Abstract
Improvements in carbon assimilation and water-use efficiency lead to increases in maximum leaf area index at elevated carbon dioxide concentration ([CO2]); however, the molecular drivers for this increase are unknown. We investigated the molecular basis for changes in leaf development at elevated [CO2] using soybeans (Glycine max) grown under fully open air conditions at the Soybean Free Air CO2 Enrichment (SoyFACE) facility. The transcriptome responses of rapidly growing and fully expanded leaves to elevated [CO2] were investigated using cDNA microarrays. We identified 1,146 transcripts that showed a significant change in expression in growing versus fully expanded leaves. Transcripts for ribosomal proteins, cell cycle, and cell wall loosening, necessary for cytoplasmic growth and cell proliferation, were highly expressed in growing leaves. We further identified 139 transcripts with a significant [CO2] by development interaction. Clustering of these transcripts showed that transcripts involved in cell growth and cell proliferation were more highly expressed in growing leaves that developed at elevated [CO2] compared to growing leaves that developed at ambient [CO2]. The 327 [CO2]-responsive genes largely suggest that elevated [CO2] stimulates the respiratory breakdown of carbohydrates, which provides increased energy and biochemical precursors for leaf expansion and growth at elevated [CO2]. While increased photosynthesis and carbohydrate production at elevated [CO2] are well documented, this research demonstrates that at the transcript and metabolite level, respiratory breakdown of starch is also increased at elevated [CO2].
By 2050, soybean (Glycine max) will grow in an atmosphere with a 50% higher carbon dioxide concentration ([CO2]) (Prentice et al., 2001). As the world's most widely grown seed legume, the physiological responses of soybean to elevated CO2 have been well characterized. Elevated [CO2] increases carbon (C) uptake, foliar carbohydrate content, plant growth, and yield, while decreasing stomatal conductance (for review, see Ainsworth et al., 2002). A Free Air CO2 Enrichment (FACE) experiment was established in one of the world's most productive soybean-growing areas, Central Illinois, in 2001. This facility allows investigation of the response of field-grown soybean to an atmosphere predicted for 2050 without alteration of the microclimate (Long et al., 2004). Across the growing season, daily integrals of leaf photosynthetic CO2 uptake increased by approximately 25%, even as midday stomatal conductance decreased by approximately 20% (Rogers et al., 2004; Bernacchi et al., 2006). Improvements in C assimilation and water-use efficiency spurred increases in maximum leaf area index (LAI) and aboveground biomass in elevated [CO2] (Morgan et al., 2005; Dermody et al., 2006). The combination of increased photosynthesis and increased LAI provided the inputs for significant increases in soybean seed yield (Ort et al., 2006).
Increased leaf growth, leading to larger individual leaf size, is one component of increased LAI measured at elevated [CO2] in field experiments (Taylor et al., 2003; Tricker et al., 2004; Dermody et al., 2006). At the molecular level, the basis for changes in LAI at elevated [CO2] is largely unknown. Both cell production rates and cell expansion have been shown to be sensitive to elevated [CO2] (Taylor et al., 1994, 2003). Transcript analysis of growing poplar (Populus spp.) leaves exposed to elevated [CO2] showed that genes involved in cell wall loosening and synthesis were up-regulated (Taylor et al., 2005; Druart et al., 2006). Therefore, at least in poplar, elevated [CO2] appears to stimulate or prolong expansion in leaves (Ferris et al., 2001; Taylor et al., 2003; Druart et al., 2006).
Leaf growth is a spatially and temporally dynamic process (for review, see Schurr et al., 2006). To understand the mechanisms controlling leaf growth in dicots, experiments must account for the spatial and diel variations in growth (Trainotti et al., 2004; Matsubara et al., 2005). Growing leaves do not expand at all times throughout the diel cycle, nor do they necessarily expand homogeneously. Tobacco (Nicotiana tabacum) leaves show a base-to-tip gradient in developmental stage of the tissue, and differential expression of genes in apical and basal tissues (Trainotti et al., 2004). While the functional maturation of dicot leaves has been described to progress with a base-to-tip gradient (Avery, 1933; Turgeon, 1989), recent experiments using digital image sequence processing revealed that some dicot species lack a base-to-tip gradient in relative expansion rates (Ainsworth et al., 2005; Matsubara et al., 2005). Prolonged cytoplasmic growth as opposed to vacuolated growth dominated leaf expansion in Populus deltoides, a species that lacks a base-to-tip gradient in leaf growth (Matsubara et al., 2005). Soybean also lacks a pronounced base-to-tip gradient in leaf growth rates (Ainsworth et al., 2005). Yet, soybean has a clear diel pattern of leaf expansion, with maximum rates occurring at night (Bunce, 1977; Ainsworth et al., 2005). How elevated [CO2] alters the dynamics of leaf expansion in soybean has not been examined to date. However, there is evidence in poplar species that altered carbohydrate status may change the fine-scale temporal and spatial patterns of growth in response to elevated [CO2] (Walter et al., 2005), and experiments with transgenic plants clearly show a link between leaf C metabolism and leaf development (Raines and Paul, 2006).
The first objective of this research was to investigate molecular changes in growing and fully expanded soybean leaves developed at elevated [CO2] under fully open-air conditions. Research has shown that the response of soybean to elevated [CO2] in the field is less than predicted from chamber studies (Long et al., 2006). One approach for maximizing future production of soybean is to increase LAI and, therefore, the potential for C uptake. Thus, the other objectives of our study were to identify transcripts that control leaf growth and elongation, and to investigate how elevated [CO2] alters the expression of those transcripts. This research provides an overview of the soybean transcriptome response to elevated [CO2] in both fully expanded and growing leaves.
RESULTS AND DISCUSSION
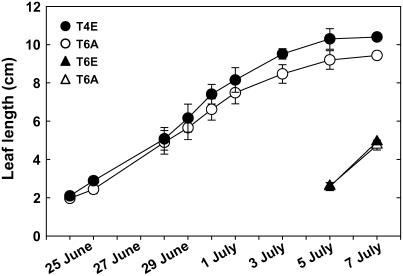
The transcriptome response of fully expanded trifoliate 4 (T4) and growing trifoliate 6 (T6) soybean leaves to elevated [CO2] was analyzed using cDNA microarrays (Fig. 1). On July 7, T4 leaflets were fully expanded and longer in elevated [CO2] compared to ambient [CO2] (Fig. 2). T6 leaflets were growing with a relative increase in length of 42% ± 6% per day in both ambient and elevated [CO2] (Fig. 2). We were specifically interested in how elevated [CO2] alters genes related to leaf development, so samples were taken between 1 and 2 am, which corresponded to the time of maximum leaf expansion rate (Ainsworth et al., 2005). Analysis of variance revealed 1,146 genes with different expression (P < 0.05) in growing leaves compared to fully expanded leaves (Supplemental Table I), 139 transcripts with a significant CO2 × development interaction (Supplemental Table II), and 327 transcripts that responded to CO2 (Supplemental Table III). The changes in transcript abundance were up to 3.5-fold in developmentally regulated transcripts (Table I) and up to 2-fold in transcripts regulated by [CO2] (Table II). This result is similar to results from two other FACE experiments where only small changes in transcript expression were detected at elevated [CO2] (Gupta et al., 2005; Taylor et al., 2005). This likely reflects the chronic nature of FACE treatment. In the FACE experiment, we analyzed transcript profiles of plants acclimated to an environmental change (a higher [CO2]) rather than observing the response of gene expression to an acute change, e.g. an herbivore attack.
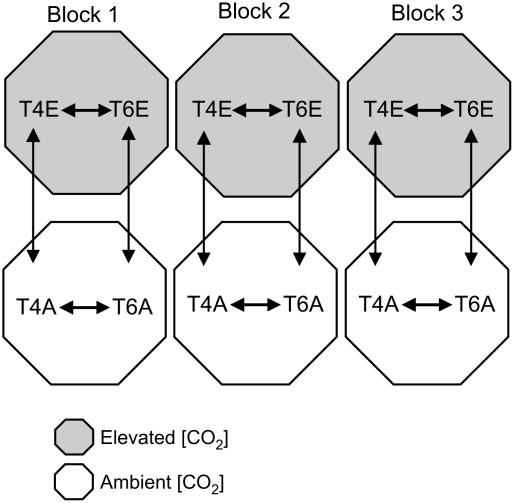
Figure 1.
Design of the cDNA microarray experiment. Each double-headed arrow represents four microarrays per library, two biological replicates and two technical replicates. Each biological replicate included pooled RNA from six individual plants. A total of 96 microarrays were analyzed. Four treatments were compared: 1, T4 versus T6 in ambient [CO2] (T4A versus T6A); 2, T4 versus T6 in elevated [CO2] (T4E versus T6E); 3, T4 in ambient [CO2] versus T4 in elevated [CO2] (T4A versus T4E); 4, T6 in ambient [CO2] versus T6 in elevated [CO2] (T6A versus T6E).
Figure 2.
Increase in length of the fourth (T4, circles) and sixth (T6, triangles) trifoliate lateral leaflets grown at ambient (A; white symbols) and elevated (E; black symbols) [CO2]. Samples for microarray analysis and leaf carbohydrates were taken on July 8, 2004, between 1 and 2 am, when T4 leaflets were fully expanded and T6 leaflets were expanding in length at 42% ± 6% d−1.
Table I.
Selection of 38 genes that were differentially expressed between growing and fully expanded leaves
We report a description based on matching BLASTX hits from sequences of both the 3′ and 5′ ends of each clone unless only the 5′ information is available (5′), relative expression in growing (T6) versus fully expanded leaves (T4), and results of the statistical analysis (F, P). The entire list of 1,146 genes that were differentially expressed between growing and fully expanded leaves is provided in Supplemental Table I.
| Clone ID | Description | T6/T4 | F | P |
|---|---|---|---|---|
| Gm-r1070-3452 | Ribosomal protein L35 (Arabidopsis thaliana) | 1.505 | 12.85 | 0.01158 |
| Gm-r1088-5863 | Ribosomal protein (Petunia x hybrida) | 1.508 | 18.25 | 0.00272 |
| Gm-r1088-5173 | 40s ribosomal protein S23 (Euphorbia esula) | 1.509 | 18.56 | 0.00259 |
| Gm-r1070-8126 | Putative ribosomal protein (Capsicum annuum) | 1.514 | 21.67 | 0.00349 |
| Gm-r1070-8058 | ADP-ribosylation factor (Hyacinthus orientalis) | 1.516 | 15.86 | 0.00726 |
| Gm-r1070-1257 | 60S ribosomal protein L7A (A. thaliana) | 1.521 | 18.69 | 0.00497 |
| Gm-r1070-8966 | Putative histone H2A protein (Oryza sativa) | 1.525 | 20.58 | 0.00395 |
| Gm-r1088-6691 | Ribosomal protein L36 (Triticum aestivum) | 1.528 | 27.57 | 0.00077 |
| Gm-r1070-7448 | α-Expansin 3 (Cicer arietinum) | 1.532 | 9.93 | 0.01979 |
| Gm-r1070-7790 | Putative 60S ribosomal protein L10A (RPL10aC) (O. sativa) | 1.538 | 18.35 | 0.00519 |
| Gm-r1088-4318 | Histone H2B (C. arietinum) | 1.541 | 20.91 | 0.00182 |
| Gm-r1070-8158 | Ribosomal protein L14-like protein (A. thaliana) | 1.576 | 9.41 | 0.02203 |
| Gm-r1070-4867 | 30S ribosomal protein S16-like (O. sativa) | 1.580 | 13.66 | 0.01013 |
| Gm-r1070-3909 | Ribosomal protein L11, putative (A. thaliana) | 1.586 | 17.14 | 0.00608 |
| Gm-r1088-164 | Putative 40S ribosomal protein (O. sativa) | 1.590 | 26.71 | 0.00086 |
| Gm-r1070-970 | Histone H2A (E. esula) | 1.624 | 7.12 | 0.03709 |
| Gm-r1070-3576 | Ribosomal protein L37 (soybean) | 1.628 | 12.90 | 0.01149 |
| Gm-r1088-7068 | 60S ribosomal protein L13E (Picea abies) | 1.639 | 7.67 | 0.02431 |
| Gm-r1070-5626 | Putative ribosomal protein L19 (O. sativa) | 1.641 | 22.99 | 0.00302 |
| Gm-r1088-4541 | Tubulin family protein (A. thaliana) | 1.646 | 10.33 | 0.01236 |
| Gm-r1088-7215 | Ribosomal protein L15 (P. x hybrida) | 1.653 | 11.23 | 0.01007 |
| Gm-r1070-8151 | 40S ribosomal protein S21, putative (O. sativa) | 1.655 | 54.66 | 0.00031 |
| Gm-r1070-8751 | Ribosomal protein L29 (Panax ginseng) | 1.660 | 45.72 | 0.00051 |
| Gm-r1070-7659 | α-Tubulin (Gossypium hirsutum) | 1.683 | 14.48 | 0.00891 |
| Gm-r1070-8444 | Ribosomal protein S26 (Pisum sativum) | 1.685 | 17.01 | 0.00619 |
| Gm-r1070-4817 | 60S ribosomal protein L7 (A. thaliana) | 1.703 | 31.44 | 0.00137 |
| Gm-r1088-4320 | Histone H2B1 (G. hirsutum) | 1.711 | 6.22 | 0.03727 |
| Gm-r1070-9008 | 40S ribosomal protein S25 (soybean) | 1.740 | 14.28 | 0.00919 |
| Gm-r1070-8414 | 40S ribosomal protein S14 | 1.760 | 33.04 | 0.00121 |
| Gm-r1070-8326 | Histone H2B-3 (Lycopersicon esculentum) | 1.780 | 17.70 | 0.00564 |
| Gm-r1070-3758 | Ribosomal protein small subunit 28 (Helianthus annuus) | 1.801 | 26.41 | 0.00214 |
| Gm-r1088-7080 | 60S ribosomal protein L34 (Solanum demissum) | 1.820 | 11.78 | 0.00893 |
| Gm-r1070-5890 | 40S ribosome protein S7 (Avicennia marina) | 1.843 | 34.44 | 0.00108 |
| Gm-r1070-7481 | Putative cytoplasmic ribosomal protein S15a (A. thaliana) | 1.862 | 10.00 | 0.01950 |
| Gm-r1070-8254 | Acidic ribosomal protein (H. orientalis) | 1.908 | 21.80 | 0.00343 |
| Gm-r1070-5505 | Expansin (Pyrus communis) | 1.941 | 6.12 | 0.04814 |
| Gm-r1070-8979 | Acidic ribosomal protein P0 (soybean) | 2.041 | 7.42 | 0.03447 |
| Gm-r1070-9002 | 40S ribosomal protein S20-like protein (A. thaliana) | 2.416 | 27.29 | 0.00197 |
Table II.
Selection of 85 genes that were differentially expressed between ambient and elevated [CO2]
We report a description based on matching BLASTX hits from sequences of both the 3′ and 5′ ends of each clone unless only the 5′ information is available (5′), relative expression in elevated [CO2]/ambient [CO2] (E/A), and results of the statistical analysis (F, P). The numbers in the final column refer to the transcripts illustrated in Figure 6. The entire list of 327 genes that were differentially expressed between ambient and elevated [CO2] is provided in Supplemental Table III.
| Clone ID | Description | E/A | F | P | Figure 6 |
|---|---|---|---|---|---|
| C Metabolism | |||||
| Gm-r1070-5613 | β-Amylase (soybean) | 1.434 | 15.39 | 0.0078 | 1 |
| Gm-r1070-7748 | Granule-bound starch synthase Ib precursor (Phaseolus vulgaris) | 1.829 | 10.60 | 0.0173 | |
| Gm-r1070-5070 | β-Amylase (soybean) | 1.360 | 16.41 | 0.0067 | 2 |
| Gm-r1070-8759 | Chain A, crystal structure of soybean β-amylase mutant (M51t) | 1.313 | 14.97 | 0.0083 | 3 |
| Gm-r1070-8484 | Chain A, crystal structure of soybean β-amylase mutant (M51t) | 1.401 | 11.88 | 0.0137 | 3 |
| Gm-r1088-4020 | myo-Inositol-1-P synthase (soybean) | 1.668 | 6.21 | 0.0375 | 4 |
| Gm-r1070-5201 | Inositol-3-P synthase (soybean) | 1.714 | 7.01 | 0.0382 | 5 |
| Gm-r1088-2682 | α-Galactosidase preproprotein (Cyamopsis tetragonoloba) | 1.476 | 5.85 | 0.0419 | |
| Gm-r1070-6564 | Phosphofructokinase (Medicago truncatula) | 1.241 | 13.13 | 0.0111 | 6 |
| Gm-r1070-4852 | Glyceraldehyde-3-P dehydrogenase (Solanum tuberosum) | 1.244 | 11.27 | 0.0153 | 7 |
| Gm-r1070-7510 | Phosphoglycerate mutase (Ricinus communis) | 1.352 | 10.75 | 0.0169 | 8 |
| Gm-r1070-6442 | Enolase, isoform 2 (Hevea brasiliensis) | 1.214 | 10.73 | 0.0169 | 9 |
| Gm-r1070-5429 | Cytosolic phosphoglycerate kinase (Pisum sativum) | 1.142 | 10.60 | 0.0173 | 10 |
| Gm-r1088-3248 | Plastidial phosphoglucomutase (P. sativum) | 1.901 | 6.49 | 0.0343 | 11 |
| Gm-r1070-7756 | Putative pyruvate dehydrogenase E1 β-subunit (Oryza sativa) | 1.322 | 7.03 | 0.0379 | 12 |
| Gm-r1070-7077 | Phosphogluconate dehydrogenase (decarboxylating) (EC 1.1.1.44) | 1.454 | 6.54 | 0.0338 | 13 |
| Mitochondrial e-Transport | |||||
| Gm-r1070-6065 | Hydrogen-transporting ATPase, rotational mechanism (Arabidopsis thaliana) | 1.198 | 6.16 | 0.0476 | |
| Gm-r1070-4318 | F1-ATP synthase δ-subunit (Ipomoea batatas) | 1.168 | 6.08 | 0.0488 | |
| Cell Wall Metabolism | |||||
| Gm-r1070-5796 | Putative NAD-dependent epimerase (A. thaliana) | 1.268 | 18.47 | 0.0051 | 14 |
| Gm-r1070-6616 | Cellulose synthase (EC 2.4.1.-) catalytic chain celA2 | 1.346 | 8.64 | 0.0260 | 15 |
| Gm-r1070-4646 | UDP-d-apiose/UDP-d-Xyl synthase (A. thaliana) | 1.303 | 8.61 | 0.0261 | 16 |
| Lipid Metabolism | |||||
| Gm-r1070-9006 | Palmitoyl-acyl carrier protein thioesterase (Gossypium hirsutum) | 1.308 | 18.55 | 0.0051 | 17 |
| Gm-r1070-7584 | Enoyl-ACP reductase (tobacco) | 1.263 | 7.71 | 0.0321 | 18 |
| Gm-r1070-3469 | Omega-3 fatty acid desaturase (EC 1.14.99.-) GMD | 1.145 | 7.02 | 0.0380 | |
| Gm-r1070-5787 | Microsomal omega-3 fatty acid desaturase (soybean) | 1.348 | 6.82 | 0.0400 | |
| Gm-r1070-5473 | Acyl-CoA thioesterase (A. thaliana) | 1.244 | 6.27 | 0.0462 | |
| N Metabolism | |||||
| Gm-r1070-5797 | P-protein precursor (S. tuberosum) | 1.417 | 15.94 | 0.0072 | |
| Gm-r1070-9038 | Putative urease accessory protein G (O. sativa [japonica cultivar group]) | 1.167 | 7.46 | 0.0342 | |
| Gm-r1088-2099 | Indole-3-glycerol phosphate synthase (A. thaliana) | 1.409 | 5.49 | 0.0472 | 19 |
| Gm-r1070-6244 | Gln synthetase precursor (soybean) | 1.328 | 8.98 | 0.0241 | 20 |
| Transport | |||||
| Gm-r1070-2965 | Ca2+/H+-exchanging protein (Vigna radiata) | 1.373 | 51.02 | 0.0004 | |
| Gm-r1070-5393 | Ca2+/H+-exchanging protein (V. radiata) | 1.616 | 40.58 | 0.0007 | |
| Gm-r1070-5594 | Vacuolar H+-ATPase B subunit (Citrus unshiu) | 1.424 | 27.29 | 0.0020 | |
| Gm-r1088-3648 | Aquaporin (Vitis vinifera) | 1.998 | 9.82 | 0.0139 | |
| Gm-r1070-5792 | Vacuolar ATPase subunit E (Phaseolus acutifolius) | 1.294 | 10.96 | 0.0162 | |
| Gm-r1070-2941 | Vacuolar H+-ATPase c subunit (C. unshiu) | 1.339 | 10.53 | 0.0176 | |
| Gm-r1070-6113 | Putative nitrate transporter NRT1-3 (soybean) | 1.734 | 8.90 | 0.0245 | 21 |
| Gm-r1070-4599 | Putative nitrate transporter NRT1-3 (soybean) | 1.259 | 6.73 | 0.0410 | 21 |
| Gm-r1070-8647 | Core protein (P. sativum) | 1.222 | 6.71 | 0.0412 | |
| Gm-r1088-1920 | ADP, ATP carrier-like protein (A. thaliana) | 1.600 | 5.75 | 0.0433 | |
| Gm-r1088-2089 | Cation diffusion facilitator 9 (Stylosanthes hamata) | 1.349 | 5.36 | 0.0494 | |
| Gm-r1088-1149 | Outer envelope protein (P. sativum) | 1.390 | 5.35 | 0.0494 | |
| Hormone Metabolism | |||||
| Gm-r1070-6109 | Lipoxygenase (P. vulgaris) | 1.293 | 25.18 | 0.0024 | |
| Gm-r1070-8913 | Auxin response factor-like protein (Mangifera indica) | 1.376 | 15.32 | 0.0079 | |
| Gm-r1070-5659 | Response reactor 4 (A. thaliana) | 1.215 | 7.72 | 0.0321 | |
| Secondary Metabolism | |||||
| Gm-r1070-1714 | Homogentisate phytylprenyltransferase (soybean) | 1.224 | 14.87 | 0.0084 | |
| Gm-r1070-3733 | 1-Deoxy-d-xylulose 5-P reductoisomerase (Pueraria montana) | 1.347 | 9.01 | 0.0240 | 22 |
| Gm-r1070-3522 | Putative caffeic acid methyl transferase (Arachis hypogaea) | 1.327 | 8.15 | 0.0290 | 23 |
| Gm-r1070-8296 | Putative cinnamoyl-CoA reductase (Solanum demissum) | 1.439 | 7.62 | 0.0328 | 24 |
| RNA Regulation of Transcription | |||||
| Gm-r1070-5468 | MYB transcription factor (A. thaliana) | 1.282 | 43.60 | 0.0006 | |
| Gm-r1070-8564 | mRNA-binding protein precursor (tobacco) | 1.316 | 24.43 | 0.0026 | |
| Gm-r1070-7770 | DNA-binding protein (A. thaliana) | 1.218 | 17.97 | 0.0054 | |
| Gm-r1070-6997 | Remorin (S. tuberosum) | 1.389 | 13.30 | 0.0107 | |
| Gm-r1070-7595 | LHY protein (P. vulgaris) | 1.287 | 11.64 | 0.0143 | |
| Gm-r1070-6170 | NAM-like protein (Prunus persica) (5′) | 1.272 | 11.64 | 0.0143 | |
| Gm-r1088-2878 | bZIP DNA-binding protein (Antirrhinum majus) | 1.643 | 8.12 | 0.0215 | |
| Gm-r1070-4141 | SEU3B protein (A. majus) | 1.370 | 9.40 | 0.0221 | |
| Gm-r1070-4704 | Double WRKY-type transfactor (S. tuberosum) | 1.389 | 8.38 | 0.0275 | |
| Gm-r1070-8883 | Nucleoid DNA-binding-like protein (A. thaliana) | 1.173 | 8.30 | 0.0280 | |
| Gm-r1070-1976 | DNA-binding protein 4 (tobacco) (5′) | 1.340 | 8.24 | 0.0284 | |
| Gm-r1070-7398 | Putative RING zinc-finger protein (A. thaliana) | 1.162 | 7.60 | 0.0330 | |
| Gm-r1070-7935 | Dc3 promoter-binding factor-3 (Helianthus annuus) | 1.228 | 7.38 | 0.0348 | |
| Gm-r1070-6989 | Nucleic acid-binding/transcription factor (A. thaliana) | 1.276 | 7.06 | 0.0377 | |
| Gm-r1070-5991 | Putative AP2-binding protein (Jatropha curcas) | 1.139 | 6.74 | 0.0409 | |
| Gm-r1070-8292 | Homeodomain-Leu zipper protein 56 (soybean) | 1.355 | 6.41 | 0.0445 | |
| Gm-r1070-3954 | MYB family transcription factor (A. thaliana) | 1.237 | 6.41 | 0.0446 | |
| Gm-r1070-7416 | LEC1-like protein (Phaseolus coccineus) | 1.136 | 6.36 | 0.0452 | |
| Gm-r1070-8125 | HMG-1-like protein gene (soybean) | 1.263 | 6.32 | 0.0456 | |
| Gm-r1070-6056 | BEL1-related homeotic protein 29 (S. tuberosum) | 1.225 | 6.25 | 0.0465 | |
| Protein Degradation | |||||
| Gm-r1070-7908 | Probable aminopeptidase F24D7.4 (imported) (A. thaliana) | 1.516 | 41.24 | 0.0007 | |
| Gm-r1070-5823 | Ubiquitin-specific protease 16 (A. thaliana) | 1.244 | 13.62 | 0.0102 | |
| Gm-r1070-8169 | Skp1 (Medicago sativa) | 1.315 | 10.24 | 0.0186 | |
| Gm-r1070-8143 | Pentameric polyubiquitin | 1.287 | 9.91 | 0.0199 | |
| Gm-r1070-3489 | Cullin 1C (tobacco) | 1.304 | 9.85 | 0.0201 | |
| Gm-r1070-5188 | Subtilisin-like protease C1 (soybean) | 1.418 | 9.21 | 0.0229 | |
| Gm-r1070-3592 | Cys proteinase (soybean) | 1.536 | 8.71 | 0.0256 | |
| Gm-r1070-4236 | Cys proteinase (soybean) | 1.250 | 8.19 | 0.0287 | |
| Gm-r1070-8885 | Aspartyl protease family protein (A. thaliana) | 1.192 | 7.72 | 0.0321 | |
| Gm-r1070-4830 | Peptidase C1A, papain; Somatotropin hormone (M. truncatula) | 1.223 | 7.61 | 0.0329 | |
| Gm-r1070-6672 | hyuC-like protein (A. thaliana) | 1.235 | 7.29 | 0.0356 | |
| Gm-r1088-4646 | Ubiquitin-conjugating enzyme E2 (Gossypium raimondii) | 1.116 | 6.33 | 0.0360 | |
| Gm-r1088-1518 | Metal-dependent hydrolase-like protein (O. sativa [japonica cultivar group]) | 1.338 | 6.22 | 0.0373 | |
| Gm-r1070-5742 | Putative PRT1 protein (A. thaliana) | 1.227 | 7.01 | 0.0381 | |
| Gm-r1070-5457 | Peptidase (A. thaliana) | 1.258 | 6.99 | 0.0384 | |
| Gm-r1070-8504 | Proteasome subunit α type 3 | 1.187 | 6.39 | 0.0448 |
Genes Associated with Leaf Development
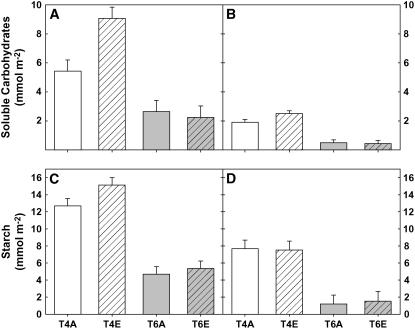
Expression of 1,146 genes was significantly different in growing versus fully expanded leaves, irrespective of growth [CO2] (for a complete list of transcripts, see Supplemental Table I). A total of 178 transcripts, encoding genes for a wide variety of functions, showed 1.5 times lower gene expression in growing leaves compared to fully expanded leaves. This group included genes involved in secondary metabolism, transport, stress and metal handling, and major and minor carbohydrate metabolism. Notably, starch phosphorylase (Gm-r1088-8633), a glucan-metabolizing enzyme (Smith et al., 2005), was expressed at lower levels in growing leaves compared to fully expanded leaves (T6/T4 = 0.595). This correlated with measured amounts of leaf carbohydrates (Fig. 3), where fully expanded, mature leaves had between 4 to 5 times the amount of starch as developing leaves (Fig. 3, C and D) and, therefore, more substrate for degradation.
Figure 3.
Leaf level contents of soluble carbohydrates (Suc, Glc, Fru; A and B) and starch (C and D) in fully expanded (T4) and growing (T6) leaves in ambient (A) and elevated (E) [CO2]. Leaves were sampled at dusk on July 7, 2004 (A and C), and between 1 and 2 am on July 8, 2004 (B and D). At dusk, there was a significant buildup of carbohydrates in mature leaves grown at elevated [CO2] (P < 0.05), but there was no significant effect of [CO2] treatment on soluble carbohydrates or starch (P > 0.05) in the middle of the night. There was a highly significant effect of development on both carbohydrate pools (P < 0.001).
A total of 132 transcripts showed at least 1.5 times higher expression in growing leaves and, therefore, represent our best estimates of control points of leaf expansion. The major family of genes that were highly expressed in growing tissues included ribosomal proteins (Table I). Some cell-cycle genes (histones) and cell wall-loosening genes (expansins) were also included in this group (Table I). Tubulin genes, necessary for regulating the direction of diffuse growth in plants (Abe et al., 2004), were also highly expressed in growing tissues (Table I). The T6 soybean leaflets in this experiment were expanding rapidly (Fig. 2). It is likely that this expansion was due to cytoplasmic growth and cell proliferation, which require significant ribosome biosynthesis (Sugimoto-Shirasu and Roberts, 2003). These findings are supported by previous work with poplar that highlighted up-regulation of ribosome biosynthesis as the primary process underlying nocturnal variations in leaf growth (Matsubara et al., 2005).
CO2 × Development Interaction
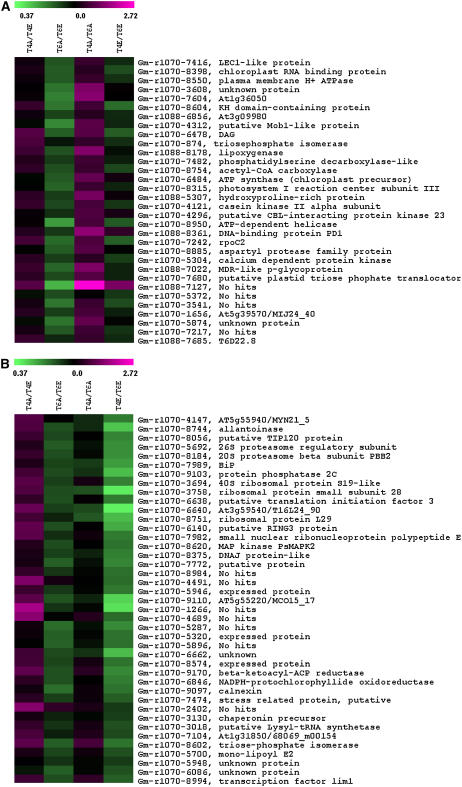
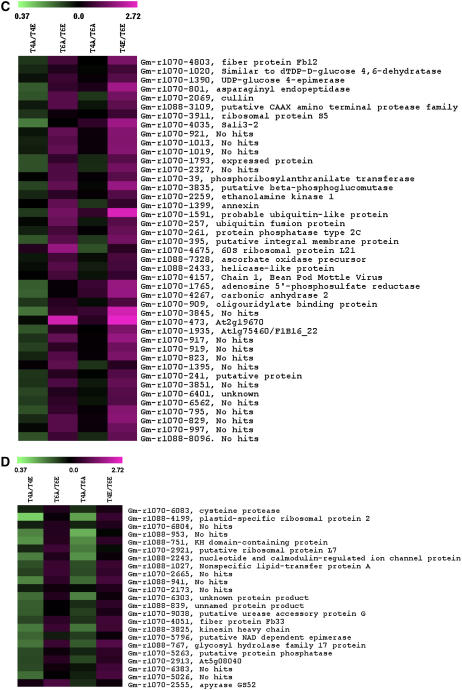
We identified 139 transcripts with a CO2 × development interaction (P < 0.05). These were of particular interest because they represent potential genes involved in growth that may be altered by [CO2] treatments. These transcripts were clustered into four groups using k-means clustering (Saeed et al., 2003). The first cluster of 32 transcripts showed lower expression in developing leaves grown at ambient [CO2] (T6A) compared to developing leaves grown at elevated [CO2] (T6E; i.e. T6A/T6E < 1), higher expression in fully expanded leaves grown at ambient [CO2] (T4A) compared to developing leaves at ambient [CO2] (T6A; i.e. T4A/T6A > 1), and no change in other comparisons (Fig. 4A). These transcripts included DAG (Gm-r1070-6478), a gene involved in chloroplast development and leaf palisade differentiation (Chatterjee et al., 1996), and a putative Mob-1 like protein, which likely functions in cell proliferation (Citterio et al., 2006). Some transcription factors and DNA-binding proteins were also included in this cluster (Fig. 4A). This provides some evidence that cell proliferation and development are increased in growing leaves at elevated [CO2] compared to growing leaves at ambient [CO2].
Figure 4.
Heat map of transcripts with significant [CO2] × trifoliate interaction (P < 0.05). Transcripts were clustered into four distinct clusters (A–D) using k-means clustering (TIGR MeV version 3.1). Values for each ratio are expressed by color intensity, where higher expression is indicated by shades of magenta and lower expression by shades of green. Comparisons between [CO2] treatments and developmental stages are described in Figure 1.
The second cluster contained transcripts with lower expression in T4E compared to T6E (Fig. 4B) and rather subtle changes in other comparisons. This group contained a number of ribosomal proteins (Gm-r1070-3758, Gm-r1070-6640, Gm-r1070-8751, Gm-r1070-3694) involved in protein synthesis, as well as a binding protein (BiP; Gm-r1070-7989), a highly conserved endoplasmic-reticulum luminal protein that functions as a molecular chaperone (Kalinski et al., 1995). Other genes involved in transcription and regulation of transcription were clustered in this group, lending further evidence to increased levels of cell proliferation and development in young leaves exposed to elevated [CO2].
The third cluster included 43 transcripts that showed higher expression in T4E compared to T6E (Fig. 4C). This cluster included genes with a wide range of functions, including amino acid synthesis and transport, carbohydrate and cell wall metabolism, protein degradation, redox, and stress response (Fig. 4C). The fourth cluster included 23 genes that showed lower expression in T4A compared to both T4E and T6A (Fig. 4D), including two genes involved in cell wall metabolism, a putative NAD-dependent epimerase and a glycosyl hydrolase family 17 protein (Gm-r1070-5796 and Gm-r1070-767). Glycosyl hydrolase family 17 proteins hydrolyze 1,3-β-glucan polysaccharides in the cell wall matrix and are involved in many stages of plant development, including cell division (Thomas et al., 2000). Previous transcriptome analysis of growing poplar leaves showed that glycosyl hydrolase was up-regulated in growing leaves at the time of maximum expansion (Matsubara et al., 2005), supporting our finding that glycosyl hydrolase was expressed at lower levels in fully expanded leaves compared to developing leaves. In general, the clustering of genes with a CO2 × development interaction led to the identification of a number of transcripts involved in growth and cell proliferation with high expression in young leaves grown at elevated [CO2].
CO2 Response
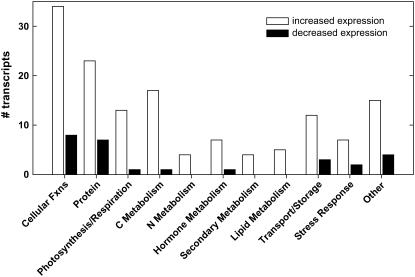
The 327 CO2-responsive genes were assigned to different functional categories (Fig. 5). Many genes with roles in cellular functions (i.e. cell cycle, RNA regulation of transcription, DNA synthesis, and cell organization) showed higher expression in elevated [CO2]. Within this category, most of the genes with higher expression in elevated [CO2] were transcription factors (Table II). While increased expression of transcription factors suggests increased protein synthesis, most transcripts in the protein category (Fig. 5) were involved in protein degradation. These included ubiquitin-specific proteases, Cys proteinases, and different proteosome subunits (Table II). Therefore, we might hypothesize that growth at elevated [CO2] accelerates protein turnover. Other categories where genes were differentially expressed in elevated [CO2] included nitrogen (N) metabolism, hormone metabolism, secondary metabolism (in particular lignin biosynthesis), and transport (Fig. 5; Table II).
Figure 5.
Categorical distribution of genes showing differential expression under elevated [CO2].
Growth of soybeans at elevated [CO2] stimulates photosynthesis during the day and results in marked and significant accumulations of soluble carbohydrates and starch at the end of the photoperiod (Fig. 3, A and C; Rogers et al., 2004). Experiments on the day preceding sampling for microarray analysis showed that soybeans grown at elevated [CO2] had increased photosynthesis and a marked end-of-day accumulation of soluble carbohydrates and starch (Fig. 3C). However, measurements of carbohydrate content made in the middle of the night showed no effect of elevated [CO2] (Fig. 3, B and D), suggesting that plants at elevated [CO2] were more rapidly utilizing the accumulated carbohydrate. This period of rapid carbohydrate utilization between dusk and the middle of the night coincided with the time of maximum leaf expansion in young leaves (Ainsworth et al., 2005). Therefore, increased carbohydrate utilization at elevated [CO2] may provide more energy and biochemical precursors to fuel leaf expansion.
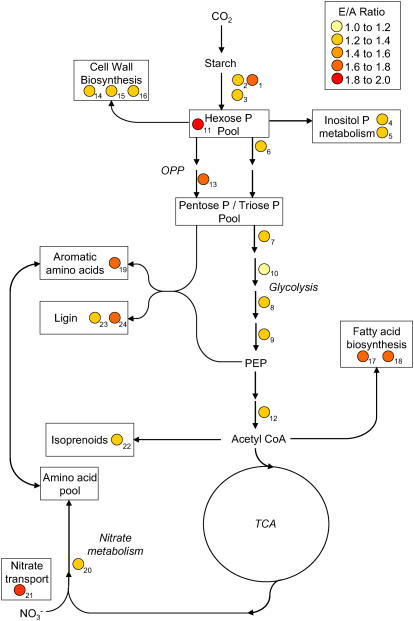
The transcripts of genes encoding enzymes of central C metabolism support this hypothesis. Figure 6 depicts a representation of central C metabolism, annotated with the steps where transcript levels indicated that they were up-regulated at elevated [CO2] (Heldt, 1997; Dennis and Blakeley, 2000). There were increased levels of β-amylase, an exoamylase involved in starch degradation (Fig. 6, nos. 1–3; see also Table II). Markedly increased levels of phosphoglucomutase (Fig. 6, no. 11; see also Table II) suggest that the hexose phosphate pool was larger in leaves grown at elevated [CO2], consistent with an increased availability of hexose from starch degradation. Moving downstream from the hexose phosphate pool, there were increased transcript levels of phosphofructokinase, the first committed step in glycolysis (Fig. 6, no. 6; see also Table II), and all of the enzymes required to make phosphoenolpyruvate from glyceraldehyde-3-P (Fig. 6, Pentose P/Triose P Pool ). Elevated [CO2] also increased transcript levels of a putative pyruvate dehydrogenase (Fig. 6, no. 12; see also Table II), which provides acetyl-CoA for the tricarboxylic acid (TCA) cycle. There was also a marked increase in phosphogluconate dehydrogenase (Fig. 6, no. 13; see also Table II) at elevated [CO2]. This enzyme is part of the oxidative pentose phosphate (OPP) pathway that is used to provide reductant for biosynthesis and pentose phosphates for nucleotide and nucleic acid biosynthesis. In short, elevated [CO2] clearly increased the transcript levels of genes encoding enzymes of glycolysis, entry to the TCA cycle, and the OPP pathway. Davey et al. (2004) found that long-term growth at elevated [CO2] led to a stimulation of foliar respiration. These data suggest one potential explanation for this observation, i.e. increased flux through glycolysis and the TCA cycle fueled by higher substrate availability (starch) at elevated [CO2]. Figure 6 and Table II also illustrate that a significant proportion of the C presumably flowing through the glycolytic pathway was diverted into secondary metabolism, in particular, cell wall, lignin, and fatty acid biosynthesis. Increases in transcripts associated with enzymes in inositol phosphate and isoprenoid biosynthesis also increased at elevated [CO2] (Fig. 6; Table II).
Figure 6.
Graphical representation of selected gene transcripts up-regulated in response to growth at elevated [CO2]. The arrows and boxes indicate metabolic steps, pathways, or metabolite pools in central C and N metabolism. The colored dots indicate that a gene encoding an enzyme for that step or pathway is significantly up-regulated at elevated [CO2]. The color of the dot indicates the degree of up-regulation at elevated [CO2] relative to ambient [CO2] controls (E to A ratio): the darker the color, the greater the up-regulation (see insert). Each spot signifies an individual gene and is coded to the complete list of clones in Table II.
While developing leaves at both ambient and elevated [CO2] had similarly high relative leaf expansion rates, mature leaves reach a larger final area at elevated [CO2] (Fig. 2). Data at the transcript level are consistent with the carbohydrate data (Fig. 3) and provide further evidence that biochemical precursors and energy from soluble carbohydrate and starch degradation may stimulate increased leaf growth and area at elevated [CO2]. Carbon from carbohydrate and starch degradation may be used along with other substrates to produce cell walls and phospholipid membranes. This was supported by the increased transcript levels of genes associated with fatty acid biosynthesis and desaturases (Fig. 6; Table II).
Soybeans get most of their N through their association with N-fixing bacteria (Ritchie et al., 1997). However, early in the season when N fixation is low, plants can be N limited and this is exacerbated at elevated [CO2] (Rogers et al., 2006). We made our measurements early in the season when soybeans at elevated [CO2] are N limited and may be more dependent on soil-borne nitrate than fixed N. Some transcript levels of genes associated with nitrate transport and assimilation (Fig. 6, nos. 20 and 21; see also Table II) were increased at elevated [CO2], and a transcript associated with aromatic amino acid biosynthesis (Fig. 6, no. 19; see also Table II) was also increased at elevated [CO2]. The reduced levels of transcripts for protein synthesis and increased levels for protein degradation, cell wall biosynthesis, lignin, and fatty acid production suggest that there may be a shift away from N-rich proteins to biosynthetic products with higher C to N ratios occurring at elevated [CO2]. This is supported by the increased levels of the OPP enzyme phosphogluconate dehydrogenase, indicating that, at elevated [CO2], C is being utilized for biosynthesis rather than simply increased energy production.
CONCLUSION
In this field study, we investigated the transcriptome response of soybean to elevated [CO2] in growing and fully expanded leaves. We tested the hypothesis that increased C assimilation in plants grown at elevated [CO2] altered pools of carbohydrates and transcripts that control growth and expansion of young leaves. It is well established that elevated [CO2] increases photosynthetic C fixation and carbohydrate synthesis (Long et al., 2004); however, this research suggests that at the transcript level, elevated [CO2] also stimulates the respiratory breakdown of carbohydrates, which likely provides increased fuel for leaf expansion and growth at elevated [CO2].
MATERIALS AND METHODS
Experimental Site
Soybeans (Glycine max cv 93B15; Pioneer Hi-Bred) were grown at the SoyFACE facility, located in Champaign, IL (40°02′N, 88°14′W, 228 m above sea level). SoyFACE was established on a tile-drained field that has been in continuous cultivation for more than 100 years. The 32-ha site has organically rich Flanagan/Drummer series soil. Following standard agronomic practice in the region, no fertilizer was applied. The crop was planted on May 28, 2004, and measurements were made on July 8, 2004, when the crop was in the vegetative growth phase (Ritchie et al., 1997). The experiment consisted of four blocks, each containing two 20-m-diameter octagonal plots. One plot was fumigated from sunrise to sunset to an elevated target [CO2] of 550 μmol mol−1, using the FACE design of Miglietta et al. (2001); the other plot provided a current ambient [CO2] control (375 μmol mol−1). In 2004, the actual elevated [CO2] averaged across the growing season was 550 μmol mol−1. One-minute averages of [CO2] within the plots were within ±20% of the 550 μmol mol−1 target 93% of the time (T. Mies, personal communication).
Leaf Growth
The length of T4 and T6 lateral leaflets was tracked with a ruler (±0.1 cm) approximately every other day from initiation of T4 until sampling of both developmental stages on July 7, 2004. Growth of 12 leaflets on six randomly selected plants per plot was followed. Leaf development in field-grown plants was similar to leaf development of plants raised in growth chambers, where a homogeneous distribution of growth along the leaf blade and a distinct diurnal rhythm of expansion were described for leaflets of a similar developmental stage (Ainsworth et al., 2005).
Leaf Carbohydrates
Leaf discs from T4 and T6 middle leaflets of three plants within each plot were sampled for analysis of carbohydrates between 1 and 2 am on July 8, 2004. Therefore, 12 leaflets per developmental stage and [CO2] treatment were sampled. Each disc (approximately 1.8 cm2) was removed from a vein-free area of a middle leaflet, wrapped in foil, and plunged immediately into liquid N. Samples were lyophilized prior to analysis.
Individual leaf discs were powdered in liquid N. Foliar contents of carbohydrates were extracted from ground leaf tissue in 80% (v/v), buffered (2 mm HEPES, pH 7.8) ethanol at 80°C. Four 20-min incubations were needed to recover the soluble carbohydrates. Glc, Fru, and Suc were determined using a continuous enzymatic substrate assay (Jones et al., 1977). For starch determination, pellets of the ethanol extraction were solubilized by heating to 95°C in 0.1 m NaOH. The NaOH solution was then acidified to pH 4.9, and starch content was determined as Glc equivalents (Hendriks et al., 2003). For the comparison of carbohydrates, a mixed-model ANOVA was performed with trifoliate and CO2 treatment as fixed effects and block as a random effect (SAS Institute).
Microarray Analysis
T4 and T6 lateral leaflets from 12 individual soybeans within each plot were harvested between 1 and 2 am. Entire leaflets were cut, wrapped in foil, plunged immediately into liquid N, and then lyophilized (Multi-Dry Lyophilizer; FTS Systems) and stored at −20°C. Total RNA was extracted from six pooled freeze-dried leaflets from each plot and developmental stage using a SDS/phenol chloroform method and lithium chloride precipitation (Wang and Vodkin, 1994). RNA content was quantified by spectrophotometry, and the integrity was confirmed using agarose gel electrophoresis (Sambrook et al., 1989). RNA was further purified using RNeasy columns (Qiagen) according to the manufacturer's instructions. Prior to labeling, purified RNA was concentrated in a Speed Vac (Savant Instruments). The cDNA synthesis, probe labeling, hybridization conditions, and slide scanning followed Vodkin et al. (2004). Microarrays from two reracked libraries, Gm-1070 and Gm-1088, were probed. The experimental design for the microarray experiment is illustrated in Figure 1. Three of the four experimental blocks in the FACE experiment were used. Each double-headed arrow represents four arrays per library, two biological samples of RNA (from six pooled leaflets), and the dye swaps (technical replicates). Therefore, a total of 96 separate hybridizations were made.
Spot intensities were quantified using Imagene 6.1 (Biodiscovery). The local background was subtracted for each spot, and spots were normalized to the median intensity of each dye on each slide. The natural log of the background-corrected median signal was used for all statistical analyses. Spots flagged by the Imagene image analysis software were removed from subsequent analyses (Prakash and Petrov, 2004). Reliability of the data was evaluated with Pearson correlation coefficients and kappa statistics on pairwise comparisons of arrays. Five slides from library Gm-1070 and three slides from library Gm-1088 had low-weighted kappa values (<0.50) and were dropped from the analysis. Gm-1070 contained 9,216 cDNA clones from various developmental stages of immature cotyledons, flowers, pods, and seed coats, and Gm-1088 contained 9,216 cDNA clones from cotyledons and hypocotyls of germinating seedlings and other plant parts subjected to various pathogens or environmental stress conditions (Vodkin et al., 2004). Transcripts that had missing data points on more than 20% of the arrays were also dropped from the analysis. Therefore, 5,314 transcripts from library Gm-1070 and 5,831 transcripts from library Gm-1088 were included in the analysis of variance.
Biological and technical replications were averaged for each plot for statistical analysis. A mixed-model ANOVA was performed, with trifoliate and CO2 treatment as fixed effects and block as a random effect. The model was tested for conformation to the assumption of normality of the residuals using the Shapiro-Wilkes Test. A Bonferroni significance level was used as an initial criterion for rejecting the null hypothesis of a significant treatment effect (0.05/5,314 for Gm-1070 and 0.05/5,831 for Gm-1088). No genes were significant at the Bonferroni level, so we used a second nominal threshold of α < 0.05 because type I and II errors are inversely related and because Bonferroni correlation is overly conservative (Kerr and Churchill, 2001; Wayne and McIntyre, 2002). If no evidence for departure from normality of the residuals was evident and the P value for the test of differences was ≤0.05, the gene was considered significant, following the methods of Li et al. (2004). All analyses were performed in SAS. Reproducibility of the hybridizations and degree of variation between technical and biological replicates and experimental blocks in the field are illustrated in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Steve Long and Tim Mies for management and maintenance of the SoyFACE facility. We thank L. McIntyre for assistance with statistical analysis of the experiment and insightful comments on an early draft of the manuscript. We thank R. Knepp, K. Gillespie, A.M. Boone, and S.I. Jones for technical help with RNA extractions, microarray protocols, and bioinformatics.
This work was supported by the Illinois Council for Food and Agricultural Research, by the Archer Daniels Midland Company, and by the U.S. Department of Agriculture/Agricultural Research Service. E.A.A. was supported by an Alexander von Humboldt postdoctoral research fellowship. A.R. was supported by the U.S. Department of Energy Office of Science contract no. DE–AC02–98CH10886 to Brookhaven National Laboratory.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Elizabeth A. Ainsworth (ainswort@uiuc.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abe T, Thitamadee S, Hashimoto T (2004) Microtubule defects and cell morphogenesis in the lefty1lefty2 tubulin mutant of Arabidopsis thaliana. Plant Cell Physiol 45: 211–220 [DOI] [PubMed] [Google Scholar]
- Ainsworth EA, Davey PA, Bernacchi CJ, Dermody OC, Heaton EA, Moore DJ, Morgan PB, Naidu SL, Ra HSY, Zhu XG, et al (2002) A meta-analysis of elevated [CO2] effects on soybean (Glycine max) physiology, growth and yield. Glob Change Biol 8: 695–709 [Google Scholar]
- Ainsworth EA, Walter A, Schurr U (2005) Glycine max leaflets lack a base-tip gradient in growth rate. J Plant Res 118: 343–346 [DOI] [PubMed] [Google Scholar]
- Avery GS (1933) Structure and development of the tobacco leaf. Am J Bot 20: 565–592 [Google Scholar]
- Bernacchi CJ, Leakey ADB, Heady LE, Morgan PB, Rogers A, Long SP, Ort DR (2006) Hourly and seasonal variation in photosynthesis and stomatal conductance of soybean grown at future CO2 and ozone concentrations for three years under fully open air conditions. Plant Cell Environ doi/10.1111/j.1365-3040.2006.01581.x [DOI] [PubMed]
- Bunce JA (1977) Leaf elongation in relation to leaf water potential in soybean. J Exp Bot 28: 156–161 [Google Scholar]
- Chatterjee M, Sparvoli S, Edmunds C, Garosi P, Finlay K, Martin C (1996) DAG, a gene required for chloroplast differentiation and palisade development in Antirrhinum majus. EMBO J 15: 4194–4207 [PMC free article] [PubMed] [Google Scholar]
- Citterio S, Piatti S, Albertini E, Aina R, Varotta S, Barcaccia G (2006) Alfalfa Mob1-like proteins are involved in cell proliferation and are localized in the cell division plane during cytokinesis. Exp Cell Res 312: 1050–1064 [DOI] [PubMed] [Google Scholar]
- Davey PA, Hunt S, Hymus GJ, DeLucia EH, Drake BG, Karnosky DF, Long SP (2004) Respiratory oxygen uptake is not decreased by an instantaneous elevation of [CO2], but is increased with long-term growth in the field at elevated CO2. Plant Physiol 134: 520–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis DT, Blakeley SD (2000) Carbohydrate metabolism. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 631–675
- Dermody O, Long SP, Delucia EH (2006) How does elevated CO2 or ozone affect the leaf-area index of soybean when applied independently? New Phytol 169: 145–155 [DOI] [PubMed] [Google Scholar]
- Druart N, Rodríguez-Buey M, Barron-Gafford G, Sjödin A, Bhalerao R, Hurry V (2006) Molecular targets of elevated [CO2] in leaves and stems of Populus deltoides: implications for future tree growth and carbon sequestration. Funct Plant Biol 33: 121–131 [DOI] [PubMed] [Google Scholar]
- Ferris R, Sabatti M, Miglietta F, Mills RF, Taylor G (2001) Leaf area is stimulated in Populus by free air CO2 enrichment (POPFACE), through increased cell expansion and production. Plant Cell Environ 24: 305–315 [Google Scholar]
- Gupta P, Duplessis S, White H, Karnosky DF, Martin F, Podila GK (2005) Gene expression patterns of trembling aspen trees following long-term exposure to interacting elevated CO2 and tropospheric O3. New Phytol 167: 129–142 [DOI] [PubMed] [Google Scholar]
- Heldt HW (1997) Plant Biochemistry and Molecular Biology. Oxford University Press, New York
- Hendriks JHM, Kolbe A, Gibon Y, Stitt M, Geigenberger P (2003) ADP-glucosepyro-phosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinski A, Rowley DL, Loer DS, Foley C, Buta G, Herman EM (1995) Binding-protein expression is similar to temporal, developmental and stress-induced regulation in terminally differentiated soybean organs. Planta 195: 611–621 [DOI] [PubMed] [Google Scholar]
- Kerr MK, Churchill GA (2001) Statistical design and the analysis of gene expression microarray data. Genome Res 77: 123–128 [DOI] [PubMed] [Google Scholar]
- Jones MGK, Outlaw WH, Lowry OH (1977) Enzymic assay of 10−7 to 10−14 moles of sucrose in plant tissues. Plant Physiol 60: 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Singh AK, McIntyre LM, Sherman LA (2004) Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp. strain PCC 6803. J Bacteriol 186: 3331–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SP, Ainsworth EA, Leakey ADB, Nosberger J, Ort DR (2006) Food for thought: lower-than-expected crop yield stimulation with rising CO2 concentrations. Science 312: 1918–1921 [DOI] [PubMed] [Google Scholar]
- Long SP, Ainsworth EA, Rogers A, Ort DR (2004) Rising atmospheric carbon dioxide: plants FACE the future. Annu Rev Plant Biol 55: 591–628 [DOI] [PubMed] [Google Scholar]
- Matsubara S, Hurry V, Druart N, Benedict C, Janzik I, Chavarría-Krauser A, Walter A, Schurr U (2005) Nocturnal changes in leaf growth of Populus deltoides are controlled by cytoplasmic growth. Planta 223: 1315–1328 [DOI] [PubMed] [Google Scholar]
- Miglietta F, Peressotti A, Vaccari FP, Zaldei A, de Angelis P, Scarascia-Mugnozza G (2001) Free-air CO2 enrichment (FACE) of a poplar plantation: the POPFACE fumigation system. New Phytol 150: 465–476 [Google Scholar]
- Morgan PB, Bollero GA, Nelson RL, Dohleman FG, Long SP (2005) Smaller than predicted increase in aboveground net primary production and yield of field-grown soybean under fully open-air [CO2] elevation. Glob Change Biol 11: 1856–1865 [Google Scholar]
- Ort DR, Ainsworth EA, Aldea M, Allen DJ, Bernacchi CJ, Berenbaum MR, Bollero GA, Cornic G, Davey PA, Dermody O, et al (2006) SoyFACE: the effects and interactions of elevated [CO2] and [O3] on soybean. In J Nösberger, SP Long, RJ Norby, M Stitt, GR Hendrey, H Blum, eds, Managed Ecosystems and CO2: Case Studies, Processes and Perspectives. Springer, Berlin, pp 71–85
- Prakash PJ, Petrov A (2004) Gene Flagging in ImaGene. Biodiscover Inc. Technical Bulletin. Biodiscover Inc., El Segundo, CA, pp 1–7
- Prentice IC, Farquhar GD, Fasham MJR, Goulden ML, Heimann M, Jaramillo VJ, Kheshgi HS, LeQuere C, Scholes RJ, Wallace DWR, et al (2001) The carbon cycle atmospheric carbon dioxide. In JT Houghton, Y Ding, DJ Griggs, M Noguer, PJ Van der Linder, X Dai, K Maskell, CA Johnson, eds, Climate Change 2001: The Scientific Basis. Contributions of Working Group I to the Third Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, New York, pp 183–239
- Raines CA, Paul MJ (2006) Products of leaf primary carbon metabolism modulate the developmental programme determining plant morphology. J Exp Bot 57: 1857–1862 [DOI] [PubMed] [Google Scholar]
- Ritchie SW, Hanaway JJ, Thompson HE, Benson GO (1997) How a Soybean Plant Develops. Special Report Number 53. Iowa State University, Ames, IA
- Rogers A, Allen DJ, Davey PA, Morgan PB, Ainsworth EA, Bernacchi CJ, Cornic G, Dermody O, Dohleman FG, Heaton EA, et al (2004) Leaf photosynthesis and carbohydrate dynamics of soybeans grown throughout their life-cycle under Free-Air Carbon dioxide Enrichment. Plant Cell Environ 27: 449–458 [Google Scholar]
- Rogers A, Gibon Y, Stitt M, Morgan PB, Bernacchi CJ, Ort DR, Long SP (2006) Increased carbon availability at elevated carbon dioxide concentration improves N assimilation in a legume. Plant Cell Environ 29: 1651–1658 [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier TM, Sturn A, et al (2003) TM4: a free, open-source system for microarray data management analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schurr U, Walter A, Rascher U (2006) Functional dynamics of plant growth and photosynthesis—from steady-state to dynamics—from homogeneity to heterogeneity. Plant Cell Environ 29: 340–352 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC, Smith SM (2005) Starch degradation. Annu Rev Plant Biol 56: 73–98 [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K (2003) “Big it up”: endoreduplication and cell-size control in plants. Curr Opin Plant Biol 6: 544–553 [DOI] [PubMed] [Google Scholar]
- Taylor G, Ranasinghe S, Bosac C, Gardner SDL, Ferris R (1994) Elevated CO2 and plant growth: cellular mechanisms and responses of whole plants. J Exp Bot 45: 1761–1774 [Google Scholar]
- Taylor G, Street NR, Tricker PJ, Sjödin A, Graham L, Skogström O, Calfapietra C, Scarascia-Mugnozza G, Jansson S (2005) The transcriptome of Populus in elevated CO2. New Phytol 167: 143–154 [DOI] [PubMed] [Google Scholar]
- Taylor G, Tricker PJ, Zhang FZ, Alston VJ, Miglietta F, Kuzminsky E (2003) Spatial and temporal effects of free-air CO2 enrichment (POPFACE) on leaf growth, cell expansion, and cell production in a closed canopy of poplar. Plant Physiol 131: 177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BR, Romero GO, Nevins DJ, Rodriguez RL (2000) New perspectives on the endo-beta-glucanases of glycosyl hydrolase Family 17. Int J Biol Macromol 27: 139–144 [DOI] [PubMed] [Google Scholar]
- Trainotti L, Pavanello A, Casadoro G (2004) Differential expression of genes in apical and basal tissues of expanding tobacco leaves. Plant Sci 167: 679–686 [Google Scholar]
- Tricker PJ, Calfapietra C, Kuzminsky E, Puleggi R, Ferris R, Nathoo M, Pleasants LJ, Alston V, de Angelis P, Taylor G (2004) Long-term acclimation of leaf production, development, longevity and quality following 3 yr exposure to free-air CO2 enrichment during canopy closure in Populus. New Phytol 162: 413–426 [Google Scholar]
- Turgeon R (1989) The sink-source transition in leaves. Annu Rev Plant Physiol Plant Mol Biol 40: 119–138 [Google Scholar]
- Vodkin LO, Khanna A, Shealy R, Clough SJ, Gonzalez DO, Philip R, Zabala G, Thibaud-Nissen F, Sidarous M, Stromvik M, et al (2004) Microarrays for global expression constructed with a low redundancy set of 27,500 sequenced cDNAs representing an array of developmental stages and physiological conditions of the soybean plant. BMC Genomics 5: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter A, Christ MM, Barron-Gifford GA, Grieve KA, Murthy R, Rascher U (2005) The effect of elevated CO2 on diel leaf growth cycle, leaf carbohydrate content and canopy growth performance of Populus deltoides. Glob Change Biol 11: 1258–1271 [Google Scholar]
- Wang C-S, Vodkin LO (1994) Extraction of RNA from tissues containing high levels of procyanidins that bind RNA. Plant Mol Biol Rep 12: 132–145 [Google Scholar]
- Wayne ML, McIntyre LM (2002) Combining mapping and arraying: an approach to candidate gene identification. Proc Natl Acad Sci USA 99: 14903–14906 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.