leaf from the bottom of 16-d-old transgenic plants was 192.5% of the average cell size from the same area of the wild-type leaf; however, these results do not exclude effects on the cell division (Fig. 5D). Examination of the root growth revealed that 16-d-old transgenic seedlings developed a significantly higher number of lateral roots (average 2.5) compared to no visibly developed lateral roots in the wild-type seedlings (Fig. 5B). In comparison to the wild-type plants, transgenic lines showed contrastingly higher root and leaf growth, which is evident from the comparison of soil grown 50-d- and 90-d-old tobacco plants (Fig. 5, C and E). Average flowering time of the CAP2-expressing plants is 85 to 90 d in comparison to 125 to 130 d for the wild-type plants in normal growth conditions. Though the expression of the transgene in CAP2L105 is much less compared to that in two other lines, no significant difference in leaf size and/or in the number of lateral roots was observed among the transgenic lines. Expression level of CAP2 in the line L105 was probably sufficient for the morphological phenotype.
Plant hormone auxin induces many developmental effects, including lateral root formation and leaf expansion, by regulating ARFs that bind to auxin-responsive element. Loss-of-function mutation of ARF7 and ARF19 together caused several phenotypes in Arabidopsis, including drastic reduction in the number of lateral roots and decrease in the leaf cell expansion. Both ARF7 and ARF19 transcripts are accumulated in response to auxin and in turn induce expression of early auxin-response genes like Aux/IAA (Wilmoth et al., 2005). Therefore, we investigated the expression of CAP2 in chickpea in response to auxin. Northern analyses of RNA from root revealed that treatment with 50 μm IAA elevated expression of CAP2 within 30 min and continued to increase up to 5 h (Fig. 5F). To investigate the activation of the auxin-response pathway in the transgenic tobacco seedlings expressing CAP2, the expressions of early auxin-response genes IAA4.2 and IAA2.5 were analyzed. Semiquantitative reverse transcription (RT)-PCR analyses of tobacco IAA4.2 and IAA2.5 clearly demonstrated that the basal expressions of these genes in the transgenic lines are more than 3- to 4-fold higher than in the wild-type plant and similar to that of an auxin-treated wild-type plant (Fig. 5G). The experiment lacking reverse transcriptase did not show any detectable band. Parallel experiment with actin-specific primers showed equal expression of the corresponding gene.
Abiotic Stress Tolerance of Transgenic Tobacco Seedlings Expressing CAP2
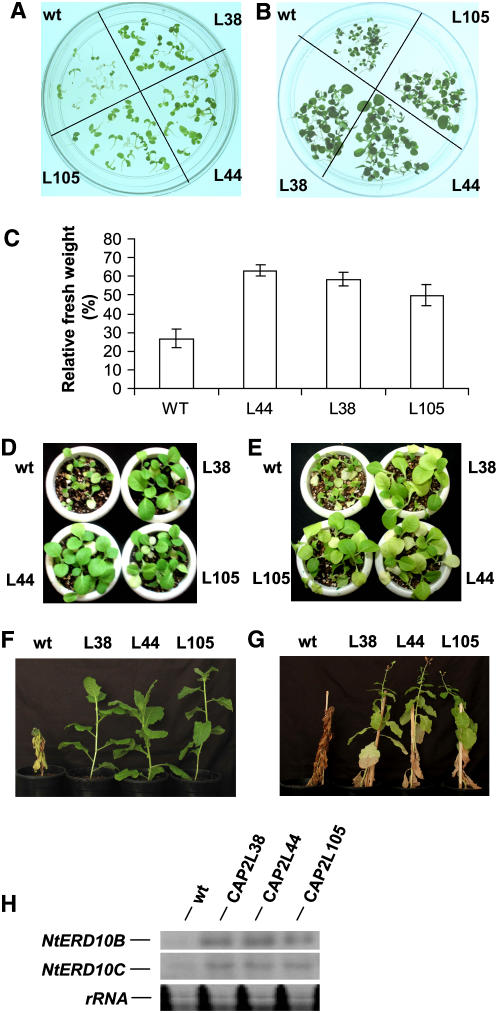
Expression of CAP2 gene increased in dehydration and exposure to high salinity, and CAP2 protein binds to DRE. T1 seedlings of the transgenic lines were therefore analyzed for salinity tolerance. Eight-day-old seedlings were transferred to medium containing 250 mm NaCl and allowed to grow for 6 d. Chlorosis was apparent in the wild-type seedlings from the second day and started appearing in transgenic seedlings from the fourth day. After 6 d, one-half of the wild-type seedlings died following complete bleaching, and the rest were showing extensive chlorosis. A total of 30% to 40% of seedlings of lines CAP2L44 and CAP2L38 and 20% of line CAP2L105 were green even after 6 d of exposure, and other seedlings showed moderate chlorosis (Fig. 6A). CAP2-expressing transgenic lines were evaluated for dehydration-stress tolerance by germinating and then growing the T1 seedlings on medium containing 0.4 m mannitol for 20 d (Fig. 6B). The germination rate for line 44 after 8 d was 78%, followed by 65% and 60% for the lines 38 and 105, respectively, while for the wild type it was only 42%. However, after 20 d, germination of the wild-type seedlings reached almost 60%. As there is a drastic difference between the fresh weights of the wild-type and the transgenic seedlings under normal growth conditions, effect of the dehydration stress was presented as percent relative fresh weight, i.e. fresh weight of the seedlings grown in experimental conditions relative to the fresh weight of the seedlings of the same line grown in control conditions (Fig. 6C). The relative fresh weights of the transgenic lines were significantly higher in comparison to that of the wild type, as reflected by the quantitative estimation. To assess the effect of CAP2 expression on stress tolerance of the greenhouse-grown seedlings, 3-week-old wild-type and transgenic seedlings were irrigated with 200 mm sodium chloride solution for 1 week and then with water for 1 week for recovery. The transgenic lines showed much better recovery than the wild-type plants (Fig. 6D). Improvement of dehydration tolerance in the greenhouse-grown transgenic seedlings was evaluated. Three-week-old seedlings were not watered for 2 weeks. The wild-type plants showed more bleaching and loss of turgor in comparison to the transgenic plants (Fig. 6E). The same experiments were performed on matured plants. The 50-d-old wild-type and transgenic plants were exposed to salt stress by irrigation of 200 mm sodium chloride solution in 4-d intervals for 16 d and then with water for 2 more weeks. For dehydration stress, 75-d-old wild-type and transgenic plants were not irrigated for 2 months. It is evident from the figures of the representative plants under the experiments (Fig. 6, F and G) that the CAP2-expressing plants are more tolerant to salt and dehydration stresses in comparison to the wild-type plants. Transfer of Arabidopsis DREB1A in tobacco under the control of constitutive 35S or inducible rd29A promoter resulted in improved drought and cold stress tolerance and constitutive expression of NtERD10, a family of genes encoding group 2 LEA proteins (Kasuga et al., 2004). We have shown that CAP2, like AtDREB1A, binds to DRE. As CAP2-expressing transgenic tobacco lines were comparatively tolerant to mannitol and salt, we have checked the expression of NtERD10B and NtERD10C, the targets of AtDREB1A, in the transgenic lines. Figure 6H shows that the transgenic lines constitutively expressed NtERD10B and NtERD10C in the control condition. Expression of these genes in line 105 was relatively lower. The reason may be the lower expression of the transgene in this line, which is also reflected in the level of tolerance in growth media containing salt and mannitol.
Figure 6.
A, Effect of salt stress on tobacco seedlings from wild type (wt) and T1 progenies of transgenic lines (CAP2L38, CAP2L44, and CAP2L105). Eight-day-old (after germination) seedlings were transferred to MSH medium supplemented with 250 mm salt and kept for 6 d. B, Effect of dehydration stress on wild-type (wt) and T1 transgenic tobacco lines. Seeds were germinated and grown on medium containing 0.4 m mannitol for 20 d. C, Fresh weight of the seedlings in stressed condition in comparison to the fresh weight of the seedlings of same line grown in control condition is presented as percent relative fresh weight. D, Three-week-old wild-type (wt) and transgenic T1 seedlings were irrigated with 200 mm salt solution for 1 week and then with water for 1 week. E, Three-week-old wild-type (wt) and transgenic T1 seedlings were not watered for 2 weeks. F, Fifty-day-old wild-type (wt) and transgenic T1 plants were irrigated with 200 mm NaCl solution in 4 d of interval for 16 d and then with water for 2 more weeks. G, Seventy-five-day-old wild-type (wt) and transgenic T1 plants were not watered for 2 more months. In F and in G, representatives of 10 plants of each line under an experiment are shown. H, Expression of abiotic stress marker genes NtERD10B and NtERD10C in wild-type (wt) and transgenic tobacco lines in normal growth condition.
DISCUSSION
AP2 defines a large family of plant-specific DNA-binding proteins that regulate diverse plant developmental processes, including stress response. We screened a cDNA library to obtain a full-length clone of a chickpea EST induced by dehydration and cloned a cDNA that encodes an AP2 family transcription factor. CAP2 ORF is relatively small in comparison to most of the well-studied AP2/ERF family members. Like CAP2, a number of small ORFs containing AP2-domain have been reported in GenBank from different plants. Two of them are AAP83131 and AAT12423 from another legume, soybean, that show high homology to CAP2 along the whole stretch of sequences. Detailed functional study on these small AP2 proteins may open up new areas in plant developmental process.
CAP2 transcription, like DREB2A of Arabidopsis and rice (Oryza sativa), is activated by dehydration and salt and not by cold, and the induction is early. But, unlike AtDREB2A, CAP2 transcription is activated by ABA. This and the fact that CAP2 transcription is also activated by auxin indicate that CAP2 and DREB2A may have different roles and are regulated by different or additional pathways. Sequence analyses of the region 5′ to the transcription initiation site of CAP2 revealed the presence of a number of abiotic stress-responsive cis-acting elements, such as ABRE, MYB2 consensus, MYC consensus, etc. Among the other significant elements, there are auxin-responsive SAUR elements (CATATG; Xu et al., 1997) and a number of AAAG motifs that are shown essential and determinant for binding of Dof domain proteins in other plants (Yanagisawa and Schmidt, 1999). Dof domain proteins are plant-specific transcription regulators with highly conserved single DNA-binding zinc finger domain. Dof domain proteins are associated with diverse plant-specific physiological roles, as suggested by the gene expression studies. They include stress responses (Zhang et al., 1995; Kang et al., 2003), light responses (Yanagisawa and Sheen, 1998), phytochrome signaling, and responses to plant hormones including auxin (DePaolis et al., 1996; Kisu et al., 1998). Tobacco Dof protein NtBBF1 has been identified as the protein involved in tissue-specific and auxin-inducible expression of rolB oncogene. It was shown that NtBBF1 binds to a target sequence ACTTTA in the promoter of rolB gene to promote auxin-responsive expression (Baumann et al., 1999). Interestingly, there is an ACTTTA motif in the CAP2 promoter region. Presence of cis-acting elements associated with abiotic stress and auxin may explain novel transcriptional regulation of CAP2. Detailed functional study of the CAP2 promoter is required to determine the pathway of CAP2 expression.
CAP2 satisfied the definition of a transcription factor by directly binding to DNA in a sequence-specific manner, trans-activating reporter genes, and being localized in nucleus. However, NtERD10B, NtERD10C, IAA4.2, and IAA2.5, the constitutively expressing genes in the transgenic lines, may not be the direct targets of CAP2, even though NtERD10B and NtERD10C are supposed to have DRE in their promoters (Kasuga et al., 2004). The development of shoot and root tissues in plants is a highly coordinated process. Leaf development occurs in conjunction with continuous development of lateral roots to extract more water and nutrients from soil. In reverse, lateral roots are believed to develop in response to shoot-derived auxin. Therefore, there is a possibility that production of auxin was increased due to increase in leaf growth. The experiments described in this report are not sufficient to define the primary effect of CAP2 expression. Chickpea mutant lacking CAP2 function is required to provide conclusive result.
CAP2 expression imparted tolerance to dehydration and salinity to the tobacco plants. The tolerance was possibly due to expression of genes like ERD10B and ERD10C coding for LEA proteins of tobacco. Constitutive expression of AtDREB1A in Arabidopsis and in tobacco caused similar expression of RD29A and NtERD10 genes, respectively, but in contrast to CAP2-transgenic plants, resulted in stunted growth (Liu et al., 1998; Kasuga et al., 2004). However, similar expression of AtDREB2 in Arabidopsis neither enhanced RD29A expression nor caused any morphological alteration (Liu et al., 1998). CAP2 possesses homology to AtDREB2 only at the AP2 domain, and its homology is much less with AtDREB1. Therefore, contrasting phenotypes of the CAP2-expressing tobacco plants and AtDREB1- and AtDREB2-expressing Arabidopsis plants may be due to the differences in their primary structures, the mode of function, and expressional regulation of these proteins. Furthermore, CAP2 may not activate all the genes that AtDREB1A does, especially those responsible for stunted growth. Additionally, CAP2 may not show its full activity in a heterologous system. However, expression of AtDREB1A in rice, though increased tolerance to abiotic stress, did not affect the growth and development of the transgenic plant (Oh et al., 2005).
Biogenesis of lateral root is affected by the interplay of various environmental and endogenous factors, including biosynthesis and distribution of phytohormones like auxin, ABA, and ethylene. It is also evident that lateral root formation is promoted by salt stress. Mutations in the signaling pathways of the phytohormones also influence the formation of lateral roots. Mutations in genes ABI3 (ABA insensitive 3) and ARF8 compromise with lateral root formation and growth (Ruegger et al., 1998; Signora et al., 2001; Brady et al., 2003; He et al., 2004). On the other hand, expression of tobacco ethylene receptors NtHK1 and NtHK2 are induced by dehydration and salt stress, and Arabidopsis plants overexpressing NtHK1 showed more sensitivity to salt stress (Zhang et al., 2001; Xie et al., 2002; Cao, 2004). Therefore, it is evident that signaling pathways of ABA, ethylene, and auxin have several common points of cross talk. Expression of Arabidopsis gene AtNAC2 is induced by salt stress, auxin, ABA, and ethylene; however, the induction under salt stress is suppressed in the mutant plants insensitive to ethylene and resistant to auxin. Interestingly, overexpression of AtNAC2, similar to CAP2, promoted lateral root formation without compromising the growth of taproot and led to a hypothesis by He et al. (2005) that salt-induced expression of AtNAC2 (and formation of lateral root) is dependent on ethylene and auxin signaling pathway. Presence of auxin and salt stress-responsive cis-acting elements in the promoter prompts us to think a similar regulation of CAP2. But unavailability of corresponding chickpea mutants is handicap for such studies.
We are aware that CAP2 expression in the transgenic tobacco was driven by 35S promoter, commonly used for strong and constitutive expression, and there may be a possibility that the phenotypes shown by the transgenic plants are due to high expression level of transgene. However, no significant morphological differences were observed in the low expressing line (CAP2L105) from the two high expressing lines. Nevertheless, CAP2, an AP2/ERF family transcription factor from chickpea, is structurally different from the other family members of other plants and shows a unique expression profile in response to different stresses and phytohormones. Expression of CAP2 in transgenic tobacco caused increase in the leaf size and number of lateral roots and simultaneously promoted tolerance to salt and osmotic stress, which highlighted its potential agronomic importance as well as provided preliminary evidences that CAP2 protein might be a common point of cross talk between growth and development and abiotic stress response.
MATERIALS AND METHODS
Plant Materials and Treatment
Chickpea (Cicer arietinum) cv BGD72 was used in this study. Seedlings were grown and treated for dehydration and with ABA, as described in Boominathan et al. (2004). Seedlings were treated with 50 μm IAA (Sigma) for auxin treatment. For transgenic lines, tobacco (Nicotiana tabacum) var Xanthi was used as wild type. For cellular localization, CAP2 ORF without translation stop codon was cloned at the XbaI site of pBI121 vector (CLONTECH) to express CAP2 protein fused with GUS at the C terminus. For functional analyses, CAP2 ORF with stop codon was cloned at the XbaI-SacI site. In both the cases, expression was driven by CaMV35S promoter. The constructs were chemically mobilized in to Agrobacterium tumefaciens strain GV3101. Agrobacterium-mediated transformation of tobacco ex-plants was carried out following standard protocol (Gelvin and Schilperoort, 1994). Integration of the transgene was tested by PCR amplification and Southern analyses of the genomic DNA. Expression of the transgene was evaluated by RNA gel-blot analyses of the total RNA extracted from leaves.
Construction of cDNA Library and Cloning of CAP2 cDNA
Chickpea cDNA library was constructed using CLONTECH SMART cDNA library kit with 4 μg of total RNA isolated from dehydrated (uprooted from soil for 5 h) chickpea seedlings (6 d after germination). The library was packaged in Packgene lambda packaging extract (Promega). The library was amplified once and obtained a titer of 1.15 × 1011 pfu. A chickpea EST (GenBank accession no. CD051361; Boominathan et al., 2004) was used as a probe to screen 2.5 × 105 pfu following the protocol described by Sambrook et al. (1989). Genome walking was done using and following methods of Genome Walker kit (CLONTECH). Analysis of cis-acting elements was done using the program PLACE.
Analyses of Transgenic Plants for Abiotic Stress Tolerance and Cellular Localization
Wild-type and T1 seeds of transgenic tobacco lines were surface sterilized, germinated, and grown essentially as described by Mukhopadhyay et al. (2004). For salt stress, the seedlings grown for 8 d after germination were transferred to half strength of Murashige and Skoog medium without Suc and organic ingredients (MSH) containing 250 mm salt for 6 d. For dehydration stress treatments, the seeds were germinated on MSH medium containing 0.4 m mannitol and grown for 20 d after germination. For quantification of relative fresh weight percent, fresh weight of 10 seedlings of each line grown in medium with or without mannitol was taken. The result from three independent experiments was presented.
For cellular localization of CAP2 protein, stem peels from transgenic tobacco plants expressing CAP2 protein fused to GUS were used. Histochemical localization of GUS activity was analyzed after incubating the samples in X-Gluc buffer (50 mm sodium phosphate buffer, pH 7, 10 mm EDTA, 0.1% Triton X 100, 5 mm potassium ferro cyanide, and 2 mg/mL 5-bromo-4-chloro-3-indolyl glucuronide) at 37°C for 12 h. The same sample was stained with Orcein to confirm the position of the nucleus.
RNA-Blot Hybridization and RT-PCR
Isolation of total RNA and RNA gel-blot hybridizations were carried out as described by Boominathan et al. (2004). Prehybridization and hybridization buffers were composed of 300 mm sodium phosphate, pH 7.2, 7% SDS, and 1 mm EDTA. Hybridized filters were washed with a solution of 0.5× SSC and 2% SDS. Hybridization and washing were performed at 60°C. An EcoRV-SspI fragment (637–1,026 bp) of the CAP2 cDNA was used to make radiolabeled probe. For semiquantitative RT followed by amplification by PCR (RT-PCR), 200 ng of total RNA was converted to cDNA with oligo(dT) primer using SuperscriptII (Life Technologies) and amplified with gene-specific primers (for IAA4.2, 5′-ACCAGAGAAACAATCTTCTTC-3′ and 5′-CATGCAAACCTTTAGCTTCAG-3′; for IAA2.5, 5′-AAGACAGAAACTGAGTGTGGAATG-3′ and 5′-TCCACAACCCAAGCCTCTTGCTTC-3′; for Actin, 5′-TTAAAGAGAAACTGGCATATGTTG-3′ and 5′-GTTGGAGACTGCAAAGAGTAGCTC-3′) for 21 cycles. For NtERD10B and NtERD10C, primers 5′-TATGGGAACCCAGTCCATCACACTGGA-3′ and 5′-CTTTAAAGGGATTTTATTGTTTGGCAG-3′, and 5′-AGGTTGAAGAGGGTAGCGCAAACG-3′ and 5′-GCCACTTCCTCTGTCTTCTTTTGC-3′, respectively, were designed from the published cDNA sequences and amplified by RT-PCR using total RNA isolated from dehydrated (5 h) tobacco seedlings as template and used for northern analyses.
Protein Expression, Site-Directed Mutagenesis, and Gel Mobility Shift Assay
CAP2 protein coding sequence was amplified by PCR with primers flanked by restriction sites for BamHI and EcoRI to clone in pGEX4T2 in frame with GST cDNA. GST-CAP2 protein was produced by inducing the Escherichia coli BL21(DE3) cells harboring pGEX4T2-GSTCAP2 with 0.5 mm isopropyl β-d-1-thiogalactopyranoside. The recombinant GST-fused CAP2 protein was purified from bacterial lysates with glutathione-Sepharose beads (Amersham). For in vitro mutagenesis, the forward and reverse primer pairs used were as follows: for V77A, 5′-GACTTGGGGAAAATGGGCCGCCGAAATTCGGGAAC-3 and 5′-GTTCCCGAATTTCGGCGGCCCATTTTCCCCAAGTC-3′; for E82D, 5′-GTGGCCGAAATTCGGGATCCAAACAGAGGGAGTAG-3′ and 5′-CTACTCCCTCTGTTTGGATCCCGAATTTCGGCCAC-3′; and for E82A, 5′-GTGGCCGAAATTCGGGCCCCAAACAGAGGGAGTAG-3′ and 5′-CTACTCCCTCTGTTTGGGGCCCGAATTTCGGCCAC-3′. Mutagenesis reactions were carried out on double-stranded plasmid DNA using Pfu Turbo DNA polymerase (Stratagene) using the following parameters: 95°C for 30 s; and 16 cycles of 95°C for 30 s, 58°C for 1 min, and 72°C for 7 min. The bacteria-derived template was digested with methylation-specific restriction enzyme DpnI, and the digested PCR product was transformed into E. coli DH5α cells. The mutation and the fidelity of the rest of the construct were confirmed by DNA sequencing. Two oligonucleotides, 5′-AATATACTACCGACATGAGTTCTAATATACTAC-3′ and 5′-AGAACTCATGTCGGTAGTATATTAGAACTCATG-3′, were mixed in equimolar ratio and elongated with Taq DNA polymerase (Invitrogen) to make double-stranded DNA probe for gel mobility shift experiment. [α-32P]-dCTP was incorporated in the elongation step to produce radiolabeled probe. Oligonucleotide pairs used for mutated probes were as follows: for M1, 5′-AATATACTACCTACATGAGTTCTAATATACTAC-3′ and 5′-AGAACTCATGTAGGTAGTATATTAGAACTCATG-3′; and for M2, 5′-AATATACTATCGACATGAGTTCTAATATACTAC-3′ and 5′-AGAACTCATGTCGATAGTATATTAGAACTCATG-3′. The substituted bases are shown in bold letters. Gel mobility shift assays were conducted as described previously (Urao et al., 1993).
Yeast One-Hybrid Assay
CAP2 protein coding sequence was cloned in yeast (Saccharomyces cerevisiae) expression vector pGBKT7 (CLONTECH) at NdeI-EcoRI site to express CAP2 protein fused to GAL4 DNA-BD. The construct was transformed into an auxotropic yeast strain PJ69-4A (Cagne et al., 2000) that contains three reporter genes, HIS3, ADE2, and β-GAL, under the control of GAL4 promoter, and plated on synthetic medium lacking His and Ade. β-Galactosidase assay of the transformed colonies was done with ortho-nitrophenyle-β-d-galactoside. Presence of CAP2 in the transformed colonies was confirmed by PCR.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ321719, AAP83131, and AAT12423.
Supplementary Material
Acknowledgments
The authors acknowledge the contribution of Dr. Manoj Prasad of the National Centre for Plant Genome Research in preparation of the manuscript.
This work was supported by the National Centre for Plant Genome Research and a grant from the Department of Biotechnology, Government of India (DBT). R.K.S. and V.T. acknowledge Council for Scientific and Industrial Research and S.R. acknowledges DBT for fellowships.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Debasis Chattopadhyay (debasis_chattopadhyay@yahoo.co.in).
The online version of this article contains Web-only data.
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Banno H, Ikeda Y, Niu QW, Chua NH (2001) Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 13: 2609–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann K, DePaolis A, Constantino P, Gualberti G (1999) The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell 11: 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boominathan P, Shukla R, Kumar A, Manna D, Negi D, Verma PK, Chattopadhyay D (2004) Long term transcript accumulation during the development of dehydration adaptation in Cicer arietinum. Plant Physiol 135: 1608–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VR, Kieft H, Onzlet T, Zhang L, Hattori J, Liu CM, Van Lammera AAM, Miki BLA, et al (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14: 1737–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743 [Google Scholar]
- Brady SM, Sarkar SF, Bonetta D, McCourt P (2003) The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved auxin signaling and lateral root development in Arabidopsis. Plant J 34: 67–75 [DOI] [PubMed] [Google Scholar]
- Cagne J, Uetz P, Fields S (2000) High throughput screening for protein-protein interactions using the two-hybrid assay. Methods Enzymol 328: 3–14 [DOI] [PubMed] [Google Scholar]
- Cao WH (2004) Ethylene receptor regulates plant response to saltstress. PhD thesis. Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing
- Cheong YH, Chang HS, Gupta R, Wang X, Zhu T, Luan S (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol 129: 661–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePaolis A, Sabatini S, DePascalis L, Constantino P, Capone I (1996) A rolB regulatory factor belongs to a new class of single zinc finger plant proteins. Plant J 10: 215–223 [DOI] [PubMed] [Google Scholar]
- Gelvin SB, Schilperoort RA (1994) Plant Molecular Biology Manual. Kluwer, Dordrecht, The Netherlands
- Guo ZJ, Chen XJ, Wu XL, Ling JQ, Xu P (2004) Overexpression of the AP2/EREBP transcription factor OPBP1 enhances disease resistance and salt tolerance in tobacco. Plant Mol Biol 55: 607–618 [DOI] [PubMed] [Google Scholar]
- He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44: 903–916 [DOI] [PubMed] [Google Scholar]
- He XJ, Zhang Z, Yan G, Zhang JS, Chen SY (2004) A salt-responsive receptor- like kinase gene regulated by the ethylene signaling pathway encodes a plasma membrane serine/threonine kinase. Theor Appl Genet 109: 377–383 [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 377–383 [DOI] [PubMed] [Google Scholar]
- Irish VF, Sussex IM (1990) Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2: 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BGW, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HG, Foley RC, Onate-Sanchez L, Lin C, Singh KB (2003) Target genes for OBP3, a Dof transcription factor, include novel basic helix-loop-helix domain proteins inducible by salicylic acid. Plant J 35: 362–372 [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2004) A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in Tobacco by gene transfer. Plant Cell Physiol 45: 346–350 [DOI] [PubMed] [Google Scholar]
- Kisu Y, Ono T, Shinofurutani N, Suzuki M, Esaka M (1998) Characterization and expression of a new class of zinc finger protein that binds to silencer region of ascorbate oxidase gene. Plant Cell Physiol 39: 1054–1064 [DOI] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer R (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophytes development is related to the floral homeotic gene APETALA2. Plant Cell 8: 137–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O (2001) Auxin signaling: the beginning, the middle, and the end. Curr Opin Plant Biol 4: 382–386 [DOI] [PubMed] [Google Scholar]
- Leyser O (2002) Molecular genetics of auxin signaling. Annu Rev Plant Biol 53: 377–398 [DOI] [PubMed] [Google Scholar]
- Liu X, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Hul AK, Kowalczyk M, Marchant A, Celenza J, Cohen JD, Sandberg G (2002) Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol Biol 50: 309–332 [DOI] [PubMed] [Google Scholar]
- Meyerowitz EM (1994) Flower development and evolution: new answers and new questions. Proc Natl Acad Sci USA 91: 5735–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A, Vij S, Tyagi AK (2004) Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci USA 101: 6309–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Song SIK, Kim YS, Jang HJ, Kim SY, Kim M, Kim YK, Nahm BH, Kim JK (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138: 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto M, Fischer RL, Goldberg RB, Nakamura K, Harada JJ (2005) Control of seed mass by APETALA2. Proc Natl Acad Sci USA 102: 3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Montagu MV, Jofuku KD (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA 94: 7076–7081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak T, Potters G, Caubergs R, Jansen MAK (2005) Complementary interactions between oxidative stress and auxins control plant growth responses at plant, organ, and cellular level. J Exp Bot 56: 1991–2001 [DOI] [PubMed] [Google Scholar]
- Reed JW (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6: 420–425 [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev 12: 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsh EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Signora L, Smet D, Foyer CH, Zhang H (2001) ABA plays a central role in mediating the regulatory effects of nitrate on rootbranching in Arabidopsis. Plant J 28: 655–662 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress response. Curr Opin Plant Biol 6: 410–417 [DOI] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM (2005) NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J 43: 118–130 [DOI] [PubMed] [Google Scholar]
- Wilson K, Long D, Swinburne J, Coupland G (1996) A dissociation insertion causes a semi dominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8: 4659–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Zhang ZG, Zhang JS, He XJ, Cao WH, He SJ, Chen SY (2002) Spatial expression and characterization of aputative ethylene receptor protein NTHK1 in tobacco. Plant Cell Physiol 43: 810–815 [DOI] [PubMed] [Google Scholar]
- Xu N, Hagen G, Guilfoyle T (1997) Multiple auxin response modules in the soybean SAUR 15A promoter. Plant Sci 126: 193–201 [Google Scholar]
- Yadav V, Malappa C, Gangappa SN, Bhatia S, Chattopadhyay S (2005) A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17: 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1997) Gene expression and signal transduction in water-stress response. Plant Physiol 115: 327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Schmidt RJ (1999) Diversity and similarity among recognition sequences of Dof transcription factors. Plant J 17: 209–214 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Sheen J (1998) Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 10: 75–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Chen W, Foley RC, Buttner M, Singh KB (1995) Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell 7: 2241–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JS, Xie C, Shen YG, Chen SY (2001) A two-component gene (NTHK1) encoding a putative ethylene receptor homolog is both development- and stress-regulated in tobacco. Theor Appl Genet 102: 815–824 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.