Abstract
Shoot apical meristems produce organs in a highly stereotypic pattern that involves auxin. Auxin is supposed to be actively transported from cell to cell by influx (AUXIN/LIKE AUXIN proteins) and efflux (PIN-FORMED proteins) membrane carriers. Current hypotheses propose that, at the meristem surface, PIN proteins create patterns of auxin gradients that, in turn, create patterns of gene expression and morphogenesis. These hypotheses are entirely based on work in Arabidopsis (Arabidopsis thaliana). To verify whether these models also apply to other species, we studied the behavior of PIN proteins during maize (Zea mays) development. We identified two novel putative orthologs of AtPIN1 in maize and analyzed their expression pattern during development. The expression studies were complemented by immunolocalization studies using an anti-AtPIN1 antibody. Interestingly, the maize proteins visualized by this antibody are almost exclusively localized in subepidermal meristematic layers. Both tassel and ear were characterized by a compact group of cells, just below the surface, carrying PIN. In contrast to or to complement what was shown in Arabidopsis, these results point to the importance of internally localized cells in the patterning process. We chose the barren inflorescence2 (bif2) maize mutant to study the role of auxin polar fluxes in inflorescence development. In severe alleles of bif2, the tassel and the ear present altered ZmPIN1a and ZmPIN1b protein expression and localization patterns. In particular, the compact groups of cells in the tassel and ear of the mutant were missing. We conclude that BIF2 is important for PIN organization and could play a role in the establishment of polar auxin fluxes in maize inflorescence, indirectly modulating the process of axillary meristem formation and development.
Plant architecture is defined by the combination of external and internal cues, which determine the expression pattern of developmental genes. The shoot apical meristem (SAM) generates the aerial part of the plant. It carries a group of totipotent stem cells, which are capable of self perpetuation and, at the same time, can differentiate as daughter cells and participate in the formation of the primordia. The SAM is formed during embryogenesis and produces modular units, called phytomers. The spatial and temporal arrangement of lateral organs around the main axis is defined as phyllotaxis. This process is highly repetitive and depends on the species and developmental stage (Traas and Vernoux, 2002; Traas and Bohn-Courseau, 2005; Carraro et al., 2006).
During vegetative growth, maize (Zea mays) produces several axillary meristems (AMs) along its stem. After the switch to the reproductive phase, plants carry a male inflorescence, called the tassel, at the tip of the main axis, and a female inflorescence, called the ear, at the axil of a leaf between the fourth and the seventh node. The tassel is highly branched with longer lateral branches at the base, produced by branch meristems (BMs), and it bears unisexuate male flowers (Irish, 1997). The female inflorescence differs by the absence of side branches at its base and carries unisexuate female flowers. The complex architecture of maize inflorescences is produced by three orders of AMs: BMs produce long lateral branches at the tassel base and later generate spikelet meristems (SMs). SMs are present along the main axis of the tassel, too, in couples, and form two spikelets, which contain two floral meristems (FMs) each (Irish, 1997; McSteen et al., 2000; McSteen and Leyser, 2005). The FM forms a male floret in the tassel and a female floret in the ear.
It has recently been demonstrated that auxin plays a major role in meristem function. In particular, polar auxin transport (PAT) is implicated in lateral organ initiation at the SAM in Arabidopsis (Arabidopsis thaliana), where it determines the position of flowers and leaves around the inflorescence stem (Reinhardt et al., 2000, 2003; Kuhlemeier and Reinhardt, 2001). This transport is supposed to be established and maintained by the members of two gene families: the PIN-FORMED (PIN) and AUXIN/LIKE AUXIN (AUX/LAX) families. The main member responsible for auxin distribution at the SAM is PIN1 (Galweiler et al., 1998; Friml et al., 2003; de Reuille et al., 2006), a putative auxin transport facilitator that localizes on the plasma membrane and acts as the auxin efflux carrier (Petrasek et al., 2006). Together with AUX1 (Bennett et al., 1996; Parry et al., 2001; Swarup et al., 2004), it regulates the PAT in the SAM. The strong alleles of pin1 do not produce lateral organs. PIN1 protein is present at the meristem surface and careful analyses of PIN1 distribution patterns have suggested that auxin transport in the surface layer L1 is a major factor in the establishment of organ position. Thus, it was proposed that auxin maxima created by PIN1 at the meristem surface are at the basis of organ initiation (Galweiler et al., 1998; Reinhardt et al., 2003). Once an organ primordium is initiated, the transport facilitator is also present at the level of the internal prevascular tissues where it supposedly removes auxin from the meristem surface. This general hypothesis was mainly established using Arabidopsis as a model system. To verify whether it also applies to monocotyledonous species, we investigated the behavior of PIN genes in maize. Although several homologs were cloned in monocotyledonous species, only one, OsPIN1, has recently been characterized in rice (Oryza sativa; Xu et al., 2005).
In this study, we analyze the expression pattern of two putative AtPIN1 orthologs, ZmPIN1a and ZmPIN1b, in the SAM, tassel, and ear of maize. We also investigate the possible role of PAT in the initiation and differentiation of maize inflorescences through localization of putative auxin efflux transporters in wild-type plants and in the barren inflorescence2 (bif2) mutant. Taken together, our results suggest that ZmPIN1a and ZmPIN1b play a fundamental role in meristem function and point to a role for the internal tissues in organ positioning. In addition, they suggest a role for internal meristematic tissues in the patterning process, in contrast to what was proposed for Arabidopsis.
RESULTS
ZmPIN1a and ZmPIN1b: Two PIN1-Like Genes from Maize
To identify maize PIN1-like genes belonging to the PIN family of putative auxin efflux carriers, we screened the National Center for Biotechnology Information (NCBI), The Institute for Genomic Research (TIGR), and Maize Genetics and Genomics Database (MaizeGDB) using the AtPIN1 (AT1G73590 for genomic DNA and AF089084 for cDNA) sequences as a query. Thirty maize tentative unique contig sequences were collected. Primers designed using the retrieved sequences were employed in PCR and reverse transcription (RT)-PCR assays to obtain maize PIN1-like (ZmPIN1) genes and cDNAs.
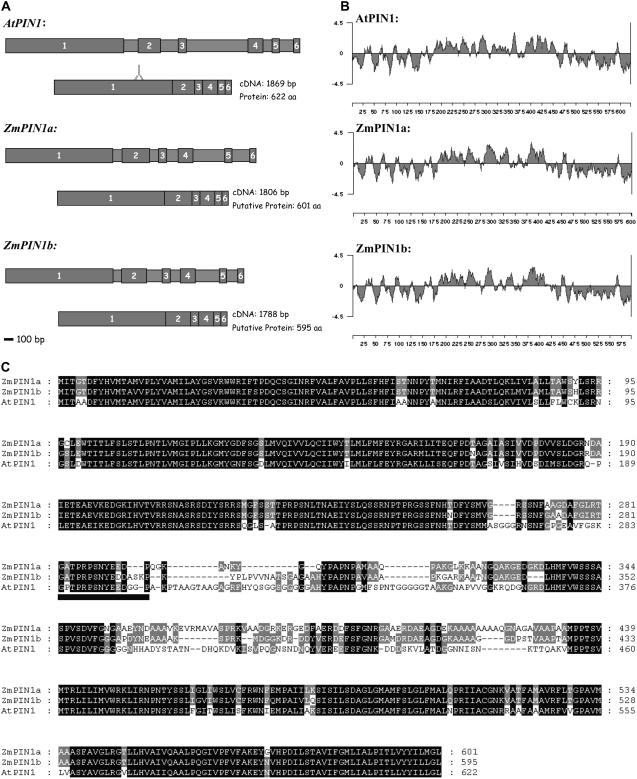
Two full-length genomic sequences of 2,649 and 2,521 bp were identified and named ZmPIN1a and ZmPIN1b, respectively. The ZmPIN1a (accession no. DQ836239) corresponding cDNA is 1,806 bp long and encodes a putative protein of 601 amino acids (65.2 kD). The ZmPIN1b (accession no. DQ836240) corresponding cDNA is 1,788 bp in length and its open reading frame (ORF) encodes a putative protein of 595 amino acids (64.5 kD; Fig. 1A). The amino acid identity between the putative maize proteins is 86.7%. The main difference in both the genomic and putative protein sequences between the two maize members is due to a 44-bp insertion at the end of the first exon in ZmPIN1b corresponding to the hydrophilic domain in the putative protein (Fig. 1C). The two maize genes showed an intron-exon structure comparable to that of AtPIN1 (Fig. 1A). The AtPIN1 protein exhibits an amino acid identity of 70.4% and 70.9% with ZmPIN1a and ZmPIN1b, respectively (Fig. 1C).
Figure 1.
Structural comparison between AtPIN1 and ZmPIN1 genes and proteins. A, Structures of the AtPIN1, ZmPIN1a, and ZmPIN1b genes (drawn to scale), with larger boxes with numbers representing exons and the smaller ones representing introns. ORFs are also depicted. The region coding for the oligopeptide recognized by the anti-AtPIN1 antibody is also reported. Scale bar = 100 bp. B, Hydropathy analysis of AtPIN1, ZmPIN1a, and ZmPIN1b proteins. The hydropathy plot was generated using Lasergene software (DNASTAR) and the method of Kyte and Doolittle, with a window size of nine amino acids. C, Amino acid sequence alignment of Arabidopsis and maize putative PIN1 proteins. AtPIN1 (At1g73590) and putative ZmPIN1a (accession no. DQ836239) and ZmPIN1b (accession no. DQ836240) were aligned using ClustalX 1.81 and then edited with Genedoc (http://www.psc.edu/biomed/genedoc). The number of amino acids is indicated on the right. Below the multiple sequence alignment, a black box indicates the oligopeptide targeted by the anti-AtPIN1 antibody.
The two maize PIN1-like proteins and AtPIN1 showed a similar hydropathy profile, with two putative lipophilic domains at the N and C ends and one central hydrophilic loop (Fig. 1B).
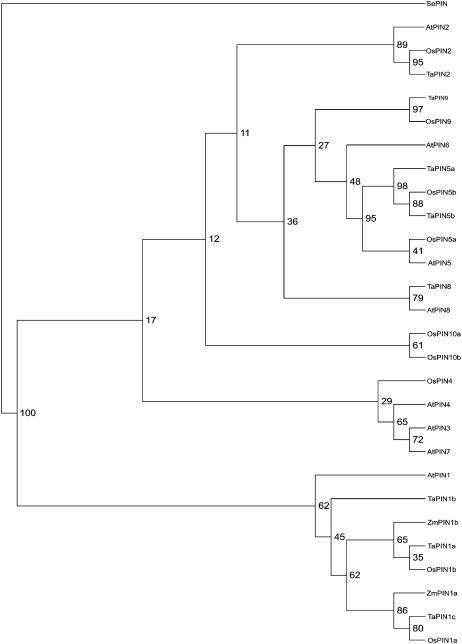
To determine the similarity of our two putative PIN sequences in relation to the PINs of Arabidopsis and other plant species, we constructed a neighbor-joining phylogenetic tree with 100 bootstrap using Phylip, including PIN-like amino acid sequences from wheat (Triticum aestivum), rice, and Arabidopsis. The resulting schematic representation was drawn using Tree-View 1.6.6 (for details, see “Materials and Methods”). ZmPIN1a and ZmPIN1b fall in the same cluster as the putative PIN1 of Arabidopsis, rice, and wheat (Fig. 2).
Figure 2.
Neighbor-joining phylogenetic tree showing the predicted relationship between ZmPIN1a, ZmPIN1b, and the Arabidopsis, rice, and wheat PIN proteins, according to Paponov et al. (2005). The tree is based on an alignment prepared using ClustalX 1.81. The phylograms were drawn using Tree-View 1.6.6 (http://darwin.zoology.gla.ac.uk/wrpage/treeviewx). Amino acid sequences for Arabidopsis were taken from http://www.Arabidopsis.org: AtPIN1, At1g73590; AtPIN2, At5g57090; AtPIN3, At1g70940; AtPIN4, At2g01420; AtPIN5, At5g16530; AtPIN6, At1g77110; AtPIN7, At1g23080; and AtPIN8, At5g15100. Data used were predicted amino acid sequences based on EST clones. The full cDNA sequences for wheat are unknown, so ESTs from the 5′ end were used. The sequences were taken from TIGR (http://www.tigr.org). Genes are named according to the cluster of the Arabidopsis PIN family to which they belong. The gene nomenclature corresponds to the following TIGR gene index: OsPIN1a, NP895789; OsPIN1b, TC250501; OsPIN2, Np897806; OsPIN4, TC259719; OsPIN5a, TC255589; OsPIN5b, TC272668; OsPIN9, TC256882; OsPIN10a, Tc260564; OsPIN10b, NP1102328; TaPIN1a, TC224207; TaPIN1b, Ck208849; TaPIN1c, Tc224208; TaPIN2, TC200857; TaPIN5a, BQ484087; TaPIN5b, CA722918; TaPIN8, CB307721; and TaPIN9, Cd895017. SEPIN (gi:57636762) from Staphylococcus epidermidis was selected as the outgroup.
Expression Patterns
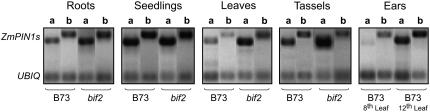
To analyze the expression of ZmPIN1a and ZmPIN1b, we performed semiquantitative RT-PCR using RNA extracted from different tissues: roots, seedlings, leaves, tassels, and ears of B73. Specific combinations of primers were used to selectively amplify ZmPIN1a or ZmPIN1b and ubiquitin as controls of the expression levels. ZmPIN1a and ZmPIN1b showed differential expression, according to the tissues and developmental stage considered. No significant differences could be observed between ZmPIN1a and ZmPIN1b expression in roots, seedlings, leaves, and tassels of B73. ZmPIN1a was not present in young ears, whereas the transcripts could be detected at a later stage of development. ZmPIN1b was also expressed in young ears and later its expression increased during development of the female inflorescence (Fig. 3).
Figure 3.
Semiquantitative RT-PCR analysis of two putative PIN1 orthologous genes from different maize tissues. B73 and bif2 show similar levels of expression in roots and seedlings, whereas in leaves ZmPIN1b displays a weaker expression than ZmPIN1a. No alteration of the expression levels is observed in bif2 roots and seedlings compared to wild type. In wild-type male inflorescences, the two genes share the same levels of expression, whereas in young ears, no ZmPIN1a expression was detected. Later, in developed female inflorescences, ZmPIN1a is expressed too. In bif2 tassels, ZmPIN1a shows high levels of expression compared to the wild type. a and b, Expression levels of ZmPIN1a and ZmPIN1b, respectively. Ubiquitin (UBIQ) was used as a control.
Expression and Localization of ZmPIN1a and ZmPIN1b in Vegetative and Reproductive Tissues
In situ hybridization with a ZmPIN1 antisense probe was employed to assess the localization of ZmPIN1 mRNA in different maize tissues. The probe was supposed to hybridize to both ZmPIN1a and ZmPIN1b mRNAs. In the mature embryo, ZmPIN1 RNA was present in a few cells positioned in the central region of the apical-basal axis. The signal was also present in the cells forming the border between the two lateral primordia of the seminal roots and the main axis of the embryo. No signal was visible in the embryonic SAM and in the embryonic leaves or cotyledon (scutellum; data not shown). In the root, the hybridization signal was mainly detected in the central cylinder and a weaker signal was detected in the cortex (data not shown).
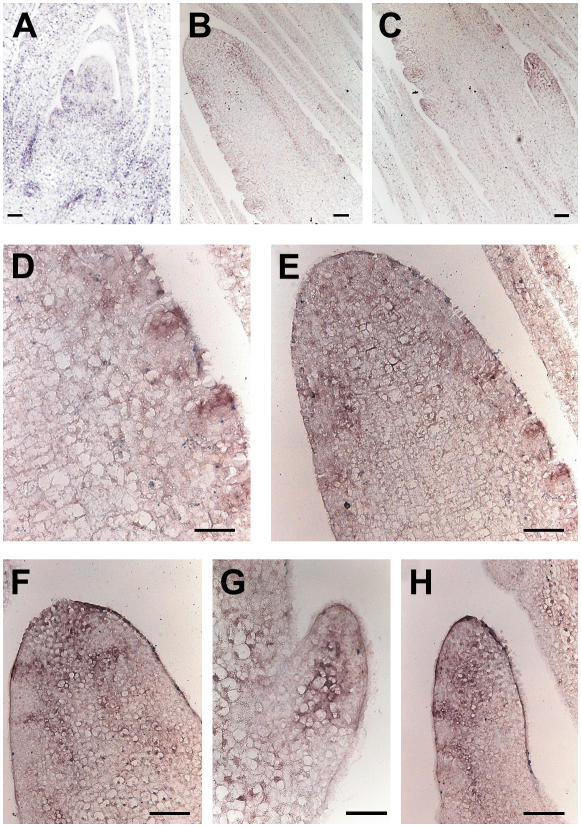
In seedlings and young maize plants, ZmPIN1 transcripts were present in inner cells of the SAM itself, in the lateral outgrowing primordia, and in the tips of young leaves (Fig. 4A). It was also strongly expressed in the vascular bundles of the apex and leaves.
Figure 4.
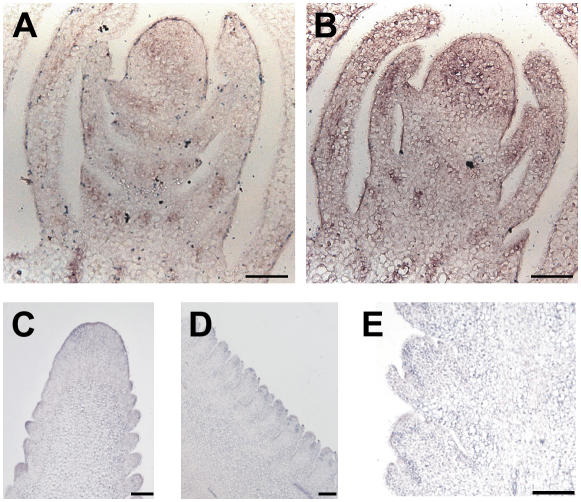
In situ hybridization of ZmPIN1 mRNA in maize apex and tassel. ZmPIN1 mRNA is expressed during maize apex development. The mRNA probe is supposed to hybridize to both ZmPIN1a and ZmPIN1b. All images represent longitudinal sections of B73 inbred line apexes. A, In the apex, the hybridization signal is mainly present in the vascular tissues; in the SAM, the signal is visible in the dome of the meristem, in the lateral outgrowing primordia, and in the tips of young leaves. B and C, In the differentiating tassel, the PIN1 probe hybridized to the corresponding mRNA in the vascular bundles and at the site of forming BM. D and E, The signal stains the whole top part of the tassel and also the upper cells of the growing SMs. G, In the lateral branches, which arise from the base of the tassel, the signal is first localized in the central part of the branch. F and H, It later follows the distichous pattern of SMs. Bars: A to E, G, and H = 100 μm; F = 50 μm.
In maize, after the transition to flowering takes place, the SAM differentiates into a tassel (Fig. 4). At the beginning of tassel differentiation, the ZmPIN1 mRNAs localized at the top of the tassel in the vascular bundles of the inflorescence axis and at the place of BM initiation (Fig. 4, B and C). When the AMs differentiated into SMs, the localization of the transcripts became restricted to the apical cells of the elongating SMs, supposedly along the differentiating provascular strands (Fig. 4, D–F). The same expression pattern was repeated in the lateral branches of the tassel, where the transcript was first detected at the top of the BM and then in the vascular bundle of the branch axis and in the main axis of the elongating SM (Fig. 4, G and H).
In developing ears, the expression was detected in lateral bracts, at the tip of bract primordia, in the vasculature and central cells of the inflorescence meristem, but not in the cells forming the outermost layer of the meristematic dome (Fig. 5, A and B). At later stages of development, the signal appeared to spread over the whole top of the upper part of the ear (the same as in the tassel) and was again localized in the AMs (Fig. 5C). Later on, the ZmPIN1 mRNA was detected in the spikelet glumes and axis of the two forming florets (Fig. 5, D and E).
Figure 5.
ZmPIN1 expression in the developing ear. In situ hybridization reveals that mRNA of ZmPIN1a and ZmPIN1b is localized in the young and adult female inflorescence. All images represent longitudinal B73 ear sections. A and B, In young ears, the signal is present in the vascular cells of the elongating axis and in the developing bracts. Also, the whole upper part, which includes the inflorescence meristem, presents diffuse staining. C, At a later stage of development, mRNA is localized in the position of growing SMs and again in the cells forming the upper part of the ear. D and E, During spikelet development, mRNA is found in the cells between the outer glumes and the differentiating SM. Bars = 100 μm.
ZmPIN1 Protein Localization
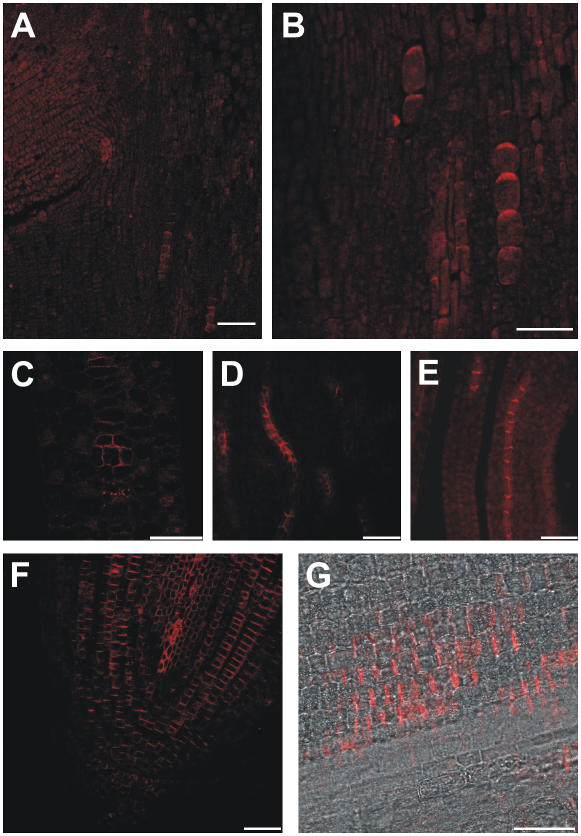
To determine the localization of ZmPIN1 proteins, we performed immunostaining experiments using an anti-AtPIN1 antibody on tissues from the B73 inbred line. This antibody has been characterized (Boutté et al., 2006; de Reuille et al., 2006) and recognizes a protein corresponding to AtPIN1 in Arabidopsis tissues and a protein with similar Mr in maize roots. In the mature embryo, the ZmPIN1 proteins localized in the main axis and were polarized in a few cells that supposedly form the provasculature. If we accept the hypothesis that localization indicates the orientation of the transport, the auxin fluxes would be directed acropetally toward the upper embryonic shoot (Fig. 6, A and B). In the leaves of seedlings and plants, the signal was clearly visible in the vascular bundles, where the PIN proteins pointed toward the inner cells (Fig. 6, C–E; data not shown). In cross sections, ZmPIN1 polarization patterns suggest that the auxin flux is directed from the outer tissues into a few or a single cell in the central part of the vascular bundle (Fig. 6C). A ladder-like staining, with the fluorescent signal localized on the basal membrane of the cells, was clearly visible in root longitudinal sections. The signal was evident both in the central vascular cylinder and in the cortex cells adjacent to the central cylinder. No signal appeared in the epidermis, external cortex, or columella apart from a few cells closely related to the root stem cells (Fig. 6, F and G).
Figure 6.
ZmPIN1 protein localization in maize vegetative tissues. The putative auxin efflux carriers are polarly localized in the vegetative organs of maize. All images portray maize B73 sections labeled with the anti-PIN1 antibody plus a secondary antibody carrying the Alexa568 fluorochrome. A to C, F, and G, Laser confocal images. D and E, Epifluorescence images acquired with a Leica DC300F camera. A and B, In the mature embryo, the anti-PIN1 antibody recognizes a polar signal in a few cells located along the embryo axis. The labeling suggests auxin fluxes directed toward the upper embryo tissues. C to E, In the leaves, putative efflux carriers are inserted into the basal membrane of vascular cell files. The labeling suggests a PAT toward the leaf base. C, In cross sections of maize vascular bundles, the signal is clearly polarized in the lateral membranes of the outer cells and indicates that auxin is loaded into the vascular bundles from the outer tissues. F and G, In the primary root, proteins are inserted into the basal membrane of the cells, suggesting an acropetal auxin flux directed toward the root tip. The labeling is present in the central cylinder and also in the inner cortex cells. Bars: A = 80 μm; B and C = 40 μm; D to G = 30 μm.
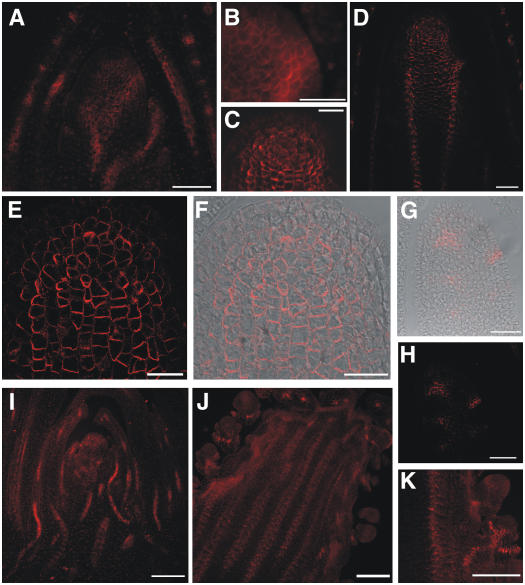
ZmPIN1 proteins were also localized in the SAM dome. Here, polar localization of the proteins appeared at the basal end of the central cells, whereas no labeling was observed in the L1 and outer layers. Furthermore, polarization was evident in the vasculature of the leaves and, most of all, at the position of young forming lateral primordia (Fig. 7, A and B). When the SAM switched from the vegetative to the reproductive phase, ZmPIN1 proteins localized in the apical region of the developing tassel, just below the surface (Fig. 7, C–F; Supplemental Fig. 2). In some of these subapical cells, the PIN orientation was not strictly basal, but other orientations were observed as well. Along the tassel axis, PIN labeling was observed in two parallel cell files. Here, the polarization pattern suggested an auxin flux directed downward toward the base of the stem (Fig. 7D; Supplemental Fig. 2). The stained cells corresponded to the provasculature. Later on, the tassel vasculature clearly differentiated and staining was present in all the vascular bundles of the tassel central axis. Cross sections (data not shown) and longitudinal sections of the young tassel indicated that the signal remained confined to the inner cells and never interested the outer layers (Fig. 7, C, E, and F). Finally, when the tassel produced SMs, the pattern appeared more complex and we detected several relocalizations of the putative ZmPIN1 (Fig. 7, G and H). The signal was evident during spikelet differentiation, first along the cells that will form the central axis of the spikelet. Successively, the two florets are produced by the FMs and ZmPIN1 proteins are visible within their central cells. At a later stage of development, ZmPIN1 proteins localized at the site of the lower spikelet glumes, pointing toward the tassel central axis (data not shown). In the young female inflorescence, the expression pattern of ZmPIN1 appeared very similar to that of the young tassel (Fig. 7I; Supplemental Fig. 3). The signal was clearly visible in the vasculature and at the place of primordia bulging from the SMs. When the SMs produced spikelets, the proteins could be detected again in the lower glumes, in the vasculature, and also in the developing florets (Fig. 7, J and K). For the negative controls of immunostaining experiments, see Supplemental Figure 1.
Figure 7.
ZmPIN1 protein localization in maize wild-type apex and reproductive structures. All images portray maize B73 longitudinal sections labeled with the anti-PIN1 antibody plus a secondary antibody carrying the Alexa568 fluorochrome. C to H, Laser confocal images. A, B, I, J, and K, Epifluorescence images acquired with a Leica DC300F camera. A, At the vegetative stage, the maize SAM shows polar labeling in cells that form the middle part of its dome, but no labeling is present in the outer layers. B, When a primordium is initiated, the auxin efflux carriers are expressed in its outer cell layers. In all cells, the signal is present in the basal or lateral membranes, suggesting an overall auxin flux directed toward the base of the plant. C to F, When the SAM switches from the vegetative to the reproductive phase, ZmPIN1 proteins are present in the inner tissues of the developing tassel and along the tassel axis in the vasculature. G and H, When the tassel forms SMs, the protein localizes in the incipient meristems, reaching the surface of the tassel. I, In the young ear, the pattern of the putative auxin efflux carriers is repeated, with labeling on the basal membranes and in the L1 at the place of outgrowing primordia. J, Again, the vascular bundles are stained in the ear. K, In the developing spikelets, the polar proteins are located in continuity with the main axis vasculature and in the place of the forming glumes. An interaction between the internal tissues of the meristems and the surface of outgrowing primordia is suggested by the immunolabeling results. The vascular tissue shows the same polarization in the tassel, ear, and leaves, where the auxin flux seems directed downward. Bars: A to C, G, and H = 50 μm; D = 75 μm; E and F = 30 μm; I to K = 100 μm.
ZmPIN1 Expression in bif2
Localization of ZmPIN1 transcripts and proteins were determined in the bif2 mutant. The most severe alleles of this mutant present extremely reduced branching, both in male and female inflorescences, although plant bodies are normally developed (McSteen and Hake, 2001).
RT-PCR experiments conducted on RNA from bif2 tissues demonstrated that the levels of expression of ZmPIN1a and ZmPIN1b are similar to those of wild-type maize in roots and seedlings, whereas ZmPIN1a is more expressed in mutant tassels (Fig. 3).
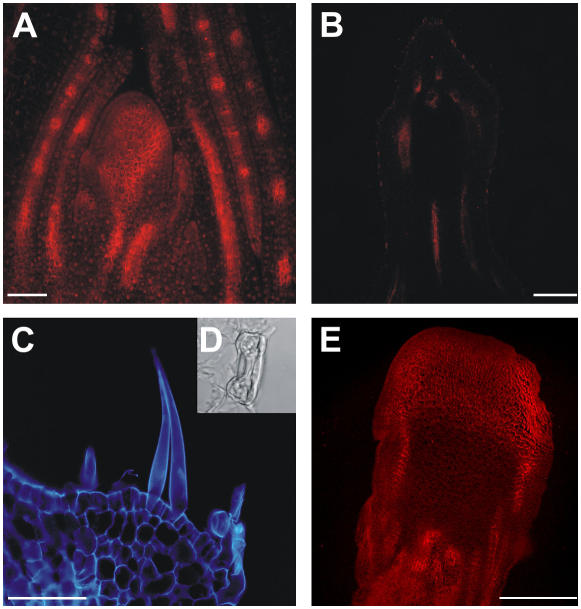
In situ hybridization experiments on bif2 vegetative apexes showed that transcript localization in the SAM resembles that of the wild-type B73 inbred line (data not shown). Moreover, immunolocalizations showed that the ZmPIN1 proteins in the mutant vegetative SAM followed a regular pattern (Fig. 8A). The anatomy of the mutant male inflorescence was analyzed in detail and the most severely affected tassels presented a highly irregular vasculature and an epidermis-like tissue forming the outermost layer. On this outer layer we found the presence of structures typical of a leaf epidermis, such as stomata and hairs (Fig. 8, C and D). Immunolocalization experiments indicated that there was no ZmPIN1 protein detection at all in the tassel of severe mutants, neither at the tip nor in the central vascular tissues (Fig. 8B). At the same time, polarization of ZmPIN1 proteins in the leaf vasculature of the same plants was evident (data not shown). Similar results were observed in the ears, with the disruption of the vasculature pattern in the central axis, but not in the leaves (Fig. 8E). Here, a few lower spikelets were formed and they presented abnormal/normal ZmPIN1 expression. Most of the ear surface did not differentiate SMs and, at the place where we expected to find these structures, an important difference was that ZmPIN1 did not appear in the central part of the SM, as observed in the wild type. A large domain, including the whole tip of the ear, presented polarized labeling, with the ZmPIN1 protein basally inserted into the plasma membrane of all cells, even in the surface layer.
Figure 8.
ZmPIN1 localization in bif2 vegetative and reproductive tissues. The bif2 mutant presents wild-type vegetative development, but abnormal reproductive structures. During the vegetative phase, bif2 plants develop a normal phyllotaxis and the putative auxin efflux carrier localization appears the same as in wild-type maize. After the switch to the reproductive phase, the tassel and ear show several developmental defects and PAT seems to be completely impaired. A, Longitudinal section of a bif2 SAM, where ZmPIN1 presents a wild-type localization. The 7-d after germination bif2 SAM section was labeled with the anti-AtPIN1 antibody coupled to an Alexa568 fluorochrome (red). B, In the fully developed tassels of severe bif2 mutants, the ZmPIN1 is completely absent. This image shows a longitudinal tassel section, which presents no polar signal; tissue autofluorescence appears red. C, Epifluorescence image of a longitudinal section detail of a mutant tassel at flowering. Structures appear blue as autofluorescence was observed with a 4′,6-diamino-phenylindole filter. In the tassel of severe bif2 mutants, the antibody does not recognize any membrane protein. The male inflorescence presents no AMs and a completely disrupted vasculature. The developmental defects include the production of leaf-like epidermis, which carries stomata (D) and trichomes. E, Epifluorescence image of a bif2 adult ear. Red color codes for the Alexa568 fluorochrome. In female reproductive structures, AMs are absent and the vascular bundles are disrupted, although the defects seem less severe than in the tassel. The anti-PIN1 antibody labels polarly localized proteins in the vasculature. Bars: A = 100 μm; B and E = 75 μm; C = 30 μm.
DISCUSSION
ZmPIN1a and ZmPIN1b Are Homologs of AtPIN1
The ORF of ZmPIN1a and ZmPIN1b encodes putative polypeptides with 71.3% and 70.9% identity to AtPIN1, respectively. Phylogenetic analysis shows that ZmPIN1a and ZmPIN1b fall into the same cluster together with the orthologous PIN1 proteins of rice, Arabidopsis, and wheat. Hydropathy profiles revealed that the PIN1 proteins contain two blocks of transmembrane segments connected by a hydrophilic region (Galweiler et al., 1998). Together, the observations suggest that ZmPIN1a and ZmPIN1b are two orthologs of AtPIN1.
Although semiquantitative RT-PCR reveals that the two genes show similar expression levels in roots, seedlings, leaves, and tassels of B73 plants, differences are observed as well. ZmPIN1a, for example, is not expressed in young ears, where only ZmPIN1b is present. This demonstrates diversification of the domains of expression, with ZmPIN1a more expressed in tissues carrying several vascular bundles as leaves and differentiated ears. At the same time, ZmPIN1b appears to be more specific for young ears and tassels.
ZmPIN1 Proteins Present a Developmental Stage-Specific Expression Pattern and May Interact with Other Proteins in the Auxin Transport Network of Maize
In the male and female inflorescences, ZmPIN1 expression patterns are strictly associated with the differentiating vascular tissues. The association of the expression pattern with the provasculature is confirmed by the localization of the ZmPIN1 protein in the mature embryo axis.
ZmPIN1 RNAs and proteins are always localized in the lateral developing primordia of the SAM, tassel, and ear and in the inner core of the meristems. During inflorescence development, ZmPIN1 is first localized in the cells corresponding to the central axis of the growing spikelet, and then it switches to the outer layers and central cells of the lower gluma. In the florets, ZmPIN1 is localized in the axis of the outgrowing organ.
In the leaf primordia and leaves and in the male and female inflorescence, the ZmPIN1 proteins are localized in the vascular cell files on the basal membrane of the cells, suggesting an auxin flux directed basipetally. This is repeated in a whole strip of cells that encircles the inflorescence tip in the tassel. If we look at the inner tissues of the upper part of the tassel, we can see that ZmPIN1 is polarized in a way that suggests auxin fluxes spreading at 360° around a small group of cells (Fig. 9A).
Figure 9.
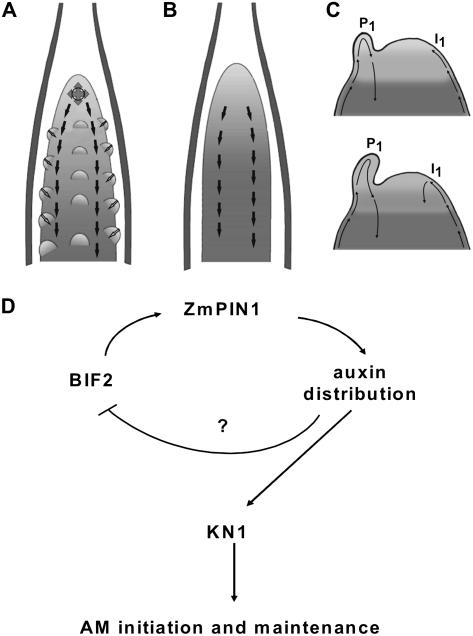
Model for the role of PAT in maize and Arabidopsis meristem differentiation. Arrows indicates polar auxin fluxes. A, Schematic representation of a maize wild-type tassel. The image was adapted from McSteen et al. (2000). ZmPIN1 is polarized in a way that suggests auxin fluxes spreading at 360° around a small group of cells at the top of the tassel and along the tassel axis in two parallel cell files. The polarization pattern suggests an auxin flux directed downward. The existence of an interaction between the internal tissues of the tassel and the surface of the outgrowing primordia is hypothesized. B, Schematic representation of a bif2 mutant tassel with alteration in AM initiation. The image was modified from McSteen et al. (2000). Whereas bif2 mutant plants still show vascular tissues with basally oriented PIN1, the subapical cells showing fluxes spreading at 360° were completely absent from the mutant tassel, which fails to initiate and maintain AMs. C, Schematic representation of two Arabidopsis SAMs in early (top) and later (bottom) stage of development during primordium formation (adapted from Reinhardt et al., 2003). The primordium P1 accumulates acropetally transported auxin, preventing accumulation on the left flank of the meristem, whereas auxin can reach the right flank (I1). Here, auxin promotes the formation of an organ initium and the establishment of a new auxin sink. D, A causal link between bif2 and ZmPIN1 is hypothesized. BIF2 could regulate the expression of ZmPIN1 and other PIN family members in the inflorescence, thus indirectly modulating auxin distribution at the site of AM formation.
The complex polar patterns of ZmPIN1 might determine auxin distribution in the different maize tissues in synergy with other polarly distributed members of the PIN family, as in Arabidopsis. PIN proteins, together with other components, seem to form an auxin transport network that mediates local auxin distribution and triggers different cellular responses, according to the organ and developmental stage. Key members of this signaling network are the P glycoproteins (PGPs), the plant homologs of human multiple drug resistance proteins. They have been identified first in Arabidopsis and shown to mediate the transport of indole acetic acid (IAA) and IAA metabolites in HeLa cells, maize protoplasts, and in planta (Arabidopsis; Geisler et al., 2005). In particular, two members of the family, PGP1 and PGP19, have been functionally characterized and shown to determine phenotypic alterations in Arabidopsis, the main one being dwarfism (Geisler et al., 2003, 2005). Recently, the PGP1 orthologous gene has been cloned in maize (brachytic2 [br2]/ZmPGP1) and in Sorghum bicolor (dwarf3/SbPGP1) and shown to be responsible for IAA transport in the stem (Multani et al., 2003). The br2 mutant presents a dwarf phenotype with very short basal internodes, altered stem anatomy, and vascular tissue defects, and has been shown to have altered auxin transport. However, br2 inflorescences develop regularly and generate fertile progeny. Thus, for the future, it would be desirable to determine the possible interaction between maize PGP and PIN proteins, for example, studying the ZmPINs pattern in the br2 mutant.
Expression Patterns Point to the Importance of Internal Tissues in Patterning
In many tissues, distribution of maize proteins resembles the one observed for PIN1 in Arabidopsis. In general, the proteins are localized in provascular and vascular tissues and, in maize roots, their localization indicates that auxin is directed toward the root tip via the mature vascular tissues, as demonstrated for Arabidopsis (Benkova et al., 2003; Blilou et al., 2005). Nevertheless, we also observed differences in labeling patterns. First, maize protein is not observed in the L1 layer of the tassel meristem, except at the level where the young primordia form. Interestingly, a pattern so far not described for the Arabidopsis SAM was also observed: in particular, the transcript and protein appear to be present in the apical region of the developing tassel in the corpus, just below the surface. The protein distribution suggests that there is a center at this level from which auxin fluxes are pointing outward into all directions. This is also the only group of cells where we identified cells in which PIN was not only localized basally. Although the precise function of this subepidermal group of cells in the apex still needs to be established, the results point to the potential importance of the internal tissues of the SAM in patterned auxin transport. This is very different from the results obtained with Arabidopsis, which precisely point to the importance of the meristem surface in the same process (Fig. 9, A–C; Heisler et al., 2005). There is the intriguing possibility that, in both species, only part of the PIN patterns was revealed so far and that both internal and surface layers play a role in both species. Alternatively, monocotyledonous and dicotyledonous species could have developed different strategies to redistribute auxin.
BIF2
The bif2 mutant presents abnormal branching of the female and male inflorescence (McSteen and Hake, 2001). ZmPIN1a and ZmPIN1b transcripts are detectable in vegetative and reproductive tissues of this mutant by RT-PCR and no differences in the expression levels were observed in bif2 roots and seedlings compared to the wild type, according to the normal vegetative aspects of these mutants. In bif2 leaves, ZmPIN1a is more expressed than in the wild type. The same can be observed in mutant tassels. By contrast, ZmPIN1b is less expressed in bif2 mutants than in the wild type. This aspect can be explained by the hypothesis of specialization in expression patterns: ZmPIN1a may be specific for the differentiation of vascular tissues, being more expressed in mutant tassels. On the other hand, ZmPIN1b may be specific for the development of primordia because it is less expressed in bif2 male inflorescences. The high levels of ZmPIN1a expression in bif2 tassels may confirm this hypothesis because the vascular tissues are formed in the male inflorescences, whereas no primordia are present.
Interestingly, whereas bif2 still shows vascular tissues with basally oriented PIN distribution, the group of cells carrying PIN just below the surface, observed in wild-type tassels and ears, was completely absent from the mutant. There are several possible explanations for this. First, BIF2 could be required to initiate this special population of cells. Their absence would subsequently lead to important changes in meristem function and organ initiation. It could also be that BIF2 has a more cellular role. It is noteworthy that we also frequently observed cells with upward-oriented PIN-labeled membranes in these cells. Because these orientations were not readily observed in the mutant, it could be, for instance, that BIF2 is required for orienting PIN in nonbasal orientations, much as PINOID in Arabidopsis (Friml et al., 2004; Furutani et al., 2004).
At the organ level, because the ZmPIN1 pattern is disrupted at the place of AM formation, it is possible that the lack of correct localization of ZmPIN1 in the provasculature indirectly causes the absence of BM, SAM, and/or SM formation and development. McSteen and Hake (2001) showed that kn1 is expressed in AM initially and this expression is completely disrupted in bif2 plants. Thus, they hypothesized that the absence of AM is due to the fact that, in bif2 mutants, AM initially fails to maintain kn1 expression (McSteen and Hake, 2001). Here, we hypothesize a causal link between BIF2 and ZmPIN1, which drives auxin distribution in both the tassel and the ear. BIF2 could regulate the expression of ZmPIN1, thus indirectly modulating auxin distribution at the site of AM formation (Fig. 9D).
Finally, possible redundancy of genes controlling branching in maize inflorescence, such as barren stalk1 (Ritter et al., 2002; Gallavotti et al., 2004), ramosa1 (Vollbrecht et al., 2005), ramosa2 (Bortiri et al., 2006), ramosa3 (Satoh-Nagasawa et al., 2006), and suppressor of sessile spikelets1 (Doebley et al., 1995), should be taken into account. Indeed, it could be that several genes take part in the interaction network, which controls auxin-responsive branching and spikelet formation in maize.
MATERIALS AND METHODS
Plant Material
The maize (Zea mays) B73 line was used for immunolocalization and in situ hybridization experiments. The maize mutant line bif2-2354 was obtained from the Maize Genetic Cooperation Stock Center (accessible online at http://www.maizegdb.org). This line was generated by ethylmethane sulfonate mutagenesis by M.G. Neuffer (Neuffer and Briggs, 1994) and characterized by McSteen and Hake (2001). Mutant seeds were germinated in the field and plants were self pollinated. F1 seeds from plants showing the most severe inflorescence phenotypes were sown and plant tissues were used to perform in situ hybridization and immunostaining experiments.
Isolation of the Full-Length ZmPIN1a and ZmPIN1b
NCBI (http://www.ncbi.nlm.nih.gov), TIGR (http://www.tigr.org/), and MaizeGDB (http://www.maizegdb.org) were screened using the AtPIN1 (AT1G73590 for genomic DNA and AF089084 for cDNA) sequence as a query. Several maize expressed sequence tag (EST) sequences were collected and those that presented the higher similarity to AtPIN1 in the domain overlapping the anti-PIN1 peptide were chosen to design primers. A first primer combination (fwd, 5′-ATGATTACGGGGACGGACTTCTA-3′ and rev, 5′-TCCAGACGAACATGTGGAGGTCCT-3′) was used to amplify a genomic fragment of 1,050 bp at the 5′ end of the gene. Sequencing revealed that the first primer combination used could amplify two different fragments. One of the two fragments contained an insertion of 44 bp at the end of the first exon. On this insertion, two forward primers (5′-GCAGGGCAAGGCGAACAAGTACGGCCAG-3′ and 5′-CTCTCCCCGTGGTGAATGCGACGTCCG-3′) were designed and used in combination with an oligo(dT)12–18 in RT-PCR amplifications. Sequencing of the amplified products allowed us to design new primers and obtain two distinct genomic clones. The primer combination (fwd, 5′-ATGATCACCGGCACGGACTTCTA-3′ and rev, 5′-CGCTGTTGGCCTGCGGGAACGAGCAGC-3′) amplified ZmPIN1a (2,770 bp), whereas the primer combination (fwd, 5′-CCACGCGTCCGGGATGGTCCAAGGAGAG-3′ and rev, 5′-CTCAAGCCATCAAACTCCGGAGGTGAGC-3′) amplified ZmPIN1b (3,300 bp).
Cloning of the PCR amplification products was performed with the pCRII-TOPO TA-cloning kit (Invitrogen) and with the pGEM-T Easy vector systems (Promega). After sequencing on both strands, sequences were blasted as queries using BLASTn, BLASTx, and tBLASTx algorithms and proteic and nucleotidic similarities were determined. Sequences were edited and aligned using EditSeq and Megalign (Lasergene DNASTAR) software and grouped to form single consensus sequences using SeqMan software (Lasergene DNASTAR). Restriction maps were created using Web Map software (available at http://pga.mgh.harvard.edu/web_apps/web_map/start). GeneSeqer software was used to excise introns from genomic sequences before alignment and assembling.
Phylogenetic Analysis
PIN sequences from Arabidopsis (Arabidopsis thaliana), wheat (Triticum aestivum), and rice (Oryza sativa) were identified through a BLAST search of The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org) and TIGR (http://www.tigr.org) databases using AtPIN genes as query (Paponov et al., 2005). Data obtained were predicted amino acid sequences based on EST clones. The full cDNA sequences were unknown for wheat; therefore, the EST at the 5′ end was used. Each EST included in this analysis was at least 288 bp long (96 amino acids). Sequences were edited and aligned using EditSeq and Megalign (Lasergene DNASTAR) software. Multiple alignments were prepared using ClustalX 1.81 according to Thompson et al. (1997). Neighbor-joining phylogenetic trees were generated using Tree-View 1.6.6 (http://darwin.zoology.gla.ac.uk/wrpage/treeviewx).
Genomic DNA, RNA Extraction, and Semiquantitative RT-PCR
Genomic DNA was extracted from maize tissues, according to the Nucleon Phytopure extraction kit (Amersham). Total RNA was extracted from maize tissues (seedlings, young roots, shoot apexes, leaves, tassels, and ears) according to the RNeasy plant mini kit (Qiagen) and subjected to on-column DNase treatment. RT-PCR was performed with the SuperScript III reverse transcriptase kit (Invitrogen), according to the manufacturer's instructions. Two micrograms of total RNA were used as a template together with 1 μL oligo(dT)12–18 (25 μg/μL). ZmPIN1a (fwd, 5′-GTCAAGGAGGTCCGCATGGCCGTCGCC-3′; rev, 5′-CGCTGTTGGCCTGCGGGAACGAGCAGC-3′) and ZmPIN1b (fwd, 5′-CTCTCCCCGTGGTGAATGCGACGTCCG-3′; rev, 5′-GAGCGTGATCGGCAGGGCGATGAGCAT-3′) specific primer combinations were used for semiquantitative amplifications of the two transcripts. Each primer combination was used in multiplex amplification reactions together with Ubi (fwd, 5′-TAAGCTGCCGATGTGCCTGCGTCG-3′) and Ubi (rev, 5′-CTGAAAGACAGAACATAATGAGCACAG-3′; Giulini et al., 2004) to amplify control ubiquitin transcripts. The PCR product lengths are 700 bp for ZmPIN1a, 863 bp for ZmPIN1b, and 210 bp for ubiquitin.
In Situ Hybridization
In situ hybridization experiments were carried out as described by Varotto et al. (2003). Plant material was fixed in 4% paraformaldehyde (Sigma) in 0.1 m phosphate buffer (pH 7.2) for 16 h at 4°C and embedded in Paraplast Plus (Sigma-Aldrich). Sections (7–10 μm) were cut using a microtome (RM 2135; Leica) and collected in xylane-coated slides. Slides were deparaffinized and treated with 10 μg/mL proteinase K. In vitro transcription of the digoxigenin-UTP-labeled (Roche) RNA sense and antisense probes was obtained using T7 and Sp6 polymerases. The probe for ZmPIN1 corresponds to a 500-bp fragment of the cDNA and is 87% identical between ZmPIN1a and ZmPIN1b; it overlaps the sequence of the peptide recognized by the anti-AtPIN1 antibody. Hybridization was performed in 50% formamide at 48°C overnight. Digoxigenin detection and signal visualization were carried out using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Roche), following the manufacturer's instructions. Slides were air dried and mounted with DPX mounting medium (Fluka Biochemika).
Immunolocalization
Immunostaining was performed on wax-embedded (9:1; PEG400 distearate:1-hexadecanol 99%; Sigma) shoots, leaves, and inflorescences. Material was previously fixed in a 4% paraformaldehyde, 1× phosphate-buffered saline (PBS) solution for 1 h, under vacuum conditions at room temperature. Embedded tissues were cut with a Leica RM2135 microtome (Leica) and stained overnight with a 1:200 anti-PIN1 1× PBS, 1% bovine serum albumin solution. The next day, sections were washed twice for 10 min in 1× PBS and stained with the secondary antibody: anti-rabbit IgG conjugated with Alexa568 or Alexa488 (Molecular Probes) and/or with fluorescein isothiocyanate (Sigma) diluted 1:400 in 1× PBS for 1 h. Sections were washed twice for 10 min with 1× PBS and mounted with VECTASHIELD mounting medium (Vector Laboratories).
Microscopy
Immunostained slides were observed with a Leica TCS SP2 laser confocal microscope (Leica Microsystems). Images were collected frame by frame with the acousto-optical tunable filter using Ag-Kr and He-Ne laser lines 488 or 543/594 nm, respectively. Images were coded red for Alexa568; green for Alexa488 and fluorescein isothiocyanate labeling; and white/blue for autofluorescence.
Histological analysis, in situ hybridization, and immunolabeled section images were acquired with a Leica DM4000B digital microscope, equipped with a Leica DC300F camera and Leica Image Manager 50 software (Leica Microsystems).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers DQ836239 and DQ836240 for ZmPIN1a and ZmPIN1b, respectively.
Supplementary Material
Acknowledgments
We thank V. Rossi for critical reading of the manuscript, C. Bonghi and E. Belles-Boix for helpful discussions, and T. Pengo for supporting experiments.
This work was supported in part by the Università Italo-Francese (grants to N.C.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Serena Varotto (serena.varotto@unipd.it).
The online version of this article contains Web-only data.
References
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Bortiri E, Chuck G, Vollbrecht E, Rocheford T, Martienssen R, Hake S (2006) ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutté Y, Crosnier MT, Carraro N, Traas J, Satiat-Jeunemaitre B (2006) The plasma membrane recycling pathway and cell polarity in plants: studies on PIN proteins. J Cell Sci 119: 1255–1265 [DOI] [PubMed] [Google Scholar]
- Carraro N, Peaucelle A, Laufs P, Traas J (2006) Cell differentiation and organ initiation at the shoot apical meristem. Plant Mol Biol 60: 811–826 [DOI] [PubMed] [Google Scholar]
- de Reuille PB, Bohn-Courseau I, Ljung K, Morin H, Carraro N, Godin C, Traas J (2006) Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc Natl Acad Sci USA 103: 1627–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley JF, Stec A, Kent B (1995) Suppressor of sessile spikelets1 (sos1)—a dominant mutant affecting inflorescence development in maize. Am J Bot 82: 571–577 [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jurgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, et al (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Furutani M, Vernoux T, Traas J, Kato T, Tasaka M, Aida M (2004) PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131: 5021–5030 [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Zhao Q, Kyozuka J, Meeley RB, Ritter MK, Doebley JF, Pe ME, Schmidt RJ (2004) The role of barren stalk1 in the architecture of maize. Nature 432: 630–635 [DOI] [PubMed] [Google Scholar]
- Galweiler L, Guan C, Muller A, Wisman E, Mendgen K, Yephremov A, Palme K (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, Titapiwatanakun B, Peer WA, Bailly A, Richards EL, et al (2005) Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J 44: 179–194 [DOI] [PubMed] [Google Scholar]
- Geisler M, Kolukisaoglu HU, Bouchard R, Billion K, Berger J, Saal B, Frangne N, Koncz-Kalman Z, Koncz C, Dudler R, et al (2003) TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol Biol Cell 14: 4238–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulini A, Wang J, Jackson D (2004) Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Irish EE (1997) Experimental analysis of tassel development in the maize mutant tassel seed 6. Plant Physiol 114: 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlemeier C, Reinhardt D (2001) Auxin and phyllotaxis. Trends Plant Sci 6: 187–189 [DOI] [PubMed] [Google Scholar]
- McSteen P, Hake S (2001) barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 128: 2881–2891 [DOI] [PubMed] [Google Scholar]
- McSteen P, Laudencia-Chingcuanco D, Colasanti J (2000) A floret by any other name: control of meristem identity in maize. Trends Plant Sci 5: 61–66 [DOI] [PubMed] [Google Scholar]
- McSteen P, Leyser O (2005) Shoot branching. Annu Rev Plant Biol 56: 353–374 [DOI] [PubMed] [Google Scholar]
- Multani DS, Briggs SP, Chamberlin MA, Blakeslee JJ, Murphy AS, Johal GS (2003) Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302: 81–84 [DOI] [PubMed] [Google Scholar]
- Neuffer MG, Briggs S (1994) Designation of bif2. Maize Newsletter 68: 28 [Google Scholar]
- Paponov IA, Teale WD, Trebar M, Blilou I, Palme K (2005) The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci 10: 170–177 [DOI] [PubMed] [Google Scholar]
- Parry G, Delbarre A, Marchant A, Swarup R, Napier R, Perrot-Rechenmann C, Bennett MJ (2001) Novel auxin transport inhibitors phenocopy the auxin influx carrier mutation aux1. Plant J 25: 399–406 [DOI] [PubMed] [Google Scholar]
- Petrasek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, et al (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Mandel T, Kuhlemeier C (2000) Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Ritter MK, Padilla CM, Schmidt RJ (2002) The maize mutant barren stalk1 is defective in axillary meristem development. Am J Bot 89: 203–210 [DOI] [PubMed] [Google Scholar]
- Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D (2006) A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441: 227–230 [DOI] [PubMed] [Google Scholar]
- Swarup R, Kargul J, Marchant A, Zadik D, Rahman A, Mills R, Yemm A, May S, Williams L, Millner P, et al (2004) Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16: 3069–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas J, Bohn-Courseau I (2005) Cell proliferation patterns at the shoot apical meristem. Curr Opin Plant Biol 8: 587–592 [DOI] [PubMed] [Google Scholar]
- Traas J, Vernoux T (2002) The shoot apical meristem: the dynamics of a stable structure. Philos Trans R Soc Lond B Biol Sci 357: 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto S, Locatelli S, Canova S, Pipal A, Motto M, Rossi V (2003) Expression profile and cellular localization of maize Rpd3-type histone deacetylases during plant development. Plant Physiol 133: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E, Springer PS, Goh L, Buckler ES IV, Martienssen R (2005) Architecture of floral branch systems in maize and related grasses. Nature 436: 1119–1126 [DOI] [PubMed] [Google Scholar]
- Xu M, Zhu L, Shou H, Wu P (2005) A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol 46: 1674–1681 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.