Abstract
Some phytohormones such as gibberellins (GAs) and cytokinins (CKs) are potential targets of the KNOTTED1-like homeobox (KNOX) protein. To enhance our understanding of KNOX protein function in plant development, we identified rice (Oryza sativa) genes for adenosine phosphate isopentenyltransferase (IPT), which catalyzes the rate-limiting step of CK biosynthesis. Molecular and biochemical studies revealed that there are eight IPT genes, OsIPT1 to OsIPT8, in the rice genome, including a pseudogene, OsIPT6. Overexpression of OsIPTs in transgenic rice inhibited root development and promoted axillary bud growth, indicating that OsIPTs are functional in vivo. Phenotypes of OsIPT overexpressers resembled those of KNOX-overproducing transgenic rice, although OsIPT overexpressers did not form roots or ectopic meristems, both of which are observed in KNOX overproducers. Expression of two OsIPT genes, OsIPT2 and OsIPT3, was up-regulated in response to the induction of KNOX protein function with similar kinetics to those of down-regulation of GA 20-oxidase genes, target genes of KNOX proteins in dicots. However, expression of these two OsIPT genes was not regulated in a feedback manner. These results suggest that OsIPT2 and OsIPT3 have unique roles in the developmental process, which is controlled by KNOX proteins, rather than in the maintenance of bioactive CK levels in rice. On the basis of these findings, we concluded that KNOX protein simultaneously decreases GA biosynthesis and increases de novo CK biosynthesis through the induction of OsIPT2 and OsIPT3 expression, and the resulting high-CK and low-GA condition is required for formation and maintenance of the meristem.
KNOTTED1-like homeobox (KNOX) proteins are encoded by knox genes and are preferentially accumulated in the indeterminate cells around the shoot apical meristem (SAM), but not in the determinate lateral organs (Jackson et al., 1994; Lincoln et al., 1994; Nishimura et al., 1999; Sentoku et al., 1999). Loss-of-function mutants shootmeristemless (stm) of Arabidopsis (Arabidopsis thaliana L. Heynh.) and knotted1 (kn1) of maize (Zea mays) show defects in SAM development or maintenance (Long et al., 1996; Kerstetter et al., 1997). The opposite phenotype, namely, formation of ectopic meristems on leaves, has been reported in transgenic plants overproducing KNOX proteins (Matsuoka et al., 1993; Sinha et al., 1993; Chuck et al., 1996; Nishimura et al., 2000; Sentoku et al., 2000). This evidence suggests that KNOX proteins play critical roles in SAM formation and maintenance as transcriptional regulators (Reiser et al., 2000).
To understand the function of KNOX proteins in plant development, it is necessary to identify the genes targeted by them and to characterize the mechanism of the transcriptional regulation of those genes. Previous studies have revealed that KNOX proteins suppress the expression of gibberellin (GA) 20-oxidase genes in the dicots tobacco (Nicotiana tabacum), Arabidopsis, and potato (Solanum tuberosum; Sakamoto et al., 2001; Hay et al., 2002; Chen et al., 2004). Because GA 20-oxidase catalyzes the rate-limiting step of bioactive GA synthesis, these findings clearly indicate that KNOX proteins in dicots play a role in maintaining the SAM through the down-regulation of GA biosynthesis. However, the decreased level of bioactive GAs cannot completely explain the altered morphologies observed in KNOX overproducers. For example, ectopic meristem formation, which is a typical abnormal phenotype of KNOX overproducers, has never been observed in GA-deficient mutants of various plant species (Sun et al., 1992; Chiang et al., 1995; Xu et al., 1995; Spray et al., 1996; Helliwell et al., 1998; Yamaguchi et al., 1998; Itoh et al., 2001, 2004; Sasaki et al., 2002; Davidson et al., 2003; Sakamoto et al., 2004).
Another candidate for regulation by KNOX proteins is cytokinin (CK) biosynthesis because production of bioactive CKs, such as trans-zeatin (tZ) and isopentenyladenine (iP), is significantly increased in KNOX overproducers (Tamaoki et al., 1997; Kusaba et al., 1998; Ori et al., 1999; Hewelt et al., 2000; Frugis et al., 2001). CKs affect many plant developmental processes, such as cell division, shoot initiation from callus, promotion of axillary bud outgrowth, direct transport of nutrients, stimulation of pigment synthesis, inhibition of root growth, and delay of senescence (Mok, 1994). The pathway for CK biosynthesis in higher plants has been established in Arabidopsis (Sakakibara, 2004, 2005). The first and rate-limiting step is prenylation of adenosine 5′ phosphates, such as ATP and ADP, at the N6-terminus with dimethylallyl diphosphate (DMAPP); this reaction is catalyzed by adenosine phosphate isopentenyltransferase (IPT). So far, plant IPT genes have been identified in dicots such as Arabidopsis (Kakimoto, 2001; Takei et al., 2001), petunia (Petunia hybrida; Zubko et al., 2002), and hop (Humulus lupulus; Sakano et al., 2004). The Arabidopsis genome encodes seven IPT genes (AtIPT1 and AtIPT3–AtIPT8; Kakimoto, 2001; Takei et al., 2001), which have different spatial expression patterns and hormone responses (Miyawaki et al., 2004; Takei et al., 2004). Recent studies demonstrated that the Arabidopsis KNOX protein STM induces expression of AtIPT7 within 2 h after induction of STM function (Jasinski et al., 2005; Yanai et al., 2005). Therefore, STM regulates expression of genes for both GA and CK biosynthesis to generate low-GA and high-CK conditions in the meristem; these conditions may be necessary to maintain meristem activity (Jasinski et al., 2005).
To elucidate the functional interaction between KNOX proteins and CK biosynthesis in monocot plants, we isolated eight IPT genes from rice. We compared transgenic rice plants overproducing OsIPT and KNOX proteins. We also examined the expression level of OsIPT genes in KNOX overproducers. We discuss the function of KNOX proteins in CK biosynthesis in rice.
RESULTS
Isolation of IPT Genes from Rice
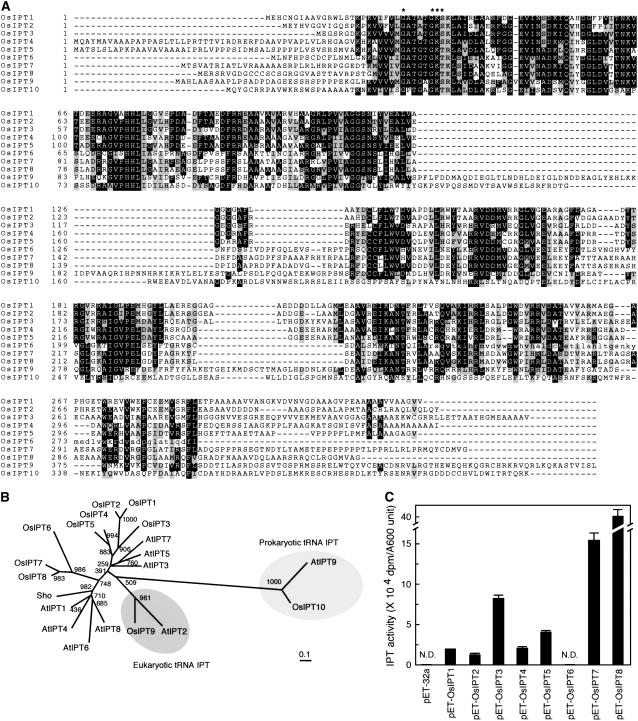
We searched for IPT genes in all available rice DNA databases, using the predicted amino acid sequences encoded by Arabidopsis IPT genes (AtIPT1 and AtIPT3–AtIPT8; Kakimoto, 2001; Takei et al., 2001) as probes. Candidate sequences detected were used reiteratively as probes for further searches. We found 10 candidates, designated OsIPT1 to OsIPT10 (Fig. 1A). The deduced open reading frames of all OsIPTs consist of one exon and no intron, with the exception of OsIPT9, which has 10 exons. The putative DMAPP-binding motif ([A, G]-X4-G-K-[S, T]) conserved in the N-terminal region of all AtIPTs was also found in all OsIPTs (Fig. 1A, asterisks). In the genome of a japonica cultivar, Nipponbare, we found that OsIPT6 has a single nucleotide substitution at Arg-236 (CGA to TGA), which generates a premature stop codon, but the indica cultivar, Kasalath, does not have this substitution. Thus, the Nipponbare OsIPT6 may be a mutant allele of the original OsIPT6 (Fig. 1A).
Figure 1.
Molecular and biochemical characterization of OsIPTs. A, Amino acid sequence alignment of OsIPTs. Exact matches are boxed in black; shaded boxes indicate conservative substitutions. Asterisks show conserved DMAPP-binding motifs. Lowercase letters indicate amino acid sequence of OsIPT6 from indica cultivar Kasalath. B, Unrooted dendrogram of IPT proteins in Arabidopsis (AtIPT1–AtIPT9), petunia (Sho), and rice (OsIPT1–OsIPT10). Bar, 0.1 amino acid substitutions per site. C, IPT activity of cell extracts. Crude extract of each transformant E. coli cell line was used to measure the IPT activity by radioisotope rapid assay. The amount of each sample for assay was equivalent to one A600 unit of cells. One A600 unit is defined as the amount of cells obtained from 1 mL of cell culture whose A600 value is 1.
Phylogenetic analysis grouped OsIPT1 to OsIPT8 with AtIPT1 and AtIPT3 to AtIPT8, and further divided this group into small subgroups (Fig. 1B). Each subgroup contained rice and Arabidopsis representatives. For instance, OsIPT1 to OsIPT5 were clustered with AtIPT3, AtIPT5, and AtIPT7. Similarly, OsIPT6, OsIPT7, and OsIPT8 were clustered with AtIPT1, AtIPT4, AtIPT6, and AtIPT8, and the petunia Sho. Pairing between rice and Arabidopsis IPTs in each subgroup leads us to speculate that each subgroup might have unique functions shared in monocots and dicots, but different from those in other subgroups. OsIPT9 and OsIPT10 were closely related to AtIPT2 and AtIPT9, respectively. AtIPT2 and AtIPT9 are considered to correspond, respectively, to eukaryotic and prokaryotic tRNA-IPTs, which catalyze prenylation of tRNA, but are not involved in CK biosynthesis (Kakimoto, 2001; Takei et al., 2001). Therefore, we predicted that OsIPT9 and OsIPT10 would also be involved in tRNA prenylation, but not in CK biosynthesis. Thus, we characterized eight genes (OsIPT1–OsIPT8).
IPT Activity of Recombinant OsIPT Proteins
To confirm the involvement of gene products in CK biosynthesis, we measured IPT activity by radioisotope rapid assay of total extract of Escherichia coli cells expressing OsIPTs. Although the activities differed among these proteins, IPT activity was detected in all cell extracts containing each recombinant OsIPT, except OsIPT6 (Fig. 1C). The result suggests that the products of OsIPTs, except OsIPT6, are involved in CK biosynthesis. The differences in IPT activity are probably due to the different efficiencies of functional protein expression, as observed in the Arabidopsis enzymes (Takei et al., 2001). Indeed, the specific activities of purified OsIPT1 and OsIPT3 (next paragraph) were 8.6 and 11.4 nmol min−1 mg−1 protein, respectively, when DMAPP and ADP were added as substrates in the reaction mixture; this indicates that the extent of IPT activity shown in Figure 1C does not always reflect the in vitro specific activity of each enzyme.
To determine the kinetic parameters of OsIPTs, we purified recombinant OsIPT1 and OsIPT3 from E. coli extracts. The Km values of both for ATP, ADP, and AMP clearly indicate that these OsIPTs prefer ATP or ADP to AMP as a substrate (Table I). Both OsIPTs utilized DMAPP as an isoprenoid side-chain donor (Table I), but hardly used hydroxymethylbutenyl diphosphate (data not shown), another candidate donor substrate (Krall et al., 2002; Sakakibara et al., 2005). Similar results were obtained with other semipurified OsIPTs, except OsIPT6 (data not shown). These results demonstrate that OsIPTs have similar substrate preferences to IPTs from Arabidopsis (Kakimoto, 2001; Sakakibara, 2004) and hop (Sakano et al., 2004), and suggest that substrate specificity is common among higher plant IPTs.
Table I.
Kinetic parametersa of OsIPT1 and OsIPT3
| Protein |
Km
|
Vmaxb | |||
|---|---|---|---|---|---|
| ATPc | ADPc | AMPc | DMAPPd | ||
| μm | nmol mg−1 min−1 | ||||
| OsIPT1 | 7.0 | 14.7 | 414 | 20.7 | 8.6 |
| OsIPT3 | 5.1 | 29.8 | 147 | 8.7 | 11.4 |
Values are means of three independent determinations.
Measured in the presence of ADP and DMAPP.
Measured with 200 μm DMAPP.
Measured with 200 μm ADP.
Expression of OsIPTs in Various Organs of Wild-Type Rice
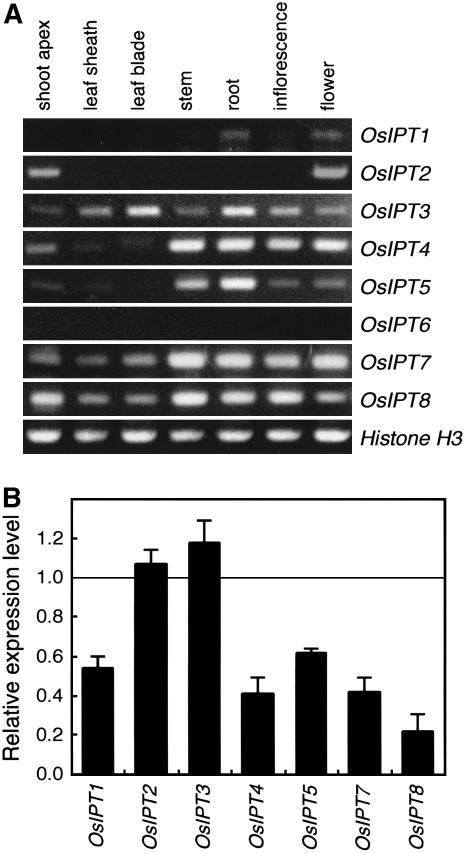
Quantitative reverse transcription (qRT)-PCR analysis revealed that seven OsIPT genes (OsIPT1–OsIPT5, OsIPT7, and OsIPT8) were expressed at different levels in various organs (Fig. 2A). Interestingly, genes grouped closely by phylogenetic analysis (Fig. 1B) showed similar expression patterns. For example, OsIPT1 transcripts were localized in the root and flower, and OsIPT2 transcripts were accumulated in the vegetative shoot apex and flower. OsIPT4 and OsIPT5 were expressed in all organs, although weakly in leaves (leaf sheath and leaf blade). OsIPT7 and OsIPT8 transcripts were broadly detected in all the organs we tested, whereas the OsIPT6 transcript was not detected in any organ. PCR without RT did not amplify any OsIPT genes (data not shown).
Figure 2.
Expression analysis of OsIPT genes. A, Expression of OsIPT genes in various organs of wild-type rice. Total RNAs were isolated from the organs listed above each lane. Histone H3 was used as a loading control. B, Feedback regulation of OsIPT genes in iP-treated wild-type plants. The value obtained without iP treatment was arbitrarily set at 1.0. qRT-PCR was performed in triplicate and the mean values with sd are shown.
Previous observations in Arabidopsis indicate that the expression of AtIPTs is regulated by the level of bioactive CKs (Miyawaki et al., 2004; Takei et al., 2004). Thus, we examined whether such feedback regulation also occurs in rice. qRT-PCR analysis revealed that iP treatment reduced the expression of OsIPT1, OsIPT4, OsIPT5, OsIPT7, and OsIPT8 (Fig. 2B). This result indicates that expression of these five genes is controlled by the CK level in a negative feedback manner, as in Arabidopsis. On the other hand, such a reduction was not observed in the expression of OsIPT2 or OsIPT3. This suggests that these genes are constitutively expressed or regulated by another mechanism (see below).
Overexpression of OsIPT Genes in Transgenic Rice
To assess the effects of overexpression of OsIPT genes and overproduction of CKs in transgenic rice, we overexpressed five OsIPT genes (OsIPT1–OsIPT4, OsIPT7) ectopically in transgenic rice under the control of the rice actin promoter (McElroy et al., 1991). All primary transformants exhibited inhibition of root formation, a typical phenotype caused by CK overproduction in mutant and transgenic dicots (Chaudhury et al., 1993; Faiss et al., 1997). The above-ground portion of primary transformants showed a range of phenotypes and some plants showed a weaker phenotype than that of a typical one (see below). Dissection of these weak phenotypes is also important to clarify the CK function on rice development. In this study, however, we focused on the typical phenotype of OsIPT transformants to simplify the discussion.
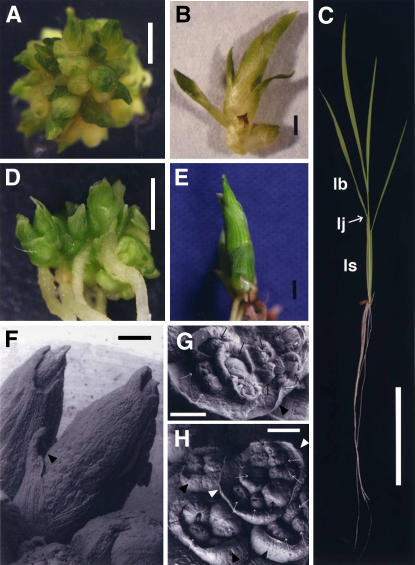
Because of the phenotypic similarity of transgenic dicots overexpressing IPT and knox, we compared the typical phenotype of transgenic rice overexpressing OsIPTs and the rice knox gene, OSH1. Most OsIPT transformants formed clumps of multiple shoots and each shoot grew to about 2 mm (Fig. 3A). Occasionally, shoots grew to about 1 cm, but they did not develop any normal leaves (Fig. 3B). The abnormal leaf-like organs of these shoots lacked the ligule, auricles, and lamina joint, which are located between the leaf blade and sheath of wild-type leaves (Fig. 3C). Most parts of the leaf-like organs seemed to derive from the leaf sheath, but we could not confirm this histologically. The typical phenotype of OSH1 transformants was similar to that of the OsIPT transformants. The above-ground portions of OSH1 transformants formed clumps of multiple shoots that grew to about 2 mm (Fig. 3D). Shoots of OSH1 transformants also occasionally grew to about 1 cm and their leaf-like organs also did not form the ligule, auricles, or lamina joint (Fig. 3E).
Figure 3.
Phenotypes of transgenic rice plants. A and B, Typical phenotype of an OsIPT overproducer 1 month after regeneration (OsIPT3 overproducer shown). C, Wild-type rice plant 1 month after germination. lb, Leaf blade; lj, lamina joint; ls, leaf sheath. D and E, Typical phenotype of OSH1 overproducer 1 month after regeneration. F, Scanning electron micrograph of shoots of OsIPT3 overproducer. Shoots were connected at their bottoms, indicating that axillary buds were successively grown. Leaf-like organs tightly overlapped each other and ectopic meristem was not observed on their adaxial surfaces (arrowhead). G and H, Scanning electron micrographs of leaf-like organs of OSH1 overproducer. Ectopic meristems (arrows) were occasionally formed on the adaxial surfaces of leaf-like organs (arrowheads). Scale bars, 2 mm (A, B, D, and E), 5 cm (C), and 250 μm (F–H).
Interestingly, OSH1 transformants developed normal roots and ectopic shoots, neither of which has been observed in OsIPT transformants. The shoot clumps of the OsIPT transformants were formed from successive development of axillary shoots, but not by ectopic shoot formation on the leaves (Fig. 3F). In addition to such successive outgrowth of axillary shoots in OSH1 transformants, ectopic shoots were formed on the adaxial surfaces of the leaf-like organs (Fig. 3, G and H). Thus, the phenotype of shoot clumps of the OSH1 transformants was caused by both successive development of axillary shoots and ectopic meristem formation on the malformed leaf-like organs.
CK Content in Transgenic Rice
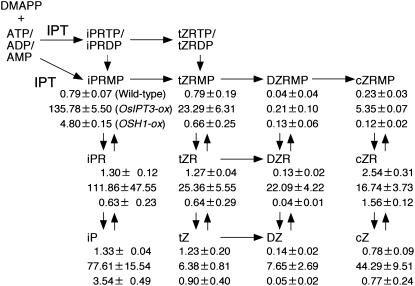
Next, we compared the endogenous levels of 12 CK species in wild-type rice and in OsIPT3 and OSH1 transformants. As shown in Figure 4, all 12 CK species examined were accumulated in very large amounts in the OsIPT3 transformants (Fig. 4, middle values), confirming that overexpression of OsIPT genes stimulates de novo CK biosynthesis. Similar results were obtained from OsIPT2 transformants (data not shown). On the other hand, levels of only three of the 12 CK species were increased in the OSH1 transformants (Fig. 4, bottom values). Although the levels of iP riboside-5′-monophosphate (iPRMP) and iP in the OSH1 transformants were 6.1 and 2.7 times those in the wild type, that of the nucleoside form, iP riboside (iPR), was about one-half that in the wild type. Levels of both tZ and cis-zeatin and of their nucleosides and nucleotides were decreased. This CK measurement analysis has revealed that overexpression of OSH1 does not cause a simple increase in de novo CK biosynthesis, but modifies CK homeostasis and consequently increases bioactive iP content, which may result in alteration of shoot development to a multiple shoot phenotype. It is noteworthy that the abundance of individual CKs was quite different between OSH1 and OsIPT3 transformants, and the enhanced level of iP caused by OSH1 overexpression (2.7-fold) was much lower than that caused by OsIPT3 overexpression (58-fold), even though ectopic shoot formation was observed only in the OSH1 transformants. This indicates that the severely abnormal phenotype of the OSH1 transformants is not caused only by CK overproduction.
Figure 4.
CK concentrations in wild-type and transgenic rice. Endogenous levels (pmol g−1 fresh weight) in wild-type (top value in each group), OsIPT3 overexpresser (middle), and OSH1 overexpresser (bottom) are shown below each product. Measurements were performed in triplicate and the mean values with sd are shown.
Endogenous OsIPT Expression in Transgenic Rice
Because IPT catalyzes the formation of iPRMP (Fig. 4), accumulation of iPRMP in the OSH1 transformants suggests that expression of one or more OsIPTs is up-regulated by the KNOX protein. To distinguish the direct effects of KNOX proteins from the various changes observed in malformed transgenic plants, we generated an artificial inducible system of OSH15 function using the human glucocorticoid receptor (GR). The steroid-binding domain of GR inactivates the function of a neighboring domain in the chimeric protein molecule in the absence of a steroid ligand, but the function is restored in the presence of the ligand, dexamethasone (DEX), even in plants (Schena et al., 1991). Using this inducible system, we have found that inhibition of GA biosynthesis via the specific suppression of GA 20-oxidase gene expression was one of the earliest events caused by the activation of tobacco KNOX protein, NTH15 (Sakamoto et al., 2001).
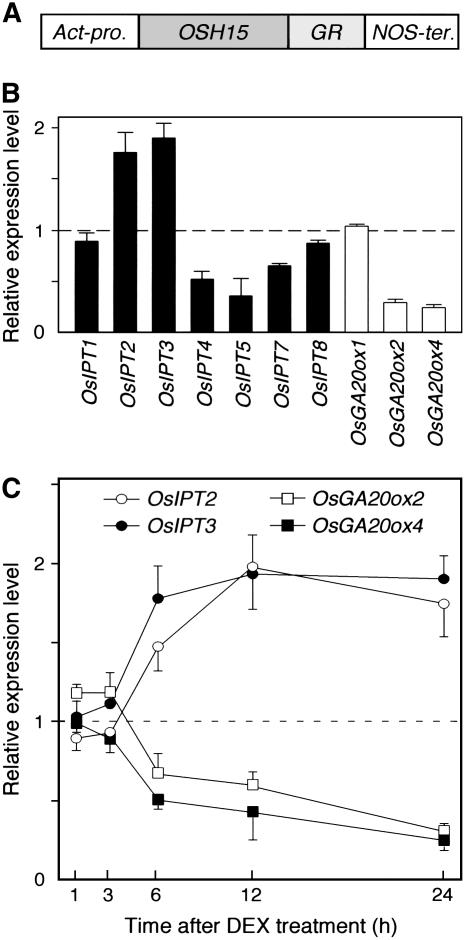
In this study, we produced the OSH15:GR fusion protein in transgenic rice plants under the control of the rice actin promoter (Fig. 5A). These transformants showed DEX-dependent induction of abnormal morphology (data not shown). Using this inducible system, we first examined the expression of GA 20-oxidase genes by qRT-PCR analysis because KNOX protein directly binds to the promoter sequence of GA 20-oxidase genes and suppresses their expression in tobacco and potato (Sakamoto et al., 2001; Chen et al., 2004). Rice has four GA 20-oxidase genes, one of which, OsGA20ox3, was specifically expressed in reproductive organs (Sakamoto et al., 2004). Therefore, we examined the expression levels of the remaining three genes in OSH15:GR transgenic seedlings at 24 h after DEX treatment. Transcripts of two genes, OsGA20ox2 and OsGA20ox4, were decreased to 30% and 27%, respectively, of the levels in DEX untreated control plants, but expression of OsGA20ox1 was not changed by the treatment (Fig. 5B). Reduction of expression of OsGA20ox2 and OsGA20ox4 occurred between 3 and 6 h after treatment (Fig. 5C). These observations indicate that suppression of GA 20-oxidase gene expression is a rapid event in the KNOX protein-targeted phenomena and is conserved between monocots and dicots.
Figure 5.
Expression of endogenous OsIPT and GA 20-oxidase genes in response to the induction of OSH15 function. A, Schematic representation of the OSH15:GR transgene. The chimeric gene consists of an in-frame fusion of the entire OSH15 cDNA and the steroid-binding domain of the human GR. This gene was driven by the rice actin promoter. B, Relative expression levels of OsIPT and GA 20-oxidase genes in 2-week-old OSH15:GR transgenic rice 24 h after 1 μm DEX treatment. C, Changes in the expression levels of OsIPT2, OsIPT3, OsGA20ox2, and OsGA20ox4 after DEX treatment. The ratio between each gene level and the histone H3 level obtained from DEX untreated control plants was arbitrarily set at 1.0. qRT-PCR was performed in triplicate and mean values with sd are shown.
Next, we examined the expression level of seven OsIPT genes in OSH15:GR transgenic seedlings 24 h after DEX treatment. Expression levels of five OsIPT genes (OsIPT1, OsIPT4, OsIPT5, OsIPT7, and OsIPT8) were slightly or greatly decreased at 24 h after treatment, whereas the levels of two genes, OsIPT2 and OsIPT3, were increased to 1.8 and 1.9 times, respectively, those in control plants (Fig. 5B). Such increased expression of both OsIPT2 and OsIPT3 occurred from 3 to 6 h after treatment, similar timing to that of the decrease in GA 20-oxidase gene expression (Fig. 5C). These observations suggest that induction of OsIPT2 and OsIPT3 is a rapid event in KNOX protein-controlled phenomena, like the down-regulation of GA 20-oxidase genes, and such IPT induction increases the endogenous CK level in KNOX overexpressers.
DISCUSSION
Many examples show that ectopic expression of KNOX proteins causes morphological alterations in transgenic plants, such as loss of apical dominance and adventitious meristem formation on leaves (Matsuoka et al., 1993; Sinha et al., 1993; Chuck et al., 1996; Tamaoki et al., 1997; Nishimura et al., 2000). KNOX overproducer phenotypes are similar to those of transformants expressing the bacterial ipt gene (Faiss et al., 1997) and the petunia IPT gene Sho (Zubko et al., 2002), and therefore the phenotypic similarity between CK overproducers and ectopic expressers of KNOX proteins suggests that KNOX proteins are involved in a CK-related pathway in plant development. Overexpression of knox genes in transgenic plants increases CK levels (Tamaoki et al., 1997; Kusaba et al., 1998; Ori et al., 1999; Hewelt et al., 2000; Frugis et al., 2001). Recent studies demonstrated that the Arabidopsis KNOX protein STM induces expression of AtIPT7 within 2 h after induction (Jasinski et al., 2005; Yanai et al., 2005). In our experiments, expression of two OsIPT genes, OsIPT2 and OsIPT3, was increased in response to the activation of a rice KNOX protein, OSH15, with similar kinetics to those of down-regulation of GA 20-oxidase genes, target genes of KNOX proteins in dicots (Fig. 5). Interestingly, expression patterns of OsIPT2 and OsIPT3 are unusual in comparison with those of other OsIPTs: These OsIPTs were not down-regulated by exogenous iP treatment in wild-type plants (Fig. 2B) or in the OSH1 overexpressers (data not shown), whereas expression of the other OsIPTs was regulated in a negative feedback manner. These results suggest that OsIPT2 and OsIPT3 have unique roles in the developmental process, which is controlled by KNOX proteins, rather than in the maintenance of bioactive CK levels in rice. Up-regulation of these IPT genes by KNOX proteins, whose kinetics were as rapid as the suppression of GA 20-oxidase gene expression (Fig. 5), also leads us to speculate that KNOX proteins may directly interact with their target sequences in the IPT genes to up-regulate expression of these genes because tobacco and potato KNOX proteins directly bind to the promoter sequences of GA 20-oxidase genes (Sakamoto et al., 2001; Chen et al., 2004). OsIPT2 contains a binding motif for OSH15 (TGTGAC; Nagasaki et al., 2001) in its 5′-flanking region at positions −2,460 to −2,455 (taking the translation initiation site as +1). Similarly, OsIPT3 contains preferable binding motifs for OSH15 (TGTCAC; Nagasaki et al., 2001) in its 5′-flanking region at positions −512 to −507 and −111 to −106. However, CK biosynthesis is not always increased by OSH1 expression. In fact, OSH1 overproducers normally develop roots, whereas root development was almost completely absent in the IPT overproducers (Fig. 3). Thus, KNOX protein is not a sufficient factor for enhanced expression of OsIPT2 and OsIPT3, and other factors may be essential.
Increased OsIPT2 and OsIPT3 expression induces de novo CK biosynthesis in KNOX overexpressers. Interestingly, the abundance of individual CKs was quite different between OSH1 and OsIPT3 overexpressers (Fig. 4). In OsIPT3 overexpressers, all CK species were greatly accumulated. In contrast, although the level of iP, the major bioactive CK in rice, was elevated about 3-fold, the level of its nucleoside, iPR, was decreased in OSH1 overexpressers. Similarly, the level of tZ, the major bioactive CK in tobacco, was increased, but the level of its nucleoside, tZR, was decreased in transgenic tobacco plants overexpressing either OSH1 or NTH15, a tobacco OSH1 homolog (Tamaoki et al., 1997; Kusaba et al., 1998). These results imply that overexpression of knox genes not only increases de novo CK biosynthesis through the induction of IPT gene expression, but also modulates CK metabolism such as the deribosylation step of iPR (or tZR in tobacco) to form the bioactive iP (or tZ). Although activation of CK is very important in the regulation of CK activity, no genes for CK nucleosidase have been identified yet. Further studies are needed to understand how knox genes function in plant development through regulation of CK biosynthesis and metabolism.
Recently, it was revealed that another type of homeodomain protein regulating stem cell fate in the SAM, WUSCHEL (WUS), directly suppresses the expression of CK-inducible type-A ARABIDOPSIS RESPONSE REGULATOR 7 (ARR7; Leibfried et al., 2005). Because type-A ARR proteins negatively regulate CK signaling (To et al., 2004), WUS and KNOX can activate CK action in different ways. However, Jasinski et al. (2005) clearly demonstrated that not only high-CK, but also low-GA, conditions are required for SAM maintenance, and KNOX protein acts as a general orchestrator by activating CK and repressing GA biosynthesis. Suppression of GA 20-oxidase gene expression by KNOX protein has been reported in various dicot plants (Sakamoto et al., 2001; Hay et al., 2002; Chen et al., 2004) and, in our experiments, expression of two GA 20-oxidase genes was rapidly down-regulated by induction of the KNOX function also in rice (Fig. 5). In addition, ectopic meristem formation was observed in KNOX overproducers, but not in OsIPT overproducers, even if they contained higher levels of bioactive CKs. These results support the possibility that rice meristems need not only high-CK, but also low-GA, conditions to maintain their activity.
Interestingly, a similar function was observed in a negative regulator of GA responses, SPINDLY (SPY). A loss-of-function mutation of SPY or GA treatment of wild-type Arabidopsis plants suppressed CK responses and CK induction of ARR5, but not ARR7 expression (Greenboim-Wainberg et al., 2005). The results indicate that Arabidopsis SPY acts as both a repressor of GA responses and a positive regulator of CK signaling. Recently, the rice SPY ortholog, OsSPY, was characterized. OsSPY also suppresses GA responses and OsSPY knockdown plants accumulate bioactive brassinosteroid (BR) and show BR-overproducing phenotypes, such as increased leaf inclination (Shimada et al., 2006). These results suggests that OsSPY functions in GA signaling and BR metabolism, whereas the effects on CK signaling and meristem maintenance are uncertain.
In conclusion, ectopic expression of KNOX proteins induces specific IPT gene expression and de novo CK biosynthesis, and this cascade is conserved in both monocots and dicots. It is noteworthy that another important function of KNOX proteins—repression of GA biosynthesis through suppression of GA 20-oxidase gene expression—is also conserved between monocots and dicots. These results indicate that plant meristems need high-CK and low-GA conditions to maintain their activity and that KNOX proteins act as central regulators to control these phytohormones at adequate levels, regardless of the differences in organization between monocots and dicots.
MATERIALS AND METHODS
Isolation of Rice IPT Genes
A BLAST search using the predicted amino acid sequences encoded by Arabidopsis (Arabidopsis thaliana) IPT genes as probes was performed against the rice (Oryza sativa) DNA databases as described (Sakamoto et al., 2004). The predicted protein sequences were initially clustered with ClustalW (Thompson et al., 1994). TreeView was used to generate graphic output (Page, 1996). Accession numbers of the sequences used are indicated in Supplemental Table I. Entire coding regions for putative rice IPT genes were amplified by PCR using rice genomic DNA. Primers were designed to generate appropriate restriction sites for constructing a translational fusion with the pET-32a expression vector (Novagen). Amplified fragments were cloned into pCR II (Invitrogen) and their nucleotide sequences were determined.
Enzyme Assays
All OsIPT genes were translationally fused to the pET-32a expression vector (Novagen) and expressed in BL21 (DE3) Escherichia coli cells (Stratagene). Detailed conditions for OsIPT expression in E. coli and measurements of IPT activity were described previously (Takei et al., 2001).
Plasmid Constructs and Plant Transformation
The entire coding region of OsIPT1, OsIPT2, OsIPT3, OsIPT4, OsIPT7, or OSH1 was inserted between the rice actin promoter and the nopaline synthase polyadenylation signal of hygromycin-resistant binary vector pAct-Hm2. This vector is modified from pBI-H1 (Ohta et al., 1990) and contains a rice actin promoter. To create the OSH15:GR fusion protein, the stop codon of OSH15 was replaced with a SmaI site by PCR and fused to the steroid-binding domain of the human GR as described (Sakamoto et al., 2001). The resulting construct was introduced into Agrobacterium tumefaciens strain EHA105, and Agrobacterium-mediated transformation of rice was performed as described (Hiei et al., 1994). Transgenic plants were selected on media containing 50 mg L−1 hygromycin.
Expression Analysis
To determine the organ specificity of OsIPT expression, total RNA was separately prepared from various organs of wild-type rice. For feedback analysis, wild-type seeds were sown on agar medium containing 5 μm iP and grown for a week, and total RNA was extracted from whole seedlings. OSH15:GR transgenic seeds were sown on agar medium and grown for 2 weeks and then transplanted to agar medium containing 10 μm DEX or the same volume of ethanol. Total RNA was extracted from whole seedlings. Single-strand cDNAs were synthesized by using an Advantage RT-for-PCR kit (CLONTECH). qRT-PCR was performed with an iCycler iQ real-time PCR system (Bio-Rad Laboratories). Expression levels were normalized against the values obtained for histone H3, which was used as an internal reference gene. Primer sequences are listed in Supplemental Table II. These primers specifically amplified the target gene sequences (data not shown).
Measurement of CK Concentrations
Wild-type seedlings and transformants (approximately 1 g) were collected and frozen at −80°C until use. CKs were extracted and fractionated from whole plants, and the resulting CK fractions were analyzed by liquid chromatography-mass spectrometry, as described previously (Takei et al., 2004).
Supplementary Material
This work was supported by the Ministry of Agriculture, Forestry, and Fisheries of Japan (Rice Genome Project IP–1010 to T.S. and Rice Genome Project IP–3003 to H.S.), by the Ministry of Education, Culture, Sports, Science and Technology of Japan (to H.S.), and by a Grant-in-Aid for the Center of Excellence from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to M.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Makoto Matuoka (makoto@nuagr1.agr.nagoya-u.ac.jp).
The online version of this article contains Web-only data.
References
- Chaudhury AM, Letham S, Craig S, Dennis ES (1993) amp1—a mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J 4: 907–916 [Google Scholar]
- Chen H, Banerjee AK, Hannapel DJ (2004) The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J 38: 276–284 [DOI] [PubMed] [Google Scholar]
- Chiang H-H, Hwang I, Goodman HM (1995) Isolation of the Arabidopsis GA4 locus. Plant Cell 7: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S (1996) KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8: 1277–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SE, Elliott RC, Helliwell CA, Poole AT, Reid JB (2003) The pea gene NA encodes ent-kaurenoic acid oxidase. Plant Physiol 131: 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faiss M, Zalubilova J, Strnad M, Schmülling T (1997) Conditional transgenic expression of the ipt gene indicates a function for cytokinins in paracrine signaling in whole tobacco plants. Plant J 12: 401–415 [DOI] [PubMed] [Google Scholar]
- Frugis G, Giannino D, Mele G, Nicolodi C, Chiappetta A, Bitonti MB, Innocenti AM, Dewitte W, Van Onckelen H, Mariotti D (2001) Overexpression of KNAT1 in lettuce shifts leaf determinate growth to a shoot-like indeterminate growth associated with an accumulation of isopentenyl-type cytokinins. Plant Physiol 126: 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D (2005) Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17: 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M (2002) The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr Biol 12: 1557–1565 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Sheldon CC, Olive MR, Walker AR, Zeevaart JAD, Reacock WJ, Dennis ES (1998) Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc Natl Acad Sci USA 95: 9019–9024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewelt A, Prinsen E, Thomas M, Onckelen HV, Meins F Jr (2000) Ectopic expression of maize knotted1 results in the cytokinin-autotrophic growth of cultured tobacco tissues. Planta 210: 884–889 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Itoh H, Tatsumi T, Sakamoto T, Otomo K, Toyomasu T, Kitano H, Ashikari M, Ichihara S, Matsuoka M (2004) A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol Biol 54: 533–547 [DOI] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M (2001) Cloning and functional analysis of gibberellin 3β-hydroxylase genes that are differently expressed during the growth of rice. Proc Natl Acad Sci USA 98: 8909–8914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S (1994) Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413 [Google Scholar]
- Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M (2005) KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Kakimoto T (2001) Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopentenyltransferase. Plant Cell Physiol 42: 677–685 [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Laudencia-Chingcuanco D, Smith L, Hake S (1997) Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124: 3045–3054 [DOI] [PubMed] [Google Scholar]
- Krall L, Raschke M, Zenk MH, Baron C (2002) The Tzs protein from Agrobacterium tumefaciens C58 produces zeatin riboside 5′-phosphate from 4-hydroxy-3-methyl-2-(E)-butenyl diphosphate and AMP. FEBS Lett 527: 315–318 [DOI] [PubMed] [Google Scholar]
- Kusaba S, Kano-Murakami Y, Matsuoka M, Tamaoki M, Sakamoto T, Yamaguchi I, Fukumoto M (1998) Alteration of hormone levels in transgenic tobacco plants overexpressing a rice homeobox gene OSH1. Plant Physiol 116: 471–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S (1994) A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Ichikawa H, Saito A, Tada Y, Fujimura T, Kano-Murakami Y (1993) Expression of a rice homeobox gene causes altered morphology of transgenic plants. Plant Cell 5: 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy D, Blowers AD, Jenes B, Wu R (1991) Construction of expression vectors based on the rice actin 1 (Act1) 5′ region for use in monocot transformation. Mol Gen Genet 231: 150–160 [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37: 128–138 [DOI] [PubMed] [Google Scholar]
- Mok MC (1994) Cytokinins and plant development—an overview. In DWS Mok, MC Mok, eds, Cytokinins—Chemistry, Activity, and Function. CRC Press, Boca Raton, FL, pp 155–166
- Nagasaki H, Sakamoto T, Sato Y, Matsuoka M (2001) Functional analysis of the conserved domains of a rice KNOX homeodomain protein, OSH15. Plant Cell 13: 2085–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura A, Tamaoki M, Sakamoto T, Matsuoka M (2000) Over-expression of tobacco knotted1-type class1 homeobox genes alters various leaf morphology. Plant Cell Physiol 41: 583–590 [DOI] [PubMed] [Google Scholar]
- Nishimura A, Tamaoki M, Sato Y, Matsuoka M (1999) The expression of tobacco knotted1-type class 1 homeobox genes correspond to regions predicted by the cytohistological zonation model. Plant J 18: 337–347 [DOI] [PubMed] [Google Scholar]
- Ohta S, Mita S, Hattori T, Nakamura K (1990) Construction and expression in tobacco of a β-glucuronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol 31: 805–813 [Google Scholar]
- Ori N, Juarez MT, Jackson D, Yamaguchi J, Banowetz GM, Hake S (1999) Leaf senescence is delayed in tobacco plants expressing the maize homeobox gene knotted1 under the control of a senescence-activated promoter. Plant Cell 11: 1073–1080 [PMC free article] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Reiser L, Sanchez-Baracaldo P, Hake S (2000) Knots in the family tree: evolutionary relationships and functions of knox homeobox genes. Plant Mol Biol 42: 151–166 [PubMed] [Google Scholar]
- Sakakibara H (2004) Cytokinin biosynthesis and metabolism. In PJ Davies, ed, Plant Hormones: Biosynthesis, Signal Transduction, Action! Springer, Dordrecht, The Netherlands, pp 95–114
- Sakakibara H (2005) Cytokinin biosynthesis and regulation. Vitam Horm 72: 271–287 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Kasahara H, Ueda N, Kojima M, Takei K, Hishiyama S, Asami T, Okada K, Kamiya Y, Yamaya T, et al (2005) Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosynthetic pathway in the host plant. Proc Natl Acad Sci USA 102: 9972–9977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Kamiya N, Ueguchi-Tanaka M, Iwahori S, Matsuoka M (2001) KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev 15: 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano Y, Okada Y, Matsunaga A, Suwama T, Kaneko T, Ito K, Noguchi H, Abe I (2004) Molecular cloning, expression, and characterization of adenylate isopentenyltransferase from hop (Humulus lupulus L.). Phytochemistry 65: 2439–2446 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al (2002) A mutant gibberellin-synthesis gene in rice. Nature 416: 701–702 [DOI] [PubMed] [Google Scholar]
- Schena M, Lloyd AM, Davis RW (1991) A steroid-inducible gene expression system for plant cells. Proc Natl Acad Sci USA 88: 10421–10425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentoku N, Sato Y, Kurata N, Ito Y, Kitano H, Matsuoka M (1999) Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell 11: 1651–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentoku N, Sato Y, Matsuoka M (2000) Overexpression of rice OSH genes induces ectopic shoots on leaf sheaths of transgenic rice plants. Dev Biol 220: 358–364 [DOI] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Sakamoto T, Fujioka S, Takatsuto S, Yoshida S, Sazuka T, Ashikari M, Matsuoka M (2006) The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and is involved in brassinosteroid synthesis. Plant J 47: (in press) [DOI] [PubMed]
- Sinha NR, Williams RE, Hake S (1993) Overexpression of the maize homeobox gene, Knotted-1, causes a switch from determinate to indeterminate cell fates. Genes Dev 7: 787–795 [DOI] [PubMed] [Google Scholar]
- Spray CR, Kobayashi M, Suzuki Y, Phinney BO, Gaskin P, MacMillan J (1996) The dwarf-1 (dt) mutant of Zea mays blocks three steps in the gibberellin-biosynthetic pathway. Proc Natl Acad Sci USA 93: 10515–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-P, Goodman HM, Ausubel FM (1992) Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell 4: 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Sugiyama T (2001) Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J Biol Chem 276: 26405–26410 [DOI] [PubMed] [Google Scholar]
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H (2004) AtIPT3, an Arabidopsis isopentenyltransferase gene, is a key determinant of macronutrient-responsive cytokinin biosynthesis. Plant Cell Physiol 45: 1053–1062 [DOI] [PubMed] [Google Scholar]
- Tamaoki M, Kusaba S, Kano-Murakami Y, Matsuoka M (1997) Ectopic expression of a tobacco homeobox gene, NTH15, dramatically alters leaf morphology and hormone levels in transgenic tobacco. Plant Cell Physiol 38: 917–927 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruere J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y-L, Li L, Wu K, Peeters AJM, Gage DA, Zeevaart JAD (1995) The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci USA 92: 6640–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Sun T-P, Kawaide H, Kamiya Y (1998) The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol 116: 1271–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O, Shani E, Dolezal K, Tarkowski P, Sablowski R, Sandberg G, Samach A, Ori N (2005) Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr Biol 15: 1566–1571 [DOI] [PubMed] [Google Scholar]
- Zubko E, Adams CJ, Machaekova I, Malbeck J, Scollan C, Meyer P (2002) Activation tagging identifies a gene from Petunia hybrida responsible for the production of active cytokinins in plants. Plant J 29: 797–808 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.