Abstract
Posttranslational protein modification by the small ubiquitin-like modifier (SUMO) is a highly dynamic and reversible process. To analyze the substrate specificity of SUMO-conjugating and -deconjugating enzymes from Arabidopsis (Arabidopsis thaliana), we reconstituted its SUMOylation cascade in vitro and tested the capacity of this system to conjugate the Arabidopsis SUMO isoforms AtSUMO1, 2, and 3 to the model substrate ScPCNA from yeast (Saccharomyces cerevisiae). This protein contains two in vivo SUMOylated lysine residues, namely K127 and K164. Under in vitro conditions, the Arabidopsis SUMOylation system specifically conjugates all tested SUMO isoforms to lysine-127, but not to lysine-164, of ScPCNA. The SUMO isoforms AtSUMO1 and AtSUMO2, but not AtSUMO3, were found to form polymeric chains on ScPCNA due to a self-SUMOylation process. In a complementary approach, we analyzed both the SUMO isopeptidase activity and the pre-SUMO-processing capacity of the putative Arabidopsis SUMO proteases At1g60220, At1g10570, and At5g60190 using the known SUMO isopeptidases ScULP1, XopD, and ESD4 (At4g15880) as reference enzymes. Interestingly, At5g60190 exhibits no SUMO protease activity but processes the pre-form of Arabidopsis Rub1. The other five enzymes represent SUMO isopeptidases that show different substrate preferences. All these enzymes cleave AtSUMO1 and AtSUMO2 conjugates of ScPCNA, whereas only the putative bacterial virulence factor XopD is able to release AtSUMO3. In addition, all five enzymes cleave pre-AtSUMO1 and pre-AtSUMO2 peptides, but none of the proteins efficiently produce mature AtSUMO3 or AtSUMO5 molecules from their precursors.
Posttranslational protein modifications enable cells to rapidly and specifically respond to endogenous or exogenous stimuli, avoiding time- and energy-consuming de novo protein synthesis. In contrast to small molecules attached to proteins as methyl, phosphate, or acetyl groups, polypeptide-like modifiers themselves are encoded in the genome and are therefore targets of duplication and diversification during evolution. Consequently, in addition to ubiquitin, several other ubiquitin-like polypeptide tags, such as Rub1 (related to ubiquitin) and SUMO (small ubiquitin-like modifier), have been identified in eukaryotic organisms (Melchior, 2000; Schwartz and Hochstrasser, 2003; Dohmen, 2004). Among the ubiquitin-like modifiers, Rub1 (Nedd8) shows the highest primary sequence similarity with ubiquitin (Schwartz and Hochstrasser, 2003). Rub1 is covalently linked to a conserved lysine residue present in the C-terminal region of most cullins through a series of steps similar to ubiquitin conjugation (Willems et al., 2004). Cullin proteins can function as subunits of the SCF (SKP1, Cullin, F-box) class of ubiquitin E3 ligases and represent the only known physiological substrates of Rub1 (Hellmann and Estelle, 2002; Willems et al., 2004). Rub1 conjugation (Neddylation) blocks the interaction with a protein called Cand1 (cullin-associated and Neddylation-dissociated) and thereby stabilizes the active SCF complex (Liu et al., 2002; J. Zheng et al., 2002; N. Zheng et al., 2002). In Arabidopsis (Arabidopsis thaliana), Neddylation of CUL1 is necessary for the normal function of SCFTIR1, an E3 ligase required for the response to the plant hormone auxin, and mutant plants deficient in Rub1 conjugation have an embryonic phenotype similar to auxin signaling mutants (Dharmasiri et al., 2003). Neddylation is a dynamic process that can be reversed by a metalloprotease activity associated with the Csn5 subunit of the Cop9 signalosome, a multiprotein complex that resembles the proteasome lid and is involved in various processes, including photomorphogenesis (Lyapina et al., 2001; Cope et al., 2002; Wei and Deng, 2003).
SUMO and ubiquitin show very similar three-dimensional structures but share only about 18% identical amino acids, and, consequently, the distribution of charged residues on the surface of both proteins differs (Bayer et al., 1998; Huang et al., 2004). SUMO is synthesized as a pre-protein that has to be processed by SUMO proteases to expose a C-terminal diglycine motif (Johnson, 2004). The mature SUMO molecule is attached to target proteins via an isopeptide linkage formed between the free carboxyl group of its C-terminal glycine residue and the ɛ-amino group of a lysine residue within the substrate protein. This ATP-dependent reaction is catalyzed by subsequent action of a SUMO-activating E1 enzyme and a SUMO-conjugating E2 enzyme. The latter is designated Ubc9 (Melchior, 2000; Dohmen, 2004). The E1 enzyme is a heteromeric complex consisting of a small SAE1 (Aos1) and a large SAE2 (Uba2) subunit (Johnson, 2004; Lois and Lima, 2005). This complex catalyzes both the formation of a SUMO adenylate as well as the transfer of this activated intermediate to one of its own cysteine sulfhydryl groups, thereby forming a thioester linkage. Subsequently, SUMO is attached to a cysteine residue of the E2 enzyme via isoenergetic thioester transfer (Johnson, 2004; Lois and Lima, 2005). In contrast to most of the ubiquitin conjugation systems, which depend on specificity-conferring E3 ligases, the SUMO E2 enzyme can directly bind and SUMOylate its substrates in vitro. Under these conditions, substrate specificity is defined by the capacity of the E2 enzyme to recognize the consensus motif ΨKxE/D (Ψ, hydrophobic amino acid; K, SUMO target lysine; D/E acidic amino acids) if this peptide stretch is exposed to the surface of the target protein (Melchior, 2000; Bernier-Villamor et al., 2002). However, their are cases in which lysine residues in nonconsensus regions are specifically modified, and recent data indicate that both secondary structure elements in the target protein itself and specificity-conferring SUMO E3 ligases can promote the SUMOylation of nonconsensus lysine residues (Hoege et al., 2002; Pfander et al., 2005; Pichler et al., 2005). SUMO E3 ligases known to date comprise members of the PIAS (protein inhibitor of activated STAT) family, including Saccharomyces cerevisiae ScSiz1 and ScSiz2, as well as Arabidopsis AtSiz1, the vertebrate nuclear pore protein RanBP2, and the polycomb group protein Pc2 (Johnson and Gupta, 2001; Pichler et al., 2002; Kagey et al., 2005; Miura et al., 2005). However, to our knowledge, only ScSiz1 has been shown to alter SUMO E2 enzyme specificity in a way that allows the recognition of nonconsensus SUMOylation motifs (Pfander et al., 2005).
SUMOylation influences stability, interaction, cellular localization, and activity of proteins and thereby regulates processes such as DNA repair (Hoege et al., 2002; Pfander et al., 2005), nuclear transport (Stade et al., 2002), transcriptional regulation (Ross et al., 2002; Sapetschnig et al., 2002), chromosome segregation (Nacerddine et al., 2005), and ion channel activity (Rajan et al., 2005). There are examples for the alternative modification of a single lysine residue by both SUMO and ubiquitin, and consequently it has been suggested that one mechanism of SUMO action in biological systems is to antagonize the effect of protein ubiquitinylation (Desterro et al., 1998; Hoege et al. 2002; Johnson, 2004). In the case of the inhibitory protein IκBα, which sequesters NF-κB in the cytoplasm, the lysine residue K21 serves as an attachment site for both SUMO and ubiquitin, and in fact SUMOylated IκBα is protected from degradation by the proteasome (Desterro et al., 1998; Ulrich, 2005). The yeast DNA polymerase processivity factor ScPCNA, which is involved in silencing, replication, and postreplicative DNA repair, can be modified by SUMO and ubiquitin at Lys-164, and it has been postulated that SUMOylation of ScPCNA inhibits ubiquitin-dependent DNA damage tolerance (Hoege et al., 2002). However, recent data indicate a more subtle scenario for the regulation of ScPCNA function with SUMO and ubiquitin acting in a cooperative manner rather than as antagonists (Pfander et al., 2005; Ulrich, 2005). Intriguingly, several transcription factors, such as Sp3, c-Jun, and c-Myb, are modified by SUMO, and in most cases this modification attenuates their activity or even induces a switch from an activator to a repressor function (Gill, 2003, 2005).
SUMOylation is a reversible and highly dynamic modification, and in several systems the continued cycling between SUMO conjugation and deconjugation seems to be more relevant than the actual steady-state level of the SUMOylated target protein (Johnson, 2004). In fact, regulation of protein function by repetitive cycles of SUMO modification has been observed with the mammalian plasma membrane ion channel K2P1 and with the excision DNA repair enzyme thymine-DNA glycosylase (Hardeland et al., 2002; Rajan et al., 2005).
Modification by SUMO is reversed by a class of cysteine proteases that shows no significant similarity with de-ubiquitinating enzymes but is distantly related to certain viral proteins (Li and Hochstrasser, 1999, 2000). S. cerevisiae contains the two SUMO isopeptidases ScULP1 and ScULP2 that execute distinct biological functions (Li and Hochstrasser, 1999, 2000, 2003). The ScULP1 protein is localized at the nuclear envelope and ScULP1 null mutants are not viable due to a defect in cell cycle progression (Li and Hochstrasser, 2003). In contrast, ScULP2 was detected within the nucleus, and ScULP2 mutants are viable but exhibit pleiotropic phenotypes, including abnormal cell morphology and decreased chromosome stability (Li and Hochstrasser, 2000). In mammals, the functional specialization of SUMO isopeptidases seems to be greater since multiple members of the gene family are encoded in these genomes, and, in at least one case, multiple peptidases with distinct subcellular localization are generated from a single gene by alternative mRNA splicing (Johnson, 2004). Interestingly, most SUMO proteases exhibit not only isopeptidase activity that recycles SUMO from conjugates, but also show endopeptidase activity that generates the mature form of SUMO from its precursor (Johnson, 2004).
Although plants seem to have a complex SUMOylation system with eight SUMO isoforms and several SUMO isopeptidases predicted for Arabidopsis, no in vivo SUMOylated plant protein has yet been reported, to our knowledge. Nevertheless, there is evidence that SUMO conjugation or deconjugation regulates hormone response, flowering time, phosphate uptake, stress adaptation, and interactions with microorganisms (Hanania et al., 1999; Orth et al., 2000; Hotson et al., 2003; Kurepa et al., 2003; Lois et al., 2003; Miura et al., 2005; Murtas et al., 2003). The conjugation of AtSUMO1 and AtSUMO2, but not AtSUMO3, to unknown target proteins increases in response to stress conditions, particularly H2O2 treatment and heat shock, suggesting a role of selected SUMO isoforms in adaptation to abiotic stress (Kurepa et al., 2003). On the other hand, artificial overexpression of AtSUMO1 and 2 has no adverse effect on plant growth, although increased levels of SUMOylated proteins were detectable (Lois et al., 2003). This increase in SUMOylated proteins was correlated with an attenuation of abscisic acid-mediated growth inhibition, indicating that abscisic acid signaling in Arabidopsis is mediated by SUMOylation (Lois et al., 2003). There are only two mutant phenotypes known so far that are connected to the plant SUMOylation system (Murtas et al., 2003; Miura et al., 2005). Inactivation of the Arabidopsis SUMO E3 ligase AtSiz1 induces phosphate starvation responses, such as anthocyanin accumulation and extensive lateral root and root hair development (Miura et al., 2005). A mutation within a reading frame that shows significant similarity to the yeast SUMO isopeptidase ScULP1 causes early flowering under short-day conditions, and biochemical analysis of the respective Arabidopsis protein confirmed its function as a SUMO peptidase (Murtas et al., 2003).
Although SUMO exists only in eukaryotes, several plant pathogenic and symbiotic bacteria encode ScULP1-like proteins, indicating that some prokaryotes are able to manipulate the SUMOylation system to reprogram host cell functions (Hotson et al., 2003; Hotson and Mudgett, 2004). In this context, it has been shown that the ScULP1-like Xanthomonas campestris protein XopD migrates to the plant nucleus after its injection by a type III secretion system and de-SUMOylates so far uncharacterized nuclear proteins (Hotson et al., 2003).
To obtain insight into the complex interplay between SUMO-conjugating and -deconjugating enzymes in plants, we reconstituted the Arabidopsis SUMOylation cascade in vitro and characterized its enzymatic properties using yeast ScPCNA as a model protein. In a complementary approach, both SUMO isopeptidase activity and pre-SUMO-processing capacity of the putative Arabidopsis SUMO proteases At1g60220, At1g10570, and At5g60190, as well as ScULP1, XopD, and ESD4 (At4g15880), were tested with various model substrates. Thereby, SUMO peptidase activity of the Arabidopsis proteins At1g60220 and At1g10570 was demonstrated. In contrast, At5g60190 exhibits no SUMO protease activity but processes the pre-form of Arabidopsis Rub1. The different capacities of the bona fide SUMO isopeptidases to cleave AtSUMO3-protein conjugates allowed the identification of a substrate quality-determining amino acid position within Arabidopsis AtSUMO3.
RESULTS
Heterologous Expression of the Arabidopsis SUMOylation Cascade
Putative enzymes of the Arabidopsis SUMOylation cascade were identified by sequence comparison with yeast and human homologs as described by others (Kurepa et al., 2003; Novatchkova et al., 2004). In all organisms analyzed so far, the SUMO E1 enzyme is a heterodimer consisting of the SAE1 subunit, which resembles the N terminus of ubiquitin E1 enzymes, and the SAE2 subunit, which corresponds to the C terminus of ubiquitin E1 enzymes and contains the active-site cysteine (Johnson, 2004; Lois and Lima, 2005). Arabidopsis encodes two highly similar proteins, namely, At4g24940 and At5g50680 that show 52% and 44% sequence similarity with yeast Aos1 (ScSAE1), respectively. In contrast, At2g21470, which shows 52% similarity to the yeast subunit Uba2 (ScSAE2), is the only SAE2 candidate protein present in Arabidopsis. We prepared the cDNAs of At5g50680 and At2g21470 from RNA derived from Arabidopsis Columbia (Col-0) wild-type plants, and found that successful expression and isolation of a functional Arabidopsis SUMO E1 complex from Escherichia coli depends on coexpression of both the 36-kD AtSAE1 subunit fused to a C-terminal hexa-His tag and the 70-kD AtSAE2 protein carrying an N-terminal hexa-His tag (for details, see “Materials and Methods”).
In fungi, invertebrates, and probably all vertebrates, the SUMO E2 enzyme Ubc9 is encoded by a single gene (Johnson, 2004). Correspondingly, only one Arabidopsis protein encoded by At3g57870 shows significant similarity (74%) with yeast Ubc9. In fact, At3g57870 has already been shown to have SUMO E2 activity and was used in this study as a hexa-His fusion protein produced in E. coli (Lois et al., 2003).
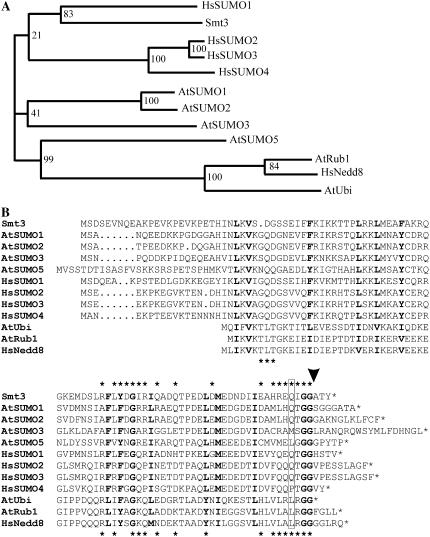
In contrast to fungi and nematodes, which contain only one SUMO variant, Arabidopsis SUMO is encoded by a small gene family that comprises eight members (Kurepa et al., 2003; Johnson, 2004; Novatchkova et al., 2004). For four of the reading frames, expressed sequence tags (ESTs) are available. The corresponding SUMO isoforms, designated AtSUMO1 to 3 and AtSUMO5, were chosen for further analysis (Kurepa et al., 2003). The cDNAs were cloned from Arabidopsis Col-0 leaf RNA, and the SUMO isoforms were expressed in their mature, conjugation-competent form in E. coli as N-terminal hemagglutinin (HA)-hexa-His double-tagged proteins. However, in the case of AtSUMO5, the expression level in E. coli was low, and as the protein could not be purified to homogeneity, this isoform was omitted from most of the experiments described below. AtSUMO1 and 2 are closely related and share 93% conserved amino acid residues, whereas AtSUMO3 shows only 53% conserved positions when compared with AtSUMO1, although it represents a putative gene duplication of AtSUMO2. With only 44% sequence conservation compared to AtSUMO1, AtSUMO5 represents the most divergent Arabidopsis SUMO isoform included in this study (Fig. 1A). As found with Arabidopsis, mammals express four SUMO isoforms (Bohren et al., 2004; Johnson, 2004). These SUMO variants can be distinguished both due to their capacity to form multimeric structures and due to their capacity to function as substrates for selected proteases (Saitoh and Hinchey, 2000; Tatham et al., 2001; Owerbach et al., 2005). However, phylogenetic analysis reveals no special relationship between selected SUMO isoforms from Arabidopsis and human, and therefore allows no prediction of the properties of AtSUMO1 to 3 and AtSUMO5 (Fig. 1A). Figure 1B shows a sequence comparison between Smt3 (ScSUMO), the four expressed Arabidopsis SUMO isoforms, the four SUMO isoforms from human, Arabidopsis ubiquitin, as well as AtRub1 and HsNedd8 before maturation to their conjugation-competent forms, and highlights the residues of Smt3 and HsNedd8 that are known interact with the proteases ScULP1 and Den1, respectively (Fig. 1B; Mossessova and Lima, 2000; Reverter et al., 2005). Interestingly, AtRub1 (At1g31340) is expressed as a naturally occurring ubiquitin-pre-AtRub1 protein fusion and therefore allows us to prove the substrate specificity of putative SUMO proteases.
Figure 1.
Phylogenetic analysis and primary sequence alignment of SUMO and Nedd8 precursors from S. cerevisiae, Arabidopsis, and mammals. A, Proteins were grouped into a phylogram using the ClustalW server at the University of Wageningen (www.bioinformatics.nl/tools/clustalw.html) with standard settings. Bootstrap values (1,000 replicates) are shown at the branches. B, Amino acid residues conserved in all aligned sequences are depicted in bold letters. The C-terminal diglycine motif that is exposed during prepeptide processing is highlighted in bold letters, and the predicted cleavage site is marked by an arrowhead. A glutamine residue known to be important for the interaction of HsSUMO1 with the protease Senp2 is boxed. Asterisks depicted above and below the alignment indicate either Smt3 or HsNedd8 residues that are known to be in direct contact with the proteases ScULP1 or Den1, respectively.
In Vitro Reconstitution and Characterization of the Arabidopsis SUMOylation Cascade
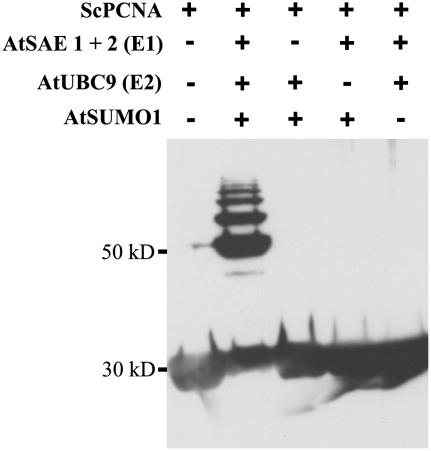
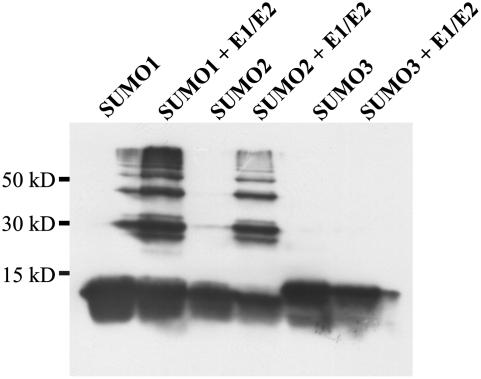
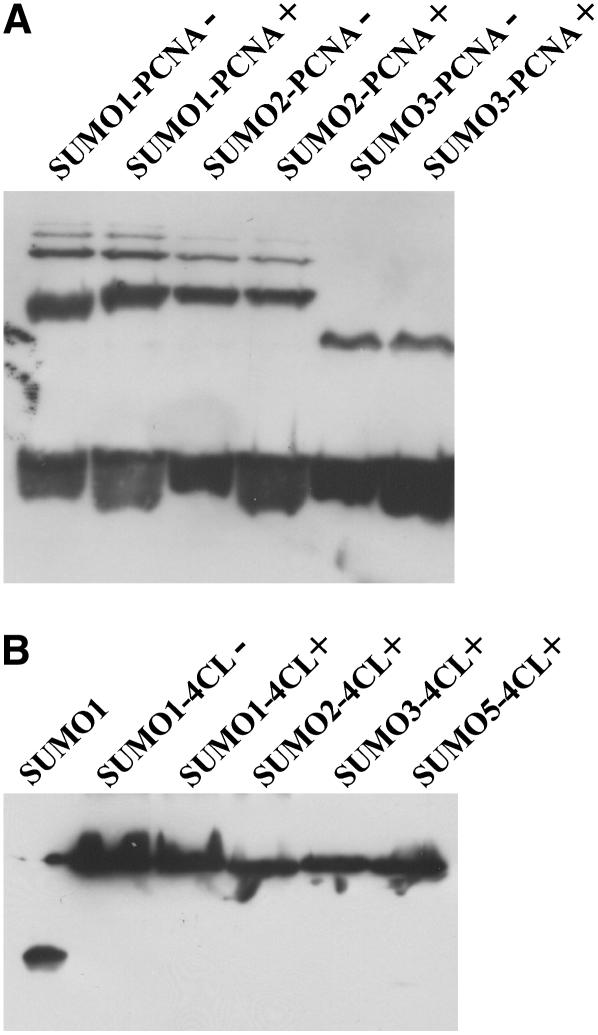
Yeast PCNA was selected as a model substrate to analyze both general enzymatic activity of the heterologously expressed Arabidopsis SUMOylation pathway components and their specificity with respect to SUMO isoform usage and modification of consensus and nonconsensus SUMOylation motifs. ScPCNA is a DNA polymerase processivity factor that contains 18 lysine residues, five of which are part of predicted SUMOylation consensus sites (www.abgent.com/doc/sumoplot). It is known that in vivo ScPCNA is SUMOylated at Lys-164, which is not part of a predicted, conserved SUMOylation motif. Additionally, ScPCNA is modified at Lys-127, a residue located within a perfect SUMOylation consensus (Hoege et al., 2002). This combination of predicted and experimentally verified SUMOylation sites makes ScPCNA a suitable model substrate to analyze both the catalytic capacity and specificity of the in vitro reconstituted Arabidopsis SUMOylation system. ScPCNA was expressed in E. coli with a Myc tag fused to its N terminus and a hexa-His tag fused to its C-terminal end. The latter modification was used for affinity purification and the Myc tag for western-blot detection of ScPCNA mobility shifts as observed after incubation with the SUMOylation cascade components (Fig. 2). When ScPCNA was incubated with the putative Arabidopsis E1 complex, AtUbc9, AtSUMO1, and Mg2+ATP, a ladder of strong modification signals was detectable (Fig. 2). The appearance of high-Mr ScPCNA species strictly depends on the presence of all the above-listed components, indicating that ScPCNA was covalently modified by AtSUMO1 and that a protein complex consisting of At5g50690 and At2g21470 in fact functions as a SUMO-activating E1 enzyme (Fig. 2). ScPCNA was also incubated with AtSUMO2, AtSUMO3, and two ubiquitin variants (Fig. 3). No conjugate formation was observed when SUMO was replaced by ubiquitin, demonstrating the specificity of the in vitro reconstituted SUMOylation cascade. As observed with AtSUMO1, AtSUMO2 and AtSUMO3 are attached to ScPCNA, although the modification pattern found with AtSUMO3 was distinguishable from that observed with the two other SUMO isoforms (Fig. 3). It is worth noting that, although SUMO has a molecular mass of approximately 11 kD, mobility shifts of SUMOylated proteins in SDS-PAGE are somewhat difficult to predict and are commonly found to range from 15 to 20 kD (Johnson, 2004). Therefore, the majority of the AtSUMO3 molecules are conjugated to a single lysine residue, whereas the modification pattern obtained after incubation with AtSUMO1 and AtSUMO2 suggests that ScPCNA is either modified at multiple sites or that AtSUMO1 and AtSUMO2 form polymeric structures due to a self-SUMOylation process (Fig. 3).
Figure 2.
SUMOylation of ScPCNA catalyzed by the Arabidopsis SUMOylation cascade. ScPCNA labeled with a Myc tag was isolated from E. coli and directly applied to the gel (lane 1), or incubated with AtSUMO1 in the presences of Arabidopsis SUMO E1 and E2 enzymes (lane 2). In lanes 3 to 5, selected components of the Arabidopsis SUMOylation machinery, as listed in the top part of the figure, were excluded from the reaction mixture. Proteins were separated on a 12% SDS-polyacrylamide gel, and ScPCNA mobility shifts were detected by western-blot analysis using a Myc peptide-specific antiserum.
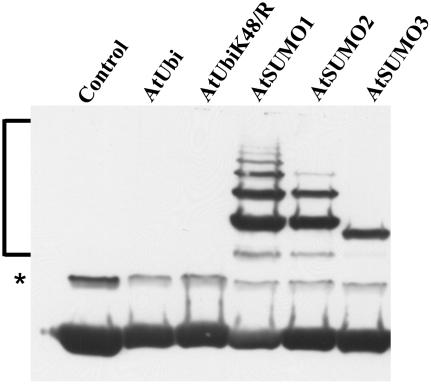
Figure 3.
Conjugation of different Arabidopsis SUMO isoforms to ScPCNA. ScPCNA labeled with a Myc tag was purified from E. coli and directly applied to the gel (control in lane 1), or incubated with the Arabidopsis E1 and E2 enzymes and either Arabidopsis ubiquitin (lane 2), the ubiquitin variant K48/R (lane 3), AtSUMO1 (lane 4), AtSUMO2 (lane 5), or AtSUMO3 (lane 6). Proteins were separated on a 12% SDS-polyacrylamide gel, and ScPCNA mobility shifts were detected by western-blot analysis using a Myc peptide-specific antiserum. SUMOylated ScPCNA species are marked by a bracket, and a signal of unclear origin is labeled with an asterisk (*).
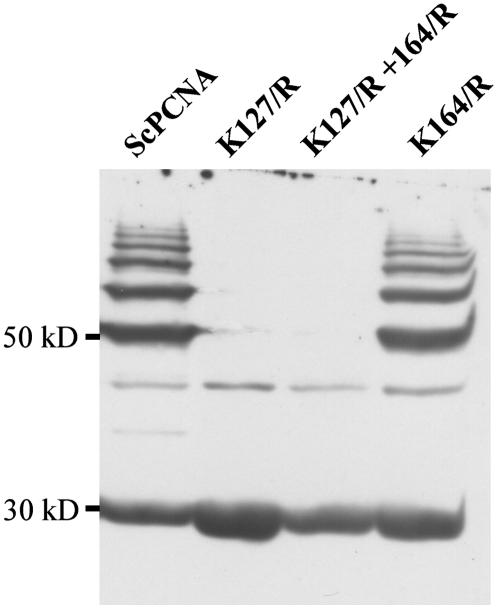
For a more detailed analysis of the recognition capacity of the Arabidopsis SUMOylation machinery, the ScPCNA variants K127/R, K164/R, as well as K127/R + K164/R were generated and used as substrates in the in vitro SUMOylation assay. When these ScPCNA variants were combined with AtSUMO1, AtSUMO2, or AtSUMO3, no SUMOylation was observed in the case of ScPCNA K127/R and ScPCNA K127/R + K164/R, whereas the ScPCNA variant K164/R was efficiently modified, demonstrating that all three Arabidopsis SUMO isoforms are conjugated to Lys-127 of ScPCNA (Fig. 4; data not shown). The observation that Lys-127 is the predominant site of modification by SUMO under in vitro conditions strongly suggests that AtSUMO1 and AtSUMO2 are able to form polymeric structures and indicates that AtSUMO3 has either no capacity to form polymeric structures or is used with reduced efficiency under in vitro conditions.
Figure 4.
Identification of SUMOylated lysine residues within ScPCNA. ScPCNA (lane 1) and the ScPCNA protein variants K127/R (lane 2), K127/R + K164/R (lane 3), as well as K164/R (lane 4) were incubated with the Arabidopsis SUMOylation machinery and AtSUMO1. ScPCNA mobility shifts were detected by western-blot analysis using a Myc peptide-specific antiserum.
Self-SUMOylation of AtSUMO1 and AtSUMO2
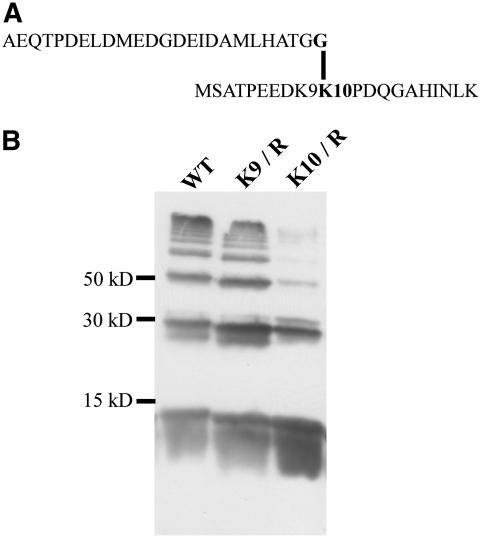
To study the self-SUMOylation of AtSUMO1 and AtSUMO2 as indicated by the mobility shifts of ScPCNA conjugates in more detail, we incubated the Arabidopsis SUMOylation cascade with AtSUMO1, 2, or 3 in the absence of any substrate protein and found a ladder of signals with the size of multimerized SUMO molecules in the case of AtSUMO1 and 2, whereas AtSUMO3 is apparently unable to form polymeric structures (Fig. 5). To identify the predicted auto-SUMOylation site within AtSUMO2, a band that corresponds in its size to a SUMO dimer was excised from a Coomassie-stained SDS gel, subjected to in-gel tryptic digestion, and analyzed by mass spectrometry (MS). MS analysis gave signals representing most of the expected peptides produced by cleavage of nonbranched AtSUMO2 at the carboxyl side of lysine and arginine residues. Additionally, one peak corresponding to a mass of 4,920.35 D was detected. This mass fits well to that predicted for the C-terminal tryptic fragment of AtSUMO2 fused to the first 20 N-terminal residues of AtSUMO2 (Fig. 6A). A more detailed analysis of MS/MS spectra confirmed this assignment and indicated that lysine residues 9 or 10 of AtSUMO2 are modified by SUMO. Thus, the weakly conserved but predictable SUMOylation motif DK9K10PD within the N-terminal region of AtSUMO2 is apparently the target of self-SUMOylation (Fig. 6A). To provide independent experimental evidence for this conclusion, both Lys-9 and Lys-10 of AtSUMO2 were individually substituted by arginine residues (Fig. 6B). Mutation of Lys-9 did not influence multimerization, whereas substitution of Lys-10 led to a strong reduction of high-Mr AtSUMO2 species, indicating that the major site of auto-SUMOylation was correctly identified by MS analysis (Fig. 6B). In agreement with these data, conjugation of AtSUMO2 K10/R to ScPCNA resulted in a clear reduction of higher molecular AtSUMO2-ScPCNA conjugates (data not shown). Although not experimentally tested, the high degree of sequence conservation between AtSUMO1 and AtSUMO2 indicates that self-SUMOylation of AtSUMO1 might also occur in the N-terminal part of the protein. The well-conserved SUMOylation consensus LK54LD present in AtSUMO3 is not located within the N-terminal region of the peptide, and therefore may not be exposed to the surface of the molecule and thus would be inaccessible to the SUMOylation machinery.
Figure 5.
Self-SUMOylation of Arabidopsis SUMO isoforms. The Arabidopsis SUMO isoforms AtSUMO1, AtSUMO2, and AtSUMO3, each labeled with an N-terminal HA tag, were heterologously expressed, purified from E. coli, and either directly applied to the gel or preincubated with a reaction mix containing the Arabidopsis SUMO E1 and E2 enzymes. Proteins were separated on a 15% SDS-polyacrylamide gel, and SUMO mobility shifts were detected by western-blot analysis using a HA peptide-specific antiserum.
Figure 6.
Analysis of AtSUMO2 multimerization by MS analysis and point mutagenesis. A, Structure of the branched AtSUMO2 peptide identified by LC-MS/MS. B, The Arabidopsis SUMO isoform AtSUMO2 labeled with an N-terminal HA tag (lane 1) and the thereof derived mutant variants K9/R (lane 2) and K10/R (lane 3) were incubated with all components of the Arabidopsis SUMOylation cascade and separated on a 15% SDS-polyacrylamide gel. AtSUMO2 mobility shifts were detected by western-blot analysis using a HA peptide-specific antiserum.
Several ScULP1-Like Genes Are Encoded in the Arabidopsis Genome
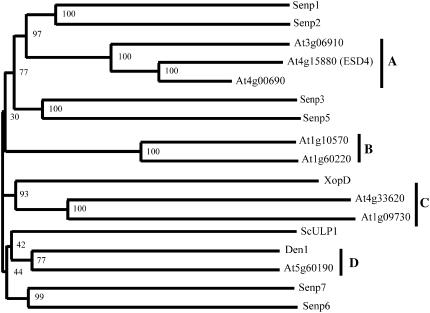
Eukaryotic organisms, some viruses, and pathogenic as well as symbiotic bacteria contain a family of cysteine proteases with similarity to the yeast SUMO isopeptidase ScULP1 (Li and Hochstrasser, 1999; Hotson and Mudgett, 2004). Typically, sequence conservation among SULP1-like peptidases is strictly limited to their C-terminal UL domains, which contain the catalytic center, whereas the large N-terminal extensions are unique for individual proteins (Li and Hochstrasser, 2003). When the UL domain of ScULP1, as defined by Li and Hochstrasser, was compared with all annotated Arabidopsis proteins, eight putative SUMO isopeptidases with at least 40% sequence similarity relative to the yeast probe were identified, and the full-length sequences were integrated into a phylogenetic tree that reveals four distinct groups (groups A–D) of putative Arabidopsis SUMO isopeptidases (Fig. 7; data not shown). All proteins identified by our BLAST search have been assigned to the C48 family of cysteine proteases (Altschul et al., 1990; Rawlings et al., 2006). However, in case of the group A reading frame At4g00690, no ESTs are available, and the predicted protein seems to be truncated as only the central part of its C-terminal catalytic domain is present. The lack of an At4g00690-encoding mRNA as suggested by the missing ESTs is in agreement with our own expression data (data not shown).
Figure 7.
Phylogram of ScULP1-like proteins. Arabidopsis proteins that exhibit at least 40% conserved residues when compared with the catalytic domain of ScULP1, as well as Senp1 to 3, Senp5 to 7, Den1, XopD, and ScULP1, were grouped into a phylogenetic tree using the ClustalW server at the University of Wageningen (www.bioinformatics.nl/tools/clustalw.html) with standard settings. Bootstrap values (1,000 replicates) are shown at the branches.
Similar to Arabidopsis, there are at least seven ScULP1-like proteins encoded in the human genome. These proteins have been designated as Senp1 to 3, Senp5 to 7, and Senp8/Den1/NEDP1 (Johnson, 2004; Reverter and Lima, 2004; Reverter et al., 2005). To identify related proteins that may represent enzymes with similar catalytic properties, a phylogenetic tree was created that comprises the ScULP1-like proteins from Arabidopsis and human, as well as ScULP1 from yeast and XopD from X. campestris (Fig. 7). This analysis reveals that most human and Arabidopsis enzymes form their own separated groups. However, At5g60190 (group D) and the human protein Senp8/Den1 group together and share 56% conserved positions on primary sequence level (Fig. 7; data not shown). It is known that Senp8 does not cleave SUMO conjugates, but represents a Nedd8-deconjugating and pre-Nedd8-processing enzyme and has therefore been designated as Den1/NEDP1 (Mendoza et al., 2003; Wu et al., 2003). In addition, cluster formation of XopD from X. campestris with the group C enzymes from Arabidopsis may also point to common catalytic properties (Fig. 7).
SUMO Isopeptidase Activity and Substrate Specificity of ScULP1-Like Enzymes from Arabidopsis, Yeast, and X. campestris
For biochemical analysis, representative members of three of the four groups of Arabidopsis ScULP1-like proteins were chosen (Fig. 7). This selection comprises At4g15880 (group A), At1g60220 and At1g10570 (group B), as well as At5g60190 (group D). We were not able to clone or express the cDNAs of any of the group C enzymes, and, consequently, At1g09730 and At4g33620 could not be analyzed. For heterologous expression in E. coli, the cDNA of At1g10570 was obtained from RIKEN (RIKEN Genomic Science Center). The At4g15880 (ESD4)-, At1g60220-, and At5g60190-encoding cDNAs were prepared from leaf RNA of 3-week-old Col-0 plants grown under short-day conditions, and sequence data obtained for At1g60220 were deposited at GenBank under the accession number DQ304543 as conflicts with the intron/exon prediction provided by the Munich Information Center for Protein Sequences (MIPS) became obvious. As reference enzymes, we expressed and purified the UL domain of yeast ScULP1 and XopD from X. campestris pv vesicatoria (Hotson et al., 2003; Li and Hochstrasser, 2003).
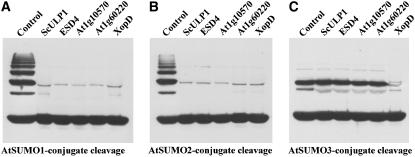
The established in vitro SUMOylation system that conjugates different SUMO isoforms to target proteins provided a useful set of model substrates to test the isopeptidase activity and substrate specificity of all of the above-listed proteins. In a first series of experiments, cleavage of AtSUMO1, 2, or 3 conjugated to ScPCNA was tested (Fig. 8). In this approach, the cleavage capacity after prolonged incubation time of 20 h was determined. Five of the six enzymes tested in this study, i.e. all enzymes except At5g60190, are able to release AtSUMO1 and 2 from ScPCNA conjugates under these conditions, demonstrating that At1g60220 and At1g10570 represent bona fide SUMO isopeptidases, while function and catalytic activity of At5g60190 remained open at this point (Fig. 8; data not shown). Interestingly, the AtSUMO3-ScPCNA conjugate was exclusively cleaved by the bacterial enzyme XopD (Fig. 8). The same enzyme preferences that were found with AtSUMO1, 2, or 3 conjugated to ScPCNA were also observed with AtSUMO1, 2, or 3 conjugated to the Arabidopsis transcription factor AtMyb30, i.e. all SUMO isopeptidases were able to release AtSUMO1 and AtSUMO2, but only XopD was able to cleave AtSUMO3-Myb30 conjugates (data not shown). This finding shows that at least under the conditions applied in this study, the attached SUMO isoform rather than the target protein represents the main determinant for recognition and cleavage by a selected protease.
Figure 8.
SUMO isopeptidase activity and substrate specificity of cysteine proteases from Arabidopsis, yeast, and X. campestris. SUMO isopeptidase activity of ScULP1, ESD4, At1g10570, At1g60220, and XopD was determined using in vitro produced AtSUMO1-ScPCNA (A), AtSUMO2-ScPCNA (B), and AtSUMO3-ScPCNA (C) conjugates. Protease-treated samples were applied as depicted. As a reference, untreated conjugates were always applied to the first lane of the respective gels. Proteins were separated on a 12% SDS-polyacrylamide gel, and cleavage of SUMO-ScPCNA conjugates was detected by western-blot analysis using a Myc peptide-specific antiserum.
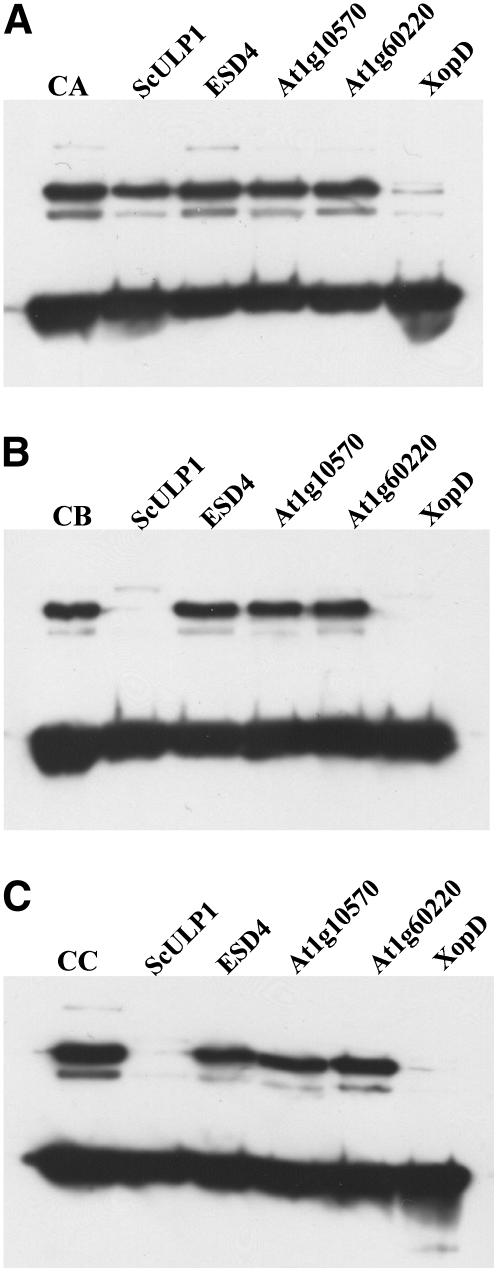
The observation that an AtSUMO3-ScPCNA conjugate is efficiently cleaved by the putative bacterial virulence factor XopD but is resistant to attack by At1g60220, At1g10570, ESD4 (At4g15880), and the catalytic domain of ScUPL1 allowed us to search for amino acid residues within the protein conjugate that hinder cleavage by the latter proteases (Figs. 8 and 9A). Since all Arabidopsis SUMO isoforms are attached to ScPCNA in the same way and at the same position, i.e. as SUMO-GG-ɛ-K127-PCNA conjugate, substrate quality-determining amino acid residues must be located within the SUMO molecules upstream of their diglycine motif. A recently resolved crystal structure of the inactivated human SUMO isopeptidase Sen2p in complex with HsSUMO1 reveals several side-chain interactions between the protease and its substrate, including the diglycine motif and the amino acids directly upstream of this motif (Reverter and Lima, 2004). In one case, a close interaction between three amino acids of the protease Sen2p and a single residue in HsSUMO1 (Gln-94) was observed, indicating that this SUMO residue might be of particular importance for substrate quality determination (Fig. 1B; Reverter and Lima, 2004). Interestingly, glutamine is found at the corresponding position within AtSUMO1 and AtSUMO2, but is substituted by methionine and leucine in AtSUMO3 and AtSUMO5, respectively (Fig. 1B). To investigate the influence of a glutamine residue at the respective position on substrate quality determination, an AtSUMO3 variant carrying a M90/Q amino acid exchange was generated and conjugated to ScPCNA. This modified conjugate was cleaved not only by XopD but also by yeast ScULP1, indicating that the selected position is in fact important for the definition of a proper protease substrate, although 17 additional contact sites between Smt3 and ScULP1 have been observed (Fig. 1B; Mossessova and Lima, 2000). In contrast, the Arabidopsis enzymes At1g60220, At1g10570, and ESD4 (At4g15880) were not able to cleave the modified AtSUMO3 conjugate, demonstrating that the selected position is important but not sufficient to define substrate quality (Fig. 9B). The crystal structure of Smt3 in complex with ScULP1 demonstrates that the SUMO residues directly upstream and downstream of the important glutamine are also in direct contact with the protease (Fig. 1B; Mossessova and Lima, 2000). Based on the sequence alignment shown in the boxed area of Figure 1B, we created the AtSUMO3 triple mutant A89/H + M90/Q + S91/T. In this AtSUMO3 variant, the amino acid composition directly upstream of the diglycine motif is identical to that found in AtSUMO1 and AtSUMO2, and the suitability of this modified AtSUMO3-ScPCNA conjugate as substrate for At1g60220, At1g10570, and ESD4 was tested (Fig. 1B). However, as observed with the AtSUMO3 M90/Q single mutant, only XopD and ScULP1, but not At1g60220, At1g10570, or ESD4, were capable to cleave the AtSUMO3 A89/H + M90/Q + S91/T-ScPCNA conjugate (Fig. 9C).
Figure 9.
Substrate quality-determining amino acid residues of AtSUMO3-ScPCNA conjugates. ScPCNA was conjugated in vitro with either AtSUMO3 wild-type protein (A; lane CA), the AtSUMO3 variant M90/Q (B; lane CB), or the AtSUMO3 triple mutant A89/H + M90/Q + S91/T (C; lane CC). Conjugates were treated with either ScULP1 (lanes 2), ESD4 (lanes 3), At1g10570 (lanes 4), At1g60220 (lanes 5), or XopD (lanes 6). Proteins were separated on a 12% SDS-polyacrylamide gel, and cleavage of AtSUMO3-ScPCNA conjugates was detected by western-blot analysis using a Myc peptide-specific antiserum.
Pre-SUMO-Processing Activity and Substrate Specificity of ScULP1-Like Enzymes from Arabidopsis, Yeast, and X. campestris
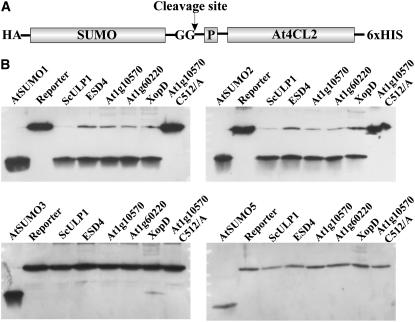
Most known SUMO proteases exhibit both isopeptidase activity, which recycles SUMO from conjugates with substrate proteins, and an endopeptidase activity, which generates the mature form of SUMO from its precursors (Johnson, 2004). To test the five SUMO proteases expressed in this study for the latter activity, a reporter construct was designed that allows detection of release of the small C-terminal SUMO prepeptide on standard SDS gels. The reporter consists of the respective pre-SUMO isoform linked to the N terminus of Arabidopsis 4-coumarate CoA ligase 2 (Fig. 10A; Stuible et al., 2000). Purification of this synthetic fusion protein from E. coli was facilitated by a C-terminal hexa-His tag, whereas cleavage of the SUMO precursor could be detected using its N-terminal HA tag (Fig. 10A). All proteases were found to be active with pre-AtSUMO1 and pre-AtSUMO2 (Fig. 10B). In the case of pre-AtSUMO3, none of the proteases except XopD showed any activity, which is reminiscent of AtSUMO3-protein conjugates to resist de-SUMOylation (Figs. 8 and 10B). However, activity of XopD with pre-AtSUMO3 was found near the detection limit, indicating that this protease interacts more readily with AtSUMO3 in the context of an isopeptide-linked conjugate (Figs. 8 and 10B). In addition, pre-AtSUMO5 was not found to represent a substrate for any of the proteases tested in this study.
Figure 10.
Pre-SUMO-processing activity and substrate specificity of cysteine proteases from Arabidopsis, yeast, and X. campestris. A, Structure of the reporter construct that was used to detect pre-SUMO-processing activity. The diglycine motif exposed in the mature SUMO molecule is depicted, and the small SUMO prepeptide is labeled with P. The expected site of protease cleavage is marked by an arrow. B, Pre-SUMO-processing activity of ScULP1 (lanes 3), ESD4 (lanes 4), At1g10570 (lanes 5), At1g60220 (lanes 6), XopD (lanes 7), and At1g10570 Cys-512/Ala (lanes 8) was determined using preAtSUMO1-4CL2, preAtSUMO2-4CL, preAtSUMO3-4CL, and preAtSUMO5-4CL reporter constructs. Mature SUMO isoforms isolated from E. coli (lanes 1) and untreated reporter fusions (lanes 2) were loaded in each case as reference probes. Proteins were separated on a 15% SDS-polyacrylamide gel, and cleavage of pre-SUMO reporter constructs was detected by western-blot analysis using a HA peptide-specific antiserum.
ScULP1 has been assigned to the C48 family of cysteine proteases, and it is known that Cys-580 of ScULP1 is directly involved in the catalytic process (Li and Hochstrasser, 2003; Rawlings et al., 2006). From the alignment of the UL domain of ScULP1 with the ScULP1-like proteases from Arabidopsis, it became obvious that the corresponding cysteine residue is highly conserved and located at position 512 within At1g10570 (data not shown). At1g10570 protease variants carrying either a C512/A or a C512/S substitution were found to be catalytically inactive, demonstrating that At1g10570 can in fact be classified as cysteine protease (Fig. 10B).
The ScULP1-Like Protein At5g60190 Represents a Rub1-Specific Protease
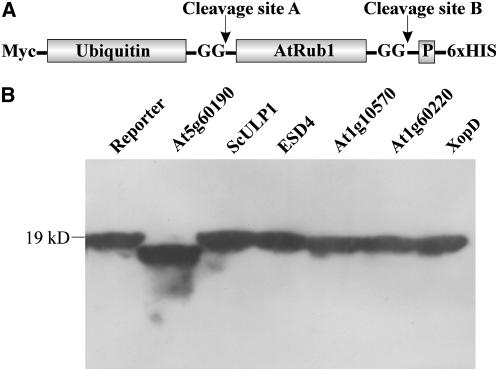
Although At5g60190 shows a clear similarity of 44% when compared with the UL domain of ScULP1, we detected neither SUMO isopeptidase activity nor pre-SUMO-processing capacity when testing this Arabidopsis enzyme with all available model substrates (Fig. 11). This observation indicated that At5g60190 does not represent a SUMO protease and, consequently, that the SUMO isopeptidase family in Arabidopsis is smaller than originally estimated (Fig. 7). The structure and domain organization of At5g60190 supports this finding. At5g60190 is only 226 amino acids long and lacks the large N-terminal noncatalytic domain typical for ScULP1-like SUMO isopeptidases (data not shown). As mentioned before, At5g60190 shares 56% conserved positions with the human de-Neddylating enzyme DEN1/NEDP1, indicating that it could represent a Nedd8 rather than a SUMO protease. To test this hypothesis, we used the naturally occurring Rub1 (Nedd8) protein fusion At1g31340 from Arabidopsis as a model substrate. This protein consists of ubiquitin fused to the N terminus of pre-AtRub1 (Fig. 12A). The cDNA of the Rub1 fusion protein was cloned from Arabidopsis Col-0 plants and expressed in E. coli with a Myc tag fused to its N terminus and a hexa-His tag fused to its C-terminal end (Fig. 12A). Treatment of the ubiquitin-preAtRub1 fusion with At1g60190 causes only a minor reduction in its Mr, indicating that cleavage occurs exclusively after the C-terminal diglycine motive of AtRub1 (Fig. 12B). This cleavage site was verified by comparative MS fingerprinting of the fusion protein before and after cleavage as well as by microsequencing of the C terminus of the cleaved protein (data not shown). Therefore, At5g60190 is not only able to cleave pre-AtRub1, but shows the capacity to discriminate between AtRub1 and ubiquitin despite the fact that these peptides share about 80% conserved residues. In contrast, all enzymes with SUMO isopeptidases activity analyzed in this study, i.e. At1g60220, At1g10570, At4g15880 (ESD4), XopD, and ScULP1, were not able to cleave the ubiquitin-preAtRub1 protein fusion at any position, indicating that the only ubiquitin-like modifier that can be recognized by these enzymes is SUMO (Fig. 12B).
Figure 11.
Analysis of SUMO isopeptidase and pre-SUMO-processing activity of At5g60190. A, The SUMO isopeptidase model substrates AtSUMO1-ScPCNA, AtSUMO2-ScPCNA, and AtSUMO3-ScPCNA were analyzed before (−) and after incubation (+) with At5g60190. Proteins were separated on a 12% SDS-polyacrylamide gel, and SUMO-ScPCNA conjugates were detected by western-blot analysis using a Myc peptide-specific antiserum. B, The pre-SUMO-4CL reporter constructs were incubated with At5g60190 as depicted. As reference probes, AtSUMO1 (lane 1) and the untreated AtSUMO1-4CL protein (lane 2) were applied. Proteins were separated on a 15% SDS-polyacrylamide gel, and the pre-SUMO reporter constructs were detected by western-blot analysis using a HA peptide-specific antiserum.
Figure 12.
Pre-Rub1 cleavage activity and substrate specificity of At1g60190. A, Structure of the reporter construct that was used to detect pre-AtRub1-processing activity. The reporter is based on a naturally occurring ubiquitin-pre-AtRub1 protein (At1g31340) that was fused with an N-terminal Myc tag and a C-terminal hexa-His tag. Two possible sites of protease cleavage are marked by arrows. The diglycine motifs that are expected to be exposed in the mature molecules are depicted, and the small prepeptide following AtRub1 is labeled with P. B, Pre-AtRub1-processing activity of At5g60190 (lane 2), ScULP1 (lane 3), ESD4 (lane 4), At1g10570 (lane 5), At1g60220 (lane 6), and XopD (lane 7) was determined. Proteins were separated on a 15% SDS-polyacrylamide gel, and cleavage of the reporter construct was detected by western-blot analysis using a Myc peptide-specific antiserum.
DISCUSSION
Although a genome survey reveals that Arabidopsis contains a comparably complex family of both SUMO isoforms and of putative SUMO isopeptidases, not much is known about the enzymology of SUMO conjugation and deconjugation in this model plant (Novatchkova et al., 2004). We started our study by in vitro reconstitution of the Arabidopsis SUMOylation cascade and analysis of the substrate specificity of its SUMO-conjugating E2 enzyme. As it is known from other eukaryotic organisms, the SUMO-activating E1 enzyme from Arabidopsis might form a heteromeric high-Mr complex, as coexpression of both At5g50690 (AtSAE1B) and At2g21470 (AtSAE2) was required to obtain E1 enzyme activity. What distinguishes Arabidopsis from most other organisms is the presence of two putative SAE1 subunits. AtSAE1A (At4g24940) and AtSAE1B (At5g50690) show 52% and 44% sequence similarity with yeast Aos1 (ScSAE1), respectively. We have verified the activity of the less conserved AtSAE1B variant, indicating that AtSAE1A may also function as subunit of a SUMO E1 enzyme, although experimental proof of its enzymatic activity is pending.
The Arabidopsis E2 enzyme is encoded by a single gene (At3g57870), which is interesting in light of the fact that eight distinct SUMO reading frames are present in the Arabidopsis genome. Our finding that conjugation of all tested SUMO variants to the model protein ScPCNA was catalyzed by this enzyme in vitro, despite significant sequence divergence between the SUMO isoforms, is consistent with the hypothesis that a single Arabidopsis E2 enzyme with relaxed substrate specificity conjugates all SUMO isoforms in vivo (Figs. 1 and 3).
Using yeast PCNA as a model protein, we found that the Arabidopsis SUMOylation system has the capacity to modify Lys-127, a residue located within a perfectly conserved SUMOylation consensus, but not Lys-164. Since the latter is located within a nonconsensus SUMOylation site, this demonstrates both the specificity and limitations of in vitro SUMOylation studies. Recently, it has been shown that in yeast the SUMO E3-ligase ScSiz1 is required for SUMOylation of ScPCNA at position 164 (Pfander et al., 2005). It has also been noted that Arabidopsis contains several proteins with sequence similarity to ScSiz1, but AtSiz1 did not allow ScPCNA modification when included in our assay (data not shown; Miura et al., 2005).
The Arabidopsis E2 enzyme shows relaxed substrate specificity and is therefore probably not a central target for the SUMOylation pathway regulation in vivo, while the different properties of the Arabidopsis SUMO isoforms that became obvious during this study may point to a regulatory function (Figs. 3 and 5). Both AtSUMO1 and AtSUMO2, but not AtSUMO3, have a tendency to form multimeric chains due to a self-SUMOylation mechanism (Figs. 3 and 5). By using specific antibodies, Kurepa et al. were able to demonstrate that conjugation of AtSUMO1 and 2, but not of AtSUMO3, to target proteins is increased during stress treatment in planta and that the cellular conjugation pattern observed with AtSUMO3 is less complex than the modification pattern observed with the other SUMO isoforms (Kurepa et al., 2003). Together, these findings correlate SUMO polychain formation with stress responses, as has been postulated for mammals (Saitoh and Hinchey, 2000; Tatham et al., 2001). The Arabidopsis SUMO isoforms cannot only be distinguished based on their capacity to form multimeric structures, but also due to their capacity to function as substrates for selected proteases. The amino acid residues neighboring the C-terminal diglycine motif in AtSUMO5 are atypical, and in fact none of the tested proteases was able to cleave a pre-AtSUMO5 reporter construct (Figs. 1B and 10B). Similarly, pre-AtSUMO3 was not processed by the four Arabidopsis peptidases tested in this study, but the detection of AtSUMO3 conjugates in vivo demonstrates that the required protease must be present in planta (Fig. 8; Kurepa et al., 2003) Astonishingly, what is found with the Arabidopsis SUMO isoforms partly resembles the situation in mammals. In mammals, SUMO is encoded by a small gene family of at least four members (Bohren et al., 2004; Johnson, 2004). Human SUMO2 and 3 form multimeric chains in vivo and in vitro and conjugation of these peptides to substrate proteins is enhanced by stress treatment, whereas HsSUMO1 shows no tendency to form multimers and is not responsive to stress stimuli (Saitoh and Hinchey 2000; Tatham et al., 2001). Based on the presence of atypical amino acid residues flanking the C-terminal diglycine motif of pre-HsSUMO4 and the protease resistance of this SUMO precursor in vitro and in vivo, it has been postulated that HsSUMO4 is never present in its mature conjugation-competent form (Owerbach et al., 2005). Therefore, HsSUMO4 may function by noncovalent interaction with its target proteins (Owerbach et al., 2005). Despite the striking correlation between the properties of human and Arabidopsis SUMO isoforms, sequence and phylogenetic comparison resulted in no informative cluster formation, i.e. all Arabidopsis and all human SUMO isoforms group together (Fig. 1A).
Not much is known about substrate specificity of SUMO isopeptidases in vivo. The observation that several organisms contain only one SUMO isoform implies either that both SUMO and the target protein within a SUMOylated complex are specifically recognized by a selected protease or that SUMO isopeptidases recognize only the SUMO residue and exhibit no defined specificity for any SUMOylated target protein. In the latter case only cellular localization would define if a SUMO conjugate can be cleaved by an isopeptidase. The observation that in yeast the nuclear pore complex associated isopeptidase ScULP1, but not the nuclear localized peptidase ScULP2, is essential for viability and the observation that both enzymes exhibit overlapping substrate specificities in fact indicate that SUMO conjugate cleavage is mainly determined by cellular localization (Li and Hochstrasser, 2000, 2003). In mammals and the model plant Arabidopsis, the situation could be more complex since multiple SUMO isoforms with distinct properties are expressed. This idea is supported by the observation that mutation of the mammalian SUMO isopeptidase Senp1 led to the accumulation of SUMO1 conjugates, whereas the overall pattern of SUMO2 and SUMO3 modified proteins was not altered (Yamaguchi et al., 2005). The importance of the attached SUMO isoform in defining a protein conjugate as substrate, i.e. the requirement for SUMO isoform-specific proteases, is strengthened by our data. All Arabidopsis SUMO isopeptidases tested were able to deconjugate both AtSUMO1 and 2 from yeast ScPCNA, whereas only XopD derived from the phytopathogenic bacterium X. campestris readily released AtSUMO3 molecules from the conjugates (Fig. 8). However, AtSUMO3 conjugates exist in planta, and, therefore, an endogenous pre-AtSUMO3-processing enzyme must be active and the presence of an endogenous AtSUMO3 protein conjugate cleaving activity is highly probable (Kurepa et al., 2003). In the case of AtSUMO5, formation of protein conjugates has not been described and, consequently, it is not clear if an AtSUMO5-specific peptidase is present in Arabidopsis. Arabidopsis SUMO variants show significant variation in their amino acid composition and at least the bacterial enzyme XopD is able to release both AtSUMO1 and AtSUMO3 residues from protein conjugates, although AtSUMO1 and AtSUMO3 share only 53% conserved positions. We therefore tested if the substrate spectrum of XopD comprises ubiquitin-like modifiers different from SUMO. However, neither XopD nor any other tested SUMO protease was able to cleave an ubiquitin-pre-AtRub1 fusion protein (Fig. 12B).
Although focusing on SUMO-conjugating and -deconjugating enzymes, we observed that At5g60190 has the capacity to process the precursor of AtRub1 but is inactive with ubiquitin (Fig. 12B). This indicates a comparably high substrate specificity since AtRub1 and Arabidopsis ubiquitin share about 80% conserved and 60% identical amino acid residues, and, focusing on the positions known to be responsible for the interaction between HsNedd8 (HsRub1) and the protease Den1, only three significant differences become obvious between AtRub1 and Arabidopsis ubiquitin (Fig. 1B; Reverter et al., 2005). It is known that Rub1 conjugation to cullins is a dynamic process that can be reversed by a metalloprotease activity associated with the Csn5 subunit of the Cop9 signalosome (Lyapina et al., 2001; Cope et al., 2002; Wei and Deng, 2003). In addition to the metalloprotease Csn5, the ScULP1-like cysteine proteases Den1 and Nep1 from Schizosaccharomyces pombe can cleave Rub1 conjugates, and it has been postulated that these proteins may regulate SCF-complex activity and protein degradation by the ubiquitin-proteasome pathway (Mendoza et al., 2003; Wu et al., 2003; Zhou and Watts, 2005). Den1/ NEDP1 is not only a Rub1-specific isopeptidase but also has the capacity to process pre-Nedd8, and, therefore, the same dual activity can be expected with At5g60190 (Mendoza et al., 2003; Wu et al., 2003). At5g60190 is similar to ScULP1 but does not group together with other ScULP1-like proteins from Arabidopsis, as is obvious from the phylogram presented in Figure 7, indicating that we have identified all putative Arabidopsis SUMO isopeptidases (Fig. 7). This assumption is in agreement with publicly available data, suggesting that Arabidopsis contains only putative SUMO isopeptidases of the C48 family, i.e. ScULP1-like enzymes, but no SUMO isopeptidases of other families such as C55 (Rawlings et al., 2006; http://merops.sanger.ac.uk). However, given the large number of uncharacterized proteases present in Arabidopsis and the uncertainty associated with the prediction of protease specificity as illustrated by the misclassification of the putative SUMO protease At5g60190, we cannot exclude that more distantly related enzymes also contribute to protein de-SUMOylation.
Our data suggest that only six ScULP1-like SUMO isopeptidases are expressed in Arabidopsis, and we presume that biological function of these enzymes is defined by three parameters, i.e. substrate and SUMO isoform specificity, cell type-specific expression level, and subcellular localization. We will address these points by extended and refined biochemical analysis, identification and characterization of protease mutants by reverse genetics, as well as expression of fluorescent-tagged protease variants to determine their subcellular localization.
MATERIALS AND METHODS
cDNA Cloning and Expression Plasmid Construction
SUMO- and Rub1-Encoding Constructs
cDNAs of the Arabidopsis (Arabidopsis thaliana) SUMO isoforms 1, 2, 3, and 5 were amplified by reverse transcription (RT)-PCR from Arabidopsis ecotype Col-0 total RNA. Synthesis of the cDNAs and subsequent PCR amplification of the full-length reading frames encoding the SUMO preproteins were performed with the Titan one-tube RT-PCR kit (Roche Diagnostics) according to the manufacturer's instructions.
For cloning and expression of mature and conjugation-competent SUMO molecules, the preprotein-encoding cDNAs were used as templates for PCR amplification of C-terminal processed constructs encoding the mature SUMO variants. A DNA region encoding a HA-hexa-His double-affinity tag was fused to the N terminus of each SUMO cDNA, and the resulting constructs were cloned into a suited vector of the pQE expression plasmid series (Qiagen). PCR primer sequences and detailed plasmid descriptions are available on request.
Point mutations were introduced into AtSUMO3 by PCR-based amplification of the entire AtSUMO3 expression plasmid, using two mutated oligonucleotide primers, each complementary to opposite strands of the vector. All components necessary for this mutagenesis procedure were included in the commercial QuikChange kit (Stratagene). Double and triple mutations were introduced in the same way by using already existing mutants as templates.
The cDNA of the Arabidopsis ubiquitin-pre-AtRub1 fusion At1g31340 was amplified by RT-PCR from Arabidopsis ecotype Col-0 total RNA. Synthesis of the cDNA and subsequent PCR amplification of the full-length reading frame were performed with the Titan one tube RT-PCR kit (Roche Diagnostics) according to the manufacturer's instructions. During a second round of PCR amplification, the SphI and BamHI sites required for cloning into a derivative of the expression plasmid pQE70 (Qiagen) were introduced. This plasmid allows the expression of cloned proteins as N-terminal Myc and C-terminal hexa-His tag fusions.
Arabidopsis SUMO E1 and E2 Enzyme-Encoding Constructs
cDNAs encoding the Arabidopsis SUMO E1 and SUMO E2 enzymes were isolated as described for the Arabidopsis SUMO variants. The cDNA of the Arabidopsis SUMO E2 enzyme was inserted into the expression vector pQE70 (Qiagen) and expressed as a C-terminal hexa-His fusion. For expression of the heteromeric Arabidopsis SUMO E1 enzyme, the cDNAs of At5g50680 and At2g21470 were used as templates for a second round of PCR amplification. In the case of At5g50680 (AtSAE1b), this second PCR reaction was used to introduce the SphI and BglII sites required for cloning into the Escherichia coli expression vector pQE70 (Qiagen). In the case of At2g21470 (AtSAE2), SphI and SalI sites were introduced in the second round of PCR amplification and the resulting product was cloned into the expression plasmid pQE30 (Qiagen). In a subsequent step, At5g50680 (AtSAE1b) was PCR amplified from the pQE70 construct together with all DNA regions necessary for proper expression in E. coli, and XbaI restriction sites were introduced at both ends of this PCR product. Finally, the AtSAE1b-encoding fragment was inserted into the unique XbaI site of the AtSAE2-encoding pQE30 construct, resulting in a plasmid that allows coexpression of both reading frames in E. coli.
SUMO Isopeptidase-Encoding Constructs
A plasmid encoding XopD from Xanthomonas campestris pv vesicatoria was kindly provided by M.B. Mudgett (Department of Biological Sciences, Stanford University). During PCR amplification of the XopD reading frame, the SphI and BamHI sites required for subcloning into the expression vector pQE70 (Qiagen) were introduced. In the case of the Saccharomyces cerevisiae SUMO isopeptidase ULP1, only the DNA region encoding the so-called UL domain, as defined by Li and Hochstrasser, was amplified from yeast genomic DNA and inserted into the expression vector pQE70 (Qiagen) using SphI and BamHI restriction sites (Li and Hochstrasser, 2003).
The cDNA encoding Arabidopsis At1g10570 was obtained from RIKEN (RIKEN Genomic Science Center, Yokohama, Japan). During PCR amplification, the SphI and NotI sites required for cloning into a derivative of the expression plasmid pQE70 (Qiagen) were introduced. Point mutations were introduced into At1g10570 by PCR-based amplification of the entire expression plasmid, using two mutated oligonucleotide primers, each complementary to opposite strands of the vector. All components necessary for this mutagenesis procedure were included in the commercial QuikChange kit (Stratagene).The cDNAs encoding At1g60220 and ESD4 (At4g15880) were amplified by RT-PCR from Arabidopsis Col-0 total RNA isolated from fully expanded leaves of 3-week-old plants grown under short-day conditions. Synthesis of the cDNAs and subsequent PCR amplification of the full-length reading frames was performed with the Titan one-tube RT-PCR kit (Roche Diagnostics) according to the manufacturer's instructions. After introduction of SphI and BamHI restriction sites, both reading frames were introduced into the expression plasmid pQE70 (Qiagen). In the case of At1g60220, cDNA sequencing revealed differences when compared with the theoretically predicted coding sequence as provided by MIPS.
SUMO Target Protein-Encoding Constructs
S. cerevisiae ScPCNA was PCR amplified from yeast genomic DNA and inserted via SphI and BamHI restriction sites into a pQE70 (Qiagen)-based plasmid called p130 that allows the expression of cloned proteins as N-terminal Myc and C-terminal hexa-His tag fusions. PCR amplification of the AtMyb30-encoding cDNA (Vailleau et al., 2002) was performed to introduce the SphI and BamHI restriction sites required for cloning into the expression vector p130.
Protein Expression and Purification
All listed proteins, i.e. AtSUMO1 (At4g26840), AtSUMO2 (At5g55160), AtSUMO3 (At5g55170), AtSUMO5 (At2g32765), AtSAE1b (At5g50680) AtSAE2 (At2g21470), AtUbc9 (At3g57870), At1g10570, AtESD4 (At4g15880), At5g60190, AtRub1 (At1g31340) ScPCNA (YBR088C), At1g60220, XopD (XCV0437), and ScULP1 (YPL020C), were heterologously expressed in E. coli DH5α carrying the repressor plasmid pREP4 (Qiagen). To enhance protein solubility, expression was performed at a reduced growth temperature of 25°C. Purification by affinity chromatography was carried out as described previously (Stuible et al., 2000). Proteins were stored in the elution buffer containing 50 mm Tris-HCl, pH 7.8, 15% glycerol, 400 mm imidazole, and 100 mm NaCl at −70°C. The purity of the enzymes was inspected by SDS gel electrophoresis. Protein concentrations were determined according to Bradford with bovine serum albumin as standard (Bradford, 1976).
In Vitro SUMOylation Assay
The standard reaction buffer contained 50 mm Tris-HCl, pH 7.8, 100 mm NaCl, 15% glycerol, 5 mm ATP, and 10 mm MgCl2. In vitro protein SUMOylation was carried out in a total volume of 50 μL at 22°C for 2 to 12 h with 4 μg of test protein, 8 μg of SUMO, 0.5 μg of E1 enzyme, and 2 μg of E2 enzyme.
MS Analysis of Branched SUMO Peptides
Tryptic in-gel digestion of self-SUMOylated Arabidopsis SUMO2 was performed as described elsewhere (Shevchenko et al., 1996). The tryptic peptides dissolved in 0.1% formic acid were loaded onto a Symmetry300 C18 precolumn of a CapLC nanobore liquid chromatography system equipped with a 75-micron × 150-mm Atlantis C18 analytical column (Waters). Peptides were eluted with a linear acetonitrile gradient ranging from 5% to 50% over 15 min. Mass spectra were recorded in positive mode with a Q-TOF2 spectrometer (Micromass) coupled to the CapLC via a picosprayer interface. The capillary voltage was set at 1,600 V and cone voltage at 40 V, and data were collected in survey scan mode. MS-TOF spectra were recorded for ions with a mass-to-charge ratio (m/z ratio) ranging from 400 to 1,800. Multiply charged ions were then fragmented in the argon collision cell with a mass- and charge-dependent collision voltage, and MS/MS spectra were recorded for fragments with a m/z ratio between 100 and 1,700. Fragmentation spectra were assigned to predicted SUMO peptides based on precursor mass and fragment ion matching. Spectra that could not be explained by the presence of linear AtSUMO2 fragments were checked for ionization products indicative of branched AtSUMO2 peptides. These diagnostic signals were established by fragmentation of the C-terminal tryptic peptide of native AtSUMO2, which revealed two high-intensity daughter b-ions at m/z 329.1 and 430.2. Since SUMO is attached to target proteins (in this case SUMO itself) via its C-terminal glycine, branched peptides must contain this tryptic SUMO peptide and should thus also display these characteristic daughter ions during fragmentation. MS/MS spectra of unexplained peptide peaks with high intensities for the diagnostic daughter ions were analyzed by hand to assign fragment ions. Thereby, two b-ion series corresponding to the two N termini of the branched SUMO peptide and one y-ion series from the C terminus of the peptide containing the SUMOylation site were observed. General data processing was carried out with the Masslynx 4.0 software package (Waters).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number DQ304543.
Acknowledgments
We thank Drs. Erich Kombrink and Paul Schulze-Lefert for critical reading of the manuscript, Dr. Mary Beth Mudgett for providing us with a XopD-encoding vector, and Dr. Kurt Stüber for helpful discussions.
This work was supported by the Max Planck Society and by grants of the Bundesministerium für Bildung und Forschung (GABI-Trilateral Project “DILEMA”; AZ 0313150) and the Deutsche Forschungsgemeinschaft (SFB 635).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hans-Peter Stuible (stuible@mpiz-koeln.mpg.de).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J (1998) Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol 280: 275–286 [DOI] [PubMed] [Google Scholar]
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD (2002) Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108: 345–356 [DOI] [PubMed] [Google Scholar]
- Bohren KM, Nadkarni V, Song JH, Gabbay KH, Owerbach D (2004) A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J Biol Chem 279: 27233–27238 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ (2002) Role of the predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298: 608–611 [DOI] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT (1998) SUMO-1 modification of IκBα inhibits NF-κB activation. Mol Cell 2: 233–239 [DOI] [PubMed] [Google Scholar]
- Dharmasiri S, Dharmasiri N, Hellmann H, Estelle E (2003) The Rub/Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J 22: 1762–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen RJ (2004) SUMO protein modification. Biochim Biophys Acta 1695: 113–131 [DOI] [PubMed] [Google Scholar]
- Gill G (2003) Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr Opin Genet Dev 13: 108–113 [DOI] [PubMed] [Google Scholar]
- Gill G (2005) Something about SUMO inhibits transcription. Curr Opin Genet Dev 15: 536–541 [DOI] [PubMed] [Google Scholar]
- Hanania U, Furman-Matarasso N, Ron M, Avni A (1999) Isolation of a novel SUMO protein from tomato that suppresses EIX-induced cell death. Plant J 19: 533–541 [DOI] [PubMed] [Google Scholar]
- Hardeland U, Steinacher R, Jiricny J, Schar P (2002) Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J 21: 1456–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann H, Estelle M (2002) Plant development: regulation by protein degradation. Science 297: 793–797 [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Hotson A, Chosed R, Shu H, Orth K, Mudgett MB (2003) Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol Microbiol 50: 377–389 [DOI] [PubMed] [Google Scholar]
- Hotson A, Mudgett MB (2004) Cysteine proteases in phytopathogenic bacteria: identification of plant targets and activation of innate immunity. Curr Opin Plant Biol 7: 384–390 [DOI] [PubMed] [Google Scholar]
- Huang WC, Ko TP, Li SS, Wang AH (2004) Crystal structures of the human SUMO-2 protein at 1.6 A and 1.2 A resolution: implication on the functional differences of SUMO proteins. Eur J Biochem 271: 4114–4122 [DOI] [PubMed] [Google Scholar]
- Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73: 355–382 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Gupta AA (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106: 735–744 [DOI] [PubMed] [Google Scholar]
- Kagey MH, Melhuish TA, Powers SE, Wotton D (2005) Multiple activities contribute to Pc2 E3 function. EMBO J 24: 108–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD (2003) The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem 278: 6862–6872 [DOI] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M (1999) A new protease required for cell-cycle progression in yeast. Nature 398: 246–251 [DOI] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M (2000) The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol 20: 2367–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M (2003) The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J Cell Biol 160: 1069–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Furukawa M, Matsumoto T, Xiong Y (2002) Nedd8 modification of Cul1 dissociates p120CAND1, an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell 10: 1519–1526 [DOI] [PubMed] [Google Scholar]
- Lois LM, Lima CD (2005) Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J 24: 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois LM, Lima CD, Chua NH (2003) Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell 15: 1347–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ (2001) Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292: 1382–1385 [DOI] [PubMed] [Google Scholar]
- Melchior F (2000) SUMO—nonclassical ubiquitin. Annu Rev Cell Dev Biol 16: 591–626 [DOI] [PubMed] [Google Scholar]
- Mendoza HM, Shen LN, Botting C, Lewis A, Chen J, Ink B, Hay RT (2003) NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J Biol Chem 278: 25637–25643 [DOI] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, Lima CD (2000) Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell 5: 865–876 [DOI] [PubMed] [Google Scholar]
- Murtas G, Reeves PH, Fu YF, Bancroft I, Dean C, Coupland G (2003) A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of small ubiquitin-related modifier conjugates. Plant Cell 15: 2308–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A (2005) The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell 9: 769–779 [DOI] [PubMed] [Google Scholar]
- Novatchkova M, Budhiraja R, Coupland G, Eisenhaber F, Bachmair A (2004) SUMO conjugation in plants. Planta 220: 1–8 [DOI] [PubMed] [Google Scholar]
- Orth K, Xu Z, Mudgett MB, Bao ZQ, Palmer LE, Bliska JB, Mangel WF, Staskawicz B, Dixon JE (2000) Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290: 1594–1597 [DOI] [PubMed] [Google Scholar]
- Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM (2005) A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Biophys Res Commun 337: 517–520 [DOI] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S (2005) SUMO modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433 [DOI] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108: 109–120 [DOI] [PubMed] [Google Scholar]
- Pichler A, Knipscheer P, Oberhofer E, van Dijk WJ, Korner R, Olsen JV, Jentsch S, Melchior F, Sixma TK (2005) SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat Struct Mol Biol 12: 264–269 [DOI] [PubMed] [Google Scholar]
- Rajan S, Plant LD, Rabin ML, Butler MH, Goldstein SA (2005) Sumoylation silences the plasma membrane leak K+ channel K2P1. Cell 121: 37–47 [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Morton FR, Barrett AJ (2006) MEROPS: the peptidase database. Nucleic Acids Res 34: D270–D272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverter D, Lima CD (2004) A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure 12: 1519–1531 [DOI] [PubMed] [Google Scholar]
- Reverter D, Wu K, Erdene TG, Pan ZQ, Wilkinson KD, Lima CD (2005) Structure of a complex between Nedd8 and the ULP/Senp protease family member Den1. J Mol Biol 345: 141–151 [DOI] [PubMed] [Google Scholar]
- Ross S, Best JL, Zon LI, Gill G (2002) SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol Cell 10: 831–842 [DOI] [PubMed] [Google Scholar]
- Saitoh H, Hinchey J (2000) Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem 275: 6252–6258 [DOI] [PubMed] [Google Scholar]
- Sapetschnig A, Rischitor G, Braun H, Doll A, Schergaut M, Melchior F, Suske G (2002) Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J 21: 5206–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DC, Hochstrasser M (2003) A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem Sci 28: 321–328 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Stade K, Vogel F, Schwienhorst I, Meusser B, Volkwein C, Nentwig B, Dohmen RJ, Sommer T (2002) A lack of SUMO conjugation affects cNLS-dependent nuclear protein import in yeast. J Biol Chem 277: 49554–49561 [DOI] [PubMed] [Google Scholar]
- Stuible H, Buttner D, Ehlting J, Hahlbrock K, Kombrink E (2000) Mutational analysis of 4-coumarate:CoA ligase identifies functionally important amino acids and verifies its close relationship to other adenylate-forming enzymes. FEBS Lett 467: 117–122 [DOI] [PubMed] [Google Scholar]
- Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT (2001) Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem 276: 35368–35374 [DOI] [PubMed] [Google Scholar]
- Ulrich HD (2005) Mutual interactions between the SUMO and ubiquitin system: a plea of no contest. Trends Cell Biol 15: 525–532 [DOI] [PubMed] [Google Scholar]
- Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylides C, Roby D (2002) A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci USA 99: 10179–10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng XW (2003) The COP9 signalosome. Annu Rev Cell Dev Biol 19: 261–286 [DOI] [PubMed] [Google Scholar]
- Willems AR, Schwab M, Tyers M (2004) A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta 1695: 133–170 [DOI] [PubMed] [Google Scholar]
- Wu K, Yamoah K, Dolios G, Gan-Erdene T, Tan P, Chen A, Lee CG, Wei N, Wilkinson KD, Wang R, et al (2003) DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper- neddylated CUL1. J Biol Chem 278: 28882–28891 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Sharma P, Athanasiou M, Kumar A, Yamada S, Kuehn MR (2005) Mutation of SENP1/SuPr-2 reveals an essential role for desumoylation in mouse development. Mol Cell Biol 25: 5171–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, Zhang H (2002) CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell 10: 1519–1526 [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al (2002) Structure of the Cul1-Rbx1-Skp1-F box Skp2 SCF ubiquitin ligase complex. Nature 416: 703–709 [DOI] [PubMed] [Google Scholar]
- Zhou L, Watts FZ (2005) Nep1, a Schizosaccharomyces pombe deneddylating enzyme. Biochem J 389: 307–314 [DOI] [PMC free article] [PubMed] [Google Scholar]