Abstract
A prototype 13-bp TATA-box sequence, TCACTATATATAG, was mutated at each nucleotide position and examined for its function in the core promoter. Specific nucleotides in the first TATA, the second TATA, as well as the flanking sequences influenced promoter function in transient transformation of tobacco (Nicotiana tabacum var Petit Havana) leaves. The effect of a given mutation on reporter gene expression in light versus dark was variable and sometimes contrasting. Some mutations, like T7 or A8→C or G, completely inactivated the expression of the minimal promoter in light but not in dark. In general, the sequence requirement for dark expression was less stringent than that for light expression. The selective effect of TATA-box mutations on light versus dark expression was exerted on core promoter function in the chromatin-integrated state also. Even in the presence of an upstream light response activator element, TATA-box mutations influenced modulation of the promoter by light. An A at the eighth position was specifically involved in the red light response of the promoter. Selectivity in gene expression was associated with a high level of transcript initiation from a site that was not active in the dark. Nuclear proteins from dark- and light-grown seedlings showed that the sequence variation within the TATA-box governs the formation of alternative transcriptional complexes. The experiments give direct evidence for the role of a core TATA-box sequence in determining the level as well as selectivity of gene expression in plants.
The minimal or core promoter, by definition, is the sequence located between the −35 to +35 region with respect to transcription start site (Smale, 2001). Eukaryotic promoters of protein-coding genes have one or more of three conserved sequences in this region (i.e. the TATA-box, initiator region, and downstream promoter element). Computational analysis of the core promoter of plants (Shahmuradov et al., 2003) suggests that core promoters are quite heterogeneous in composition for different conserved cis-regulatory elements. The TATA-box, found commonly in eukaryotic promoters, is typically a T/A-rich sequence, located about 25 to 30 bp upstream of the transcription start site. Breathnach and Chambon (1981) analyzed a number of eukaryotic genes, most of those from invertebrates and vertebrates, and identified TATAAA as the consensus TATA-box. Joshi (1987) did computational analysis of 79 plant promoters and suggested TCACTATATATAG as the consensus sequence in the TATA region. Our analysis (Sawant et al., 1999) attributed this sequence preferentially to highly expressed plant genes rather than plant genes in general. By examining the effect of mutations on in vitro transcription, a duplicated TATA sequence in the TATA-box was reported to enhance transcription (Mukumoto et al., 1993). Mutational studies (Zhu et al., 1995) have also established the importance of the TATA-box in the correct selection of a transcription initiation site.
Although the minimal promoter is generally believed to determine the rate of transcription and the point of its initiation (for review, see Roeder, 1996), there is some evidence suggesting its role in promoter selectivity also. Two different TATA-boxes in the human globin gene determine the regulation of γ-globin gene expression in embryogenesis and adult stage (Duan et al., 2002). Human myoglobin enhancer activates TATAAAA and not TATTTAT, although the latter sequence functions in the SV40 early promoter (Wefald et al., 1990). Heterogeneous composition of TFIID complexes (Martinez, 2002) associated with different promoters also suggests that sequence variations within the core promoter may govern alternative transcription complexes needed in different physiological settings. The diversity observed in the sequence of minimal promoters can also be interpreted as related to combinatorial regulation by activators bound to distal sites because such transcription factors often function through contacts with factors associated with specific TATA-box sequences (Butler and Kadonaga, 2001). However, the effect of specific mutations in the core TATA-box on the in vivo selectivity in eukaryotic gene expression has not been examined in detail.
Mukumoto et al. (1993) and Yamaguchi et al. (1998) carried out mutational analysis and examined the effect of changes at each position in a consensus plant TATA element on in vitro transcription, using the HeLa- and tobacco (Nicotiana tabacum)-based transcription systems, respectively. This article presents results on the effect of mutations at each position in a 13-nucleotide-long prototype TATA-box on promoter function in leaf tissue. The effect of various mutations in light-regulated expression was examined in tobacco leaves after transient, as well as stable, transformation using β-glucuronidase (gusA) as the reporter gene. The results suggest that the sequence architecture in the TATA region may be important not only in modulating the level of gene expression, but also in determining selectivity of promoter function in response to the absence or presence and quality of light.
RESULTS
Several Single Mutations in the TATA-Box Region Are Tolerated in in Vivo Gene Expression
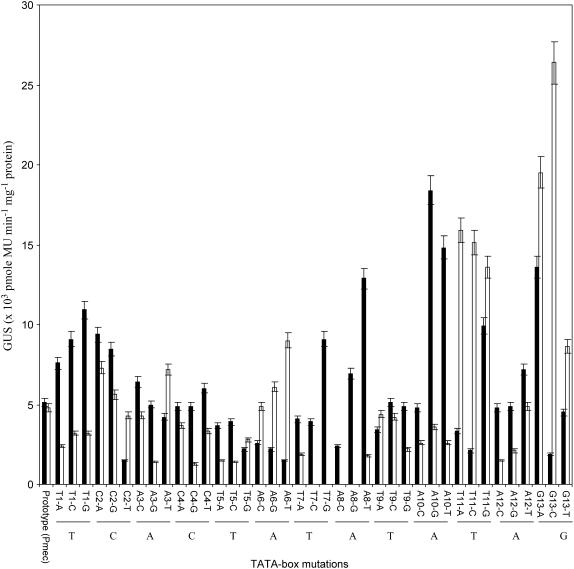
The effect of single mutations in the TATA-box on transient gene expression from reporter gene constructs was observed by determining the GUS activity after incubating the leaf discs for 48 h in continuous light. The GUS activity expressed by the prototype TATA-box, TCACTATATATAG, was compared with the activity of the mutated derivatives following incubation of the excised leaves in light. As seen in Figure 1, although several mutations had no significant effect on the activity of the basal promoter, a majority of them resulted in a decrease or an increase in promoter expression as compared to the prototype promoter that contained the parent TATA sequence. For example, a 5-fold increase in light was obtained when G at the thirteenth position was mutated to C (G13→C). In some cases, the results are in contradiction to those published earlier on plant promoter mutants in HeLa-based (Mukumoto et al., 1993) and tobacco-based (Yamaguchi et al., 1998) in vitro transcription systems. A decline in transcription was reported in the case of most of the mutations in those studies. Our experiments on transient transformation of tobacco leaves show that, in contrast to in vitro transcription, mutations in the core TATA sequence are more tolerated in vivo. In the case of mutations like T9→A, no significant effect was observed by us, which is in agreement with that reported in an in vitro tobacco-based transcription system (Yamaguchi et al., 1998). Several mutations, especially those at the fifth, seventh, eighth, and tenth positions, and others like A3→G, C4→G, A12→C, etc., decreased transcription substantially. However, others, especially at the eleventh and thirteenth positions, had a substantial increase in activity in light. The effect of mutations on gene expression suggested the importance of both TATATATA as well as the flanking sequences (i.e. the TCAC and G) located, respectively, before and after it, in determining the level of in vivo transcription.
Figure 1.
Light-related effect of the TATA-box mutations on promoter function. Each bar represents reporter gene (gusA) expression in the absence (black bars) and presence (white bars) of light. The x axis identifies the mutations for which GUS activities are shown. Each reading is an average of 18 to 24 leaf disc bombardments over three to four independent experiments.
Mutations that were not tolerated at all in gene expression in leaves exposed to continuous light were the substitutions to C and G at the seventh and eighth positions (i.e. the latter TA of the first TATA sequence). The promoters containing a TATA-box with these mutations failed to activate gusA expression in leaves exposed to continuous light (Fig. 1). The results suggest that certain positions in the prototype TATA-box have very little or no sequence preference for in vivo promoter function, although others could be critical to the level of gene expression.
Nucleotide Sequence in the TATA-Box Determines Promoter Selectivity in Gene Expression
We examined whether specific mutations in the TATA-box sequence influenced promoter strength in general or whether the mutated promoters behaved differently, depending upon other physiological cues. In other words, is the sequence requirement of the TATA-box the same for all promoters or only if the TATA-box sequence reflects specific sequence signatures related to the needs of gene expression specificity? To address this question, the expression of gusA, driven by each of the above-mentioned mutated promoters, was compared between the leaf discs incubated in light or dark. The dark versus light data are included in Figure 1. In general, the expression in dark was affected less substantially and varied from 41% to 214% of the expression driven by the prototype promoter. The results suggest that a given TATA-box cannot be considered as the optimal signal for transcription initiation. Depending upon the physiological cues, fine differences in the TATA-box sequence may modulate promoter expression differently.
The effect of some of the mutations in the first TATA (i.e. T7→C, T7→G, A8→C, and A8→G) were most noticeable in light-activated promoter expression. Whereas these mutations made the promoter completely nonfunctional in light, they continued to express at a high level in dark. In fact, dark activity was not reduced to zero in any of the 40 promoters. In some cases, as in the case of C2→T and A6→T, dark activity declined by about 3-fold as compared to the prototype TATA-box sequence. In other cases, such as T1→G, A8→T, A10→G, A10→T, and G13→A, expression from the core promoter in dark increased 2- to 3.6-fold. The mutations that stimulated dark transcription did not necessarily give a similar effect in light. For example, among the aforementioned five mutations that increased promoter activity in dark, only G13→A gave a comparable increase, whereas the others had little effect or resulted in a decline in light-modulated promoter function. Other mutations, like T11→A, T11→C, and G13→C, gave a 3- to 5.5-fold increase in light-dependent activity but not in dark.
Multiple Mutations in the TATA-Box Cause General Loss of Promoter Function
To examine the extent to which sequence distortions could be tolerated by the transcription machinery in dark, double mutations were constructed in a few selected cases. Table I shows that dark transcription, although less sensitive to single mutations, fell off completely in some of the double mutations. The A6→G mutation in the first TATA reduced dark-mediated expression by about 2-fold, whereas it gave a small increase in light-mediated promoter function. The second accompanying mutation at the third, seventh, or eighth position resulted in near-complete or complete failure of promoter function in both light and dark. The results suggest that expression from the promoter used in this study is dependent on the TATA-box for expression in both light and dark. The results also point to the role of the previously unreported flanking motif TCAC in promoter function and its selectivity in combination with the TATA-box sequence. As noted in Table I, the A3→G mutation in TCAC by itself had no effect on dark expression, although it inhibited light expression. However, when accompanied by the A6→G mutation, the A3→G resulted in 10-fold lower and no expression, respectively, in dark and light.
Table I.
Synergistic effect of the nucleotides at different positions in the TATA-box on the specificity of promoter expression
GUS values are averages ± sd of data from three independent experiments comprising six to nine replicates each. The variation in GFP values expressed from the 35S promoter in the internal control were 34.05 ± 4.56 × 103 RFU mg protein−1.
| Promoter Mutation | GUS Expression
|
||
|---|---|---|---|
| Dark | Light | Ratio (Dark/Light) | |
| 103 pmol min−1 mg protein−1 | |||
| Pmec | 5.40 ± 1.10 | 5.00 ± 1.10 | 1.08 ± 0.22 |
| A6→G | 2.40 ± 0.45 | 6.49 ± 1.29 | 0.36 ± 0.06 |
| A3→G | 4.87 ± 1.11 | 1.52 ± 0.33 | 3.20 ± 0.62 |
| T7→G | 9.15 ± 1.92 | 0 | ∞ |
| A8→C | 2.60 ± 0.50 | 0 | ∞ |
| A6→G; A3→G | 0.48 ± 0.09 | 0 | ∞ |
| A6→G; T7 →G | 0 | 0 | 0 |
| A6→G; A8→C | 0 | 0 | 0 |
The Seventh and Eighth Positions in the TATA-Box Are Critical to Transcription in Light
The actual level of the transcript was measured in a few representative cases by real-time PCR. The T7→C, T7→G, A8→C, and A8→G mutations were examined for the level of gusA transcript in light and dark vis-à-vis that for ubiquitin as an internal control. As seen in Table II, in light-incubated leaves, nearly no gusA transcript was formed in the case of promoters with any of the four single mutations. On the other hand, none of these mutations led to nonexpression of the gusA transcript when the leaf was incubated in dark. The results on transcript quantitation in the above four representative mutations therefore confirm that the relative levels of glucuronidase activity measured after microprojectile bombardment (Fig. 1) are an authentic reflection of the actual transcription.
Table II.
Relative levels of gusA transcript in selected promoter mutations
Ratios of the transcripts are the average of three independent experiments, where gusA expression was normalized by internal ubiquitin gene expression. gfp gene expression was taken as a control for the bombardments. The values in the column on relative enzyme level have been derived from the data in Figure 1.
| Serial No. | Promoter Mutation | Incubation Condition | Relative Transcript Level | Relative Enzyme Activity |
|---|---|---|---|---|
| Expression in Mutations at T7 in the TATA-Box | ||||
| 1 | Pmec | Light | 1.00 | 1.00 |
| Dark | 1.04 ± 0.20 | 1.06 | ||
| 2 | T7→C | Light | 0.01 ± 0.00 | ∞ |
| Dark | 0.79 ± 0.16 | 0.81 | ||
| 3 | T7→G | Light | 0.02 ± 0.00 | ∞ |
| Dark | 2.04 ± 0.40 | 1.89 | ||
| Expression in Mutations at A8 in the TATA-Box | ||||
| 1 | Pmec | Light | 1.00 | 1.00 |
| Dark | 0.91 ± 0.18 | 1.06 | ||
| 2 | A8→C | Light | 0.01 ± 0.00 | ∞ |
| Dark | 0.42 ± 0.08 | 0.5 | ||
| 3 | A8→G | Light | 0.02 ± 0.00 | ∞ |
| Dark | 1.68 ± 0.32 | 1.44 | ||
Modulation of Gene Expression in Light by a TATA-Box Sequence in the Chromatin-Integrated State
It was important to establish whether the TATA-box sequence regulated gene expression selectively under light versus dark conditions even when the promoter was integrated in the chromatin. For this purpose, stable transgenic lines of tobacco were developed using minimal promoter-reporter constructs representing the selected mutations.
Pmec-Mediated Transcription in Light as Well as Dark Is Primarily TATA Dependent
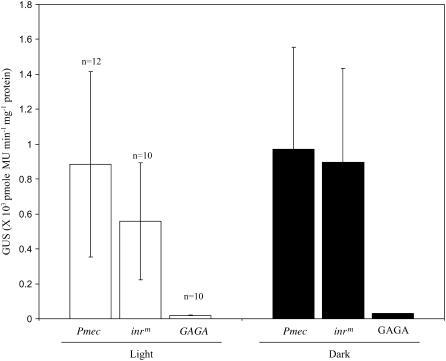
The minimal promoter sequence Pmec used in this study is a typical core promoter having both the consensus TATA and a putative initiator sequence PyTCANTPyPy (Nakamura et al., 2002). To establish the contribution of these elements in transcription regulation of Pmec, we mutated both the TATA sequences in TCACTATATATAG to GAGA in one case, and the initiator-like motif TTGTCATTTCTC to AAGACAAAACAC (inrm) in another case. The constructs Pmec-GAGA and Pmec-inrm were then transformed in tobacco to obtain several independent transgenic lines. gusA expression was monitored in transgenic seedlings grown either continuously in dark or in a 16-h-light/8-h-dark cycle for a period of 11 d. As compared to the prototype promoter Pmec, transgenic lines with Pmec-GAGA showed almost no activity either in light- or dark-grown seedlings (Fig. 2). In contrast to the GAGA mutation, transgenic seedlings expressing Pmec-inrm showed about a 36% decline in gusA activity in light-grown seedlings. Dark-grown seedlings with inrm showed promoter activity similar to that of Pmec. The results indicate that Pmec used in this study is primarily a TATA-dependent promoter, although the inr-like element also contributes to light-mediated transcription.
Figure 2.
Role of TATA-box and inr-like element in light-regulated expression of Pmec GUS activity in 11-d-old light-grown (white bars) or dark-grown (black bars) transgenic seedlings containing minimal promoter Pmec with the prototype sequence and its derivatives mutated in the inr (inrm) and TATA-box (GAGA) regions. The values above the bars indicate the sd of three independent experiments. The numbers of independent transgenic lines (n) used in the analysis are indicated.
Light-Specific Influence of the TATA-Box Sequence on Transcription Is Maintained in the Chromatin-Integrated State
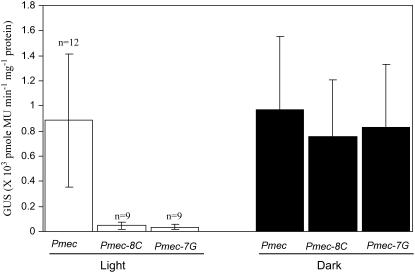
Figure 3 presents data on the expression of gusA in the leaves of several independent transgenic lines transformed with constructs carrying the prototype promoter Pmec or promoter mutations (i.e. T7→G or A8→C). Transgenic seedlings were grown either in continuous dark or a 16-h light cycle before estimation of GUS activity. Whereas Pmec showed no difference in the expression of gusA in light and dark, the mutants gave strikingly different results. In a majority of transgenic plants, the T7→G as well as T8→C mutations resulted in complete inhibition of promoter function in light. On the other hand, dark expression of the promoter was not affected significantly by any of the two mutations (Fig. 3). The results show that light-specific regulation of transcription by the TATA-box sequence was not overridden by the neighborhood genomic sequences in any of the 12 integration events. Thus, selective regulation of gene expression by TATA-box sequence per se exerts itself in spite of the chromatin structure and nonspecific sequences in the neighborhood in the integrated state. The above results suggested that certain sequences should preferentially be present in the TATA-box of promoters in genes that express in light.
Figure 3.
Role of the seventh and eighth positions in the TATA-box in light-mediated expression of Pmec. GUS activities in 11-d-old light-grown (white bars) or dark-grown (black bars) transgenic seedlings containing the minimal promoter with prototype TATA (Pmec), mutation A8C (Pmec-8C), or T7G (Pmec-7G). The values above the bars indicate the sd of three independent experiments.
TATA-Box-Dependent Modulation of Transcription by Light Is Not Overridden by the Light-Responsive Activator Element
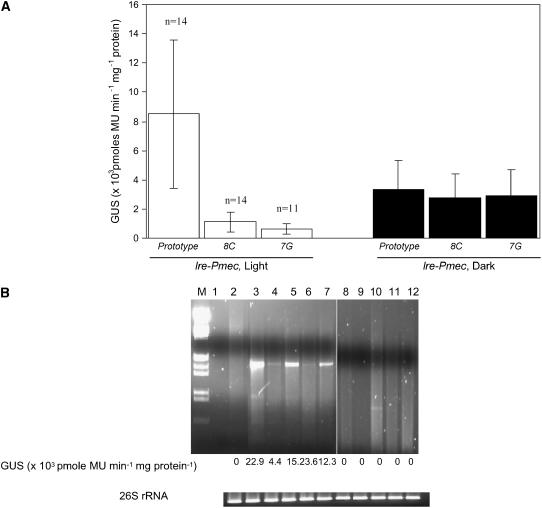
To evaluate the contribution of the TATA-box sequence in the presence of the light-specific activator motif, we placed the 52-bp light-responsive activator element (lre) fragment of the small subunit of Rubisco (rbcS) 8B gene of tobacco (Martinez-Hernandez et al., 2002) upstream of Pmec and some selected mutants. The 52-bp lre has been reported to confer light-specific activation of transcription to even heterologous promoters (Martinez-Hernandez et al., 2002). As expected, the lre fragment conferred typical light-dependent activation on the Pmec promoter in transgenic lines. The lre increased expression from Pmec severalfold in both light and dark, the increase being greater in light (compare Figs. 3 and 4A). lre-Pmec transgenic lines showed 2- to 3-fold higher expression in light as compared to the seedlings grown in dark (Fig. 4A).
Figure 4.
Role of the seventh and eighth positions in the TATA-box in light-dependent, lre-activated transcription from Pmec. A, GUS activities in 11-d-old light-grown (white bars) or dark-grown (black bars) seedlings containing lre-Pmec with the prototype TATA-box (lre-Pmec) and its variants with mutations A8C and T7G. The values above the bars indicate the sd from the means of three independent experiments. B, Semiquantitative RT-PCR carried out on five light-grown transgenic lines of lre-Pmec (lanes 3–7) and lre-Pmec-8C (lanes 8–12). Lanes 1 and 2 represent nontemplate control and semiquantitative RT-PCR of nontransformed tobacco, respectively. The values below the lanes indicate GUS activities in respective transgenic lines. The 26S rRNA levels detected by semiquantitative PCR are shown for comparison.
The relationship between the activation by lre and the sequence of the TATA-box was further analyzed by placing the lre fragment upstream of Pmec with the 7G and 8C mutations in transgenic tobacco lines. Expression studies were carried out on dark etiolated seedlings and those grown for 11 d in a 16-h light cycle. As depicted in Figure 4A, both mutations resulted in a 7- to 14-fold decrease in the expression of gusA specifically in light-grown seedlings. However, expression of gusA was unaffected in dark-grown seedlings regardless of the two mutations. Semiquantitative reverse transcription (RT)-PCR was performed on several independent transgenic lines carrying lre-Pmec with the 7G or 8C mutations to establish a correlation between reporter gene expression and the level of transcripts. The results in Figure 4B show that the lre-Pmec (lanes 2–7) results in formation of the gusA transcript, whose levels are in proportion to reporter enzyme activity in the seedlings. In contrast, the mutant lre lines showed no transcript in semiquantitative PCR (Fig. 4B, lanes 8–12) in agreement with the very poor GUS activity. Thus, the seventh and eighth positions in the canonical TATA-box sequence influence transcription in light even in the presence of lre. In contrast, these positions do not significantly affect transcription from Pmec in the dark.
Light-Related Transcription Regulation by TATA-Box Sequence Is Mainly Phytochrome Specific
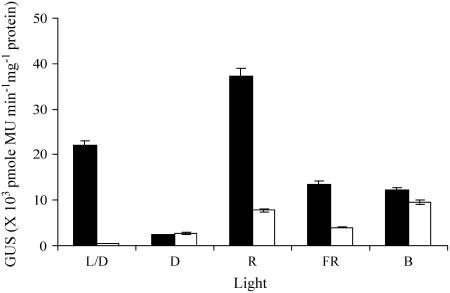
Specificity of TATA-box sequence-dependent transcription regulation was examined further in monochromatic light. The results for one transgenic line of lre-Pmec and of lre-Pmec 8C are shown in Figure 5. A typically contrasting expression was noticed in light and dark (Fig. 5, L/D). Seedlings were germinated either in continuous dark or in a 16-h light regime. For the monochromatic study, plants were germinated in dark for 9 d and exposed to specific monochromatic light, namely, red, far-red, or blue light for 2 d before determining GUS activity. As expected (Martinez-Hernandez et al., 2002), lre responds to various monochromatic light by showing activation (Fig. 5). As compared to the mutant lre-Pmec 8C transgenic line, the lre-Pmec transgenic line showed 55-, 4.7-, and 3.3-fold higher expression in white, red, and far-red light, respectively. However, expression of both lines was comparable in dark (0.87-fold) and blue (1.27-fold) light. Activation by red light was most inhibited in the presence of the 8C mutation in Pmec. The inhibitory effect was not seen in blue light. Thus, TATA-box sequence-dependent transcription regulation is more specific to red/far-red light and thus indicative of phytochrome-specific transcription regulation.
Figure 5.
Effect of quality of light in TATA-box-dependent gene expression from Pmec GUS activities in the transgenic line of lre-Pmec (black bars) and lre-8C (white bars) constructs grown in dark for 9 d and then exposed for 2 d of different light conditions (i.e. 16 h white light/8 h dark [L/D], continuous dark [D], continuous red [R], continuous far-red [FR], and continuous blue [B] light). Intensities of the WL, R, FR, and B irradiation were 80, 10, 0.86, and 15 μmol m−2 s−1, respectively.
Light-Dependent Nucleoprotein Complex Formation on the Core Promoter Is Inhibited by Specific TATA-Box Mutations
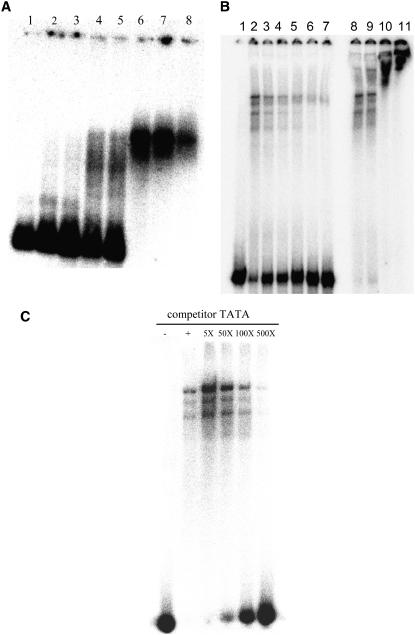
To study the mechanism by which mutations at the seventh and eighth positions may inhibit transcription in light (and not in dark), electrophoretic mobility shift assays were carried out using nuclear protein extracted from either light-grown (NEL) or dark-grown (NED) seedlings. At the optimal DNA to NEL ratio, the oligonucleotide containing the prototype TATA-box sequence made a discrete nucleoprotein complex (Fig. 6A). The discrete complex formation occurred as we increased the protein concentration by 5-fold more than the starting protein concentration (lanes 6–8). A further increase in NEL resulted in the disappearance of the specific complex because, under such conditions, larger nonspecific complexes that did not enter the agarose gel were formed. That the discrete complex seen was a functional transcriptional complex was established by various approaches. First, under incubation conditions, in vitro transcription gave a single major transcript (Supplemental Fig. 1). Second, the distinct promoter-protein complex disappeared on competition with molar excess of Pmec oligo (Fig. 6B), resulting in the reappearance of the free oligonucleotide. Titration with random nucleotides, poly(dG-dC), poly(dA-dC), or salmon sperm DNA did not result in such dissociation (data not shown). To establish that the complex formed was indeed TATA-box dependent, we completed the complex with the oligo containing only the canonical TATA-box sequence (multimer of TCACTATATATAG). Somewhat unexpectedly, the concentration of the complex increased with the increase in competitor TATA from 5- to 50-fold (Fig. 6C). However, when the competitor oligo was increased further, it completely abolished the complex at 500-fold molar excess (Fig. 6C). The results established that the complex formed on Pmec was a TATA-box-dependent complex. The addition of polyclonal anti-TATA-binding protein (TBP) antibodies to the incubation mixture resulted in a supershift (Fig. 6B, lanes 9–11) of the complex. Thus, the nucleoprotein complex seen on the agarose gel represented a transcriptionally relevant TATA-box/TBP-containing complex.
Figure 6.
Autoradiograph of 2% agarose (A) and 5% PAGE (B and C) showing nucleoprotein complex formation on 32P-labeled 120-bp Pmec oligo. A, In the presence of 0, 5, 10, 50, 100, 500, and 1,000 ng of NEL protein per 20 μL of incubation mixture (lanes 1–8). B, In the presence of 500 ng of NEL at 0-, 2-, 5-, 10-, 50-, and 100-fold molar excess of unlabeled 120-bp Pmec oligo (lanes 2–7). Lanes 8 to 11 show a supershift of the complex following addition of 0.5, 1.0, 2.0, and 5.0 μL of anti-TBP antibody. C, In the presence of 500 ng of NEL at 0-, 5-, 50-, 100-, and 500-fold molar excess (lanes 2–6) of unlabeled multimerized TATA-box sequence. Lane 1 shows the position of the free 32P-labeled 120-bp Pmec oligo.
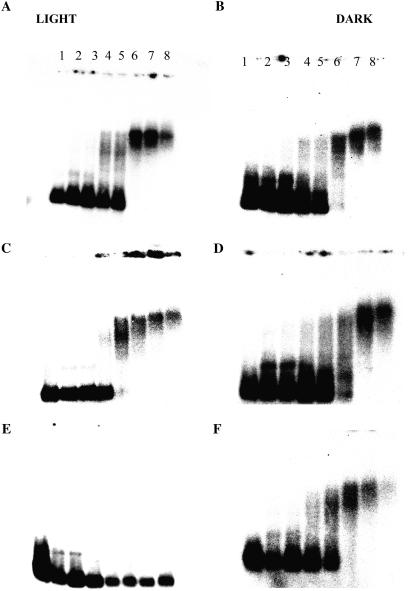
The results of titration of the NEL and NED with the promoter containing the prototype TATA-box are shown in Figure 7, A and B, respectively. In both cases, a discrete TBP-TATA complex was formed and maintained because the nuclear extract was increased by about 5-fold. However, contrasting results on the complex formation were obtained when promoter mutations were used. Nuclear extract from light-grown seedlings showed weak complex formation with the TATA-box oligonucleotide containing the T7→C mutation (Fig. 7C). A diffused complex was formed, which disappeared into nonspecific larger complexes that do not enter the agarose gel at higher concentrations of NEL. With the oligo containing the A8→G mutation, the NEL failed to form a specific complex (Fig. 7E). On the other hand, in the case of the dark-grown extract NED, a specific nucleoprotein complex was formed with the TATA-box containing either of the two mutations (Fig. 7, D and F). The results suggest that the nuclear preparations from the seedlings grown in the absence or presence of light have different sequence requirements for the formation of transcriptional complexes on the TATA-box.
Figure 7.
Light-related specificity of the TATA-box sequence in complex formation with nuclear extract. The formation of complexes using 0 to 1,000 ng of nuclear protein (lanes 1–8) prepared from light-grown (left) and dark-grown (right) seedlings is shown. Shown is the mobility shift in the promoter oligo containing the prototype TATA-box without T7C (A and B) and with T7C (C and D) and A8G (E and F) mutations.
Transcription Activation by Light Operates from a Novel Initiation Site
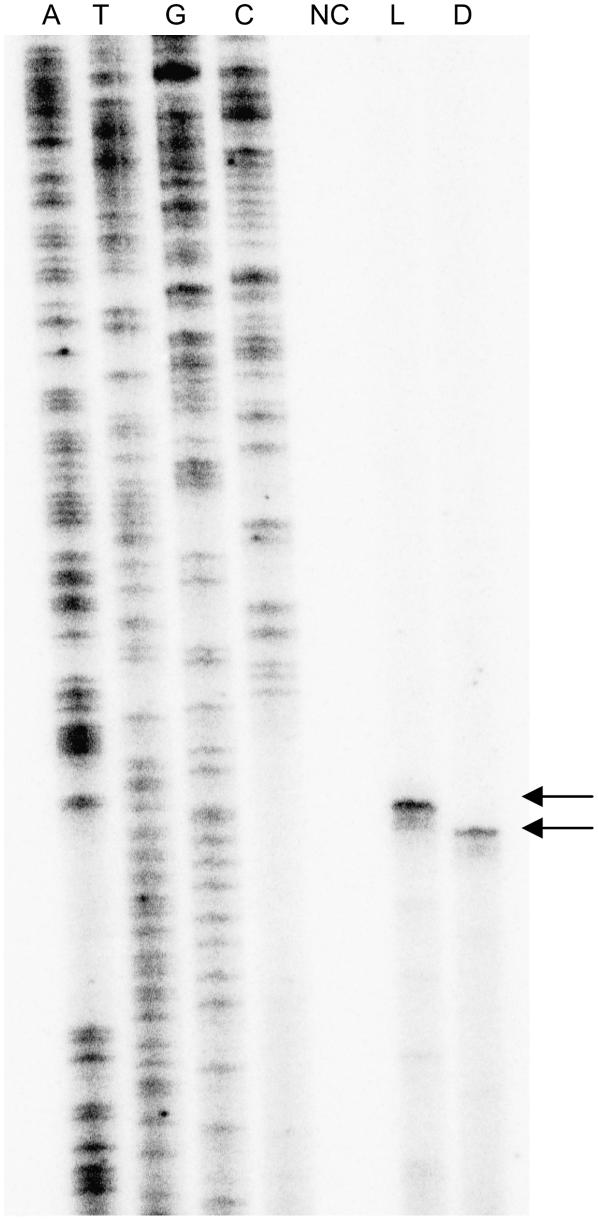
To probe further into mechanistic differences operating on the minimal promoter during transcription in light and dark, we mapped the transcription start site. We carried out RNA ligation-mediated (RLM)-RACE to map the transcription start site in several lre-Pmec transgenic lines grown in light or dark. A major transcription start site (Fig. 8, arrow) was obtained in light-grown seedlings. In the case of etiolated seedlings, we obtained another transcript two nucleotides downstream of transcription site (*) obtained in light-grown seedlings.
Figure 8.
Mapping of point of transcript initiation from lre-Pmec. Shown is primer extension analysis conducted with 32P-labeled PE1 primer using RLM-RACE. Lanes A, T, G, and C represent sequencing ladders with the primer. Lanes L and D represent primer extension of the transcripts formed in the lre-Pmec transgenic line grown in light and dark, respectively. Lane NC represents the control lane (i.e. nontransgenic plant).
To evaluate whether differences in transcription site in the case of light and dark conditions is a general mechanism operating on light-activated promoters, we mapped a transcription start site in the rbcS 8B gene of tobacco from light-grown and etiolated seedlings. In contrast to the results obtained on lre-Pmec, the rbcS 8B gene showed a single major transcript in light and a weak transcript at the same position in etiolated seedlings (data not shown). The results suggest that the mechanism that operates during light and dark might be context dependent and might selectively operate on some, but not all, light-regulated promoters.
DISCUSSION
Our study shows that several single mutations in the prototype TATA-box (inclusive of the immediately flanking sequences) are largely tolerated in vivo by leaf transcriptional machinery. Transcription in the dark was particularly more tolerant to several mutations that resulted in substantial to complete failure in reporter gene expression in light. This is in contrast to earlier studies based on in vitro transcription (Mukumoto et al., 1993; Yamaguchi et al., 1998), which suggested that the function of the TATA-box region was highly vulnerable to single mutations. Differences in the results may be because certain factors involved in the stabilization of TATA-TBP interaction in vivo may be absent in in vitro transcription systems. Intracellularly, TBP is maintained in a dynamic state of equilibrium with other protein complexes, like TFIID (Coleman et al., 1999), where it is associated with a variety of TBP-associating factors (TAFs). Some of the TAFs stabilize TATA-TBP interactions directly or indirectly in association with other general transcription factors (Wassarman and Sauer, 2001). Complexes like TFIID-TATA are rate limiting for transcription in vivo (Colgan and Manley, 1992) and are likely to be limiting or absent in nuclear extracts used for in vitro transcription. Hence, the results on in vitro and in vivo transcription could be different, as noted in this study.
The results establish that the prototype minimal promoter Pmec is a typical TATA-box-dependent promoter. None of the 10 transgenic lines with the TATA-box mutated to GAGA showed significant GUS activity. Thus, GUS activity expressed from the variants containing the TATA-box mutations reflects true behavior of the TATA-box in the promoters. The inr-like sequence also contributed to light-mediated expression of Pmec but failed to activate transcription significantly in the absence of a TATA-box. This is in agreement with Nakamura et al. (2002), who reported the importance of the inr region in light-mediated regulation of psaDb in tobacco.
High vulnerability of transcription in light-incubated leaf to mutations in the first TATA-box largely agrees with the above in vitro studies and with other mutational and TATA-TBP interaction studies in yeast (Saccharomyces cerevisiae; Steven et al., 2002). Light-dependent transcription from the core promoter appears to require TATA-box architecture different from that required by the transcriptional machinery that functions in the dark. Some of the substitutions (T11A, T11C, G13C) in the flanking positions showed light-specific stimulation of transcription. Others, like G13A, resulted in general improvement in promoter function. Footprinting studies using in vitro and in vivo systems in eukaryotes have shown that the points of contact of the TFIID complex extend well beyond the core TATA region (Nakajima et al., 1988; Purnell and Gilmour, 1993). Some of the TAFs have been reported to interact with the sequences flanking TATA and thus stabilize transcription. For example, TFIIB interacts with the TATA-box 5′-flanking sequences (Wolner and Gralla, 2001) and stabilizes the TBP-TFIIA complex on the TATA-box (Pan et al., 2000).
Our results suggest that the TATA-box flanking sequences are involved in in vivo stability of the pre-initiation complex (PIC) as well as specificity of gene expression. Double mutations, including the sequences flanking the TATA-box, reduced transcription synergistically. Hence, a larger context in the TATA-box region determines the performance of the core promoter. For example, as seen in Table I, TGTA in a sequence context, TCGCTGTA, is more deleterious to promoter function than that in a different context, like TCACTGTA. Earlier, TGTA has been reported to inactivate promoter function in Arabidopsis (Arabidopsis thaliana; Pan et al., 2000). In that study, the TATA-box of the cauliflower mosaic virus 35S promoter was mutated from CCTCTATA to CCTCTGTA. However, a mutated TATA-box binding protein, TBPm3, was reported to recognize the promoter containing the TGTA motif (Strubin and Struhl, 1992). Our results show that a change in the context of the TATA-box can also alter the specificity and functional interactions of the core promoter with components of the transcriptional complex. Thus, given a different context, the core promoter containing TGTA can also be functionally active.
Light-specific failure in expression of the minimal promoter Pmec with C and G at the seventh or eighth positions was neither affected by chromatin context nor the light-specific activator motif placed upstream. The results were completely reproducible in independent transgenic lines. Although GUS activity varied significantly in different transgenic lines due to the positional effect, the promoter-specific differential transcription in light versus dark conditions was noticed in all cases. A similar case occurred when we cloned the lre from the rbcS 8B promoter upstream of either the prototype Pmec or the mutants 7G and 8C. As expected, the lre rendered light activation to Pmec. The activation was abolished when the lre was cloned upstream of the TATA-box with 7G or 8C mutations. Our results suggest that the expression of Pmec in light and dark happens by different mechanisms operating on the minimal promoter.
The light-specific response of the TATA-box mutations seems to be phytochrome A (PhyA) and phytochrome B (PhyB) dependent because expression from the 8C mutation was substantially compromised in red and far-red light, but not in blue light. The results reflect that the TATA-box-specific expression obtained with Pmec is primarily PhyA and PhyB dependent and may involve synergistic action because inhibition was greater in white light (composed of both red and far-red components) as compared to the individual monochromatic light. Such cooperative inhibition due to PhyA and PhyB has been reported for light-dependent promoters like TUB1 and lhcb in the phyA phyB double-mutant background in Arabidopsis (Leu et al., 1995).
Our results suggested that certain sequences containing a G or a C at the third and fourth positions in the TATA-box of promoters with double TATA may preferentially be present in genes whose expression is suppressed in light. To test this hypothesis, we generated an Arabidopsis minimal promoter database containing 30,975 entries. This database was searched for the occurrence of TATA variants TAGATATA, TACATATA, TATGTATA, and TATCTATA within the first 50 nucleotides upstream of the reported transcription start site. The above variants in TATATATA were present in 166 Arabidopsis genes (Supplemental Sequence Data 1). However, only 44 of these 166 genes contain the variant TATA as the only identifiable TATA-box. In all other cases, more than one putative TATA-box was identified. In the case of none of these 44 genes, clearly interpretable light versus dark expression data are available in the literature. Thus, TATA variants of the kind noticed to make distinct transcriptional complexes in light versus dark in this study are present naturally in the Arabidopsis genome. However, unfortunately, sufficient expression data are currently unavailable to validate the general applicability of the conclusions arrived in our study.
More stringent sequence requirements of the TATA-box region for transcription in light than in dark may be due to the involvement of light-specific general transcription factors or their modifications that recognize specific sequences in the TATA-box region. Our results on promoter-nuclear protein complex formation suggest the presence of such factors. Evidence suggesting the presence of such a general transcription factor in Arabidopsis has recently been published (Bertrand et al., 2005), where the involvement of a TAF1 in light-induced gene expression has been established. Tissue-specific TAFs, TBP-related factors (TRFs), or TBP-like factors (TLFs) have previously been discussed in yeast, Drosophila, and humans. The molecular details of their selectivity for sequences in the TATA-box are not known. Such factors may interact directly with sequences in the TATA-box or do so indirectly through components of chromatin and complexes like TFIID (Martinez, 2002). Some of the known cell-specific TAFs reported from nonplant systems are hTAFII105, which expresses at a higher level in B cells (Dikstein et al., 1996), although its function is not clear (Freiman et al., 2002); TAFII135 in neuronal differentiation (Metsis et al., 2001); and the cell type- and stage-specific TRF2 in mouse spermatogenesis (Zhang et al., 2001). In the case of plant cells, Pan et al. (2000) reported that activation of promoters driven by a single activation element (and not complex promoters with multiple cis-motifs) requires functional interaction between the cognate transcription factor and the PIC. By mutagenesis, interaction between the C-terminal stirrup of TBP and TFIIB was reported as essential for the function of three different transcription factors. Because the stability of such complexes is dependent on the TATA-box sequence (Stewart and Stargell, 2001), the specificity of promoter expression will be expected to be influenced by the TATA-box sequence per se. Differential levels of activation by the transcription factor IE62 of Varicella zoster virus can be conferred by sequence variations within the TATA motif in mammalian cells (Perera, 2000). The differential level of transcriptional activation through different TATA-box motifs can therefore be accomplished by direct and indirect interactions of conformationally variable complexes of TBP, TBP-TFIIA/B, TBP-TAFs, TF-PIC, etc.
Our results establish the effect of a TATA-box-mediated sequence-specific modulation of gene expression in plants in response to light. A variety of factors, including cis-motifs in the neighborhood, long-distance positional effects, and chromatin structure can modify the function of the core promoter in the integrated state. The results on stable transgenic lines show that the TATA-box continued to exert its light-related sequence-specific influence despite the nonspecific positional effects that would normally be associated with the expression of transgenes inserted at different positions in the chromatin. Selectivity in light-mediated gene expression by the TATA-box sequence was substantially maintained in the presence of the light-regulated upstream activation element.
MATERIALS AND METHODS
Construction of Mutations in the TATA-Box Region
We have previously reported (Sawant et al., 1999) TCACTATATATAG as the most commonly present TATA-box sequence in highly expressed plant genes. The functional validity of this sequence has been established previously by demonstrating that an upstream region activates transcription dependent upon the core promoter containing this TATA-box sequence. It functioned at a high level in a number of plant species and in different tissues (Sawant et al., 2000). In this study, mutations at each of the 13 positions in the prototype TATA-box, TCACTATATATAG, were created by PCR using degenerate primers. A total of 13 degenerate primer sets were synthesized. A given primer set represented the three possible nucleotide substitutions at a specific position. The PCR products were cloned in a pUC19 (New England Biolabs) vector. A total of 10 random clones were picked for each primer and sequenced. Finally, a total of 39 mutations in the 13-bp-long prototype TATA-box sequence were selected. A few representative double mutations were also created by a second cycle of PCR mutagenesis, wherever described.
Construction of Cassettes and Transient Expression Studies
Each of the 40 variants of the TATA-box was placed in a 139-nucleotide-long minimal promoter sequence, Pmec, that contained a transcription initiation site and translation initiation context followed by gusA, as reported previously (Sawant et al., 2001). The nucleotide sequence of the gusA promoter cassette showing the prototype TATA-box and the transcription initiation site is 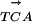 CTATATATAGGAAGTTCATTTCATTTGGAATGGACACGTGTTGTCATTTCTCA
CTATATATAGGAAGTTCATTTCATTTGGAATGGACACGTGTTGTCATTTCTCA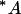 C
C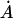 ATTACCAACAACAACAAACAACAAACAACATTATACAATTACTATTTACAATTACATC
ATTACCAACAACAACAAACAACAAACAACATTATACAATTACTATTTACAATTACATC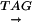 ATAAACAATGGCTTCCTCC-gusA.
ATAAACAATGGCTTCCTCC-gusA.
The prototype TATA-box and the initiator sequences (inr) are underlined. The transcription initiation sites, light-dependent (*) and dark-dependent (·), are indicated. The horizontal arrows indicate the 120-bp sequence used in mobility shift studies described later.
The 40 Pmec-gusA cassettes containing one mutation at a time in the prototype TATA-box sequence were examined for core promoter function in tobacco (Nicotiana tabacum var Petit Havana) leaves, following both transient and stable transformation. A 52-bp LRE, called CMA5 (Martinez-Hernandez et al., 2002), was synthesized and inserted immediately upstream of the minimal promoters Pmec, Pmec7G, and Pmec8C to obtain respective constructs under the control of lre.
The effect of promoter mutations was studied by delivering onto tobacco leaves different promoter cassettes coated on gold microparticles. A highly reproducible protocol, published previously for microprojectile bombardment, was used (Sawant et al., 2000). An internal control comprising green fluorescent protein (gfp) expressed from the cauliflower mosaic virus 35S promoter was included in some experiments to establish reproducibility of the delivery of gold particles in leaves.
The controls performed with the gusA cassette devoid of the first TATA-box motif in the TATA-box gave very low background glucuronidase activity under otherwise similar experimental conditions. Activity of the prototype minimal promoter (4.8 ± 0.9 × 103 pmol methylumbelliferone [MU] released min−1 mg−1 soluble leaf protein after subtracting the background) used in the study was about 3-fold higher than the background.
RNA Extraction and RT
Total RNA was isolated from the bombarded leaves by TRIzol LS reagents (Invitrogen Life Technologies). After DNase treatment, the integrity of RNA was tested by electrophoresis. Total RNA (1 μg) was used for first-strand cDNA synthesis. A 20-μL reaction mixture consisting of 2 μL 10× incubation buffer, 2.5 μm primer [poly(dT)], 500 μm dNTPs, 5.5 mm MgCl2, 0.4 units RNase inhibitor, and 1.25 units Multiscribe reverse transcriptase (TaqMan reverse transcription reagents kit; PE Biosystems) was incubated at 42°C for 1 h, heat inactivated at 70°C for 10 min, and used for real-time PCR.
Real-Time PCR
Real-time PCR was carried out using an ABI Prism 7000 sequence detection system (TaqMan; Perkin-Elmer/PE Biosystems). Each multiplex PCR mixture contained 12.5 μL of TaqMan master mixture, an amount of cDNA equivalent to 200 ng total RNA, 20 pm gusA forward and reverse primers, 12.5 pm ubiquitin forward and reverse primers, 5.6 pm gusA, and 0.7 pm ubiquitin probes. The oligonucleotides and probes were supplied by PE Biosystems. The relative values for quantitation of RNA are expressed at 2−ΔΔCt, where ΔCt represents the difference between the threshold cycle (Ct; reflects the cycle number at which the fluorescence generated within a reaction crosses the threshold) value of a sample and that of ubiquitin (endogenous control) in the same sample, and ΔΔCt is the difference between the ΔCt value of a sample and that of the prototype promoter.
Plant Material and Transient Transformation Assay to Study Promoter Expression
Leaves from the third or fourth node of 9- to 10-week-old tobacco plants were used to study expression of the reporter gusA gene from different promoter cassettes. These were surface sterilized with 0.1% HgCl2, washed, and incubated on Murashige and Skoog agar medium (Murashige and Skoog, 1962) at 25°C in dark or continuous light (irradiance 80 μmol m−2 s−1) provided by cool-white fluorescent tube lights for 24 h before bombardment with DNA-coated gold microprojectiles. The DNA was bombarded at 1,100 lbs cm−3 pressure using a PDS 1000 He machine (Bio-Rad). Ten micrograms of DNA were coated on 1.5-mg gold particles and used for the bombardment of three discs of tobacco leaves taken as replicates for each treatment. Each experiment was repeated at least three times. After bombardment, the leaves were placed on fresh Murashige and Skoog agar medium and incubated in continuous light or dark as desired for a total period of 48 h before estimating the expression of GUS using the fluorogenic substrate MUG (Jefferson and Wilson, 1991). Total soluble protein in leaf extracts was quantified using Bradford reagent (catalog no. 500-0006; Bio-Rad Laboratories). Enzyme activity was expressed as pmol MU released min−1 mg−1 soluble protein.
Tobacco Transformation
The desired TATA-box-gusA cassettes or their corresponding counterparts with lre were cloned at the BamHI site in T-DNA in the binary vector pBI101. The recombinant vector plasmids were mobilized into Agrobacterium tumefaciens strain LBA4404 by electroporation. Leaf-disc explants were transformed following the protocol of Horsch et al. (1985). Several independently transformed plants were developed with each construct. Transgenic plants were grown in a 16-h light/dark cycle. The number of independent transgenic lines (n) tested in a given experiment are specified in the figures.
Growth Conditions and GUS Assays
After transformation, seeds of the R0 generation were collected and homozygous lines were obtained through kanamycin selection. Seeds of homozygous lines from the R1 generation were used for further study. For dark versus light experiments, seeds were grown in a 16-h-dark/8-h-light cycle or in complete darkness for 11 d. For light induction experiments, 9-d-old dark-grown seedlings were exposed to white light (80 μmol m−2 s−1), far-red light (0.86 μmol m−2 s−1), red light (10 μmol m−2 s−1), and blue light (15 μmol m−2 s−1) for 2 d before measurement of GUS activity.
Nuclear Protein Preparation
Nuclear protein from tobacco leaves was prepared from plants grown in a 16-h light/dark cycle. The plants were then incubated for 72 h in continuous light (NEL) or dark (NED). Following isolation of intact nuclei, protein was extracted in high ionic strength buffer and concentrated by ammonium sulfate precipitation. A procedure modified from Green et al. (1987) was used.
Tobacco leaf tissue was washed thoroughly with chilled distilled water. It was powdered in a mortar and pestle in liquid nitrogen and extracted in homogenization buffer (1 m hexylene glycol, 10 mm PIPES/KOH, pH 7.0, 10 mm MgCl2, 0.5% [v/v] Triton X-100, 5 mm 2-mercaptoethanol, and 0.8 mm phenylmethylsulfonyl fluoride [PMSF]). The homogenized extract was filtered sequentially through four layers of muslin cloth, followed by 80-, 60-, 40-, and 20-μm nylon mesh. Nuclei were pelleted at 3,000g for 10 min and resuspended gently with a soft paintbrush in wash buffer (0.5 m hexylene glycol, 10 mm PIPES/KOH, pH 7.0, 10 mm MgCl2, 5 mm 2-mercaptoethanol, 0.5% [v/v] Triton X-100, and 0.8 mm PMSF). Nuclei were pelleted at 3,000g for 15 min, the pellet was resuspended in 5% Percoll buffer (0.5 m Suc, 25 mm Tris, pH 8.0, 10 mm MgCl2, 0.8 mm PMSF, 0.5% Triton X-100, 5 mm 2-mercaptoethanol, 5% Percoll) and loaded on to 40% to 80% Percoll step gradient containing a 2 m Suc pad at the bottom. The nuclei were collected from the 80% Percoll-2 m Suc interface, washed in the wash buffer without Triton X-100, and resuspended in lysis buffer (110 mm KCl, 15 mm HEPES/KOH, pH 7.5, 5 mm MgCl2, 1 mm dithiothreitol [DTT], and 5 μg mL−1 leupeptin). One-tenth volume of 4 m ammonium sulfate was added and placed on a rocker for 30 min. Chromatin and particulate matter were pelleted at 100,000g for 45 min. Ammonium sulfate powder was added at 0.3 g mL−1 to the supernatant to precipitate the protein. The protein pellet was suspended in nuclear extract buffer (40 mm KCl, 250 mm HEPES/KOH, pH 7.5, 0.1 mm EDTA, 10% glycerol, 1 mm DTT, 5 μg mL−1 leupeptin) and subjected to dialysis for 2 h in the same buffer.
Electrophoresis Mobility Shift Assay
Electrophoresis mobility shift assays with nucleoprotein prepared from tobacco leaves were performed using 120-bp-long double-stranded oligonucleotides (Pmec without the coding sequence of gusA, as shown in the Construction of Cassettes section). The binding reactions were performed in a buffer containing 20 mm HEPES/KOH, pH 7.5, 40 mm KCl, 1 mm EDTA, 10% glycerol, and 0.5 mm DTT. As nonspecific carrier DNA, 5 μg of poly(dG-dC) was included in the binding reaction. Seventy-three fmol of the desired 32P-labeled double-stranded oligonucleotide were incubated with 5 ng to 1 μg of tobacco nucleoprotein at 25°C for 1 h, loaded onto 2% agarose gel or 4% PAGE, and electrophoresed in 0.5× Tris-borate/EDTA buffer at 200 V or 25 W, respectively, at 4°C.
Generation of Polyclonal Anti-Tobacco TBP Antibody
Total RNA was isolated from tobacco leaves, reverse transcribed using SuperScript II reverse transcriptase (GIBCO-BRL) to obtain cDNA, which was used to amplify the tobacco TBP gene. The TBP was cloned in pET 19b expression vector and expressed in Escherichia coli strain BL-21 as per standard protocol (Novagen PET manual). Most of the TBP expressed in this way formed inclusion bodies. The protein was solubilized using 8 m urea in the buffer. After solubilization, the protein was loaded onto nickel-nitrilotriacetic acid agarose column. The eluted protein was dialyzed to remove urea using urea step gradient. After the final step of dialysis, some protein still remained soluble and was removed by centrifugation. Two milligrams of purified protein were given to Bangalore Genei Pvt. Ltd. for generation of polyclonal antibodies in rabbit. The recognition of tobacco-TBP by antibody was confirmed through western blotting and ELISA assay using tobacco nuclear extract (data not shown).
RLM-RACE to Map gusA Transcripts of lre-Pmec Transgenic Lines in Light and Dark Conditions
Total RNA of lre-Pmec transgenic seedlings was used to perform RLM- RACE using a kit from Ambion. PCR was performed on cDNAs using the gene-specific primer GSP1 (5′-TAGTCTGCCAGTTCAGTTCGFT-3′) and the 5′-RACE outer primer supplied with the kit. A second round of PCR was performed with the product of the first PCR using a nested gene-specific primer (5′-GCTCCATCACTTCCTGATTATT-3′) coupled with the 5′-RACE inner primer supplied with the kit. The products of RACE reaction were confirmed with three sets of primers. Primer extension was then performed using PCR products of the second RACE reaction with 32P-labeled primer (from +94 to +1 of the gusA gene). The 5′ ends were identified by electrophoresis of the primer extension product alongside sequencing fragments obtained with the same primer.
Supplementary Material
Acknowledgments
The work presented in Figure 5 was performed in the laboratory of Dr. Jitendra Khurana at Delhi University (South Campus). Computational analysis was done by using software developed by Coral GeneCraft (arunsharma@genecraft.in).
This work was supported by a grant from the Council of Scientific and Industrial Research, Government of India, under the New Millennium Indian Technology Leadership Initiative program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Rakesh Tuli (rakeshtuli@hotmail.com).
The online version of this article contains Web-only data.
References
- Bertrand C, Benhamed M, Li YF, Ayadi M, Lemonnier G, Renou JP, Delarue M, Zhou DX (2005) Arabidopsis HAF2 gene encoding TATA-binding protein (TBP)-associated factor TAF1 is required to integrate light signals to regulate gene expression and growth. J Biol Chem 280: 1465–1473 [DOI] [PubMed] [Google Scholar]
- Breathnach R, Chambon P (1981) Organization and expression of eukaryotic split genes coding for proteins. Annu Rev Biochem 50: 349–383 [DOI] [PubMed] [Google Scholar]
- Butler JEF, Kadonaga JT (2001) Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev 15: 2515–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RA, Taggart AK, Burmam S, Chicca JJ II, Pugh BF (1999) TFIIA regulates TBP and TFIID dimers. Mol Cell 4: 451–457 [DOI] [PubMed] [Google Scholar]
- Colgan J, Manley JL (1992) TFIID can be rate limiting in vivo for TATA-containing, but not TATA-lacking, RNA polymerase II promoters. Genes Dev 6: 304–315 [DOI] [PubMed] [Google Scholar]
- Dikstein R, Zhou S, Tjian R (1996) Human TAFII105 is a cell type specific TFIID subunit related to hTAFII130. Cell 67: 137–146 [DOI] [PubMed] [Google Scholar]
- Duan ZJ, Fang X, Rohde A, Han H, Stematoyannopoulos G, Li Q (2002) Developmental specificity of recruitment of TBP to the TATA-box of the human γ-globin gene. Proc Natl Acad Sci USA 99: 5509–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiman RN, Albright SR, Chu LE, Zheng S, Hong-Ert L, Liang Sha WC, Tjian R (2002) Redundant role of tissue-selective TAFII 105 in lymphocytes. Mol Cell Biol 22: 6564–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PJ, Kay SA, Chua N-H (1987) Sequence-specific interactions of pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J 6: 2543–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Wilson KJ (1991) The GUS gene fusion system. In S Gelvin, R Schilperoot, eds, Plant Molecular Biology Manual, Vol B 14. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–33
- Joshi CP (1987) An inspection of the domain between putative TATA-box and translation start site in 79 plant genes. Nucleic Acids Res 15: 6643–6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu W, Cao X, Wilson T, Snustad DP, Chua N-H (1995) Phytochrome A and phytochrome B mediate the hypocotyl-specific downregulation of TUB1 by light in Arabidopsis. Plant Cell 7: 2187–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E (2002) Multi-protein complexes in eukaryotic gene transcription. Plant Mol Biol 50: 925–947 [DOI] [PubMed] [Google Scholar]
- Martinez-Hernandez A, Lopez-Ochoa L, Arguello-Astroga G, Herrera-Esterella L (2002) Functional properties and regulatory complexity of a minimal RBCS light-responsive unit activated by phytochrome, cryptochrome, and plastid signals. Plant Physiol 128: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsis M, Branhorst A, Neuman T (2001) Cell-type-specific expression of the TFIID component TAF(II)135 in the nervous system. Exp Cell Res 269: 214–221 [DOI] [PubMed] [Google Scholar]
- Mukumoto F, Hirose S, Imaseki H, Yamazaki KI (1993) DNA sequence requirement of a TATA element binding protein from Arabidopsis for translation in vitro. Plant Mol Biol 23: 995–1003 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised method for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Nakajima N, Horikoshi M, Roeder RG (1988) Factors involved in specific transcription by mammalian RNA polymerase II: purification, genetic specificity, and TATA-box-promoter interactions of TFIID. Mol Cell Biol 8: 4028–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Tsunoda T, Obokata J (2002) Photosynthesis nuclear genes generally lack TATA-boxes: a tobacco photosystem I gene responds to light through an initiator. Plant J 29: 1–10 [DOI] [PubMed] [Google Scholar]
- Pan S, Verner EC, Gurley WB (2000) Role of the TATA binding protein transcriptional factor IIB interaction in supporting basal and activated transcription in plant cells. Plant Cell 12: 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera LP (2000) The TATA motif specifies the differential activation of minimal promoter by Varicella zoster virus immediate-early regulatory protein IE62. J Biol Chem 275: 487–496 [DOI] [PubMed] [Google Scholar]
- Purnell BA, Gilmour DS (1993) Contribution of sequences downstream of the TATA element to protein-DNA complex containing the TATA-binding protein. Mol Cell Biol 13: 2593–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci 21: 327–335 [PubMed] [Google Scholar]
- Sawant SV, Kiran K, Singh PK, Tuli R (2001) Sequence architecture downstream of the initiator codon enhances gene expression and protein stability in plants. Plant Physiol 126: 1630–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawant SV, Singh PK, Gupta SK, Madanala R, Tuli R (1999) Conserved nucleotide sequence in highly expressed genes in plants. J Genet 78: 123–131 [Google Scholar]
- Sawant SV, Singh PK, Tuli R (2000) Pretreatment of microprojectiles to improve the delivery of DNA in plant transformation. Biotechniques 29: 246–248 [DOI] [PubMed] [Google Scholar]
- Shahmuradov IA, Gammerman AJ, Hancock JM, Bramley HP, Solovyev YV (2003) Plant Prom: a database of plant promoter sequences. Nucleic Acids Res 31: 114–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST (2001) Core promoters: active contributors to combinatorial gene regulation. Genes Dev 15: 2503–2508 [DOI] [PubMed] [Google Scholar]
- Steven LS, Krassimira AG, Weil PA (2002) Molecular characterization of Saccharomyces cerevisiae TFIID. Mol Cell Biol 22: 6000–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JJ, Stargell LA (2001) The stability of the TFIIA-TBP-DNA complex is dependent on the sequence of the TATAAA element. J Biol Chem 276: 30078–30084 [DOI] [PubMed] [Google Scholar]
- Strubin M, Struhl K (1992) Yeast and human TFIID with altered DNA-binding specificity for TATA elements. Cell 68: 721–730 [DOI] [PubMed] [Google Scholar]
- Wassarman DA, Sauer F (2001) TAF(II) 250: a transcription tool box. J Cell Sci 114: 2895–2902 [DOI] [PubMed] [Google Scholar]
- Wefald FC, Devlin BH, Williams RS (1990) Functional heterogeneity of mammalian TATA-box sequences relived by interaction with a cell specific enhancer. Nature 344: 260–262 [DOI] [PubMed] [Google Scholar]
- Wolner BS, Gralla JD (2001) TATA-flanking sequences influence the rate and stability of the TATA-binding protein and TFIIB binding. J Biol Chem 276: 6260–6266 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Itoh Y, Takeda Y, Yamazaki KI (1998) TATA sequence requirements for the initiation of transcription for an RNA polymerase II in vitro transcription system from Nicotiana tabacum. Plant Mol Biol 38: 1247–1252 [DOI] [PubMed] [Google Scholar]
- Zhang DPT, Morris PL, Roeder RG (2001) Cell- and stage-specific high level expression of TBP-related factor 2 (TRF2) during mouse spermatogenesis. Mech Dev 106: 203–205 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Dabi T, Lamb C (1995) TATA-box and initiator functions in the accurate transcription of a plant minimal promoter in vitro. Plant Cell 7: 1681–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.