Abstract
Virus-induced gene silencing (VIGS) is a plant RNA-silencing technique that uses viral vectors carrying a fragment of a gene of interest to generate double-stranded RNA, which initiates the silencing of the target gene. Several viral vectors have been developed for VIGS and they have been successfully used in reverse genetics studies of a variety of processes occurring in plants. This approach has not been widely adopted for the model dicotyledonous species Arabidopsis (Arabidopsis thaliana), possibly because, until now, there has been no easy protocol for effective VIGS in this species. Here, we show that a widely used tobacco rattle virus-based VIGS vector can be used for silencing genes in Arabidopsis ecotype Columbia-0. The protocol involves agroinfiltration of VIGS vectors carrying fragments of genes of interest into seedlings at the two- to three-leaf stage and requires minimal modification of existing protocols for VIGS with tobacco rattle virus vectors in other species like Nicotiana benthamiana and tomato (Lycopersicon esculentum). The method described here gives efficient silencing in Arabidopsis ecotype Columbia-0. We show that VIGS can be used to silence genes involved in general metabolism and defense and it is also effective at knocking down expression of highly expressed transgenes. A marker system to monitor the progress and efficiency of VIGS is also described.
In the past, plant biologists relied almost exclusively on forward genetics; that is, the identification of a mutant and the subsequent cloning of the mutated gene to identify the wild-type sequence responsible for the process being investigated. The past several years have seen the complete sequencing of two plant genomes and the generation of large databases of sequence information from several other plant species. The availability of these large sets of genome sequences means that alternative approaches to traditional forward genetics can be implemented to identify the genes involved in a process of interest. An important alternative approach made possible by the availability of genome sequences is reverse genetics. Reverse genetics investigates the function of a gene or DNA sequence directly by altering the expression of the sequence of interest and then identifying the mutant phenotype that is produced. Most reverse genetics approaches described in plants to date rely on posttranscriptional gene silencing (PTGS; Watson et al., 2005).
PTGS is an RNA silencing-based approach used to reduce the level of expression of a gene of interest. It is also described as quelling in fungi (Cogoni et al., 1996) and RNA interference in animals (Fire et al., 1998). The mechanism of PTGS involves the sequence-specific degradation of RNA and several different techniques have been developed to harness PTGS phenomena for investigation. One of these is virus-induced gene silencing (VIGS; Baulcombe, 1999; Dinesh-Kumar et al., 2003). The observation that plants could overcome infection by viruses and then be rendered resistant to subsequent infection by closely related viruses was the first suggestion that PTGS was an innate antiviral defense in plants (Lindbo et al., 1993; Ratcliff et al., 1997; Soosaar et al., 2005). VIGS takes advantage of this defense system to silence endogenous RNA sequences that are homologous to a sequence engineered into the viral genome, which generates the double-stranded RNA that mediates silencing.
Several viral genomes have been modified to produce VIGS vectors (for review, see Burch-Smith et al., 2004). The majority of these have been based on RNA viruses that can infect several plant species used in scientific investigations. The most widely used VIGS vectors are based on the Tobacco rattle virus (TRV; Ratcliff et al., 2001; Liu et al., 2002b). TRV-based VIGS vectors have been used to silence genes in a number of Solanaceous plant species, including Nicotiana benthamiana (Ratcliff et al., 2001; Liu et al., 2002b), tomato (Liu et al., 2002a), pepper (Capsicum annuum; Chung et al., 2004), potato (Solanum tuberosum; Brigneti et al., 2004), and petunia (Petunia hybrida; Chen et al., 2005). VIGS using TRV-derived vectors is also effective in opium poppy (Papaver somniferum), a basal eudicot (Hileman et al., 2005). One distinct advantage of using TRV for VIGS is the ability of the virus to infect the meristem of its hosts (Ratcliff et al., 2001) and it has been used to study flowering in N. benthamiana (Liu et al., 2004) and petunia (Chen et al., 2004), in addition to fruit development in tomato (Fu et al., 2005). The distinct advantage of TRV-based VIGS in Solanaceous species is the ease of introduction of the VIGS vector into plants. This is usually mediated by Agrobacterium tumefaciens with the VIGS vector placed between T-DNA borders (Ratcliff et al., 2001; Liu et al., 2002b). A. tumefaciens can then be easily introduced into plant tissues by a variety of techniques, including infiltration with a needleless syringe, direct inoculation of bacterial colonies, agrodrenching, or vacuum infiltration (Lu et al., 2003b; Burch-Smith et al., 2004; Ryu et al., 2004; Hileman et al., 2005; Wang et al., 2006).
In addition to RNA viruses, DNA viruses have also been adapted for use as VIGS vectors. One of the more interesting of these is derived from the bipartite Cabbage leaf curl geminivirus (CbLCV) to perform VIGS in the model plant species Arabidopsis (Arabidopsis thaliana; Turnage et al., 2002). However, this vector has seen limited use for VIGS in Arabidopsis. This may be due in part to the difficulty in introducing the VIGS vector into the plant through particle bombardment (Turnage et al., 2002), a relatively tedious process. Another reason for the CbLCV vector's restricted use may be the limited insert size accommodated by the viral genome. An insert of up to 800 bp can be used in the CbLCV vector for Arabidopsis VIGS (Muangsan and Robertson, 2004).
Besides CbLCV as a VIGS vector in this model species, only TRV has been reported to be effective for transient VIGS in Arabidopsis. This is the same TRV VIGS vector described by Ratcliff and coworkers (Ratcliff et al., 2001) and used extensively for VIGS in N. benthamiana. However, the protocol used for silencing in Arabidopsis requires that the TRV vector first be introduced into N. benthamiana to produce virions and then the virions are used secondarily to infect Arabidopsis (Lu et al., 2003b). The recently described alternative approach to the technique described by Lu and coworkers uses vacuum infiltration to introduce the Agrobacterium inoculum into Arabidopsis plants (Wang et al., 2006). Both of these procedures are time consuming and tedious, especially in large-scale functional studies. In an effort to generate a more useful set of tools for VIGS in this model dicotyledonous species, we have used our TRV-based VIGS vector (Liu et al., 2002b) and optimized its delivery to and effectiveness in Arabidopsis. Here, we show that our TRV-based VIGS can be introduced into Arabidopsis ecotype Columbia-0 (Col-0) by agroinfiltration using the same technique employed with Solanaceous species. We found that the age and growth conditions of the Arabidopsis plants to be silenced were the most important factors determining the effectiveness of TRV VIGS. We demonstrate that our TRV-based VIGS method can be easily and reliably used for silencing a combination of genes, and we have also developed a silencing marker system for use in Arabidopsis.
RESULTS
Optimal Conditions for TRV VIGS in Arabidopsis
To investigate the optimal conditions under which TRV-based VIGS in Arabidopsis ecotype Col-0 might be effective, we attempted to silence the Arabidopsis Phytoene desaturase (AtPDS) gene. The silencing of PDS has been used as a marker for the effectiveness of VIGS in several instances (Ratcliff et al., 2001; Liu et al., 2002b; Turnage et al., 2002). The silencing of PDS produces a typical white color that is the result of photobleaching, which occurs in the absence of the gene product. We first used the TRV vector alone to see what symptoms, if any, would be produced in the Arabidopsis seedlings. We observed no visible symptoms of TRV infection in these plants and they were indistinguishable from wild-type, uninfiltrated seedlings (compare Fig. 1, A and B).
Figure 1.
Silencing of the PDS gene using TRV-based VIGS. A, Wild-type Arabidopsis Col-0 plant. B, Arabidopsis Col-0 plant 12 dpi with empty vector TRV VIGS at OD600 = 1.5. The plant shows no symptoms of viral infection. C, Arabidopsis Col-0 plants 12 dpi with TRV VIGS vector carrying a PDS insert. The white patches are caused by photobleaching that occurs due to reduced PDS levels. All plants infiltrated exhibit the bleaching indicative of PDS silencing. D, Silencing in the growing points of the plant. The cauline leaves and flowers are white due to PDS silencing. Photograph taken 4 weeks after infiltration with TRV AtPDS.
We examined whether the growing conditions of the Arabidopsis seedlings affected TRV VIGS. We compared the number of plants showing the pds phenotype after growth under long-day (16/8-h photoperiod) to those grown under short-day (8/16-h photoperiod) conditions. For the seedlings grown in 16-h light, 90% to 100% of the plants displayed photobleaching. Only 10% of those grown under short-day conditions exhibited AtPDS VIGS. After testing seedlings of different ages, we found that silencing of AtPDS was most effective in seedlings inoculated at the two- to three-leaf stage. When we used seedlings at the four- to five-leaf stage, the number of plants displaying the pds phenotype decreased by 50%. We observed an even more drastic reduction in the number of plants displaying the pds phenotype when we used older plants that contained many rosette leaves. The number of plants showing photobleaching decreased by 90% when compared to the number of two- to three-leaf-stage plants exhibiting VIGS. Therefore, younger plants are better for TRV-based silencing in Arabidopsis.
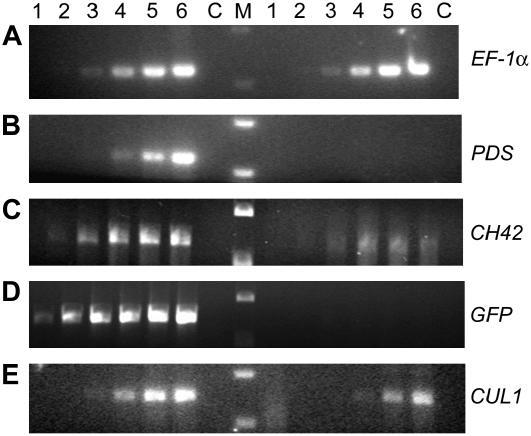
After establishing the growth conditions and the age of the seedlings that were most conducive to TRV VIGS, we investigated whether the concentration of the A. tumefaciens cultures used to introduce the VIGS vectors had any effect on the outcome of silencing. We found that the effectiveness of silencing of PDS was somewhat dependent on the concentration of the cultures used for agroinfiltration, and for all our future investigations we used cultures resuspended to OD600 = 1.5, compared to OD600 = 1.0 as described for VIGS in N. benthamiana (Liu et al., 2002b). Thus, we found that Arabidopsis seedlings inoculated at the two- to three-leaf stage and grown under 16-h light displayed the photobleaching phenotype indicative of PDS silencing in almost 100% of the cases examined (Fig. 1C). Indeed, when we allowed these seedlings to grow older, we observed photobleaching of the cauline leaves and flowers of the bolt (Fig. 1D). This is consistent with the ability of TRV to infect the growing points of its hosts (Ratcliff et al., 2001) and with the use of TRV VIGS to study flowering in petunia (Chen et al., 2004) and N. benthamiana (Liu et al., 2004). Semiquantitative reverse transcription (RT)-PCR analyses indicate that PDS transcript levels in silenced plants are reduced by 95% (Fig. 3B).
Figure 3.
VIGS effect on PDS, CH42, and CUL1 transcripts. Semiquantitative RT-PCR was used to determine the degree of silencing. Lanes 1 to 6 represent PCR cycles 15, 18, 21, 24, 27, and 30, respectively. C is the no RT control and M is the size marker. PCR products on the left of the marker are the nonsilenced controls. Those on the right are from the silenced plants. EF-1α is used as an internal control (A). Reductions of transcript levels are 95% for PDS (B), 89% for CH42 (C), 92% for GFP (D), and 79% for CUL1 (E).
VIGS of Endogenous Transgenes and Essential Genes in Arabidopsis
We wanted to investigate the efficacy of TRV VIGS on silencing other Arabidopsis genes. We chose the Chlorata42 (CH42) gene because silencing produces a visible phenotype: yellow color due to inhibition of chlorophyll biosynthesis (Kjemtrup et al., 1998). It has also been used as a marker for silencing in Arabidopsis (Turnage et al., 2002) and other systems (Kjemtrup et al., 1998). A fragment of the CH42 gene was cloned into TRV RNA2 and two- to three-leaf seedlings were infiltrated with cultures containing the silencing construct. At 10 to 12 d postinfiltration (dpi), the yellow sulfur phenotype of ch42 was visible in 100% of silenced plants (Fig. 2B). We monitored the level of silencing by semiquantitative RT-PCR and found an 89% reduction in CH42 transcript levels in yellow, silenced leaves (Fig. 3C).
Figure 2.
Silencing of CH42, GFP, and Cullin 1 using TRV-based VIGS. A, Arabidopsis Col-0 plant 12 dpi with empty vector TRV VIGS. B, Silencing of AtCH42 results in the yellow color of the sulfur phenotype of the plants. The AtCH42 phenotype was observed in all plants tested. C, Wild-type Arabidopsis Col-0 plant gives red fluorescence under a UV light (image 1), whereas a transgenic Col-0∷GFP plant expressing GFP appears green (image 2). Col-0∷GFP plants infiltrated with empty vector TRV VIGS remain green under UV light 12 dpi (image 3, left). Infiltration of Col-0∷GFP plants with TRV-GFP leads to loss of GFP and red fluorescence under UV light 12 dpi (image 3, right, and image 4). D, Silencing of an essential gene, AtCUL1, produces chlorosis of leaves, stunting, and eventually kills the plant. Photographs were taken 4 weeks after infiltration with TRV AtCUL1.
The ability of TRV VIGS to silence a green fluorescent protein (GFP) transgene in Arabidopsis was also examined. Wild-type Arabidopsis plants appear red under UV light due to chlorophyll autofluorescence, whereas transgenes containing GFP appear green (compare Fig. 2C, first two sections). We agroinfiltrated seedlings with cultures containing TRV RNA2 carrying a GFP fragment. We observed the plants under UV light after 12 dpi. Plants infiltrated with the TRV VIGS vector alone retained their green color, whereas those infiltrated with TRV RNA2-GFP were mostly red, with only small patches of green fluorescence still visible at the edge of leaves (Fig. 2C). This indicates that TRV-mediated silencing of a transgene was effective in Arabidopsis. The silencing observations were confirmed by analysis of GFP transcript levels by semiquantitative RT-PCR of RNA derived from a red leaf from Figure 2C, section 4. The silenced plants showed a 92% reduction in GFP transcript levels (Fig. 3D). Thus, TRV VIGS can also be used to silence highly expressed transgenes in Arabidopsis.
One of the most useful applications of VIGS is in studying genes whose traditional knockout phenotype is lethal. In Arabidopsis, one such gene is Cullin 1 (CUL1), a component of the Skp1/Cullin/F-box (SCF)-type E3 ubiquitin ligases (Shen et al., 2002). cul1 mutants are embryonic lethal with an early arrest in development that precedes division of the endosperm (Shen et al., 2002). This suggests that CUL1 has an essential role in early embryogenesis that may be due to its ability to form a variety of SCF complexes containing different F-box proteins. The availability of cul1 mutant tissue would allow an investigation of other roles of CUL1 in SCF function and in plant development. Following the approach used for silencing PDS, CH42, and GFP, we silenced AtCUL1. This produced a pleiotropic phenotype (Fig. 2D) that includes yellowing of leaves, severe stunting, and reduced bolting. We determined that the phenotypes observed coincided with a 79% reduction in AtCUL1 transcript levels (Fig. 3E).
TRV VIGS as a Tool for Studying Disease Resistance
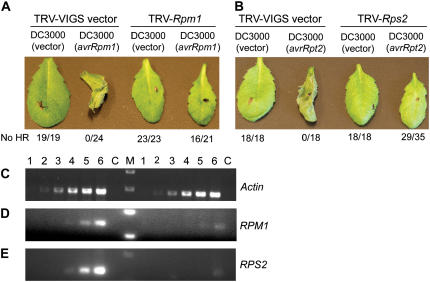
We wanted to demonstrate that TRV VIGS could be used to investigate a variety of biological questions and so we chose to examine the effectiveness of silencing Arabidopsis disease resistance (R) genes. RPM1 confers resistance to Pseudomonas syringae carrying either the AvrRpm1 or AvrB effector proteins (Grant et al., 1995). Arabidopsis Col-0 leaves infected with P. syringae carrying AvrRpm1 or AvrB initiate programmed cell death (PCD) that is part of the typical R-gene-mediated response, called the hypersensitive response (HR; Fig. 4A). We used TRV VIGS to silence RPM1 and then infected plants with P. syringae carrying AvrRpm1. In nonsilenced controls, 24 of 24 plants showed HR PCD. In contrast, 16 of 21 RPM1-silenced plants gave no HR PCD (Fig. 4A). Thus, the silenced plants show a loss of resistance to P. syringae carrying AvrRpm1. We assessed the degree of reduction of RPM1 transcript levels and found that the silenced plants have 92% lower levels of transcripts (Fig. 4D). Similarly, we examined the effect of silencing another Arabidopsis R gene, RPS2. Infection of RPS2-containing Arabidopsis with P. syringae carrying AvrRpt2 produces HR PCD (Bent et al., 1994; Mindrinos et al., 1994). Consistent with this, all plants infiltrated with TRV VIGS vector alone showed HR PCD when infected with P. syringae carrying AvrRpt2. In contrast, 29 of 35 RPS2-silenced plants failed to produce HR PCD after introduction of AvrRpt2 (Fig. 4B). We confirmed the silencing of RPS2 by semiquantitative RT-PCR and found a 95% reduction in transcript levels (Fig. 4E). Thus, TRV VIGS is effective at silencing R genes and can be used as a tool for investigating plant innate immunity.
Figure 4.
VIGS to study disease resistance in Arabidopsis. A, RPM1 was silenced in wild-type Col-0 plants by infiltration with TRV-RPM1. Control plants infected with P. syringae not containing AvrRPm1 do not show HR (19 of 19 plants tested), whereas HR is observed in all plants in the presence of AvrRpm1. RPM1-silenced plants do not show HR in the absence of AvrRpm1 (23 of 23 plants tested); 16 of 21 RPM1-silenced plants do not produce HR in response to AvrRpm1, indicating a loss of RPM1 function. All observations were made 14 h postinoculation (hpi). B, Silencing of RPS2 results in loss of HR to AvrRpt2 in 29 of 35 plants tested. Observations were made 22 hpi. C and D, Semiquantitative RT-PCR was used to determine the degree of RPM1 and RPS2 silencing. Lanes 1 to 6 represent PCR cycles 15, 18, 21, 24, 27, and 30, respectively. C is the no RT control and M is the size marker. PCR products on the left of the marker are the nonsilenced controls and those on the right are from the silenced plants. Actin is used as an internal control (C). Reductions of transcript levels are 92% for RPM1 (D) and 95% for RPS2 (E).
Development of a VIGS Marker System
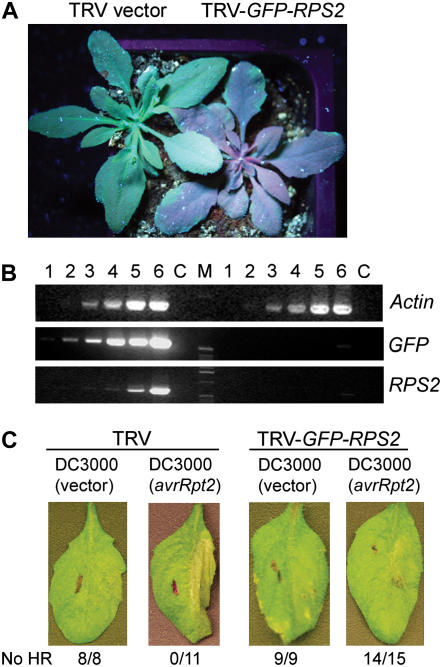
In most instances, the efficacy of silencing of a gene of interest cannot be visibly assessed. We wanted to develop a system that contained a marker that could be easily observed to indicate whether silencing had been effective and the approximate degree of silencing. For this, we generated a TRV RNA2 vector that contains a fragment of the GFP gene upstream of a multiple cloning site. This allows the insertion of sequence of a gene of interest into the plasmid to facilitate the simultaneous silencing of both genes in transgenic Arabidopsis containing the GFP transgene. To test our system, we inserted fragments of GFP and RPS2 in tandem into TRV RNA2. Two weeks after agroinoculation of seedlings, we observed the plants under UV light. As expected, we observed red fluorescence in the GFP-RPS2-silenced plants (Fig. 5A).
Figure 5.
A VIGS reporter system for Arabidopsis. The TRV VIGS reporter system couples the silencing of GFP in a transgenic Col-0∷GFP plant with the silencing of an endogenous gene of interest. A, GFP is silenced after infiltration with TRV-GFP-RPS2 and plants appear red when observed under UV light. B, The level of GFP and RPS2 transcript reduction was monitored by semiquantitative RT-PCR. Lanes 1 to 6 represent PCR cycles 15, 18, 21, 24, 27, and 30, respectively. C is the no RT control and M is the size marker. PCR products on the left of the marker are the nonsilenced controls and those on the right are from the silenced plants. Actin is used as a control. C, Fourteen of 15 plants tested did not produce an HR in response to AvrRpt2 after infiltration with TRV-GFP-RPS2 indicating silencing of RPS2.
To confirm that silencing of the GFP transgene overlaps with silencing of the endogenous gene, RPS2, we performed quantitative RT-PCR analyses of relative GFP and RPS2 mRNA levels in the tissue showing loss of UV fluorescence. Our analyses indicate that GFP and RPS2 transcript levels are reduced by 95% compared to the control TRV vector alone in infected nonsilenced plants (Fig. 5B). These results clearly illustrate that GFP (transgene) silencing overlaps with RPS2 (endogenous) gene silencing at the molecular level.
We tested the ability of the GFP-RPS2 double-silenced plants to respond to P. syringae carrying AvrRpt2. We found that 14 of 15 plants tested did not produce HR PCD in response to AvrRpt2, indicating that RPS2 function had been compromised (Fig. 5B). Thus, the marker system coupling GFP to a gene of interest allows effective silencing of the target gene of interest as well as a visible marker for silencing.
DISCUSSION
We have optimized a TRV-based VIGS protocol for use in the model dicotyledonous species, Arabidopsis. The availability of the genome sequence of the Col-0 ecotype of Arabidopsis has been a valuable tool for the identification and characterization of many mutants in a broad range of processes. Despite this, there are still challenges to working in this species. One problem is embryonic lethality of some mutants. As we have demonstrated for CUL1, VIGS can be used to examine the effects of loss of the product of the essential gene in adult tissues. Another problem is the possible functional redundancy of members of gene families. It is difficult and tedious to generate traditional T-DNA insertion mutants in multiple members of a gene family to tease apart the function of those genes. VIGS can be used to silence multiple members of a family by using a highly conserved region for silencing (He et al., 2004). This provides an idea of the processes in which these genes function. In addition, the directed nature of VIGS allows the targeted knock down of expression of any gene of interest. This is important when traditional knockouts of a gene of interest are unavailable due to the limitations of the kind of mutations introduced by chemical mutagenesis or the preference of T-DNA insertion sites. Thus, VIGS represents an important tool to complement traditional forward genetics tools in Arabidopsis.
The previously described protocols for VIGS in Arabidopsis have been time consuming and difficult. Indeed, they have seen limited use for study of gene function. There has been a single report using the CbLCV-based VIGS vector to study plant development (Fan et al., 2005). The CbLCV VIGS vector is introduced into Arabidopsis seedlings by particle bombardment, a process that requires care in the preparation of microprojectile particles, the risk of carryover between experiments, and the somewhat unpredictable introduction of the silencing vector (Turnage et al., 2002; Muangsan and Robertson, 2004). The previously described TRV-based VIGS protocol required that the vector first be introduced into N. benthamiana to produce virions (Lu et al., 2003b). The purification of the crude N. benthamiana sap to produce the viral inoculum for Arabidopsis also involves several different steps. The other recently described TRV-based protocol uses vacuum infiltration of the whole plant that is submerged in Agrobacterium cultures (Wang et al., 2006). Not only is this process tedious, but also it requires relatively large culture volumes (100 mL) for infiltration. None of these protocols are therefore suitable for large-scale approaches to gene function analysis in Arabidopsis.
In contrast, our protocol allows direct introduction of the TRV VIGS vector into Arabidopsis seedlings by simple agroinfiltration. Given the small size of seedling leaves, it takes less than a minute to inoculate a plant. With the modifications we have described here, previously published protocols for VIGS in N. benthamiana or tomato can be used for silencing in Arabidopsis (Lu et al., 2003b; Burch-Smith et al., 2004). This protocol provides an avenue for large-scale functional genomic screens in Arabidopsis, as has been performed in N. benthamiana (Lu et al., 2003a; Liu et al., 2005). Thus, TRV-based VIGS holds promise as a powerful tool for genetic analysis in this indispensable model organism.
MATERIALS AND METHODS
Plasmid Construction
pTRV1 (pYL192) and pTRV2 (pYL156) vectors have been described in Liu et al. (2002b). The pYL170 TRV2 vector was derived by cloning a PstI-blunt-DraIII fragment of pYL156 into EcoRI-blunt-DraIII-cut pCAMBIA3301. This vector is identical to pYL156, except for a plant selection marker. To generate pTRV2-AtPDS, a cDNA fragment was PCR amplified using Arabidopsis (Arabidopsis thaliana) ecotype Col-0 cDNA and primers 5′-CGCGAATTCTGCGGCGAATTTGCCTTATCAAAACG-3′ and 5′-CGCTCTAGAAACTCTTAACCGTGCCATCGTCATTGAG-3′. The resulting PCR product was cloned into XbaI-EcoRI-cut pTRV2. To generate pTRV2-AtCH42, the Arabidopsis CH42 cDNA fragment was PCR amplified from Arabidopsis cDNA using primers 5′-CGACGACAAGACCCTGGCGTCTCTTCTTGGAACATCTTC-3′ and 5′-GAGGAGAAGAGCCCTCGCAATAACAGGAACTTGCTCTC-3′ and cloned into pTRV2. To generate pTRV2-GFP, the GFP fragment was PCR amplified from the tobacco mosaic virus-GFP plasmid using primers CGGTCTAGAGGTACCCTTGTTAATCGTATCGAG-3′ and 5′-CCGTCTAGAGCTCATCCATGCCATGTGT-3′. The resulting PCR product was cloned into XbaI-cut pYL170. To generate pTRV2-AtCUL1, the Arabidopsis Cullin1 cDNA fragment was PCR amplified from Arabidopsis cDNA using primers 5′-CGGGAATTCGAGCGCAAGACTATTGACTTGGAGC-3′ and 5′-CGGGAATTCTTGCAAACACAACCAGCAATTCATG-3′. The resulting PCR product was cloned into EcoRI-cut pTRV2. To generate pTRV2-AtRPM1, the Arabidopsis RPM1 cDNA fragment was PCR amplified using primers 5′-CGGGAATTCTGATCGCAACTGCAAGCATTGAGAAGCT-3′ and 5′-CGGGAATTCAGATGAGAGGCTCACATAGAAAGAGCC-3′. The resulting PCR product was cloned into EcoRI-cut pTRV2. To generate pTRV2-AtRPS2, the Arabidopsis RPS2 cDNA fragment was PCR amplified using primers 5′-CGGGAATTCGAACTCCTCTACTTCAATCTCCCATC-3′ and 5′-CGGGAATTCGGAACAAAGCGCGGTAAATAACAAAG-3′. The resulting PCR product was cloned into EcoRI-cut pTRV2. To generate pTRV2-GFP-AtRPS2, the RPS2 cDNA fragment was PCR amplified using primers 5′-CGACGACAAGACCCTGAACTCCTCTACTTCAATCTCCCATC-3′ and 5′-GAGGAGAAGAGCCCTGGAACAAAGCGCGGTAAATAACAAAG-3′. The resulting PCR product was cloned into pYL989, a pTRV2-GFP derivative (pTRV2-GFP). All PCR amplification was performed using Taq DNA polymerase (New England Biolabs).
Plant Growth, Agroinfiltration, and GFP Imaging
Wild-type Arabidopsis ecotype Col-0 and GFP-expressing transgenic Arabidopsis Col-0 plants were grown in pots at 23°C in a growth chamber under a 16/8-h photoperiod with 60% humidity. Two- to three-leaf seedlings were used for VIGS, approximately 15 to 17 d after seed germination. For the VIGS assay, pTRV1 or pTRV2 and its derivatives were introduced into Agrobacterium tumefaciens strain GV3101. A 5-mL culture was grown overnight at 28°C in 50 mg/L gentamycin and 50 mg/L kanamycin. The next day, the culture was inoculated into 50-mL of Luria-Bertani medium containing antibiotics, 10 mm MES, and 20 μm acetosyringone. The culture was grown overnight in a 28°C shaker. A. tumefaciens cells were harvested and resuspended in infiltration media (10 mm MgCl2, 10 mm MES, and 200 μm acetosyringone), adjusted to an OD600 of 1.5, and left at room temperature for 3 to 4 h. Agroinfiltration was performed with a needleless 1-mL syringe into two leaves of two- to three-leaf-stage plants, infiltrating the entire leaf. Plants were left covered overnight. GFP imaging was performed using UV illumination and photographs were taken using an Olympus Camedia E10 digital camera.
HR Assay in Arabidopsis Plants
Pseudomonas syringae DC3000 containing empty vector pVSP61, AvrRpm1, or AvrRpt2 were grown from glycerol stocks for 36 to 48 h on King's B solid medium containing 100 mg/L rifampicin and 25 mg/L kanamycin. Cells were scraped into 10 mm MgCl2 and diluted to OD600 = 0.2 and infiltrated using a 1-mL needleless syringe into the silenced and control Arabidopsis plants. RPM1-mediated and RPS2-mediated HR cell death were observed and photographed at 14 and 22 h postinfiltration of P. syringae strains, respectively.
RNA Isolation and RT-PCR Analysis
Total RNA was extracted from pooled tissue samples of two to three silenced and nonsilenced Arabidopsis plants using the RNeasy plant minikit, including an RNase-free DNase treatment step (Qiagen). First-strand cDNA was synthesized using 1 μg of total RNA, oligo d(T) primer, and SuperScript reverse transcriptase (Invitrogen). Semiquantitative RT-PCR was performed as described in Liu et al. (2002b). For RT-PCR, primers that anneal outside the region targeted for silencing were used to ensure that the gene of interest was silenced. The intensities of PCR-generated fragments were analyzed and quantified using Gel Doc 2000 and Quantity One, version 4.3 (Bio-Rad).
Acknowledgments
We thank Brian Staskawicz for providing P. syringae strains and Dominique Robertson for providing GFP-expressing Arabidopsis Col-0.
This work was supported by a National Science Foundation Plant Genome grant (grant no. DBI–0211872).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: S.P. Dinesh-Kumar (savithramma.dinesh-kumar@yale.edu).
References
- Baulcombe DC (1999) Fast forward genetics based on virus-induced gene silencing. Curr Opin Plant Biol 2: 109–113 [DOI] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ (1994) RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265: 1856–1860 [DOI] [PubMed] [Google Scholar]
- Brigneti G, Martin-Hernandez AM, Jin H, Chen J, Baulcombe DC, Baker B, Jones JD (2004) Virus-induced gene silencing in Solanum species. Plant J 39: 264–272 [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP (2004) Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J 39: 734–746 [DOI] [PubMed] [Google Scholar]
- Chen JC, Jiang CZ, Gookin TE, Hunter DA, Clark DG, Reid MS (2004) Chalcone synthase as a reporter in virus-induced gene silencing studies of flower senescence. Plant Mol Biol 55: 521–530 [DOI] [PubMed] [Google Scholar]
- Chen JC, Jiang CZ, Reid MS (2005) Silencing a prohibitin alters plant development and senescence. Plant J 44: 16–24 [DOI] [PubMed] [Google Scholar]
- Chung E, Seong E, Kim YC, Chung EJ, Oh SK, Lee S, Park JM, Joung YH, Choi D (2004) A method of high frequency virus-induced gene silencing in chili pepper (Capsicum annuum L. cv. Bukang). Mol Cells 17: 377–380 [PubMed] [Google Scholar]
- Cogoni C, Irelan JT, Schumacher M, Schmidhauser TJ, Selker EU, Macino G (1996) Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J 15: 3153–3163 [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Anandalakshmi R, Marathe R, Schiff M, Liu Y (2003) Virus-induced gene silencing. Methods Mol Biol 236: 287–294 [DOI] [PubMed] [Google Scholar]
- Fan J, Quan S, Orth T, Awai C, Chory J, Hu J (2005) The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol 139: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Fu DQ, Zhu BZ, Zhu HL, Jiang WB, Luo YB (2005) Virus-induced gene silencing in tomato fruit. Plant J 43: 299–308 [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846 [DOI] [PubMed] [Google Scholar]
- He X, Anderson JC, del Pozo O, Gu Y-Q, Tang X, Martin GB (2004) Silencing of subfamily I of protein phosphatase 2A catalytic subunits results in activation of plant defense responses and localized cell death. Plant J 38: 563–577 [DOI] [PubMed] [Google Scholar]
- Hileman LC, Drea S, Martino G, Litt A, Irish VF (2005) Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy). Plant J 44: 334–341 [DOI] [PubMed] [Google Scholar]
- Kjemtrup S, Sampson KS, Peele CG, Nguyen LV, Conkling MA (1998) Gene silencing from plant DNA carried by a geminivirus. Plant J 14: 91–100 [DOI] [PubMed] [Google Scholar]
- Lindbo JA, Silva-Rosales L, Proebsting WM, Dougherty WG (1993) Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell 5: 1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Nakayama N, Schiff M, Litt A, Irish VF, Dinesh-Kumar SP (2004) Virus induced gene silencing of a DEFICIENS ortholog in Nicotiana benthamiana. Plant Mol Biol 54: 701–711 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002. a) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP (2002. b) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Lu R, Malcuit I, Moffett P, Ruiz MT, Peart J, Wu AJ, Rathjen JP, Bendahmane A, Day L, Baulcombe DC (2003. a) High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J 22: 5690–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC (2003. b) Virus-induced gene silencing in plants. Methods 30: 296–303 [DOI] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu GL, Ausubel FM (1994) The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78: 1089–1099 [DOI] [PubMed] [Google Scholar]
- Muangsan N, Robertson D (2004) Geminivirus vectors for transient gene silencing in plants. Methods Mol Biol 265: 101–115 [DOI] [PubMed] [Google Scholar]
- Ratcliff F, Harrison BD, Baulcombe DC (1997) A similarity between viral defense and gene silencing in plants. Science 276: 1558–1560 [DOI] [PubMed] [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC (2001) Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 25: 237–245 [DOI] [PubMed] [Google Scholar]
- Ryu CM, Anand A, Kang L, Mysore KS (2004) Agrodrench: a novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse Solanaceous species. Plant J 40: 322–331 [DOI] [PubMed] [Google Scholar]
- Shen WH, Parmentier Y, Hellmann H, Lechner E, Dong A, Masson J, Granier F, Lepiniec L, Estelle M, Genschik P (2002) Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol Biol Cell 13: 1916–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soosaar JL, Burch-Smith TM, Dinesh-Kumar SP (2005) Mechanisms of plant resistance to viruses. Nat Rev Microbiol 3: 789–798 [DOI] [PubMed] [Google Scholar]
- Turnage MA, Muangsan N, Peele CG, Robertson D (2002) Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J 30: 107–117 [DOI] [PubMed] [Google Scholar]
- Wang C, Cai X, Wang X, Zheng Z (2006) Optimisation of tobacco rattle virus-induced gene silencing in Arabidopsis. Funct Plant Biol 33: 347–355 [DOI] [PubMed] [Google Scholar]
- Watson JM, Fusaro AF, Wang M, Waterhouse PM (2005) RNA silencing platforms in plants. FEBS Lett 579: 5982–5987 [DOI] [PubMed] [Google Scholar]