Abstract
The early light-induced proteins (Elips) in higher plants are nuclear-encoded, light stress-induced proteins located in thylakoid membranes and related to light-harvesting chlorophyll (LHC) a/b-binding proteins. A photoprotective function was proposed for Elips. Here we showed that after 2 h exposure of Arabidopsis (Arabidopsis thaliana) leaves to light stress Elip1 and Elip2 coisolate equally with monomeric (mLhcb) and trimeric (tLhcb) populations of the major LHC from photosystem II (PSII) as based on the Elip:Lhcb protein ratio. A longer exposure to light stress resulted in increased amounts of Elips in tLhcb as compared to mLhcb, due to a reduction of tLhcb amounts. We demonstrated further that the expression of Elip1 and Elip2 transcripts was differentially regulated in green leaves exposed to light stress. The accumulation of Elip1 transcripts and proteins increased almost linearly with increasing light intensities and correlated with the degree of photoinactivation and photodamage of PSII reaction centers. A stepwise accumulation of Elip2 was induced when 40% of PSII reaction centers became photodamaged. The differential expression of Elip1 and Elip2 occurred also in light stress-preadapted or senescent leaves exposed to light stress but there was a lack of correlation between transcript and protein accumulation. Also in this system the accumulation of Elip1 but not Elip2 correlated with the degree of PSII photodamage. Based on pigment analysis, measurements of PSII activity, and assays of the oxidation status of proteins we propose that the discrepancy between amounts of Elip transcripts and proteins in light stress-preadapted or senescent leaves is related to a presence of photoprotective anthocyanins or to lower chlorophyll availability, respectively.
The chlorophyll (chl)-binding proteins in photosynthetic organisms are located either in core complexes of PSI and PSII or form antenna systems (Green and Durnford, 1996). The antenna systems of higher plants consist of nuclear-encoded chl a/b-binding (Cab) proteins whose primary function is the absorption of light through chl excitation and transfer of absorbed energy to photochemical reaction centers.
The Cab superfamily in Arabidopsis (Arabidopsis thaliana) consists of 20 different proteins (Jansson, 1999), out of which six are associated with PSI (Lhca1–6) and 14 with PSII (Lhcb1–6 and their isomers). The Cab proteins associated with PSII are organized in minor (Lhcb4–6) and major (Lhcb1–3) antenna systems. The major antenna is either tightly associated with PSII core complex or forms an outer pool at more distant locations relative to PSII that is loosely bound (Bassi and Dainese, 1992; Drepper et al., 1993; Vink et al., 2004; Consoli et al., 2005). This pool can reversibly dissociate/reassociate with PSII during the process called state transition (Allen and Forsberg, 2001; Haldrup et al., 2001; Wollman, 2001). Recent crystallization of spinach (Spinacia oleracea) light-harvesting complex from PSII (LHCII) revealed that the basic structural and functional unit of the major antenna is the trimer (Liu et al., 2004). However, it still remains uncertain whether the mobile pool consists of monomers (mLhcb) or trimers (tLhcb; Kouril et al., 2005). The monomerization of tLhcb was also reported during assembly (Dreyfuss and Thornber, 1994) or disassembly of LHCII prior to its degradation upon excess of light (Garab et al., 2002).
In the past years several distant relatives of Cab protein superfamily with conserved chl-binding residues and a transient expression pattern were found in PSI and PSII of pro- and eukaryota. These distant relatives belong to the early light-induced protein (Elip) family of light stress proteins (Montané and Kloppstech, 2000; Adamska, 2001). Three-helix Elips, two-helix stress-enhanced proteins, and one-helix one-helix proteins (Ohps) were described from Arabidopsis (Heddad and Adamska, 2000; Jansson et al., 2000; Andersson et al., 2003).
The Elip family members in higher plants and green algae are nuclear-encoded proteins that accumulate in thylakoid membranes in response to various abiotic stresses, when the expression of Cab proteins is down-regulated (Montané and Kloppstech, 2000; Adamska, 2001). Therefore, it was proposed that Elip family members have a photoprotective role under light stress conditions either by transient binding of released chls and preventing the formation of free radicals and/or by participating in energy dissipation (Montané and Kloppstech, 2000; Adamska, 2001).
Previous studies showed that Elip from pea (Pisum sativum) is located in the nonappressed regions of the thylakoid membrane in the vicinity of PSII (Adamska and Kloppstech, 1991) and is associated into a multisubunit complex together with unidentified polypeptides of 24 to 26 kD (Adamska et al., 1999). However, no detailed localization studies were performed for these proteins.
Here, we investigated the localization of Elips in green light-stressed leaves of Arabidopsis and compared their expression in light stress-preadapted or senescing leaves exposed to low or high intensity light. Two closely related Elip1 and Elip2 (81.05% identity at the amino acid level) are present in Arabidopsis, thus raising the question about their redundant physiological function. Alternatively, Elip1 and Elip2 may perform their function at different intramembrane locations or during different stages of light stress. We demonstrated that although both proteins were found in isolated mLhcb and tLhcb under light stress conditions they coisolated with different LHCII subpopulations. Interestingly, during the first 2 h of light stress the amount of Elips in relation to the amount of tLhcb and mLhcb was almost equal, but a longer exposure to light stress resulted in higher Elip amounts in tLhcb as compared to mLhcb. We showed further that both proteins were differentially expressed in response to light stress in green, light stress-preadapted and senescing leaves and their expression was controlled at transcriptional and/or transcriptional/posttranslational levels. However, it is not known whether the steady-state level of Elip transcripts is controlled by their transcription and/or degradation rates. While the accumulation of Elip1 occurred in parallel to the inactivation and damage of PSII, a massive accumulation of Elip2 was assayed when approximately 40% of PSII was photodamaged and lost their D1 protein. This suggests that Elip1 and Elip2 function is related to different phases of light stress and occurs in different LHCII subpopulations.
RESULTS
Coisolation of Elip1 and Elip2 with PSII Antenna
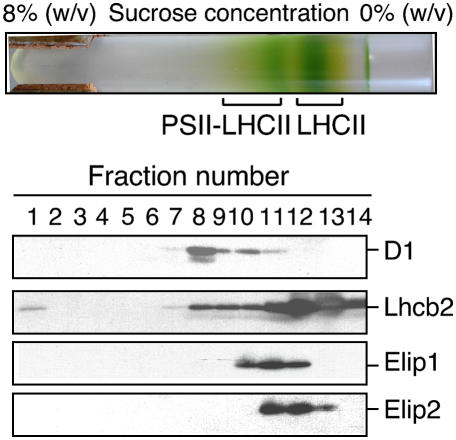
Cross-linking studies indicated that Elip in pea is located in the vicinity of PSII reaction center (Adamska and Kloppstech, 1991). We explored the location of Elip1 and Elip2 in an isolated PSII-LHCII supercomplex from thylakoid membranes of light-stressed green Arabidopsis leaves using a single-step detergent solubilization and Suc density gradient centrifugation as described (Eshaghi et al., 1999). Two green bands were visible in the gradient after centrifugation (Fig. 1, top section). Previous studies (Eshaghi et al., 1999) showed that the top green band contained LHCII and the bottom band PSII-LHCII supercomplex. To confirm the identity of these complexes the immunoblot analyses were performed using collected gradient fractions. The results revealed (Fig. 1, bottom section) that approximately 60% of the total amount of D1 protein used as a marker for PSII reaction center was present in the fraction 8 and 40% was detected in fractions 9 to 11. Immunoblot with anti-Lhcb2 antibodies used for the detection of LHCII complex showed that 76% of the total amount of Lhcb2 was present in fractions 11 to 13 and the remaining 24% was distributed onto fractions 8 to 10 and 14.
Figure 1.
Localization of Elip1 and Elip2 in LHCII of Arabidopsis. The PSII-LHCII supercomplex was released from light stress-treated thylakoid membranes with DM and separated by a Suc density gradient centrifugation (top section). The protein composition in collected gradient fractions was analyzed by immunoblotting using polyclonal antibodies raised against Elip1, Elip2, the D1 protein of PSII reaction center (D1), and the major Cab protein of PSII (Lhcb2).
To investigate the distribution of Elips in collected gradient fractions anti-Elip1 or anti-Elip2 antibodies were raised and used for such analysis. The results revealed that both proteins were located mainly in fractions 11 and 12 (83.5% and 90% of the total amount of Elip1 and Elip2, respectively) with 16.5% of the total amount of Elip1 detected in the fraction 10 and 10% of the total amount of Elip2 in the fraction 13. Using discontinuous 0.5% to 10% (w/v) Suc gradients (1 mL of 0.7% steps of increasing Suc concentration) more drastic differences in the distribution of Elip1 and Elip2 were observed (data not shown). These data indicate that Elip1 and Elip2 colocalize with LHCII but their different distribution in the gradient suggests their association with different LHCII subpopulations.
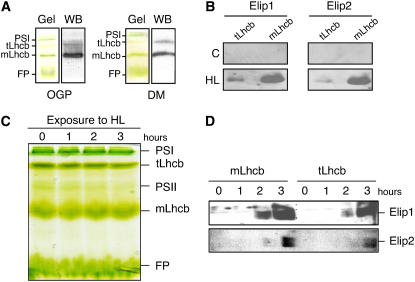
When isolated from thylakoid membranes Lhcb1 to 3 proteins were mainly organized into homo- or hetero-tLhcb or mLhcb (Caffarri et al., 2004). To test whether Elip1 and Elip2 coisolate with mLhcb and/or tLhcb populations we isolated both Lhcb forms using nondenaturating green gel electrophoresis and tested the presence of Elips by immunoblotting. Detergents, such as Triton X-100 in combination with lithium dodecyl sulfate (LDS; data not shown), n-octyl-β-d-glucopyranoside (OGP), and n-dodecyl β-d-maltoside (DM) were tested for the optimal solubilization of light-stressed thylakoid membranes and the separation of mLhcb and tLhcb populations. The results revealed that independent of the detergent used PSI, tLhcb, mLhcb, and free pigments were well separated in the gel and visible as green bands, while PSII core complexes were almost invisible in some preparations and their amounts depended on the solubilization conditions (Fig. 2, A and C). To confirm the position of mLhcb and tLhcb in the gel we performed immunoblot analysis using anti-Lhcb2 antibodies. The results revealed (Fig. 2A) that the amounts of isolated tLhcb and mLhcb varied depending on the detergent used. A higher amount of tLhcb was obtained after the solubilization of thylakoid membranes with DM as compared with OGP (Fig. 2A) or Triton X-100:LDS (data not shown). Therefore, DM was used routinely for further analysis (see “Materials and Methods” for details). To investigate the presence of Elip1 and Elip2 in mLhcb and tLhcb populations under control and light stress conditions we excised corresponding green bands from the gel and subjected them to the denaturating gel electrophoresis followed by immunoblotting. In agreement with previous reports (Heddad and Adamska, 2000; Andersson et al., 2003), Elip1 and Elip2 were not detected in control leaves kept under low light conditions but accumulated in both Lhcb populations in response to light stress (Fig. 2B). Much higher amounts of Elip1 and Elip2 (80% to 95% of the total amount) were detected in the mLhcb as compared to the tLhcb population (5% to 20% of the total amount). However, the amount of Elips calculated per Lhcb content was almost equal for mLhcb and tLhcb (compare Fig. 2, A and B).
Figure 2.
Localization of Elip1 and Elip2 in tLhcb and mLhcb populations. A, Thylakoid membranes were isolated from high light-treated (3 h at 1,800 μmol m−2 s−1) leaves and solubilized with OGP or DM to release pigment-protein complexes prior to their separation on native green gels (Gel). Green bands corresponding to PSI complex, tLhcb, mLhcb, and free pigments (FP) are marked. The identity of tLhcb and mLhcb was confirmed by immunoblot (WB) with anti-Lhcb2 antibody. B, Pigment-protein complexes were isolated from low light-treated control (C; 3 h at 20 μmol m−2 s−1) or high light-treated (HL; 3 h at 1,800 μmol m−2 s−1) leaves as described in A using DM as detergent. Green bands containing tLhcb and mLhcb were excised from the gel, incubated in sample buffer (Laemmli, 1970) for 30 min at 50°C, and loaded onto 15% denaturated SDS-PAGE gels prior to immunoblotting with anti-Elip1 and anti-Elip2 antibodies. C, Green leaves were exposed to high light (1,800 μmol m−2 s−1) for 0 to 3 h, the pigment-protein complexes were solubilized with DM, and separated on native green gels as described in A. D, Bands containing tLhcb and mLhcb were excised from the gels and treated as described in B prior to immunoblotting.
It was reported that the tLhcb pool dissociates into the mLhcb population upon light stress (Garab et al., 2002). To investigate whether the distribution of Elip1 and Elip2 in mLhcb and tLhcb populations changes during the duration of light stress we isolated pigment-protein complexes at various time points of light stress exposure. The results revealed (Fig. 2C) that no significant changes in amounts of PSI and mLhcb were observed during the exposure of leaves to light stress for 3 h. In contrast, the amounts of tLhcb or PSII core complexes decreased almost linearly with the time of exposure to light stress and reached after 3 h of illumination 50% and 55% of the initial values, respectively. Immunoblot analysis showed that Elip1 and Elip2 were not detected in control leaves or leaves exposed to light stress for 1 h but their amounts increased drastically during a longer illumination period (Fig. 2D). After 2 h of illumination approximately 30% and 17% of the maximal amount of Elip1 present in 3 h illuminated leaves and 10% and 14% of Elip2 were detected in mLhcb and tLhcb, respectively. Similar to data shown in Figure 2B, the amounts of Elips in relation to amounts of tLhcb or mLhcb were comparable after 2 h of illumination (Fig. 2D). However, after 3 h of the high light exposure approximately 40% of the total Elips amount was detected in tLhcb as compared to 60% in mLhcb but due to a strong decrease of the tLhcb amount (50% of the initial value) the ratio of Elip:tLhcb was much higher (40%:50%) than Elip:mLhcb (60%:100%).
Expression of Elip1 and Elip2 in Green Arabidopsis Leaves Exposed to Light Stress
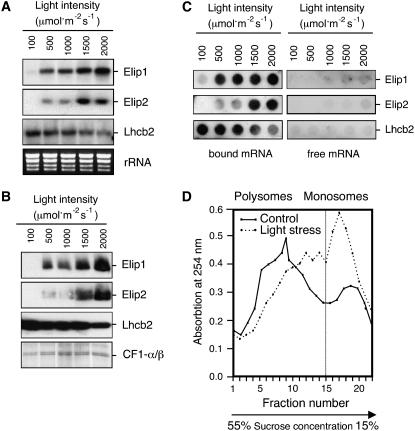
To test whether Elip1 and Elip2 are differentially expressed under light stress conditions we analyzed the accumulation of both proteins and their transcripts in leaves exposed to increasing light intensities from 100 μmol m−2 s−1 (low light conditions) to 2,000 μmol m−2 s−1 (severe light stress conditions). Northern-blot analysis revealed (Fig. 3A) that although the level of transcript accumulation for these two genes increased in a light intensity-dependent manner, different accumulation profiles were observed for each of these mRNAs. While Elip1 transcripts accumulated almost linearly with an increasing light intensity starting from 500 μmol m−2 s−1 and reaching the maximal level at 2,000 μmol m−2 s−1, a comparable amount of Elip2 transcripts was induced at 500 μmol m−2 s−1 and 1,000 μmol m−2 s−1 (Fig. 3A). Exposure of leaves to 1,500 μmol m−2 s−1 resulted in a 6-fold increase of Elip2 transcripts but higher light intensities did not significantly influence this accumulation level. In agreement with reports published for barley (Hordeum vulgare; Pötter and Kloppstech, 1993) also in Arabidopsis the level of transcript accumulation of Lhcb2 decreased almost linearly with increasing light intensities (Fig. 3A).
Figure 3.
Light intensity-dependent accumulation of Elip1 and Elip2 in green Arabidopsis leaves exposed to light stress. Mature green leaves were exposed to increasing light intensities for 3 h prior to the isolation of total RNA, polysome-bound and -free mRNAs, or total membrane proteins. A, Northern blot. As a reference, the rRNA pattern in the gel, visualized by staining with ethidium bromide, is shown. B, Immunoblot analysis. As a reference, the α- and β-subunits of the CF1-ATP-synthase complex (CF1-α/β) are shown. C, Distribution of Elip1, Elip2, and Lhcb2 mRNAs between polysome-free and polysome-bound fractions assayed by dot-blot hybridization. D, Analysis of translationally active (polysomes) and inactive (monosomes) ribosomes isolated after exposure of leaves to low light (control, 20 μmol m−2 s−1) or high light (light stress, 1,800 μmol m−2 s−1) conditions for 3 h. Polysomes and monosomes were separated on a linear 5% to 55% Suc density gradient and their distribution was monitored by the measurement of absorption spectra at 254 nm.
Immunoblotting analysis revealed that Elips accumulated in thylakoid membranes with kinetics resembling the accumulation pattern of their transcripts (Fig. 3B). Similarly, the amount of Lhcb2 protein followed the level of transcript amount and decreased with an increasing light intensity (Fig. 3B). The amounts of α/β-subunits of the CF1-ATP-synthase complex (CF1-α/β) assayed as a control did not change significantly during the light stress treatment (Fig. 3B).
To assess how the increasing irradiance influences the translational activity of Elip1 and Elip2 transcripts we analyzed the distribution of these mRNAs between polysome-bound and -free fractions. During the first step of protein synthesis in the cytoplasm, mRNAs are integrated into the ribosomal complex and polysomes are formed. Analysis of the mRNA content of polysomes is assumed to be indicative of active protein synthesis. Our results revealed that over 99% of Elip1 and Elip2 mRNAs were detected in the polysomal fraction, indicating their simultaneous translation into corresponding proteins. The distribution of Lhcb2 transcripts, assayed for comparison, demonstrated (Fig. 3C) that also these mRNAs were actively translated. Less than 1% of the total amount of Elip1, Elip2, and Lhcb2 transcripts were present in polysome-free fractions under all light intensities tested.
Interestingly, exposure of leaves to light stress resulted in a drastic decrease of polysomal complexes followed by the enrichment in monosomes (Fig. 3D). While under low light conditions 71% of ribosomes formed polysomes and only 29% were present as monosomes, under light stress conditions the polysome amount was reduced to 55% and monosomes increased to 45%. This suggests a selective translation of induced Elip1 and Elip2 mRNAs under light stress conditions.
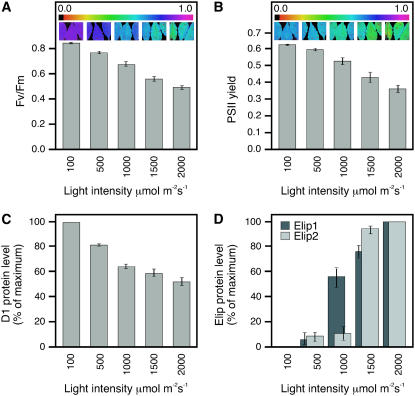
It was reported that the accumulation of the carotenoid biosynthesis-related (Cbr) protein, a homolog of Elip in the green alga Dunaliella salina, occurs in parallel with the accumulation of photodamaged PSII reaction centers (Jin et al., 2001, 2003). To test whether the accumulation of Elip1 and Elip2 in Arabidopsis correlates with the photoinactivation and photodamage of PSII we exposed leaves to increasing light intensities and measured the PSII activity by the assay of chl fluorescence (Fig. 4, A and B), the loss of D1 protein (Fig. 4C), and the accumulation of Elips (Fig. 4D) using quantitative immunoblotting. The data revealed that Fv/Fm values representing a maximal efficiency of PSII (Fig. 4A) and PSII yield (Fig. 4B) in the dark-adapted state decreased almost linearly with increasing light intensities. The Fv/Fm ratio assayed for leaves exposed to 100 (control) and 2,000 μmol m−2 s−1 (light stress) was 0.840 ± 0.008 and 0.478 ± 0.047, respectively (Fig. 4A). The PSII yield was 0.624 ± 0.009 and 0.356 ± 0.060 for control and light stress-treated leaves, respectively (Fig. 4B). Both parameters indicate that exposing leaves to 2,000 μmol m−2 s−1 inactivated 43% of PSII reaction centers.
Figure 4.
Comparison of the Elip protein accumulation with photoinactivation and photodamage of PSII under increasing light intensities. Mature green leaves were exposed to increasing light intensities for 3 h prior to images of chl fluorescence parameters and isolation of total membrane proteins. A, Fv/Fm values expressing a maximal efficiency of PSII shown as a bar graph and leaf images. B, PSII yield expressing photon use efficiency of PSII shown as a bar graph and leaf images. C, Densitometric quantification of the D1 protein amount assayed by immunoblotting. The amount of D1 protein detected at 100 μmol m−2 s−1 was set as 100%. D, Densitometric quantification of Elip1 and Elip2 protein amounts assayed by immunoblotting. The amount of Elips detected at 2,000 μmol m−2 s−1 was set as 100%.
As reported before (Andersson and Aro, 2001), the exposure of leaves to light stress led to reduced amounts of D1 protein, probably when the degradation rate exceeded the rate of its de novo synthesis (Fig. 4C). The loss of D1 protein occurred in parallel to the inactivation of PSII, and the steady-state level of D1 in leaves exposed to 2,000 μmol m−2 s−1 was reduced to approximately 50% as compared to the amount present in control leaves. Quantifications of Elip signals in immunoblots revealed (Fig. 4D) that Elip1 accumulated almost linearly with increasing light intensities and its level of accumulation correlated with increasing photoinactivation (Fig. 4, A and B) and photodamage (Fig. 4C) of PSII reaction centers. In contrast, the level of Elip2 accumulation was low (7%–13% of the maximal value) in leaves exposed to 100 to 1,000 μmol m−2 s−1 and a massive accumulation (92%–100%) of these proteins occurred only at 1,500 to 2,000 μmol m−2 s−1, when at least 40% of PSII reaction centers were photodamaged (Fig. 4C).
Differential Expression of Elip1 and Elip2 in Light Stress-Preadapted or Senescent Arabidopsis Leaves Exposed to Light Stress
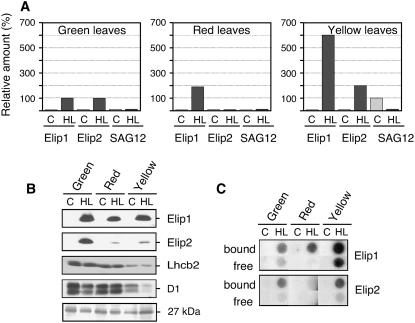
We investigated the expression pattern of Elip1 and Elip2 transcripts and proteins in Arabidopsis leaves preadapted to moderate light stress conditions and in senescent leaves exposed to low or high light conditions. Exposure of low light-acclimated leaves to moderate light stress conditions was accompanied by a massive accumulation of anthocyanins (Merzlyak and Chivkunova, 2000) in leaf vacuoles (red leaves). During natural senescence a dramatic loss of chls occurred and leaves turned yellow due to remaining carotenoids (Hörtensteiner, 1999; Matile, 2001) that were previously masked by chls (yellow leaves). For comparison, the expression of Elips in mature green leaves was assayed under the same experimental conditions. The results revealed that no significant amounts of Elip1 or Elip2 transcripts were detected in green, red, and yellow leaves kept at low light intensity (Fig. 5A). The exposure of green leaves to light stress resulted in the accumulation of comparable amounts of Elip1 and Elip2 transcripts, thus confirming our previous data (Fig. 3A; Heddad and Adamska, 2000). In red leaves exposed to light stress the level of mRNA accumulation for Elip1 was 2-fold higher as compared to green leaves but only traces of Elip2 transcripts were detected in such leaves (Fig. 5A). An approximately 6- or 2-fold higher level of Elip1 or Elip2 transcript accumulation was observed in light-stressed yellow leaves as compared to green leaves, respectively (Fig. 5A).
Figure 5.
Differential expression of Elip1 and Elip2 in green, red, and yellow Arabidopsis leaves. Mature green, light stress-preadapted (red), or senescent (yellow) leaves were exposed to low light (control [C], 20 μmol m−2 s−1) or high light (HL; 1,800 μmol m−2 s−1) conditions for 3 h. A, Quantification of the transcript level assayed by dot-blot hybridization. The amount of Elip1 and Elip2 transcripts detected in green leaves exposed to HL was set as 100%. As a molecular marker for natural senescence the accumulation of SAG12 (encoding a Cys protease) transcripts was assayed under the same conditions and the maximal value was set as 100%. The quality and quantity of isolated RNAs was tested on the agarose gel prior to dot blotting (data not shown). B, Immunoblot analysis using thylakoid membranes isolated from green, red, and yellow leaves. For comparison, the level of the major Lhcb2 and the D1 protein of PSII reaction center (D1) are shown. The level of 27 kD unknown protein stained with Coomassie blue confirmed an equal gel loading. C, Analysis of polysome-bound (bound) and -free (free) RNAs for Elip1 and Elip2 assayed by dot-blot hybridization. Signals obtained with 0.25 μg RNAs are shown in this section.
The expression of senescence-associated gene 12 (SAG12) was assayed as a molecular marker for naturally induced senescence. This gene encodes a Cys protease (Lohman et al., 1994) and is expressed as a consequence of natural aging of tissues (Noh and Amasino, 1999). As expected, a high level of SAG12 transcript accumulation was detected in yellow leaves under low light conditions but their amounts were strongly reduced after the exposure of such leaves to light stress (Fig. 5A). The SAG12 transcripts were neither detected in red nor in green leaves, both under low light or light stress conditions (Fig. 5A).
Immunoblot analysis revealed that the accumulation of Elip1 and Elip2 in thylakoid membranes followed the induction of corresponding transcripts only in green leaves exposed to light stress but not in red or yellow leaves (compare Fig. 5, A and B). In red and yellow leaves exposed to light stress amounts of Elip1 were reduced to 70% and 90%, respectively, as compared to green leaves, although its transcripts were induced to a higher extent. Only 5% and 13% of Elip2 amounts present in green leaves were detected in thylakoid membranes of red and yellow leaves exposed to light stress, respectively, and these amounts did not correlate with the high level of Elip2 transcripts induced in yellow leaves (compare Fig. 5, A and B). As expected, no Elip1 and Elip2 were detected in green, red, or yellow leaves exposed to low light intensity.
For comparison, the levels of Lhcb2 and D1 protein from PSII reaction center were assayed in green, red, and yellow leaves under the same experimental conditions. The results revealed (Fig. 5B) that the amount of Lhcb2 decreased to a different extent in red and yellow leaves as compared with green leaves. While red leaves still contained 80% of the Lhcb2 amount present in green leaves, only 15% this protein was assayed in yellow leaves. No significant differences in the amount of Lhcb2 were detected in low or high light-treated green, red, and yellow leaves (Fig. 5B). Comparable amounts of D1 protein were detected in green and red leaves but its level was reduced to 18% in yellow leaves under low light conditions as compared to amounts present in green leaves (Fig. 5B). In yellow leaves exposed to light stress the amount of D1 protein was reduced to 7% (Fig. 5B) probably due to a disturbed balance between the rates of protein synthesis and degradation (Andersson and Aro, 2001). Surprisingly, the amount of D1 protein present in low light-exposed red leaves was unaffected by the light stress treatment.
The discrepancy between transcript and protein levels for Elip1 and Elip2 in red and yellow leaves suggests the existence of a posttranscriptional control in the expression of both genes. This posttranscriptional regulation can occur at translational or posttranslational levels. To investigate whether Elip1 and Elip2 transcripts induced in red and yellow leaves by light stress are actively translated we analyzed the distribution of mRNAs between polysome-bound and -free fractions (Fig. 5C). For comparison, polysomal fractions from green leaves exposed to light stress were also analyzed. Our results revealed (Fig. 5C) that 75% and 97% of total Elip1 and 80% and 70% of total Elip2 mRNAs induced by light stress in green and red leaves, respectively, were found to be bound to polysomes and to be actively translated. The remaining amounts of these transcripts were detected in the polysome-free fractions. However, in yellow leaves 40% of the amount of Elip1 mRNAs were found as free RNAs.
Analysis of Pigments, PSII Activity, and Oxidation of Proteins in Light Stress-Preadapted and Senescent Leaves
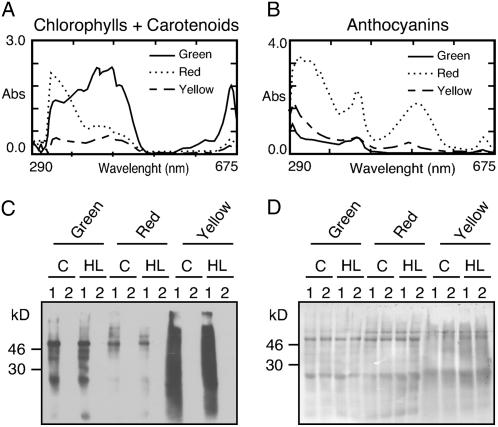
It was shown (Adamska et al., 2001) that the insertion of Elips into isolated membranes from barley depended on chl a. Thus, a low chl content in yellow leaves could explain low amounts of Elip1 and Elip2 as compared to enhanced levels of their transcripts under light stress conditions (Fig. 5, A and B). It was also reported that anthocyanins act as a pigment screen protecting the photosynthetic apparatus from excess light energy (Feild et al., 2001). This could be a reason for the low Elip2 content in red leaves. To test these two possibilities we performed pigment analysis in green, red, and yellow leaves, measured the maximal PSII efficiency, and assayed the oxidation status of proteins as an indication for photoprotection (Fig. 6, A–D). Pigments (chls, carotenoids, and anthocyanins) were isolated from control leaves and their identity was confirmed by the measurement of absorption spectra in 80% acetone (chls and carotenoids) or in a methanol-based solvent (anthocyanins; Fig. 6, A and B). Quantitative analysis of pigments demonstrated (mean values for three independent experiments are given below) that green leaves contained 18.5 μg cm−2 chl a, 9.39 μg cm−2 chl b, 2.53 μg cm−2 carotenoids, and 0.02 μg cm−2 anthocyanins. The pigments in red leaves consisted of 3.98 μg cm−2 chl a, 1.79 μg cm−2 chl b, 1.18 μg cm−2 carotenoids, and 1.42 μg cm−2 anthocyanins. Yellow leaves contained 2.03 μg cm−2 chl a, 1.19 μg cm−2 chl b, 0.83μg cm−2 carotenoids, and 0.21 μg cm−2 anthocyanins. This demonstrated that in red leaves chl and carotenoid content was reduced to approximately 20% and 45%, respectively, as compared to values assayed in green leaves. Even more drastic reduction of the pigment content was measured in yellow leaves, where only 10% of chl and 30% of carotenoid contents were assayed. Also the anthocyanin content differed between green, red, and yellow leaves. While green leaves contained almost nondetectable amounts of anthocyanins, their content in red leaves was very high (set as 100%) and yellow leaves contained approximately 14% of the amount measured in red leaves.
Figure 6.
Pigment analysis and oxidation status of proteins in green, red, and yellow Arabidopsis leaves. A, Absorption spectra of chl and carotenoids. B, Absorption spectra of anthocyanins. Total pigments were extracted from leaf discs (12 mm diameter) excised from mature green, light stress-preadapted (red), or senescent (yellow) leaves and incubated in the dark for 24 h at 4°C in 1 mL of 80% acetone (chls and carotenoids) or 3 m mixture of HCl:water:MeOH, 1:3:16 (v/v/v; anthocyanins). C, Oxyblot showing the oxidation level of proteins in green, red, and yellow leaves exposed to low light (control [C], 20 μmol m−2 s−1) or high light (HL; 1,800 μmol m−2 s−1) conditions for 3 h. Protein extracts were treated with an oxidation detection reagent (lane 1) or a control reagent (lane 2) included into the Oxyblot kit according to the manufacturer's protocol prior to the separation by a 14% SDS-PAGE and immunoblotting. D, Coomassie blue staining of the corresponding membrane as described in C.
To test whether red leaves showed a better protection of photosynthetic apparatus against light stress due to the accumulation of anthocyanins we measured the photosynthetic activity before and after the exposure to light stress in correlation to amounts of D1 protein and Elips. The similar measurements were performed for green and yellow leaves for comparison. The results revealed (Table I) that the photosynthetic activity assayed as changes in chl fluorescence induction kinetics and expressed as Fv/Fm was reduced to 58% and 72% after exposure of green and yellow leaves to light stress, respectively. This photoinactivation correlated with decreased amounts of D1 protein whose level was reduced to 55% and 80% in green and yellow leaves. Interestingly, only a minor reduction of Fv/Fm (93%) and the D1 amount (93%) was assayed in red leaves. Similar to results shown in Figure 4 also in this experimental system the amount of Elip1 correlated with the degree of photoinactivation and photodamage of PSII. The highest amount of Elip1 accumulated in green leaves (this value was set as 100%) followed by yellow (90%) and red (70%) leaves. In contrast, Elip2 was strongly induced only in green leaves (this value was set as 100%) but not in red (7%) or yellow (15%) leaves, which were less photodamaged.
Table I.
Accumulation of Elips compared to photoinactivation and photodamage of PSII reaction centers in green, light stress-preadapted (red) or senescent (yellow) leaves exposed to light stress
Leaves were exposed to low light (20 μmol m−2 s−1) or high light (1,800 μmol m−2 s−1) for 3 h and the maximal efficiency of PSII (Fv/Fm) and amounts of Elip1, Elip2, and D1 protein were measured. LL, Low light; HL, high light.
| Parameter | Green Leaves
|
Red Leaves
|
Yellow Leaves
|
|||
|---|---|---|---|---|---|---|
| LL | HL | LL | HL | LL | HL | |
| Fv/Fm | 0.810 ± 0.010 | 0.470 ± 0.037 | 0.620 ± 0.017 | 0.578 ± 0.027 | 0.298 ± 0.007 | 0.215 ± 0.012 |
| Fv/Fm (%) | 100 ± 0 | 58 ± 6 | 100 ± 0 | 93 ± 2 | 100 ± 0 | 72 ± 2 |
| Amount of D1 (%) | 100 ± 0 | 55 ± 10 | 100 ± 0 | 93 ± 7 | 100 ± 0 | 80 ± 11 |
| Amount of Elip1 (%) | 0 ± 0 | 100 ± 0 | 0 ± 0 | 70 ± 9 | 0 ± 0 | 90 ± 5 |
| Amount of Elip2 (%) | 0 ± 0 | 100 ± 0 | 0 ± 0 | 7 ± 5 | 0 ± 0 | 15 ± 5 |
Protein oxidation results mainly in introduction of carbonyl groups, which can be easily detected. We measured the oxidation status of proteins in green, red, or yellow leaves that were exposed at low or high light conditions (Fig. 6, C and D). The results revealed that several oxidized proteins were detected in green leaves and their content increased dramatically in yellow leaves. Several additional oxidized proteins were detected in green leaves under high light conditions (Fig. 6C). Interestingly, only few oxidized proteins were detected in red leaves and their content did not change significantly after the light stress treatment. A Coomassie blue-stained immunoblot membrane, shown as a control, confirmed that the equal amount of proteins was loaded in each line (Fig. 6D). The diffused protein pattern in samples isolated from yellow leaves results from their modification by photooxidation.
DISCUSSION
Differential Expression of Elip1 and Elip2 in Green Arabidopsis Leaves Exposed to Light Stress Is Regulated at the Transcript Level and Related to the Degree of PSII Photodamage
We showed that the accumulation of Elip1 and Elip2 transcripts and proteins was induced in a light intensity-dependent manner but the induction kinetics differed for both genes. While Elip1 transcripts and proteins accumulated almost linearly with increasing light intensities, the induction of Elip2 transcripts and proteins occurred stepwise. Furthermore, much higher photon fluency rates were required for elip2 than elip1 genes for a comparable expression level. While significant amounts of Elip1 transcripts and proteins were induced already at 500 μmol m−2 s−1, a comparable amount of Elip2 transcripts and proteins were detected only at 1,500 μmol m−2 s−1 (Fig. 3). The induced Elip1 and Elip2 transcripts were simultaneously translated into correspondent proteins that accumulated in thylakoid membranes. Interestingly, the amount of accumulated Elip1 but not Elip2 correlated with the degree of photoinactivation and photodamage of PSII reaction centers. While Elip1 accumulation and the degree of photoinactivation and photodamage of PSII reaction centers increased almost in parallel in green (Fig. 4), red, and yellow leaves (Table I), the comparable amounts of Elip2 were induced only when approximately 40% of PSII reaction centers were damaged. Thus, the expression pattern of Elip1 resembles that reported for its Cbr homolog in Dunaliella, where the induction of Cbr followed the accumulation of photodamaged D1 protein in 160 kD complex (Jin et al., 2001, 2003). We expect that the physiological role of Elip2 is required under light stress conditions, where the accumulation of Elip1 alone did not offer a sufficient protection and an additional protection from Elip2 might ensure an adequate light stress defense.
The differential expression of four high light-induced A to D (Hli) proteins, related to higher plants Ohps, was reported for the cyanobacterium Synechocystis PCC 6803 (He et al., 2001) and for Elip1 and Elip2 transcripts during photomorphogenesis of Arabidopsis seedlings (Harari-Steinberg et al., 2001). The HliA, HliB, and HliC proteins rapidly accumulated in high light, but while the level of HliC protein remained high for at least 24 h, the level of HliA and HliB proteins began to decline already after 9 to 12 h of light stress exposure. The HliD protein was transiently expressed in high light and was not detected 24 h after beginning of illumination (He et al., 2001). Thus, the differential expression of Elip family members under light stress conditions seems to be very conserved for prokaryotic and eukaryotic organisms, thus indicating a different physiological role of each Elip/Hlip species (see below).
A differential expression of elip1 and elip2 genes might be connected with the differences in their promoter regions. It was reported that the transcription factor HY5 promotes the light induction of elip1 but not elip2 gene during greening of etiolated Arabidopsis seedlings (Harari-Steinberg et al., 2001). HY5 is a basic Leu zipper transcription factor that binds directly to G box in DNA sequences of light-responsive promoters (Chattopadhyay et al., 1998). Using the Plant Cis-Acting Regulatory Elements database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) we performed searches for cis-acting elements present in promoters of elip1 and elip2 genes that have been identified as components of light-responsive elements in photosynthetic genes. It was postulated that the combination of at least two elements is required to confer light responsiveness (Martínez-Hernández et al., 2002). We found the light-responsive element was composed of three GATA motifs located between the CCAAT (−875 bp) and TATA (−538 bp) boxes in elip2 but not in elip1 promoter region, which might explain different light stress responses assayed for both genes.
Differential Expression of Elip1 and Elip2 in Light Stress-Preadapted or Senescent Arabidopsis Leaves Exposed to Light Stress Is Independently Regulated at the Level of Transcript and Protein Accumulation
We demonstrated that the level of transcript accumulation for Elip1 in red and yellow leaves exposed to light stress increased 2- or 6-fold, respectively, as compared with green leaves, while the protein amount was down-regulated. Also the enhanced level of Elip2 transcript accumulation present in yellow leaves exposed to light stress was not accompanied by the accumulation of the correspondent protein. Since the induced Elip RNAs were actively translated as shown by their association with polysomes the reduced amounts of proteins could be a result of lower rates of translation, import into plastids, insertion into plastid membranes, or an increased rate of the degradation.
It was shown in the past that the chl a availability is crucial for the stable insertion of Elips into etioplast membranes of barley (Adamska et al., 2001). Furthermore, the amount of inserted Elip increased almost linearly with chl concentration. Thus, it is possible that a strongly reduced chl content in red or yellow leaves limited the accumulation of Elip1 and Elip2 in thylakoid membranes and promoted their degradation.
Investigation of the global gene expression pattern in naturally senescing leaves of poplar (Populus tremula) trees revealed that transcripts for Elips, together with those for metallothioneins and Cys proteases, were the most abundant (Bhalerao et al., 2003). Also, significant amounts of Elip transcripts, but not proteins, accumulated in senescing leaves of some pea cultivars under low light conditions and the exposure to light stress resulted in a massive accumulation of Elip transcripts and proteins (Norén et al., 2003). Furthermore, the accumulation of Elip transcripts and proteins was reported also in artificially induced senescence of detached tobacco (Nicotiana tabacum) leaves kept at low light intensities (Binyamin et al., 2001). In contrast to these reports no Elip1 or Elip2 transcripts or proteins were detected in senescent yellow leaves of Arabidopsis under low light conditions. This suggests that the presence of Elip transcripts during senescence is not always of a physiological relevance, as is the case for Arabidopsis.
Our data revealed that red leaves contained very high amounts of anthocyanins as compared with green or yellow leaves. Although the role of anthocyanins is not very well understood, it was postulated that they could be involved in several protective mechanisms, including the modification of the quantity and quality of light (Barker et al., 1997), the protection from photoinhibition (Gould et al., 1995; Dodd et al., 1998), and the scavenging of reactive oxygen intermediates under stressful environments (Yamasaki, 1997; Sherwin and Farrant, 1998). It is also believed that anthocyanins protect shade-adapted chloroplasts from brief exposures to high intensities of sunlight (Gould et al., 2000). Our results demonstrated that the high level of anthocyanins in red leaves protected the photosynthetic machinery from light stress damage. A much lower photoinactivation of PSII was assayed by measurements of changes in variable chl fluorescence after exposure of such leaves to light stress as well as a lower degree of photodamage of D1 protein of PSII reaction center. Also the photooxidation status of proteins was much lower in red leaves as compared with green or yellow leaves.
Anthocyanins absorb mainly light between 270 and 290 nm (UV-B), 310 and 320 nm (UV-A), and 500 and 550 nm (green light; Barker et al., 1997). It was previously reported that the induction of Elips in pea leaves exposed to light stress is controlled by a blue light receptor of cryptochrome-type absorbing blue and UV-A light (Adamska et al., 1992). A massive accumulation of Elips was reported also in pea plants exposed to low levels of UV-B (Sävenstrand et al., 2004). Thus, we can assume that low amounts of Elip2 transcripts in red leaves as compared with green or yellow leaves were a consequence of a reduced photon fluency rate reaching a photoreceptor that was sufficient for induction of elip1 but not for elip2 gene (see Fig. 3).
Coisolation of Elip1 and Elip2 with mLhcb and tLhcb Populations
There is only very limited information available on the intrathylakoid location of Elip family members. It was demonstrated that Elip from pea is located in PSII (Adamska and Kloppstech, 1991), while Ohp2 from Arabidopsis is associated with PSI (Andersson et al., 2003). A high molecular mass Elip complex of 100 kD with unidentified thylakoid membrane components was reported to exist in barley under combined light and cold stress conditions (Montané et al., 1999). A similar high molecular mass Elip complex composed of several unidentified polypeptides of 24 to 26 kD was found in pea leaves exposed to light stress (Adamska et al., 1999). Here we demonstrated that Elip1 and Elip2 in Arabidopsis coisolate with LHCII and are distributed with an equal ratio between mLhcb and tLhcb as based on the Lhcb content. On the basis of these data we can expect that the previously reported Elip complex might represent Elips bound to tLhcb.
It is still unclear whether Elips are attached to the preexisting homo- or hetero-tLhcb and/or form heterotrimers with Lhcb1, Lhcb2, or Lhcb3. Mutation analysis has shown that Trp-16, Tyr-17, and Arg-21 residues within the N-terminal domain of the mature Lhcb1 are needed for trimerization of LHCII complexes (Hobe et al., 1995). These residues are conserved in all Lhcb1 to 3 but not in Elips, thus speaking in favor of the first possibility.
Different distribution of Elip1 and Elip2 in the gradient fractions suggests their coisolation with different LHCII subpopulations. Native LHCII trimers are not homogeneous but consist of homo- and heterotrimers in different combinations of the various Lhcb1 to 3 isoforms (Larsson et al., 1987; Jackowski et al., 2001; Caffarri et al., 2004; Standfuss and Kühlbrandt, 2004). Depending of the composition of LHCII trimers a different role in photosynthesis, light adaptation, or photoprotection was proposed (Standfuss and Kühlbrandt, 2004). Thus, it is possible that Elip1 and Elip2 colocalize with LHCII trimers of different composition and function.
Based on past expression and localization studies it was proposed that Elips might be involved in binding of chls released during proteolytic degradation of photodamaged D1 protein from PSII (Adamska and Kloppstech, 1991) and/or proteolysis of LHCII (Lindahl et al., 1997). Both the outer population of LHCII and damaged PSII migrate from the grana stacks to the stroma lamellae of the thylakoid membrane and this is in agreement with the location of Elips in nonappressed regions of thylakoid membranes (Adamska and Kloppstech, 1991). It was proposed that Cbr in Dunaliella may participate in PSII repair process and may by critical for the protection of PSII during the process of degrading and replacing nonfunctional D1 reaction center protein (Jin et al., 2001, 2003). This hypothesis is also consistent with the accumulation pattern of Elip1 and Elip2 in Arabidopsis that amounts correlated with the degree of photoinactivation and photodamage of PSII reaction center.
It was shown that high intensity light induces the monomerization of tLhcb (Garab et al., 2002) prior to its degradation (Lindahl et al., 1995). It has been proposed that the trimeric form of Lhcb is protected from degradation due to a close interaction between the N-terminal parts of the three mLhcb, which hide the protease recognition site (Yang et al., 2000). Thus coisolation of Elips with mLhcb (subjected to degradation) and tLhcb (photodamaged) is consistent with the function proposed for these proteins. This scenario would also explain a higher content of Elips in relation to tLhcb during the prolonged light stress treatment.
Based on our data we can assume that the function of Elip1 and Elip2 in Arabidopsis is not redundant but connected to various locations within LHCII at different stages of the light stress treatment.
MATERIALS AND METHODS
Growth of Plants and Stress Conditions
Arabidopsis plants (Arabidopsis thaliana L. cv Columbia and cv Wassilewskaja) were grown in a growth chamber at 20°C at a photon flux density of 100 μmol m−2 s−1 under the light regime of 8-h dark/16-h light. Plants were cultivated either hydroponically (Norén et al., 2004) or on soil for 40 to 55 d prior to the collection of mature green leaves. The hydroponic system was shown to produce 5 times as much leaf material compared to plants grown on soil (Norén et al., 2004). For experiments shown in Figures 1 to 3, 5, and 6 both hydroponically and soil-grown plants were used in parallel for comparisons. Since no significant differences in experimental data were detected, experiments shown in Figure 4 and the replicas of all experiments were performed only with soil-grown plants.
For preadaptation to higher light intensities, 50- to 60-d-old, low light-grown plants were transferred to a moderate light intensity of 300 μmol m−2 s−1 for 4 d. A massive accumulation of anthocyanins occurred during this period (Fig. 6B). For natural senescence, plants were grown for 80 to 110 d under conditions described above prior to collection of senescing leaves. During this period a massive loss of chls occurred (Fig. 6A). Detached mature green leaves, anthocyanins containing red leaves, and naturally senescing yellow leaves floating on water were exposed to light stress for 3 h at a photon flux density of 1,800 μmol m−2 s−1 provided by white fluorescent lamps (Osram Power star HQI-E 250W/D). The spectrum of the lamp covered a visible light region from 380 to 720 nm. The temperature of the water was kept constant between 22°C and 25°C. Photon fluency rates were measured with a photometer (Skye, Techtum Laboratory AB). Control leaves were incubated at 20 μmol m−2 s−1 for the same time. After treatment leaves were frozen in liquid nitrogen and stored at −80°C for further analysis.
For light intensity-dependent induction of Elips (transcripts and proteins) and the association of mRNAs with polysomes leaves were exposed to increasing light intensities from 100 to 2,000 μmol m−2 s−1 for 3 h at 25°C. For isolation of polysomes and monosomes leaves were exposed to low light (20 μmol m−2 s−1) or high light (1,800 μmol m−2 s−1) for 3 h at 25°C.
Isolation and Assay of RNA
The total RNA was isolated with a RNeasy kit (Qiagen, GmbH), spotted on the Hybond-N+ membrane (Amersham Biosciences) at four different concentrations, 5.0, 2.5, 1.25, and 0.63 μg (data shown in Fig. 3), or 1.0, 0.5, and 0.25 μg (data shown in Fig. 5) with a dot-blot apparatus (Schleicher & Schuell), and the membrane was used for hybridization as described (Heddad and Adamska, 2000). The cDNA probe was labeled with [32α]dCTP using a megaprime DNA-labeling kit (Amersham Biosciences). The signals on the filter were analyzed with a Phosphorimager FLA3000 (Fujifilm) or x-ray film (Cronex 5, Agfa). For quantification, signals linear in intensity with exposure time (A600 > 0.8) were scanned at 600 nm (Personal Densitometer, Molecular Dynamics) using the Image-Quant 3.3 program.
For northern blots 5 μg RNA was separated in 1.2% agarose gel and transferred to Hybond-N+ membrane prior to the hybridization as described (Heddad and Adamska, 2000).
Isolation of Monosomes and Polysomes
The frozen plant material (5 g) was ground in liquid nitrogen and the resulting tissue powder was resuspended at 4°C in 50 mL polysome buffer containing 400 mm KCl, 50 mm Tris-HCl, pH 8.3, 10 mm Mg acetate, 250 mm Suc, 2% (w/v) Triton X-100, and 0.005% (v/v) β mercaptoethanol. The suspension was filtrated through Miracloth (Calbiochem) and centrifuged at 15,000g for 10 min at 4°C. The supernatant was loaded onto a two-step gradient containing 5 mL of 0.7 m and 7 mL of 1.7 m Suc in polysome buffer and gradients were centrifuged for 17 h at 200,000g and 4°C. The supernatant (containing free RNA) was collected and used for RNA isolation as described above. The pellet (containing polysomes) was resuspended in 2 mL of polysome buffer and centrifuged at 200,000g for 30 min at 4°C over a 0.5 mL 1.7 m Suc cushion. The pellet was used for isolation of polysome-bound RNA as described above.
For investigations of ribosomal profiles 1 to 2 g of frozen leaves were ground in liquid nitrogen and resuspended in five volumes of buffer A (200 mm Tris-HCl, pH 8.9, 200 mm KCl, 35 mm MgCl2, 0.6 m sorbitol, 12.5 mm EGTA, and 15 mm dithiothreitol). After differential centrifugation at 10,000g and 30,000g for 10 min, homogenate was filtrated through Miracloth (Calbiochem). After adding Triton X-100 to a final concentration of 2% (w/v), ribosomes were pelleted by centrifugation at 160,000g for 3.5 h at 4°C over a 1.5 m Suc cushion in buffer B (40 mm Tris-HCl, pH 8.9, 20 mm KCl, 10 mm MgCl2, 5 mm EGTA, and 5 mm dithiothreitol). The pellets were gently resuspended in buffer C (10 mm Tris-HCl, pH 7.6, 25 mm KCl, and 5 mm MgCl2) and centrifuged at 8,000g in the Eppendorf centrifuge. The polysomal suspension was loaded onto a linear 15% to 55% (w/v) Suc density gradient prepared in buffer C and centrifuged at 260,000g for 70 min. Gradient fractions containing polysomes and monosomes were collected by monitoring the absorption at 254 nm.
Isolation and Assay of Proteins
Leaves frozen in liquid nitrogen were homogenized in extraction medium containing 300 mm sorbitol, 20 mm HEPES-NaOH pH 7.4, 5 mm MgCl2, 2.5 mm EDTA, and 10 mm KCl. The homogenate was filtrated through Miracloth (Calbiochem), mixed (1:1, v/v) with 3× concentrated sample buffer (Laemmli, 1970), proteins denatured at 70°C for 5 min, and separated by SDS-PAGE according to Laemmli (1970) using 14% polyacrylamide gels and a Hoefer mini gel system. The gels were loaded on an equal protein basis.
Polyclonal antibodies were raised against synthetic peptides of Elip1 and Elip2 selected shortly after the predicted N-terminal cleavage sites. These peptide sequences were unique for Arabidopsis Elip1 and Elip2 and consisted of amino acids 48 to 60 for mature Elip1 (EGGPTNEDSSPAP) and amino acids 45 to 57 for mature Elip2 (QGDPIKEDPSVPST; AgriSera AB). The peptides were conjugated to keyhole limpet hemocyanin carrier protein prior to immunization of rabbits. The IgG fraction was purified by affinity chromatography on the G-protein-sepharose column (AgriSera AB). These antibodies were highly specific for Elip1 or Elip2 and did not show cross-reactivity with a second Elip form or with any other thylakoid membrane protein. The polyclonal D1 and Lhcb2 antibodies were purchased from Agrisera AB and the antibody against α/β-subunits of the CF1-ATP-synthase complex was kindly provided by Dr. Ralf Oelmüller (University of Jena, Germany).
Immunoblotting was carried out according to Towbin et al. (1979) using a polyvinylidene diflouride PLUS transfer membrane with 45-μm pores (Micron Separations) and an enhanced chemiluminescence assay (ECL, Amersham Biosciences) as the detection system. For quantification, signals linear in intensity with exposure time were scanned as described above.
For isolation and assay of oxidized proteins, leaves frozen in liquid nitrogen were homogenized in extraction medium as described above, supplemented with 2% (v/v) β-mercaptoethanol, and the protein oxidation was assayed using an OxyBlot protein oxidation detection kit (Intergen Company) after SDS-PAGE and immunoblotting. Oxidative modifications of proteins introduced carbonyl groups into protein side chains that were immunodetected with antibodies supplied in the OxyBlot kit.
Fractionation of Thylakoid Membranes and Isolation of Photosynthetic Complexes
Thylakoid membranes were prepared from fresh or frozen leaf material as described (Tidholm et al., 2002). For isolation of photosynthetic complexes on Suc density gradients, thylakoid membranes (0.5 mg chl mL−1 corresponding to approximately 3.0 mg protein) were disrupted by six strokes of a glass homogenizer in the presence of 20 mm DM (Glycon Biochemicals) in 25 mm MES pH 6.0, 10 mm NaCl, 5 mm MgCl2, and 2 m Gly betaine (MNMβ buffer) as described in Eshaghi et al. (1999). The homogenate (700 mL) was then loaded onto continuous Suc density gradients. The Suc density gradients were prepared by freezing and thawing of 8% (w/v) Suc in 25 mm MES pH 5.7, 10 mm NaCl, 5 mm CaCl2, 2 m Gly betaine (MNCβ buffer), and 0.03% (w/v) DM. The mixture was kept in SW-28 ultracentrifuge tubes at −80°C for at least 2 h. The thawing of the frozen mixture was initiated at room temperature using a water bath covering 2 to 3 cm of the tube bottom for 5 to 10 min. The tubes were then transferred to the cold room until the thawing was completed and a 0% to 8% Suc gradient was formed. Centrifugation was carried out at 4°C using a SW40 rotor (Beckman Instruments) at 27,000 rpm for 12 to 13 h. The gradient fractions (0.7 mL from a total gradient volume of 11 mL) were carefully collected using a peristaltic pump and the polypeptide composition in each fraction was analyzed on 15% SDS-PAGE according to Laemmli (1970).
To establish optimal conditions for the isolation of pigment-protein complexes from light-stressed leaves on green native gels (0.5 mg chl/mL) were solubilized either with Triton X-100:LDS mixture, 15 mm DM, or 15 mm OGP. For the solubilization with OGP or DM (data shown in Fig. 2, A and B) leaves were homogenized in medium containing 300 mm sorbitol, 20 mm HEPES, pH 7.4, 5 mm MgCl2, 2.5 mm EDTA, and 10 mm KCl and an aliquot of homogenate corresponding to 20 μg chl was mixed (10:1, v/v) with 1 m Tris, pH 6.6, and 30% glycerol, and supplemented with DM or OGP. Solubilization was carried out at room temperature for 5 min in the dark, with brief vortexing and samples corresponding to 2 to 4 μg chl separated on native green gels as described below. For the solubilization with Triton X-100:LDS mixture (10% stock solutions were mixed at the ratio 12:1, v/v) samples were treated as previously described (Peter and Thornber, 1991; Dreyfuss and Thornber, 1994).
For routinely used isolation of pigment-protein complexes (shown in Fig. 2, C and D) 2% (v/v) DM was used and the solubilization took place on ice for 10 min as described for greening barley (Hordeum vulgare) leaves (Peter and Thornber, 1991; Dreyfuss and Thornber, 1994). An aliquot of sample corresponding to 15 μg chl was loaded on a 10% native green gel (Camm and Green, 1980) and electrophoresis was carried out for 2 h at 4°C in the dark. Green bands containing mLhcb and tLhcb were excised from the gel, incubated in Laemmli buffer for 30 min at 50°C, and loaded onto a 15% denaturated SDS-PAGE (Laemmli, 1970) as described above.
Isolation and Assay of Pigments
For extraction of pigments two leaf discs with 12 mm diameter were agitated gently in the dark for 24 h at 4°C in 1 mL of 80% (v/v) acetone (chls and carotenoids) or 3 m mixture of HCl:water:MeOH (1:3:16, v/v/v, anthocyanins) according to Gould et al. (2000). Absorption spectra were measured at A647 and A663 for chl a and b or at A470 for carotenoids using a spectrophotometer (Beckman DU 640) and the concentration of pigments was calculated according to Lichtenthaler (1987). The anthocyanin level was calculated using an equation A530 − 0.24 A653 according to Murray and Hackett (1991). The anthocyanins absorbed maximally at 530 nm and the subtraction of 0.24 A653 compensated for the small overlap in absorbency with chls (Murray and Hackett, 1991).
Measurements of the Photosynthetic Activity
Chl fluorescence induction kinetics were measured at room temperature on detached leaves using either an imaging/pulse-amplitude modulation fluorimeter (Walz GmbH) for data shown in Figure 4 or a FlurCam fluorimeter (PSI Instruments) for data shown in Figure 5. Leaves were preadapted in the dark for 15 min and then exposed to a saturating 1 s light flash. The minimal fluorescence (F0) in the absence of actinic light and maximal fluorescence (Fm) after a saturating light flash were measured and the variable fluorescence (Fv = Fm − F0) was calculated as described (Butler and Kitajima, 1975). The photochemical yield of open PSII reaction centers, commonly known as the relative variable fluorescence, was calculated as Fv/Fm. This value reflects the maximal efficiency of PSII that is measured in dark-adapted tissues.
The effective quantum yield of PSII photochemistry (PSII yield) was calculated as described (Maxwell and Johnson, 2000; ΔF/Fm = [Fm′ − Ft]/Fm′), where Fm′ represented the maximal fluorescence in the light and Ft was the steady-state fluorescence yield in the light measured immediately before the saturating light flash. The PSII yield represents the proportion of the light absorbed by chl associated with PSII that is used in photochemistry.
Statistical Analysis
Results shown in Figures 1 and 2 are the average of five to seven independent experiments using 10 to 15 leaves of the same age collected from three to five plants. Results shown in Figure 3 are the average of three independent experiments using 8 to 10 (sections A and B), 50 to 60 (section C), and 10 to 20 (section D) leaves per point collected from three, or eight to 10 plants, respectively. Data shown in Figure 4 are the average of three independent experiments using three to four leaves per point. The measurement of Fv/Fm and PSII yield were performed on three leaves per point on six different leaf areas. Data shown in Figures 5 and 6 are the average of three independent experiments using 20 leaves per point collected from eight to 10 plants.
Sequence data from this article can be found in the GenBank under the following accession numbers: At3g22840 (Elip1) and At4g14690 (Elip2).
Acknowledgments
We thank Dr. Ralf Oelmüller (University of Jena, Germany) for providing polyclonal antibodies against α/β-subunits of the CF1-ATP-synthase complex and Amine Heddad (Stockholm) for the assisting with the promoter analysis. We would like also to thank Lars Hiertas Minne (Sweden) for the sponsoring peptide antibodies against Elip1 and Elip2 (to M.H.).
This work was supported by research grants from the Swedish Research Council, the Swedish Strategic Foundation and the Carl Tryggers Foundation (to I.A. and B.A.), the Deutsche Forschungsgemeinschaft (grant nos. AD92/7–1 and AD92/7–2 to I.A.), and the Konstanz University grant (to I.A.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Iwona Adamska (iwona.adamska@uni-konstanz.de).
References
- Adamska I (2001) The Elip family of stress proteins in the thylakoid membranes of pro- and eukaryota. In EM Aro, B Andersson, eds, Advances in Photosynthesis and Respiration-Regulation of Photosynthesis, Vol 11. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 487–505
- Adamska I, Kloppstech K (1991) Evidence for an association of the early light-inducible protein (ELIP) of pea with photosystem II. Plant Mol Biol 16: 209–223 [DOI] [PubMed] [Google Scholar]
- Adamska I, Kruse E, Kloppstech K (2001) Stable insertion of the early light-inducible proteins into etioplast membranes requires chlorophyll a. J Biol Chem 276: 8582–8587 [DOI] [PubMed] [Google Scholar]
- Adamska I, Ohad I, Kloppstech K (1992) Synthesis of early light-inducible protein is controlled by blue light and related to light stress. Proc Natl Acad Sci USA 89: 2610–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamska I, Roobol-Boza M, Lindahl M, Andersson B (1999) Isolation of pigment-binding early light-inducible proteins from pea. Eur J Biochem 260: 453–460 [DOI] [PubMed] [Google Scholar]
- Allen JF, Forsberg J (2001) Molecular recognition in thylakoid structure and function. Trends Plant Sci 6: 317–326 [DOI] [PubMed] [Google Scholar]
- Andersson B, Aro EM (2001) Photodamage and D1 protein turnover in photosystem II. In EM Aro, B Andersson, eds, Advances in Photosynthesis and Respiration-Regulation of Photosynthesis, Vol 11. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 377–393
- Andersson U, Heddad M, Adamska I (2003) Light stress-induced one helix protein of the chlorophyll a/b-binding family associated with photosystem I. Plant Physiol 132: 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DH, Seaton GGR, Robinson SA (1997) Internal and external photoprotection in developing leaves of the CAM plant Cotyledon orbiculata. Plant Cell Environ 20: 617–624 [Google Scholar]
- Bassi R, Dainese P (1992) A supramolecular light-harvesting complex from chloroplast photosystem II membranes. Eur J Biochem 204: 317–326 [DOI] [PubMed] [Google Scholar]
- Bhalerao R, Keskitalo J, Sterky F, Erlandsson R, Björkbacka H, Birve SJ, Karlsson J, Gardeström P, Gustafsson P, Lundeberg J, et al (2003) Gene expression in autumn leaves. Plant Physiol 131: 430–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binyamin L, Falah M, Portnoy V, Soudry E, Gepstein S (2001) The early light-induced protein is also produced during leaf senescence of Nicotiana tabacum. Planta 212: 591–597 [DOI] [PubMed] [Google Scholar]
- Butler WL, Kitajima M (1975) Energy transfer between photosystem II and photosystem I in chloroplasts. Biochim Biophys Acta 396: 72–85 [DOI] [PubMed] [Google Scholar]
- Caffarri S, Croce R, Cattivelli L, Bassi R (2004) A look within LHCII: differential analysis of the Lhcb1-3 complexes building the major trimeric antenna complex of higher plant photosynthesis. Biochemistry 43: 9467–9476 [DOI] [PubMed] [Google Scholar]
- Camm EL, Green BR (1980) Fractionation of thylakoid membranes with the nonionic detergent octyl β-d-glucopyranoside. Plant Physiol 66: 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consoli E, Croce R, Dunlap DD, Finzi L (2005) Diffusion of light-harvesting complex II in the thylakoid membranes. EMBO Rep 6: 782–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I, Critchley C, Woodall GS, Stewart GR (1998) Photoinhibition in differently coloured juvenile leaves of Syzygium species. J Exp Bot 49: 1437–1445 [Google Scholar]
- Drepper F, Carlberg I, Andersson B, Haehnel W (1993) Lateral diffusion of an integral membrane protein: Monte Carlo analysis of the migration of phosphorylated light-harvesting complex II in the thylakoid membrane. Biochemistry 32: 11915–11922 [DOI] [PubMed] [Google Scholar]
- Dreyfuss BW, Thornber JP (1994) Assembly of the light-harvesting complexes (LHCs) of photosystem II. Plant Physiol 106: 829–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi S, Andersson B, Barber J (1999) Isolation of a highly active PSII-LHCII supercomplex from thylakoid membranes by a direct method. FEBS Lett 446: 23–26 [DOI] [PubMed] [Google Scholar]
- Feild SF, Lee DW, Holbrook M (2001) Why leaves turn red in autumn: the role of anthocyanins in senescing leaves of red-osier dogwood. Plant Physiol 127: 566–574 [PMC free article] [PubMed] [Google Scholar]
- Garab G, Cseh Z, Kovacs L, Rajagopal S, Varkonyi Z, Wentworth M, Mustardy L, Der A, Ruban AV, Papp E, et al (2002) Light-induced trimer to monomer transition in the main light-harvesting antenna complex of plants: thermo-optic mechanism. Biochemistry 41: 15121–15129 [DOI] [PubMed] [Google Scholar]
- Gould KS, Kuhn DN, Lee DW, Oberbauer SF (1995) Why leaves are sometimes red. Nature 378: 241–242 [Google Scholar]
- Gould KS, Markham KR, Smith RH, Goris JJ (2000) Functional role of anthocyanins in the leaves of Quintinia serrata A. Cunn. J Exp Bot 51: 1107–1115 [DOI] [PubMed] [Google Scholar]
- Green BR, Durnford DG (1996) The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 47: 685–714 [DOI] [PubMed] [Google Scholar]
- Haldrup A, Jensen PE, Lunde C, Scheller HV (2001) Balance of power: a view of the mechanism of photosynthetic state transitions. Trends Plant Sci 6: 301–305 [DOI] [PubMed] [Google Scholar]
- Harari-Steinberg O, Ohad I, Chamovitz DA (2001) Dissection of the light signal transduction pathways regulating the two early light-induced protein genes in Arabidopsis. Plant Physiol 127: 986–997 [PMC free article] [PubMed] [Google Scholar]
- He Q, Dolganov N, Björkman O, Grossman AR (2001) The high light-inducible polypeptides in Synechocystis PCC6803: expression and function in high light. J Biol Chem 276: 306–314 [DOI] [PubMed] [Google Scholar]
- Heddad M, Adamska I (2000) Light stress-regulated two-helix proteins in Arabidopsis thaliana related to the chlorophyll a/b-binding gene family. Proc Natl Acad Sci USA 97: 3741–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobe S, Förster R, Klingler J, Paulsen H (1995) N-proximal sequence motif in light-harvesting chlorophyll a/b binding protein is essential for the trimerization of light-harvesting chlorophyll a/b complex. Biochemistry 34: 10224–10228 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S (1999) Chlorophyll breakdown in higher plants and algae. Cell Mol Life Sci 56: 330–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowski G, Kacprzak K, Jansson S (2001) Identification of Lhcb1/Lhcb2/Lhcb3 heterotrimers of the main light-harvesting chlorophyll a/b-protein complex of photosystem II (LHCII). Biochim Biophys Acta 1504: 340–345 [DOI] [PubMed] [Google Scholar]
- Jansson S (1999) A guide to the identification of the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci 4: 236–240 [DOI] [PubMed] [Google Scholar]
- Jansson S, Andersson J, Kim SJ, Jackowski G (2000) An Arabidopsis thaliana protein homologous to cyanobacterial high-light-inducible protein. Plant Mol Biol 42: 345–351 [DOI] [PubMed] [Google Scholar]
- Jin ES, Polle JWE, Melis A (2001) Involvement of zeaxanthin and of the Cbr protein in the repair of photosystem II from photoinhibition in the green alga Dunaliella salina. Biochim Biophys Acta 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Jin ES, Yokthongwattana K, Polle JEW, Melis A (2003) Role of the reversible xanthophyll cycle in the photosystem II damage and repair cycle in Dunaliella salina. Plant Physiol 132: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouril R, Zygadlo A, Arteni AA, der Wit CD, Dekker JP, Jensen PE, Scheller HV, Boekema EJ (2005) Structural characterization of a complex of photosystem I and light-harvesting complex II of Arabidopsis thaliana. Biochemistry 44: 10935–10940 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Larsson UK, Anderson JM, Andersson B (1987) Variations in the relative content of the peripheral and inner light-harvesting chlorophyll a/b-protein complex (LHCII) subpopulations during thylakoid light adaptation and development. Biochim Biophys Acta 894: 69–75 [Google Scholar]
- Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Lindahl M, Funk C, Webster J, Bingsmark S, Adamska I, Andersson B (1997) Expression of Elips and PSII-S protein in spinach during acclimative reduction of the photosystem II antenna in response to increased light intensities. Photosynth Res 54: 227–236 [Google Scholar]
- Lindahl M, Yang DH, Andersson B (1995) Regulatory proteolysis of the major light-harvesting chlorophyll a/b-protein of photosystem II by a light-induced membrane-associated enzymic system. Eur J Biochem 231: 503–509 [DOI] [PubMed] [Google Scholar]
- Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An X, Chang W (2004) Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428: 287–292 [DOI] [PubMed] [Google Scholar]
- Lohman K, Gan S, John M, Amasino RM (1994) Molecular analysis of natural senescence in Arabidopsis thaliana. Physiol Plant 92: 322–328 [Google Scholar]
- Martínez-Hernández A, López-Ochoa L, Argüello-Astorga G, Herrera-Estrella L (2002) Functional property and regulatory complexity of a minimal RBCS light-responsive unit activated by phytochrome, cryptochrome, and plastid signal. Plant Physiol 128: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile P (2001) Senescence and cell death in plant development: chloroplast senescence and its regulation. In EM Aro, B Andersson, eds, Advances in Photosynthesis and Respiration-Regulation of Photosynthesis, Vol 11. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 277–296
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence: a practical guide. J Exp Bot 51: 659–668 [DOI] [PubMed] [Google Scholar]
- Merzlyak MN, Chivkunova OB (2000) Light stress-induced pigment changes and evidence for anthocyanin photoprotection in apples. J Photochem Photobiol B 55: 155–163 [DOI] [PubMed] [Google Scholar]
- Montané MH, Kloppstech K (2000) The family of light-harvesting-related proteins (LHCs, ELIPs, HLIPs): was the harvesting of light their primary function? Gene 258: 1–8 [DOI] [PubMed] [Google Scholar]
- Montané MH, Petzold B, Kloppstech K (1999) Formation of early light-inducible complexes and status of xanthophyll levels under high light and cold stress in barley (Hordeum vulgare L.). Planta 208: 1287–1290 [Google Scholar]
- Murray JR, Hackett WP (1991) Dihydroflavonol reductase activity in relation to differential anthocyanin accumulation in juvenile and mature phase Hedera helix. Plant Physiol 97: 343–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh YS, Amasino RM (1999) Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol Biol 41: 181–194 [DOI] [PubMed] [Google Scholar]
- Norén H, Svensson P, Andersson B (2004) A convenient and versatile hydroponic cultivation system for Arabidopsis thaliana. Physiol Plant 121: 343–348 [Google Scholar]
- Norén H, Svensson P, Stegmark R, Funk C, Adamska I, Andersson B (2003) Expression of the early light-induced protein but not the PsbS protein is influenced by low temperature and depends on the developmental stage of the plant in field-grown pea cultivars. Plant Cell Environ 26: 245–253 [Google Scholar]
- Peter GF, Thornber JP (1991) Biochemical composition and organization of higher plant photosystem II light-harvesting pigment-proteins. J Biol Chem 266: 16745–16754 [PubMed] [Google Scholar]
- Pötter E, Kloppstech K (1993) Effect of light stress on the expression of early light-inducible proteins in barley. Eur J Biochem 214: 779–786 [DOI] [PubMed] [Google Scholar]
- Sävenstrand H, Olofsson M, Samuelsson M, Strid Å (2004) Induction of early light-inducible protein gene expression in Pisum sativum after exposure to low levels of UV-B irradiation and other environmental stresses. Plant Cell Rep 22: 532–536 [DOI] [PubMed] [Google Scholar]
- Sherwin HW, Farrant JM (1998) Protection mechanisms against excess light in the resurrection plants Craterostigma wilmsii and Xerophyta viscosa. Plant Growth Regul 24: 203–210 [Google Scholar]
- Standfuss J, Kühlbrandt W (2004) The three isoforms of the light-harvesting complex II: spectroscopic features, trimer formation, and functional roles. J Biol Chem 279: 36884–36891 [DOI] [PubMed] [Google Scholar]
- Tidholm E, Lindström V, Tisser C, Robinson C, Schröder WP, Funk C (2002) Novel approach reveals localization and assembly pathway of the PsbS and PsbW proteins into photosystem II dimer. FEBS Lett 513: 217–222 [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink M, Ter H, Alumot N, Gaathon A, Niyogi K, Herrmann RG, Andersson B, Ohad I (2004) Light-modulated exposure of the light-harvesting complex II (LHCII) to protein kinase(s) and state transition in Chlamydomonas reinhardtii xanthophyll mutants. Biochemistry 43: 7824–7833 [DOI] [PubMed] [Google Scholar]
- Wollman FA (2001) State transitions reveal the dynamics and flexibility of the photosynthetic apparatus. EMBO J 20: 3623–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H (1997) A function of color. Trends Plant Sci 2: 7–8 [Google Scholar]
- Yang DH, Paulsen H, Andersson B (2000) The N-terminal domain of the light-harvesting chlorophyll a/b-binding protein complex (LHCII) is essential for its acclimative proteolysis. FEBS Lett 466: 385–388 [DOI] [PubMed] [Google Scholar]