Abstract
Chromoplastogenesis during flower development and fruit ripening involves the dramatic overaccumulation of carotenoids sequestered into structures containing lipids and proteins called plastid lipid-associated proteins (PAPs). CHRC, a cucumber (Cucumis sativus) PAP, has been suggested to be transcriptionally activated in carotenoid-accumulating flowers by gibberellin (GA). Mybys, a MYB-like trans-activator identified here, may represent a chromoplastogenesis-related factor: Its expression is flower specific and parallels that of ChrC during flower development; moreover, as revealed by stable ectopic and transient-expression assays, it specifically trans-activates ChrC promoter in flowers accumulating carotenoids and flavonoids. A detailed dissection of ChrC promoter revealed a GA-responsive element, gacCTCcaa, the mutation of which abolished ChrC activation by GA. This cis-element is different from the GARE motif and is involved in ChrC activation probably via negative regulation, similar to other GA-responsive systems. The GA responsiveness and MYBYS floral activation of the ChrC promoter do not overlap with respect to cis-elements. To study the functionality of CHRC, which is activated in vegetative tissues similar to other PAPs by various biotic and abiotic stresses, we employed a tomato (Lycopersicon esculentum) plant system and generated RNAi-transgenic lines with suppressed LeCHRC. Transgenic flowers accumulated approximately 30% less carotenoids per unit protein than controls, indicating an interrelationship between PAPs and flower-specific carotenoid accumulation in chromoplasts. Moreover, the transgenic LeCHRC-suppressed plants were significantly more susceptible to Botrytis cinerea infection, suggesting CHRC's involvement in plant protection under stress conditions and supporting the general, evolutionarily preserved role of PAPs.
Carotenoid-accumulating plastids, the chromoplasts, are responsible for the yellow, orange, and red colors of flower parts, fruits, old leaves, and some roots in various plant species. The pigment accumulates in structures composed of lipids and proteins, allowing sequestration/stabilization of the large amounts of carotenoids. Although any plastid can be converted to chromoplasts, they are usually derived from chloroplasts during fruit maturation and flower development. Studies on the molecular organization of carotenoid-overaccumulating structures have led to the identification and characterization of several nuclear genes encoding carotenoid-associated proteins (Deruere et al., 1994; Bartley and Scolnik, 1995; Vishnevetsky et al., 1999a; Murphy, 2004). The first gene involved with carotenoid storage, termed fibrillin (Fib), was identified in red fruit chromoplasts of Capsicum annuum (Newman et al., 1989; Deruere et al., 1994). In floral tissues, a 35-kD carotenoid-associated protein (CHRC) was characterized in chromoplasts of yellow cucumber (Cucumis sativus) corollas (Vainstein et al., 1994; Vishnevetsky et al., 1996). At both the protein and transcript levels, Fib and ChrC accumulation was shown to parallel carotenoid accumulation and fibril development in fruits and corollas (Deruere et al., 1994; Vishnevetsky et al., 1996). In fruit and flower tissues, spatial and temporal expression of these genes is regulated essentially at the transcriptional level. In leaves, their expression under standard greenhouse conditions is very low (Deruere et al., 1994; Chen et al., 1998; Vishnevetsky et al., 1999b). In floral tissue, GA3 plays a critical role in chromoplastogenesis: It leads to enhanced carotenoid accumulation as well as to transcriptional activation of ChrC expression. The response to GA was localized to a 290-bp fragment within the ChrC promoter that does not contain known GA-responsive elements (GARE/CARE; Gubler et al., 1999; Vishnevetsky et al., 1999b; Sutoh and Yamauchi, 2003).
Modulation of the composition and amount of carotenoids accumulating in chromoplasts affects the expression of ChrC. Nevertheless, its flower-specific transcriptional activation is not strictly dependent on accumulated carotenoids, but rather on an unidentified regulatory factor related to chromoplastogenesis. This proposal was based on the observation that in chromoplast-lacking flavonoid-accumulating flowers, ChrC promoter is not active, as revealed by transient-expression experiments (Vishnevetsky et al., 1999b).
Numerous CHRC/Fib homologs have been identified in different plant systems, as well as in plastids other than chromoplasts, and have therefore been collectively termed plastid lipid-associated proteins (PAPs; Pruvot et al., 1996; Kessler et al., 1999; Kim et al., 2001). Recently, using bioinformatics/genomic tools, several Arabidopsis (Arabidopsis thaliana) homologs have been identified and phylogenetically characterized (Laizet et al., 2004). The expression of PAPs in nonchromoplastogenic tissues led to the suggestion that they are involved not only in the storage of carotenoids but also in the general sequestration of hydrophobic compounds such as lipids. Supporting this hypothesis, Ting et al. (1998) and Hernandez-Pinzon et al. (1999) showed that CHRC homologs are components of tapetal lipid bodies in Brassica napus. Within the chloroplast of pea (Pisum sativum), the CHRC homolog PG1 was identified on the surface of lipid-containing structures—plastoglobules, which are the origin of carotenoid-accumulating structures in chromoplasts (Kessler et al., 1999).
The accumulation of PAPs in plastids as well as the biogenesis of structures that sequester hydrophobic compounds are accelerated by various stresses (Murphy, 2004). For example, the expression of Fib is transcriptionally up-regulated by both biotic and abiotic stresses (Chen et al., 1998; Kuntz et al., 1998; Manac'h and Kuntz, 1999; Langenkamper et al., 2001). Furthermore, gene expression and protein accumulation of the potato (Solanum tuberosum) PAP homolog CDSP34 in leaf chloroplasts are induced by different osmotic and oxidative stress conditions (Pruvot et al., 1996; Gillet et al., 1998; Langenkamper et al., 2001). This PAP has been implicated in the modulation of photosynthetic efficiency and in the dissipation of excess absorbed light energy (Monte et al., 1999). Similarly, the expression of B. napus and Arabidopsis PAPs is regulated by various abiotic stresses (Kim et al., 2001; Laizet et al., 2004). Moreover, Rey et al. (2000) have shown that overexpressing Fib in tobacco (Nicotiana tabacum) improves plant performance under stress conditions. Hence, it is apparent that in addition to chromoplastogenesis, PAPs play an important general role in the sequestration of hydrophobic molecules, a process that may be essential for plant survival under stress.
Here, we identified a MYB-like factor, MYBYS, which specifically trans-activates ChrC promoter in floral tissue irrespective of chromoplastogenesis, and dissected the ChrC promoter to identify a GA-responsive element, gacCTCcaa, involved in ChrC activation, probably via negative regulation. To characterize CHRC's role in vegetative tissues as well, we generated transgenic tomato (Lycopersicon esculentum) plants with RNAi-suppressed LeCHRC. Using these plants, we showed the involvement of CHRC not only in flower-specific carotenoid accumulation in chromoplasts but also in susceptibility to Botrytis cinerea infection.
RESULTS
MYB-Like Factor MYBYS Activates the ChrC Promoter
We previously found that in floral tissues, expression of ChrC depends on a factor related to chromoplastogenesis and that it is transcriptionally up-regulated by the hormone GA3 (Vishnevetsky et al., 1999b). With the aim of identifying this factor, we screened a cDNA expression library from cucumber flowers with a 137-bp segment of the ChrC promoter that is responsive to chromoplastogenesis and floral signals. A MYB-like transcription factor, having a typical DNA-binding domain containing the helix-turn-helix conserved R2 and R3 repeats, was identified and termed MYBYS (MYB-like, GenBank DQ311672). C-terminal to the DNA-binding domain, MYBYS contains a motif of amino acids (Gln and Pro) that is frequently associated with activation domains (Fig. 1A). Analyses of mybys expression at the RNA level in floral tissue revealed that it accumulates in parallel to flower development, up to anthesis, at which stage no transcript was revealed. No expression was detected in leaf tissues. This spatial and temporal pattern of expression is essentially identical to that of ChrC (Fig. 1B). To assess whether the identified MYBYS can activate ChrC promoter in floral tissue irrespective of chromoplastogenesis, we cobombarded mybys, under the regulation of cauliflower mosaic virus (CaMV) 35S (35S:MYBYS), with 137ChrC:β-glucuronidase (GUS) or ChrC:GUS (137 bp or 3,500 bp of the ChrC promoter fused to GUS, respectively) into petunia (Petunia hybrida) corollas. Trans-activation of ChrC promoter by MYBYS was revealed through the expression of GUS in chromoplast-lacking petunia corollas following cobombardment. No GUS expression was observed in petunia corollas following bombardment with 137ChrC:GUS or ChrC:GUS alone, or when they were cobombarded with an unrelated MYB driven by CaMV 35S (35S:PAP or 35S:Lc; Ben-Meir, 2003; Fig. 2A). Nor was GUS expression observed when 35S:MYBYS was cobombarded with a minimal TATA-box promoter fused to GUS or with the unrelated promoter construct glutathione S-transferase:GUS (Zenvirt, 2000). The same results, i.e. trans-activation of 137ChrC:GUS or ChrC:GUS specifically by MYBYS, were obtained when other flowers that do not accumulate chromoplasts, carnation (Dianthus caryophyllus) and gypsophila (Gypsophila paniculata), were used in the transient-expression assays (data not shown). In leaves, MYBYS was not sufficient for ChrC:GUS trans-activation.
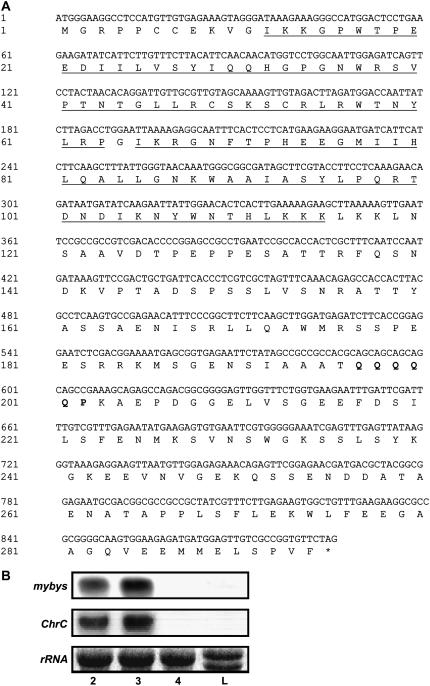
Figure 1.
A, Nucleotide and predicted amino acid sequences of mybys. The conserved R2/R3 DNA-binding domains at the 5′ end of the mybys (GenBank DQ311672) sequence are underlined and a typical activation domain at the 3′ end is in bold. The terminal codon is marked with an asterisk. B, Temporal and spatial regulation of mybys transcript levels in cucumber tissues. Total RNA extracted from cucumber leaves (L) and corollas at different developmental stages (stages 2–4) was probed with a radiolabeled fragment of mybys specific to the 3′ end of the gene. The same RNA blot was rehybridized with radiolabeled ChrC.
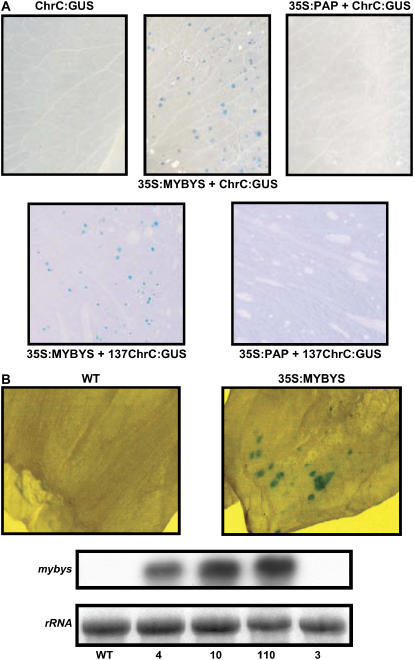
Figure 2.
MYBYS transcription factor specifically activates the ChrC promoter. A, Histochemical visualization of GUS activity in petunia flowers bombarded with ChrC:GUS alone, or ChrC:GUS or 137ChrC:GUS (containing GUS driven by 3,500 or 137 bp of the ChrC promoter, respectively) cobombarded with 35S:MYBYS. In control cobombardment experiments, MYBYS was replaced by another MYB factor, PAP, which regulates the anthocyanin pathway (35S:PAP). B, Histochemical visualization of GUS activity in young green transgenic tomato flowers constitutively expressing 35S:MYBYS (line 10) following bombardment with ChrC:GUS. As a control, nontransgenic flowers (WT) were also bombarded with ChrC:GUS and histochemically analyzed (top). Accumulation of mybys in 35S:MYBYS-transgenic tomato flowers (independent transgenic lines 4, 10, and 110) is shown in the bottom section. RNA-blot analysis was performed using radiolabeled 3′ mybys as a probe. A nontransgenic line (WT) and a transgenic line with no expression of mybys (line 3) were used as controls.
To further evaluate the activity of MYBYS in planta, we generated transgenic tomato plants expressing it under the CaMV 35S promoter. Tomato plants were used as they are highly amenable to transformation (see also below). Expression of mybys in young corollas of transgenic (T2-generation) plants, as compared to control nontransgenic ones, was confirmed by RNA-blot analysis (Fig. 2B, bottom). Bombardment of young green flower corollas with 137ChrC:GUS or ChrC:GUS yielded GUS expression in mybys-transgenic flowers but not in control plants (Fig. 2B). It should be noted that at this early stage of flower development, endogenous PAP is not yet expressed (Vishnevetsky, 1999).
GA-Responsive Element within the ChrC Promoter
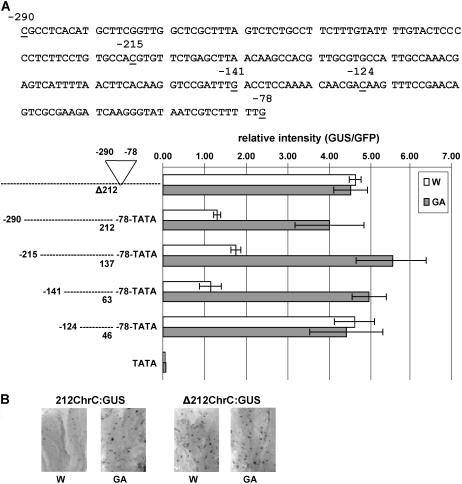
GA3 has been shown to promote chromoplastogenesis and CHRC accumulation in floral tissue (Vainstein et al., 1994). The accumulation of CHRC was shown to be regulated at the RNA level (Vishnevetsky et al., 1997). To detail GA3-responsive cis-elements in the ChrC promoter, we constructed a series of vectors (Fig. 3A) aimed at characterizing the 212-bp region shown to be responsible for the response to GA3 (Vishnevetsky et al., 1999b). One construct contained the ChrC promoter (3,500 bp) without the 212-bp region, fused to a GUS reporter gene (Δ212ChrC:GUS). The others contained 212 (from −290 to −78), 137 (−215 to −78), 63 (−141 to −78), or 46 (−124 to −78) bp of ChrC promoter fused via a minimal TATA-box promoter to the GUS reporter gene. All constructs were delivered by particle bombardment to young cucumber flower buds subjected to GA3 or water treatment. Construct containing CaMV 35S promoter fused to green fluorescent protein (GFP) was cobombarded and used to normalize the transient GUS expression results. Whereas the 212-, 137-, and 63-bp fragments of ChrC promoter were responsive to GA3, showing 3- to 4-fold higher GUS activity in GA3 versus water treatment, the 46-bp fragment was not affected by GA3 (Fig. 3A). Interestingly, this 46-bp fragment drove high GUS expression in control, water-treated corollas, suggesting that there may be a GA3-responsive repressor acting via a cis-element within the 18-bp region at the 5′ end of the 63-bp promoter region (between −141 and −124). In addition, GUS expression was similarly high in both water- and GA3-treated corollas when the vector with the deletion of the entire 212-bp region of ChrC promoter was used, further supporting repressor-mediated regulation of ChrC promoter by GA3 (Fig. 3, A and B).
Figure 3.
Identification of GA3-responsive cis-elements in the ChrC promoter region. A, ChrC promoter fragments fused to GUS via the 35S minimal promoter (TATA) were cobombarded with 35S:GFP into stage 1 cucumber corollas grown in vitro without (W) or with GA3 (GA). GUS expression was normalized to the GFP signal using ImageJ software. The results of five replicates ±se are presented. B, Histochemical visualization of GUS activity in stage 1 cucumber corollas grown in vitro without (W) or with GA3 (GA) following bombardment with ChrC promoter lacking 212 bp (−290 to −78) fused to GUS (Δ212ChrC:GUS) or the 212-bp fragment of the promoter (−290 to −78) fused to GUS via an 35S minimal promoter (212ChrC:GUS).
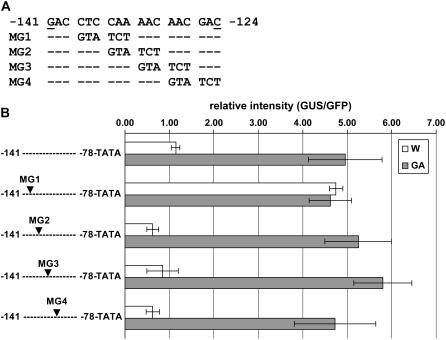
To further evaluate the GA-responsive cis-elements, we generated four mutations in the 18-bp region by replacing six bases, with three-base gaps between each of the four replacements (Fig. 4A). One construct (MG1) containing a mutation between −138 and −133, upon delivery to young flower corollas, yielded similarly high GUS expression in both water- and GA3-treated corollas, whereas other constructs, including the one with a mutation between −135 to −130 (MG2), were still responsive to GA3 in a manner similar to that of the control nonmutated promoter (Fig. 4B). These results point to the three bases, CTC, between −138 and −136 as necessary elements for the response of ChrC promoter to GA3 activation. It should be noted that none of these analyzed mutations in the 18-bp region affected activation of the ChrC promoter by MYBYS, suggesting that this trans-factor acts through different cis-elements present in the promoter.
Figure 4.
Effect of mutations on GA3 responsiveness of the ChrC promoter. A, Mutated region of the ChrC promoter. The 6-bp sequence GTA TCT was used to replace the original sequence of the promoter, with three-base gaps between each of the four (MG1–MG4) mutations. B, ChrC promoter fragments, original and mutated, fused to GUS via an 35S minimal promoter (TATA) were cobombarded with 35S:GFP into stage 1 cucumber corollas grown in vitro without (W) or with GA3 (GA). GUS expression was normalized to the GFP signal using ImageJ software. The results of five replicates ±se are presented.
Dual Role of CHRC: Carotenoid Accumulation and Stress Responses
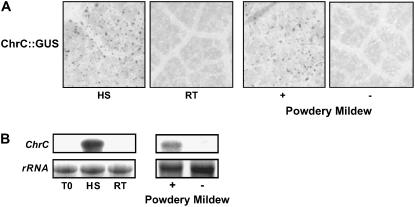
Various stresses, as shown in several plant systems, affect the accumulation of PAPs. For example, while under normal growth conditions PAP expression is specific for carotenoid-overaccumulating organs, biotic and abiotic stress conditions have been shown to activate their expression in additional tissues (Murphy, 2004). Also, analyses of ChrC promoter activity, using a ChrC:GUS transient-expression assay, revealed that it is induced in heat shock-treated cucumber leaves (Fig. 5A). Transcriptional activation of ChrC expression was also revealed following bombardment of powdery mildew-infected leaves. No GUS expression was detected in control, room temperature-treated or uninfected leaves (Fig. 5A). Moreover, endogenous transcript levels of ChrC in stressed leaves, both heat shock-treated and powdery mildew-infected, were strongly up-regulated (Fig. 5B).
Figure 5.
Induction of ChrC expression in cucumber leaves by biotic and abiotic stresses. A, Activation of ChrC promoter by heat shock and fungal inoculation. Cucumber leaves were cultured in vitro for 4 h at 42°C (HS) or room temperature (RT). In addition, leaves from plants infected (+) with powdery mildew Sphaerotheca fuliginea (Oidium sp.) were compared to control uninfected leaves (−). Following bombardment with ChrC:GUS, leaves were histochemically analyzed for GUS expression. B, Effect of heat shock and fungal inoculation on ChrC transcript levels. Total RNA was extracted from leaves treated as in A, and following blotting probed with radiolabeled ChrC. T0, Detached leaves prior to the in vitro culture.
To allow functional analyses of CHRC, which is responsive to numerous developmental and environmental cues, we cloned its tomato homolog with the aim of generating transgenic plants with suppressed CHRC expression. Tomato plants provide a useful research system because in addition to bearing chromoplasts accumulating CHRC homolog (Pozueta-Romero et al., 1997), their expressed sequence tag (EST) databases are elaborate and their transformation protocols well defined. Furthermore, in the tomato system, Fib promoter and endogenous PAP homolog have been shown to be activated by stresses in a manner similar to that of ChrC in cucumber tissues (Fig. 5; Kuntz et al., 1998; Langenkamper et al., 2001). Based on the partial sequence of the tomato homolog (Pozueta-Romero et al., 1997), we cloned the complete cDNA sequence of LeChrC (Fig. 6; GenBank DQ310151). Analyses of LeChrC expression at the RNA level revealed spatial and temporal regulation, mimicking that of ChrC in cucumber flowers and leaves (data not shown). It should be noted that LeChrC BLAST search in The Institute for Genomic Research (TIGR) database revealed one homolog, TC162898, putatively coding for PAP with 37% identity at the amino acid level.
Figure 6.
Comparison of CHRC homolog amino acid sequences. Multiple sequence alignment was performed with ClustalW (Thompson et al., 1994). GenBank accession numbers of the CHRC homologs are as follows: tomato LeChrC, DQ310151; C. annuum Fib, CAA50750; and cucumber CHRC, AAD05165. The transit peptide of the CHRC sequence is underlined. Identical residues in the column are marked with an asterisk (*), and conserved (:) and semiconserved (.) substitutions are indicated as well.
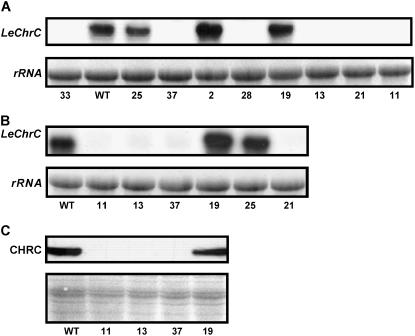
The RNAi approach was employed to generate transgenic tomato plants with suppressed LeCHRC. To this end, we used a 5′ partial sequence of LeChrC; when analyzed by BLAST against all tomato ESTs in the TIGR database (default parameters), this fragment did not reveal the second TC162898. Following transformation, the regenerated plants surviving selection on kanamycin were screened by PCR and characterized using RNA-blot analysis to evaluate suppression of LeChrC (Fig. 7A). The suppression of LeCHRC in T2-generation plants was further confirmed using RNA- and western-blot analyses (Fig. 7, B and C). Analysis of pigment level in corollas of independent T2 lines with suppressed LeCHRC revealed an approximately 30% reduction in carotenoid level per unit protein as compared to their level in control corollas, nontransgenic lines, or transgenic lines with no suppression of LeCHRC (Table I), indicating an effect of PAP on pigment accumulation. It should be noted that carotenoid levels per unit protein in leaves were essentially the same in transgenic and control plants (Table I).
Figure 7.
Transgenic tomato plants with suppressed LeCHRC. A and B, Molecular analysis of transgenic tomato flowers with suppressed LeChrC generated via the RNAi approach. Total RNA from stage 2 corollas was extracted and probed with radiolabeled LeChrC. RNA-blot analysis of RNAi LeChrC-transgenic plants (independent lines 11, 13, 21, 28, 33, and 37) versus control transgenic lines with no suppression (independent lines 2, 19, and 25) and control nontransgenic (WT) tomato plants is presented. Analyses were performed with both T0 (A) and T2 (B) generation plants. C, Western-blot analyses of CHRC levels in stage 2 corollas of RNAi LeChrC-transgenic plants (independent lines 11, 13, and 37) versus control transgenic lines with no suppression (line 19) and control nontransgenic (WT) tomato plants (T2 generation). Equal loading, as revealed by Ponceau-S red staining of the membrane prior to incubation with affinity-purified polyclonal antibodies against CHRC, is shown in the bottom section.
Table I.
Carotenoid levels in transgenic tomato with suppressed LeChrC expression versus control plants
Carotenoid content in stage 3 flowers and young leaves of T2 plants were measured in LeChrC RNAi (independent lines 11, 13, 21, and 37), control transgenic with no suppression (line 19), and control nontransgenic (WT) tomatoes. Means of four replicates ±se are presented.
| Line | Carotenoid in Corollas | Carotenoid in Leaves |
|---|---|---|
| ng/μg protein | ng/μg protein | |
| 21 | 4.6 ± 0.1 | 0.21 ± 0.02 |
| 11 | 4.4 ± 0.1 | 0.22 ± 0.01 |
| 37 | 4.3 ± 0.2 | 0.21 ± 0.02 |
| 13 | 4.1 ± 0.3 | 0.22 ± 0.01 |
| 19 | 5.8 ± 0.2 | 0.22 ± 0.02 |
| WT | 6.0 ± 0.3 | 0.23 ± 0.02 |
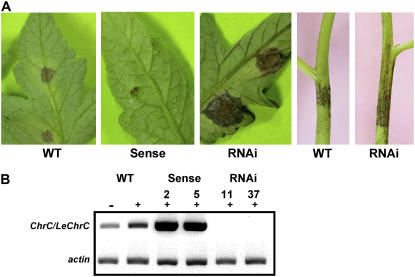
Over the course of several years of plant propagation, we observed that transgenic tomato lines with suppressed LeCHRC are highly susceptible to B. cinerea infection as compared to either control nontransgenic tomato or tomato transgenic for different, unrelated genes. To evaluate the susceptibility of LeCHRC-suppressed plants to B. cinerea infection, leaves were detached from these and control nontransgenic plants and inoculated with the conidia. Disease severity in leaves of plants with suppressed LeCHRC expression was 2- to 3-fold higher than that in control plants (Table II). To further evaluate this susceptibility, a conidial suspension was applied to leaves and stems of 1-month-old plants. Inoculated plants were then grown in a growth chamber (Table III; Fig. 8A). Three days after inoculation, there was no significant difference in the size of the necrotic lesions on leaves of transgenic LeCHRC-suppressed versus control plants. However, during the following 3 d of growth, lesions on the transgenic leaves increased rapidly, reaching approximately twice the size of controls. A significant difference was also observed in the necrotic lesions developed on stems of transgenic LeCHRC-suppressed plants versus control nontransgenic ones: Whereas the lesions on the control plants remained restricted in size, from days 6 to 8 after inoculation those on LeCHRC-suppressed plants spread, covering up to 60% more area (versus controls) 8 d postinoculation. Mock-inoculated plants did not generate lesions on either stems or leaves. Moreover, tomato transgenic lines characterized by Vishnevetsky et al. (1999b) that overaccumulate CHRC were significantly less susceptible to B. cinerea infection, as compared to both control nontransgenic plants and LeCHRC-suppressed plants (Table III), further indicating the involvement of LeCHRC in the plant's resistance to the fungus. As expected, following inoculation with B. cinerea, no expression of LeChrC was detected in RNAi-transgenic plants, whereas in control plants LeChrC levels increased (Fig. 8B).
Table II.
Susceptibility of detached transgenic tomato leaves with modulated ChrC expression levels to B. cinerea infection
Detached leaves from transgenic tomato overexpressing ChrC (sense), control nontransgenic (WT), and RNAi-suppressed leaves (independent lines 11, 13, and 37) were infected with B. cinerea. Disease severity on leaves, measured 3 and 6 d after inoculation, was determined by estimating the size of the necrotic area on a scale of 0% to 100%, where 100% severity = a lesion of 20 mm2. Numbers followed by a common letter are not significantly different (t test, P ≤ 0.05).
| Line | Disease Severity on Leaves
|
|
|---|---|---|
| 3 d | 6 d | |
| % | ||
| Sense | 0.6 c | 3.6 c |
| 11 | 1.6 b | 19.0 a |
| 13 | 1.2 b | 15.0 a |
| 37 | 1.9 a | 19.0 a |
| WT | 0.9 c | 6.5 b |
Table III.
Susceptibility of transgenic tomato plants with modulated ChrC expression levels to B. cinerea infection
Transgenic tomato overexpressing ChrC (sense), control nontransgenic (WT), and RNAi-suppressed plants (independent lines 11, 13, and 37) were infected with B. cinerea. Disease severity on leaves, measured 3 and 6 d after inoculation, was determined by estimating the size of the necrotic area on a scale of 0% to 100%, where 100% severity = a lesion of 20 mm2. Stem infection, 6 and 8 d after inoculation, was evaluated by measuring the length of the lesion. Numbers followed by a common letter are not significantly different (t test, P ≤ 0.05).
| Line | Disease Severity on Leaves
|
Lesion Size on Stem
|
||
|---|---|---|---|---|
| 3 d | 6 d | 6 d | 8 d | |
| % | mm | |||
| Sense | 3.5 b | 21.5 c | 12.6 d | 21.7 d |
| 11 | 6.4 b | 89.8 a | 35.5 a | 43.7 ab |
| 13 | 3.4 b | 83.5 a | 29.3 b | 47.3 a |
| 37 | 5.0 b | 86.8 a | 22.2 c | 37.0 b |
| WT | 4.6 b | 54.8 b | 28.7 b | 28.7 c |
Figure 8.
Susceptibility of transgenic tomato plants with modulated ChrC expression levels to B. cinerea infection. Transgenic tomato overexpressing ChrC (sense lines 2 and 5), control nontransgenic (WT), and RNAi LeChrC-suppressed plants (RNAi lines 11 and 37) were infected with B. cinerea. A, B. cinerea disease symptoms in leaves and stems 6 and 8 d after inoculation, respectively. B, RT-PCR analysis of ChrC/LeChrC (and actin as a control) expression in control and transgenic leaves prior to infection (−) or 3 d after infection (+) with B. cinerea.
DISCUSSION
From evolutionary and phylogenetic standpoints, PAPs appear to be a very old and conserved group of genes (Vishnevetsky et al., 1999a). Their role in sequestering overaccumulated carotenoids within chromoplasts of angiosperm reproductive organs may be a rather recent development, whereas their original function may have been to aid in the sequestration of hydrophobic molecules and protect against various stresses. Indeed, PAPs are up-regulated by various stresses, e.g. drought, mechanical wounding, treatments generating active oxygen species, and biotic stresses (Pruvot et al., 1996; Chen et al., 1998; Gillet et al., 1998; Manac'h and Kuntz, 1999; Langenkamper et al., 2001). Numerous stress conditions, such as heat shock at 42°C and inoculation with different viruses (Tomato mosaic virus, Potato virus Y, and Tomato yellow leaf curl virus) and fungi (Oidium sp. and B. cinerea), resulted in the induction of ChrC and LeChrC in leaves of cucumber and tomato, respectively (Fig. 5B; data not shown). Using ChrC promoter fused to GUS, we further showed (Fig. 5) that similar to Fib, activation by both biotic and abiotic stresses occurs at the transcriptional level. Monte et al. (1999) and Rey et al. (2000) have shown that suppression of the PAP CDSP34 leads to increased sensitivity of potato to photooxidative stress, and overexpression of Fib in tobacco improves plant performance under high-light conditions. Oxidative events are also strongly implicated in the interaction of B. cinerea with plants (Malolepsza, 2005). This pathogen has a broad host range, causing gray mold on more than 200 plant species, including tomato (Elad et al., 2004). Using a tomato plant system, we show that LeCHRC expression is needed for resistance to B. cinerea infection. Transgenic plants with suppressed LeCHRC were significantly more susceptible to infection in both in vitro experiments with detached leaves and in the growth chamber with intact leaves and stems (Tables II and III).
The production of active oxygen species during infection of plants by B. cinerea has been well documented, and there is growing evidence that the fungus exploits this situation to aid its development in planta (Lyon et al., 2004). There is now evidence (Urbanek et al., 1996; Von Tiedemann, 1997) that the generation of active oxygen species assists in the colonization of plant tissues by necrotrophic organisms, such as B. cinerea. One of the most obvious and consistent effects of B. cinerea infection is a large increase in the electron paramagnetic resonance free-radical signal in rotted tissue compared to controls; similar results have been obtained with leaves and fruit (Deighton et al., 1999; Muckenschnabel et al., 2001). LeCHRC, similar to other PAPs, may assist in contending with the oxidative stress: Its suppression may lower the plant's ability to withstand the oxidative stress associated with B. cinerea attack, hence leading to increased susceptibility to infection.
The involvement of LeCHRC in resistance to biotic stresses further supports the hypothesized general, evolutionarily preserved role of PAPs in plant protection. Conservation of PAPs is also evident from the fact that even the sequence of a Synechocystis homolog shares approximately 30% identity with plant PAPs (Vishnevetsky et al., 1999a). It is reasonable to assume that during their evolution, PAPs were harnessed for diverse cellular activities. Originally, PAPs were proposed to play a role in the organization of carotenoid-accumulating structures in flower and fruit chromoplasts. This was mainly based on their intrachromoplast location and on the correlation between their expression pattern and carotenoid levels in chromoplasts during flower/fruit development (Deruere et al., 1994; Vishnevetsky et al., 1999a). Here, we further evaluated the role of CHRC in carotenoid accumulation within chromoplasts in floral tissues, using transgenic tomato plants with suppressed LeCHRC. Analyses of carotenoid levels in flowers with down-regulated LeCHRC relative to controls revealed that the former accumulate up to 32% less carotenoids per unit protein, strengthening the notion of an interrelationship between PAPs and flower-specific carotenoid accumulation in chromoplasts.
The functional characterization of PAPs by RNAi is strongly hindered by the presence in plastids of several members of this family, as has been shown in various plant systems (Laizet et al., 2004; Murphy, 2004; Ytterberg et al., 2006). In Arabidopsis, for example, 13 genes encoding members of this family have recently been identified, some of which appear to be the result of recent gene duplication (Laizet et al., 2004). The functional divergence of PAPs is also evidenced by their different patterns of expression, as revealed by both proteome and transcriptome analyses, in chromoplastogenic versus nonchromoplastogenic tissue (Laizet et al., 2004; Murphy, 2004; Ytterberg et al., 2006). A search in the National Center for Biotechnology Information and TIGR databases for PAPs reveals only one contig in cucumber and two homologs (TC162898 and TC161992, the latter characterized here) in tomato. The homology between the two tomato open reading frames is low: 44.5% at the nucleotide level with 37% identity at the amino acid level. The LeChrC characterized here is 95.7% identical in amino acids with Fib, whereas the second homolog (TC162898) is identical in only 37.4% of its amino acids. Clearly, we cannot exclude the occurrence of additional PAP homologs with redundant or overlapping functions, as no genome sequence is currently available for the plant species studied here. To evaluate the role of LeCHRC, we applied an RNAi approach using a nonconserved 5′ gene region. This in itself does not ensure specificity of suppression for individual family members (Valencia-Sanchez et al., 2006). However, since only two LeChrC homologs were identified in the databases and the fragment used for RNAi was generated from a divergent region, and since the promoter of ChrC is responsive to both chromoplastogenesis and stress signals, we are tempted to propose that the characterized phenotype is due to LeChrC suppression.
Regulation at the RNA level has been shown to control PAPs in diverse processes/tissues, i.e. developmental and hormonal expression and induction by biotic and abiotic stresses of both ChrC and Fib (Fig. 5; Kuntz et al., 1998; Vishnevetsky et al., 1999b). While carotenoid type/amount exerts a posttranscriptional effect on ChrC expression, isoprenoid pathway-related, flower-specific trans-factors were proposed to modify its expression at the transcriptional level (Vishnevetsky et al., 1999b). The MYBYS characterized here may represent such a ChrC-activating trans-factor. Its expression is spatially controlled with no detectable levels in leaves. In floral tissue, it is temporally regulated, mimicking the increase in ChrC levels during flower development (Fig. 1B). The ability of MYBYS to specifically trans-activate ChrC promoter was shown by two alternative approaches (Fig. 2). Ectopic expression of MYBYS was sufficient for activation of ChrC promoter in young green tomato flowers. Using cobombardment of ChrC:GUS and 35S:MYBYS, GUS expression was also revealed in floral tissues of different plants that accumulate anthocyanin and not carotenoids, such as petunia and carnation. No GUS expression was observed when MYBYS or ChrC promoter was replaced with other transcription factors/promoters. In leaves, no trans-activation of ChrC promoter was observed, indicating that MYBYS is not sufficient to activate ChrC promoter in tissues other than flowers. It should be noted that the ability of MYBYS to specifically activate ChrC promoter was recently harnessed to develop a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants (Tzfira et al., 2005).
In addition to isoprenoid pathway-related trans-factor MYBYS, also GA3, a product of the pathway, has been shown to rapidly activate ChrC expression in floral tissues in parallel to chromoplastogenesis. Activation of ChrC by GA, localized to a 290-bp region of the promoter, was characterized as a primary response (Vishnevetsky et al., 1997, 1999b), in contrast to some other well-established, GA-regulated systems, such as the flavonoid pathway in flowers and activation of the α-amylase gene in aleurone cells, which are slow-response processes that depend on de novo protein synthesis (Weiss et al., 1992; Huttly and Phillips, 1995). A detailed characterization of mainly GA-responsive mutants in various plants allowed the identification of DELLA proteins functioning as negative regulators in the GA-signaling cascade (Gomi and Matsuoka, 2003). In barley (Hordeum vulgare), GAMyb, a transcriptional activator of α-amylase gene expression that is rapidly regulated by GA, was shown to be the target, although not necessarily a direct one, for this class of proteins (Gubler et al., 2002). However, cis-elements responsible for its GA responsiveness, in contrast to GARE/CARE elements of the slow-response α-amylase genes, were not characterized (Gubler et al., 1999; Sutoh and Yamauchi, 2003). Dissection of 290 bp of the ChrC promoter revealed an 18-bp motif that was necessary for the GA responsiveness. Further scanning of this motif allowed the identification of three nucleotides (gacCTCcaa), the mutation of which abolished GA responsiveness. Interestingly, this new motif is partially similar to the GA-responsive CARE regulatory element (CAACTC) in the promoters of rice (Oryza sativa) and barley Cys proteinases (REP-1 and EPB1) and the rice α-amylase gene (RAmy1A; Sutoh and Yamauchi, 2003). Deletion of the 18 bp or mutation of three nucleotides in the GA-responsive motif of the ChrC promoter (Figs. 3 and 4) led to its increased activity, to a level similar to that of the native promoter following GA activation. This suggests that negative regulation, similar to other GA-responsive systems, underlies the response of ChrC to GA. The observation that responsiveness of the promoter to GA and MYBYS, both related to the isoprenoid pathway, does not overlap with respect to cis-elements brings additional support to the notion that throughout evolution, plants have integrated diverse functions into PAPs. To this end, their involvement in resistance to stresses/diseases further emphasizes the complexity of the machinery governing the multitasking of plant components.
MATERIALS AND METHODS
Plant Material and in Vitro Flower Bud Culture
Cucumber (Cucumis sativus L. cv Shimshon) and tomato (Lycopersicon esculentum Mill. cv Adi) plants were grown under standard greenhouse conditions (Vishnevetsky et al., 1999b). Flower development was divided into stages: In cucumber, stages 1, 2, and 3 represent flowers 120, 72, and 24 h before anthesis, respectively, and stage 4 flowers are at anthesis; in tomato, stages 1, 2, and 3 represent flowers 84, 48, and 12 h before anthesis, respectively, stage 4 flowers being at anthesis.
In vitro culture of flower buds was performed as described previously (Vishnevetsky et al., 1997). Stage 1 flower buds were cultured in double distilled water for 12 h and then transferred to fresh double distilled water or 100 μm GA3 (Sigma-Aldrich) for another 12 h prior to bombardment.
Isolation of the MYBYS Regulatory Factor
A λZap-cDNA expression library from stage 3 flower corollas (Vishnevetsky et al., 1996) was screened with radiolabeled promoter region of ChrC (137-bp fragment, positions −215 to −78) to identify cDNA clones encoding ChrC-DNA-binding protein. Approximately 106 bacteriophages were screened in the presence of an excess of nonspecific competitor calf thymus DNA (Ausubel et al., 2001), and, following two rounds of rescreening, potential recombinants of interest were isolated and sequenced.
Constructs for Transient Expression and Particle Bombardment
The mybys gene was introduced into a PCD vector (Broido et al., 1991) 3′ to the CaMV 35S promoter, to generate 35S:MYBYS plasmid. The ChrC:GUS construct (pGEM3Z/201.2; Vishnevetsky et al., 1999b), containing 3,500 bp of ChrC promoter upstream of the GUS gene, was digested with EheI and EcoRI to generate the deletion within the ChrC promoter region between positions −290 and −78 (Δ212ChrC:GUS). The 212-bp fragment (position −290 to −78) released from the ChrC promoter region was introduced 5′ to a minimal (−46 to +8) TATA-box promoter fused to a GUS reporter gene, creating 212ChrC:GUS. This plasmid was used in all further ChrC promoter 5′ deletions. Plasmid containing a promoter fragment (−215 to −78) fused to TATA:GUS (137ChrC:GUS) was generated following digestion of 212ChrC:GUS with PmaCI and ApaI and blunt ligation. To generate plasmids containing promoter fragments −141 to −78 and −124 to −78 fused to TATA:GUS, PCR fragments of the relevant promoter regions were used. Primers used for the generation of the former fragment were F-63 (5′-GACCTCCAAAACAACGACA-3′) and R (5′-TCACGGGTTGGGGTTTCTAC-3′), and for the latter fragment primers were F-46 (5′-CAAGTTTCCGAACAGTCGCG-3′) and R. Mutagenesis of the four adjacent regions (six nucleotides each) within the −141 to −124 promoter fragment was generated using primers containing a GTATCT replacement of the promoter sequence: MG1 (5′-GTATCTAACAACGACAAGTTTCCGAA-3′), MG2 (5′-CTCGTATCTAACGACAAGTTTCCGAA-3′), MG3 (5′-CTCCAAGTATCTGACAAGTTTCCGAA-3′), and MG4 (5′-GACCTCCAAAACGTATCTAAGTTTCCGAA-3′).
Transient expression of the described constructs in leaves and petals was evaluated following bombardment using the Biolistic PDS 1000/He system (Bio-Rad) at a pressure of 1,350 dpi (effector to reporter taken in 1:1 molar ratio), as described by Vishnevetsky et al. (1999b). In MYBYS transient-expression experiments, the transcriptional regulator Pap1 (production of anthocyanin pigment 1) fused to CaMV 35S (35S:PAP vector; Ben-Meir, 2003) was used as a control MYB factor. Following bombardment, tissue was incubated for a few hours at 37°C in a 0.1% (w/v) X-Gluc (5-bromo-4-chloro-3-indolyl β-d-GlcUA; Duchefa) solution containing 0.1 m sodium phosphate buffer, pH 7.0, 10 mm EDTA, and 0.1% (w/v) Triton X-100 (Vishnevetsky, 1999). In GA experiments, EGFP driven by CaMV 35S (pEGFP-PL vector; Ben-Nissan et al., 2004) was cobombarded as reference, and GFP expression was monitored using a fluorescence binocular (480/40 nm excitation filter and 510 nm barrier filter; MZ FLIII; Leica) equipped with a DC300FX camera (Leica), prior to transfer of the tissue to the X-Gluc solution. GUS expression was normalized to GFP using ImageJ software (Bezanilla et al., 2003). All experiments were repeated at least five times.
LeChrC and MYBYS Vectors and Tomato Transformation
Based on the tomato EST database, the ChrC homolog was identified (TC98907) and cloned (LeChrC, 775 bp) from a tomato petal cDNA library using two primers: F47 (5′-TGCTCTTTCTCTGTTTCACTCTGA-3′) and R822 (5′-TTGTCCCAAGAATTCAACGTTC-3′). A 5′ LeChrC fragment (530 bp) generated using PCR primers 5′-ATGGCTTCCATCTCTTCTCTCA-3′ and 5′-TCGAACCAGAAGCAGATTGC-3′ was cloned into pRNA69 (Waterhouse et al., 1998) 3′ to the CaMV 35S promoter in an antisense and sense orientation before and after the intron, respectively. The resultant plasmid was digested with NotI, and the fragment was inserted into the binary vector pART27 to create the RNAi construct (Gleave, 1992). For the MYBYS sense construct, 35S:MYBYS was released from 35S:MYBYS plasmid using BamHI and XbaI and transferred to the binary vector pCGN1559 (Comai et al., 1990). Binary vectors were electroporated into Agrobacterium tumefaciens strain AGLO and used for transformation of tomato cv Adi as described by Vishnevetsky et al. (1999b).
RNA-Blot, RT-PCR, and Western-Blot Analyses
Total RNA, isolated as described previously (Vishnevetsky et al., 1996), was used in either blot or RT-PCR analysis. For the former, 10 μg of total RNA was fractionated through a 1.6% formaldehyde gel and transferred to a Hybond-N+ filter (Amersham Biosciences). cDNA clones of mybys (371–885 nucleotide fragment, upstream of the conserved R2/R3 DNA-binding sites), ChrC, and LeChrC served as specific probes following labeling with 32P using a random priming kit (Rediprime; Amersham Biosciences). The blots were hybridized as described by Vishnevetsky et al. (1996). Hybridization was at 60°C, and the membranes were washed twice in 2× SSC, 0.1% (w/v) SDS at 60°C for 20 min each and exposed to x-ray film (Fuji) at −70°C. For RT-PCR, total RNA was treated with RNase-free DNase (Promega) and transcribed using oligo(dT)15 primer (Promega) and M-MLV reverse transcriptase (Promega) according to the manufacturer's recommendations. For PCR amplification of LeChrC, primers F47 and R822 were used. The reaction was performed in the presence of an additional two primers, 5′-GGTTTTGCTGGGGATGC-3′ and 5′-CATTGAATGTCTCAAACAGTATTTGAGTC-3′, allowing amplification of reference actin cDNA.
Proteins were extracted from stage 3 tomato corollas, fractionated by 12.5% SDS-PAGE (50 μg per lane), and analyzed following western blotting using affinity-purified polyclonal antibodies against CHRC and a chemiluminescence detection kit (Amersham Biosciences; Vishnevetsky et al., 1999b). Membranes were then exposed to x-ray film (Fuji). Prior to incubation with antibodies, membrane was stained with Ponceau-S red (Sigma-Aldrich).
Botrytis Infection and Measurements
Botrytis cinerea (isolate BcI16; Guetsky et al., 2002) was maintained and grown for infection experiments on potato dextrose agar. Conidia were harvested from the cultures by agitating small pieces of 14-d-old agar bearing mycelium and conidia in a glass tube containing 2 mL of tap water and 0.01% (w/v) Tween-80. The suspension was filtered through a double layer of cheesecloth to screen out mycelium plugs, and the conidial concentration was calibrated by means of a hemacytometer and adjusted to 5 × 105 cells/mL. Glc (0.05%, w/v) and KH2PO4 (0.05%, w/v) were added to the conidial suspension. Detached leaves were infected by placing 8- to 20-μL drops of the suspension on each of six leaves from each plant line. The leaves were then placed on a plastic grid that was laid over moist paper, all of which was then placed in a box covered with transparent polyethylene to ensure high humidity. In experiments with whole plants, four leaflets of each of four leaves on six plant replicates (1-month-old tomato plants) were infected with 20 μL of conidial suspension and covered with a polyethylene bag: Care was taken to avoid contact between the polyethylene and the inoculum drop. Stem infection was carried out with 5-mm-diameter mycelium discs that originated from the edge of a 4-d-old potato dextrose agar culture of B. cinerea. The mycelium disc was placed on the stem surface between the second and third nodes. Leaves and plants were incubated in a walk-in growth chamber set at 20°C ± 2°C and a 12-h photoperiod.
Disease severity on leaves in all experiments was determined by estimating the size of the necrotic area developed from each suspension drop on a scale of 0% to 100%, where 100% severity = a lesion of 20 mm2 (Guetsky et al., 2002). Severity of stem infection was evaluated by measuring the length of the lesion.
Analytical Methods
Carotenoid contents were determined as described by Lichtenthaler (1987). Protein content was determined using a detergent-compatible protein assay (Bio-Rad).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ311672 and DQ310151.
This work was supported by the Israel Science Foundation (grant no. 466/01).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Alexander Vainstein (vain@agri.huji.ac.il).
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (2001) Current Protocols in Molecular Biology. John Wiley and Sons, New York
- Bartley G, Scolnik P (1995) Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant Cell 7: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Meir H (2003) Characterization of the flavonoid biosynthetic pathway in Gypsophila flowers. PhD thesis. Hebrew University of Jerusalem, Jerusalem
- Ben-Nissan G, Lee J-Y, Borohov A, Weiss D (2004) GIP, a Petunia hybrida GA-induced cysteine-rich protein: a possible role in shoot elongation and transition to flowering. Plant J 37: 229–238 [DOI] [PubMed] [Google Scholar]
- Bezanilla M, Pan A, Quatrano RS (2003) RNA interference in the moss Physcomitrella patens. Plant Physiol 133: 470–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broido S, Loyter A, Vainstein A (1991) Expression of plant genes in transfected mammalian cells: accumulation of recombinant preLHC IIb protein within cytoplasmic inclusion bodies. Exp Cell Res 192: 248–255 [DOI] [PubMed] [Google Scholar]
- Chen HC, Klein A, Xiang MH, Backhaus RA, Kuntz M (1998) Drought- and wound-induced expression in leaves of a gene encoding a chromoplast carotenoid-associated protein. Plant J 14: 317–326 [Google Scholar]
- Comai L, Moran P, Maslyar D (1990) Novel and useful properties of a chimeric plant promoter combining CaMV 35S and MAS elements. Plant Mol Biol 15: 373–381 [DOI] [PubMed] [Google Scholar]
- Deruere J, Romer S, d'Harlingue A, Backhaus RA, Kuntz M, Camara B (1994) Fibril assembly and carotenoid overaccumulation in chromoplasts: a model for supramolecular lipoprotein structures. Plant Cell 6: 119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deighton N, Muckenschnabel I, Goodman BA, Williamson B (1999) Lipid peroxidation and the oxidative burst associated with infection of Capsicum annuum by Botrytis cinerea. Plant J 20: 485–492 [DOI] [PubMed] [Google Scholar]
- Elad Y, Williamson B, Tudzynski P, Delen N (2004) Botrytis spp. and diseases they cause in agricultural systems—an introduction. In Y Elad, B Williamson, P Tudzynski, N Delen, eds, Botrytis: Biology, Pathology and Control. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–8
- Gillet B, Beyly A, Peltier G, Rey P (1998) Molecular characterization of CDSP 34, a chloroplastic protein induced by water deficit in Solanum tuberosum L. plants, and regulation of CDSP 34 expression by ABA and high illumination. Plant J 16: 257–262 [DOI] [PubMed] [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organization structure conducive to efficient integration of cloned DNA into plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Gomi K, Matsuoka M (2003) Gibberellin signaling pathway. Curr Opin Plant Biol 6: 489–493 [DOI] [PubMed] [Google Scholar]
- Gubler F, Chandler PM, White RG, Llewellyn DJ, Jacobsen JV (2002) Gibberellin signaling in barley aleurone cells. Control of SLN1 and GAMYB expression. Plant Physiol 129: 191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV (1999) Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J 17: 1–9 [DOI] [PubMed] [Google Scholar]
- Guetsky R, Elad Y, Shtienberg D, Dinoor A (2002) Improved biocontrol of Botrytis cinerea on detached strawberry leaves by adding nutritional supplements to a mixture of Pichia guillermondii and Bacillus mycoides. Biocontrol Sci Technol 12: 625–630 [Google Scholar]
- Hernandez-Pinzon I, Ross JHE, Barnes KA, Damant AP (1999) Composition and role of tapetal lipid bodies in the biogenesis of the pollen coat of Brassica napus. Planta 208: 588–598 [DOI] [PubMed] [Google Scholar]
- Huttly AK, Phillips AL (1995) Gibberellin-regulated plant genes. Physiol Plant 95: 310–317 [Google Scholar]
- Kessler F, Schnell D, Blobel G (1999) Identification of proteins associated with plastoglobules isolated from pea (Pisum sativum L.) chloroplasts. Planta 208: 107–113 [DOI] [PubMed] [Google Scholar]
- Kim HU, Wu SH, Ratnayake C, Huang AHC (2001) Brassica rapa has three genes that encode proteins associated with different neutral lipids in plastids of specific tissues. Plant Physiol 126: 330–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz M, Chen HC, Simkin AJ, Romer S, Shipton CA, Drake R, Schuch W, Bramley PM (1998) Upregulation of two ripening-related genes from a non-climacteric plant (pepper) in transgenic climacteric plant (tomato). Plant J 13: 351–361 [Google Scholar]
- Laizet Y, Pontier D, Mache R, Kuntz M (2004) Subfamily organization and phylogenetic origin of genes encoding plastid lipid-associated proteins of the fibrillin type. J Genome Sci Tech 3: 17–26 [Google Scholar]
- Langenkamper G, Manac'h N, Broin M, Cuine S, Becuwe N, Kuntz M, Rey P (2001) Accumulation of plastid lipid-associated proteins (fibrillin/CDSP34) upon oxidative stress, ageing and biotic stress in Solanaceae and in response to drought in other species. J Exp Bot 52: 1545–1554 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Lyon GD, Goodman BA, Williamson B (2004) Botrytis cinerea perturbs redox processes as an attack strategy in plants. In Y Elad, B Williamson, P Tudzynski, N Delen, eds, Botrytis: Biology, Pathology and Control. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 119–141
- Malolepsza U (2005) Spatial and temporal variation of reactive oxygen species and antioxidant enzymes in o-hydroxyethylorutin-treated tomato leaves inoculated with Botrytis cinerea. Plant Pathol 54: 317–324 [Google Scholar]
- Manac'h N, Kuntz M (1999) Stress induction of a nuclear gene encoding for a plastid protein is mediated by photo-oxidative events. Plant Physiol Biochem 37: 859–868 [DOI] [PubMed] [Google Scholar]
- Monte E, Ludevid D, Prat S (1999) Leaf C40.4: a carotenoid-associated protein involved in the modulation of photosynthetic efficiency. Plant J 19: 399–410 [DOI] [PubMed] [Google Scholar]
- Muckenschnabel I, Goodman BA, Deighton N, Lyon GD, Williamson B (2001) Botrytis cinerea induces the formation of free radicals in fruits of Capsicum annuum at positions remote from the site of infection. Protoplasma 218: 112–116 [DOI] [PubMed] [Google Scholar]
- Murphy DJ (2004) The roles of lipid bodies and lipid-body proteins in the assembly and trafficking of lipids in plant cells. In Proceedings of the 16th International Plant Lipid Symposium. Hungarian Scientific Society for Food Industry and Budapest University of Economic Sciences and Public Administration Faculty of Food Science Budapest, Budapest, pp 55–62
- Newman L, Hadjeb N, Price C (1989) Synthesis of two chromoplast-specific proteins during fruit development in Capsicum annuum. Plant Physiol 91: 455–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozueta-Romero J, Rafia F, Houlne G, Cheniclet C, Carde JP, Schantz ML, Schantz R (1997) A ubiquitous plant housekeeping gene, PAP, encodes a major protein component of bell pepper chromoplasts. Plant Physiol 115: 1185–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruvot G, Cuine S, Peltier G, Rey P (1996) Characterization of a novel drought-induced 34-kDa protein located in the thylakoids of Solanum tuberosum L. plants. Planta 198: 471–479 [DOI] [PubMed] [Google Scholar]
- Rey P, Gillet B, Romer S, Eymery F, Massimino J, Peltier G, Kuntz M (2000) Over-expression of a pepper plastid lipid-associated protein in tobacco leads to changes in plastid ultrastructure and plant development upon stress. Plant J 21: 483–494 [DOI] [PubMed] [Google Scholar]
- Sutoh K, Yamauchi D (2003) Two cis-acting elements necessary and sufficient for gibberellin-upreguated proteinase expression in rice seeds. Plant J 34: 635–645 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JTL, Wu SSH, Ratnayake C, Huang AHC (1998) Constituents of the tapetosomes and elaioplasts in Brassica campestris tapetum and their degradation and retention during microsporogenesis. Plant J 16: 541–551 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Tian G-W, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in planta. Plant Mol Biol 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Urbanek H, Gajewska E, Karwowska R, Wielanek M (1996) Generation of superoxide anion and induction of superoxide dismutase and peroxidase in bean leaves infected with pathogenic fungi. Acta Biochim Pol 43: 679–685 [PubMed] [Google Scholar]
- Vainstein A, Halevy A, Smirra I, Vishnevetsky M (1994) Chromoplast biogenesis in Cucumis sativus corollas. Plant Physiol 104: 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R (2006) Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20: 515–524 [DOI] [PubMed] [Google Scholar]
- Vishnevetsky M (1999) Chromoplast biogenesis during flower development. PhD thesis. Hebrew University of Jerusalem, Jerusalem
- Vishnevetsky M, Ovadis M, Itzhaki H, Levy M, Libal-Weksler Y, Adam Z, Vainstein A (1996) Molecular cloning of a carotenoid-associated protein from Cucumis sativus corollas: homologous genes involved in carotenoid sequestration in chromoplasts. Plant J 10: 1111–1118 [DOI] [PubMed] [Google Scholar]
- Vishnevetsky M, Ovadis M, Itzhaki H, Vainstein A (1997) CHRC, encoding a chromoplast-specific carotenoid-associated protein, is an early gibberellic acid-responsive gene. J Biol Chem 272: 24747–24750 [DOI] [PubMed] [Google Scholar]
- Vishnevetsky M, Ovadis M, Vainstein A (1999. a) Carotenoid sequestration in plants: the role of carotenoid-associated proteins. Trends Plant Sci 4: 232–235 [DOI] [PubMed] [Google Scholar]
- Vishnevetsky M, Ovadis M, Zuker A, Vainstein A (1999. b) Molecular mechanisms underlying carotenogenesis in the chromoplast: multilevel regulation of carotenoid-associated genes. Plant J 20: 423–431 [DOI] [PubMed] [Google Scholar]
- Von Tiedemann A (1997) Evidence for a primary role of active oxygen species in induction of host cell death during infection of bean leaves with Botrytis cinerea. Physiol Mol Plant Pathol 50: 151–166 [Google Scholar]
- Waterhouse PM, Graham MW, Wang MB (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense. Proc Natl Acad Sci USA 95: 13959–13964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D, van Blokland R, Kooter JM, Mol JNM, van Tunen AJ (1992) Gibberellic acid regulates chalcone synthase gene transcription in the corolla of Petunia hybrida. Plant Physiol 98: 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ytterberg AJ, Peltier J-B, van Wijk KJ (2006) Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol 140: 984–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenvirt S (2000) Development of transformation system for Gypsophila (Gypsophila paniculata L.). MSc thesis. Hebrew University of Jerusalem, Jerusalem