Abstract
Under normal field growth conditions, canola (Brassica napus) seeds produce chloroplasts during early seed development and then catabolize the photosynthetic machinery during seed maturation, producing mature seeds at harvest that are essentially free of chlorophyll (Chl). However, frost exposure early in canola seed development disrupts the normal programming of Chl degradation, resulting in green seed at harvest and thereby significantly devaluing the crop. Pheophorbide a oxygenase (PaO), a key control point in the overall regulation of Chl degradation, was affected by freezing. Pheophorbide a, the substrate of PaO, accumulated during late stages of maturation in seeds that had been exposed to freezing during early seed development. Freezing interfered with the induction of PaO activity that normally occurs in the later phases of canola seed development when Chl should be cleared from the seed. Moreover, we found that the induction of PaO activity in canola seed was largely posttranslationally controlled and it was at this level that freezing interfered with PaO activation. The increased accumulation of PaO transcript and protein levels during seed development was not altered by the freezing episode, and the increase in PaO protein was small compared to the increase in PaO activity. We found that PaO could be phosphorylated and that phosphorylation decreased with increasing activity, implicating PaO dephosphorylation as an important posttranslational control mechanism for this enzyme. Two PaO genes, BnPaO1 and BnPaO2, were identified in senescing canola leaves and during early seed development, but only BnPaO2 was expressed in maturing, degreening seeds.
Canola (Brassica napus) is an important oil seed crop grown extensively in North America and northern Europe with annual yields exceeding 7 million metric tons. Canola is the world's third-most important vegetable oil crop, in significant part due to the low levels of erucic acid and glucosinolates in canola oil (Levadoux et al., 1987; Zhang et al., 2004). However, the chlorophyll (Chl) content is significantly higher than that found in other major vegetable oils and is the biggest quality impediment in the canola oil industry.
During the early stages of seed development, photosynthetically produced carbohydrate is transferred from the leaves and silique walls to the seeds for the synthesis of oil and other storage products. In canola seeds, the conversion of sugars to fatty acids is the primary metabolic flux with more than 60% of carbon stored as oil (Schwender et al., 2004b). There is evidence that developing embryos are capable of significant rates of photosynthesis directly associated with fatty acid biosynthesis of the developing seed (Eastmond et al., 1996; Willms et al., 1999; Ruuska et al., 2004). However, the low light levels able to reach the seeds through the silique walls made it difficult to explain the significance of photosynthesis in oil biosynthesis within the seed. This conundrum was explained by the recent discovery that Rubisco in developing canola embryos can use the energy of photosynthesis while operating in a novel pathway that produces acetyl-CoA with much greater efficiency than the previously described glycolytic pathway (Schwender et al., 2004a). As the seed matures and the rate of oil synthesis declines, the need for photosynthesis declines and chloroplasts are degraded, resulting in seeds at harvest free of Chl. Although canola is in general a cold hardy plant, a freezing episode early in seed development can disrupt the normal program of Chl degradation, resulting in a green seed at harvest and significantly devaluing the crop (Johnson-Flanagan and Thiagarajah, 1990). This so-called green seed problem is a high priority seed quality issue within the canola industry.
The key reactions in Chl degradation are catalyzed by chloroplast localized enzymes (Matile et al., 1999; Hörtensteiner, 2006). Chl is removed from the Chl binding proteins within the thylakoid membranes by a yet undescribed process. Once free from the membrane, the initial step in Chl degradation is the removal of the phytol tail by chlorophyllase (Chlase), which catalyzes hydrolysis of the ester linkage of the phytol chain to the porphyrin macrocycle (Jacob-Wilk et al., 1999; Matile et al., 1999; Tsuchiya et al., 1999). In Arabidopsis (Arabidopsis thaliana), there are two known Chlase genes. AtCLH1 appears to be induced in response to wounding and pathogen attack (Kariola et al., 2005), whereas AtCLH2 expression is constitutive. Chlase activity is at least in some cases latent prior to the onset of senescence (Benedetti and Arruda, 2002), but in other cases, high Chlase activity has been associated with high rates of Chl synthesis (Roca and Minguez-Mosquera, 2003), making it unlikely that Chlase is a central controlling step in Chl degradation.
Magnesium-dechelatase, for which the gene and protein are yet to be identified, is responsible for removing the magnesium ion from the tetrapyrrole producing the chlorin molecule pheophorbide (Pheide) a (Shioi et al., 1996). Although the release of Mg2+ could in principle occur spontaneously, Mg-dechelatase activity has been demonstrated with the artificial substrate chlorophyllin for which spontaneous Mg2+ removal is unlikely (Shioi et al., 1996). Removal of the magnesium ion from the macrocycle prepares it for Pheide a oxygenase (PaO), which opens the macrocycle of Pheide a, resulting in the final disappearance of the green color (Rodoni et al., 1997).
PaO, a nonheme iron monooxygenase localized to the inner envelope of maturing gerontoplasts, opens the porphyrin macrocycle by adding two oxygen atoms (Matile and Schellenberg., 1996). Pheide a has been shown to be an efficient substrate in PaO activity measurements, whereas Pheide b acts as a competitive inhibitor (Hörtensteiner et al., 1995; Pružinská et al., 2003). In Arabidopsis, AtPaO belongs to a five-member gene family encoding nonheme iron oxygenases defined by the presence of a Rieske-type domain in addition to a mononuclear iron-binding domain. This gene family also includes Chl a oxygenase, choline monoxygenase, Tic55, and Ptc52 (Gray et al., 2004). The Arabidopsis cell death mimic mutant accelerated cell death 1 (acd1) is an AtPaO mutant allele (Pružinská et al., 2003) and is orthologous to lethal leaf spot 1 (lls1) of maize (Zea mays; Gray et al., 2004). The mutant leaves of both plants accumulate Pheide a, making them highly photosensitive and thereby producing the cell death mimic phenotype.
The conversion of Pheide a to the colorless primary fluorescent Chl catabolite (pFCC) is a complex step that involves not only PaO but also red Chl catabolite reductase (RCCR), a stromal protein, and is the causal gene in acd2 mutant of Arabidopsis. The opening of the Pheide a macrocycle by PaO produces a red colored catabolite (RCC), an intermediary product, which in turn is reduced by RCCR in a reaction requiring ferredoxin to form pFCC, a colorless compound that is detected by its distinctive blue fluorescence (Wüthrich et al., 2000). The RCCR gene is expressed in most tissues, even roots (Mach et al., 2001; Yao and Greenberg, 2006), and is constitutively active throughout leaf development, including senescence. Moreover, RCCR protein levels do not change during senescence or pathogen attack (Mach et al., 2001), removing it from consideration as a significant control step in Chl degradation. The final steps of Chl degradation involve the hydroxylation and conjugation of the pFCC tetrapyrroles. Fluorescent Chl catabolites (FCCs) are exported from the gerontoplasts for further modification in the cytosol. The modified FCCs are imported to the vacuole, eventually leading to the formation of noncolored catabolites, which are not phototoxic and stored in the vacuoles (Oberhuber et al., 2003).
The major objective of this study was to identify those steps in Chl degradation in maturing canola seeds that are disrupted by exposure to freezing temperatures early in seed development. The results show that freezing interfered with the induction of PaO activity that normally occurs in the later phases of canola seed development. Moreover, we found that the regulation of PaO activity was largely posttranslational and it was at this level that freezing interfered with PaO activation in canola seeds.
RESULTS
Nonlethal Freezing Exposure Prevented the Complete Clearing of Chl from Mature Canola Seeds
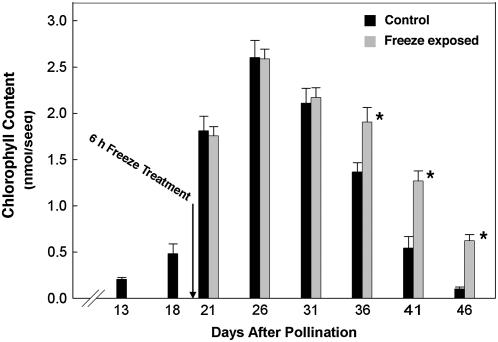
At 20 d after pollination (DAP), when seeds had attained about 45% of final dry weight (Fig. 1A) and 60% of maximum Chl content (Fig. 2), canola plants were cooled in the dark at 5°C/h until reaching −4°C, where the temperature was held for 6 h followed by rewarming at 5°C/h back to the growth temperature. The Chl content of seeds collected at intervals from 13 to 46 DAP was measured spectrophotometrically in N, N-dimethylformamide using the specific absorption coefficients of Porra and Grimme (1974) and expressed on the basis of seed fresh weight. The rate of Chl accumulation increased over the first 26 DAP with the greatest increase occurring between 18 and 21 DAP, during which time there was a nearly 4-fold increase in Chl content per seed (Fig. 2). The freeze exposure at 20 DAP had no significant affect on maximum Chl content (Fig. 2) or the fresh (data not shown) or dry weight (Fig. 1A) of the developing seeds. Net Chl degradation was initiated sometime after 26 DAP in both control and freeze-exposed samples with Chl degraded to trace amounts by 46 DAP in control canola seeds. The freezing-induced delay in Chl loss became evident at 36 DAP, and thereafter the rate of net degradation in freeze-exposed seeds proceeded more slowly. Chl degradation stopped when the seed moisture content dropped below approximately 40% (compare Figs. 1B and 2). The freezing-induced delay in Chl degradation resulted in mature seeds with approximately 600% higher Chl content than control (Fig. 2).
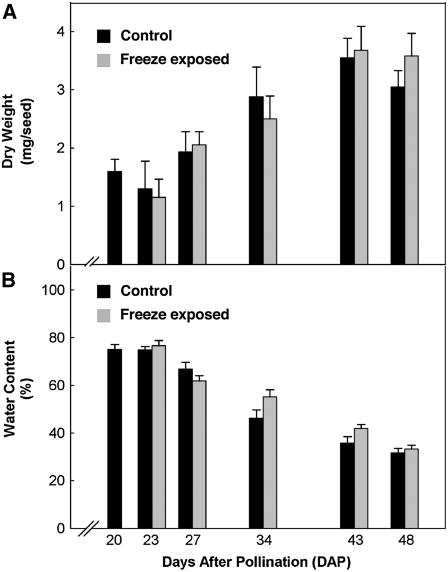
Figure 1.
Freezing exposure did not affect canola seed dry weight (A) or water content (B). At 20 DAP, canola plants were cooled 5°C/h until reaching −4°C, where the temperature was held for 6 h followed by rewarming at 5°C/h back to the growth temperature. The freeze-exposed plants were compared to control plants. The dry weight of the seeds was determined by incubating the seeds overnight at 75°C. Canola seed water content during development was calculated by (fresh weight − dry weight)/fresh weight. To separate the effects of freeze exposure from seed moisture loss on seed degreening, plants were maintained at high humidity during the freeze treatment and the recovery period.
Figure 2.
A 6-h freeze exposure at 20 DAP caused Chl to subsequently be retained in mature canola seed. The amount of Chl retained in the seeds of freeze-treated plants was statistically greater (P < 0.05 [*]) on 36 DAP and beyond.
The Effects of Freeze Exposure on Chl Loss Was Direct and Not Mediated by Differential Effects of Freezing on Seed Moisture Content
Although the interference with developmentally programmed Chl degradation in maturing canola seeds by freeze exposure is well established (Johnson-Flanagan and Thiagarajah, 1990), it has been suggested that the effect is indirectly mediated by acceleration of seed water loss from freeze-exposed plants (Green et al., 1998). Work was done to attempt to separate the direct effects of freeze exposure from ancillary effects of water loss on Chl degradation by maintaining high humidity during the freeze exposure and recovery period. Seed water content was calculated as (fresh weight − dry weight)/fresh weight, and the dry weight of the seeds was determined following overnight incubation at 75°C (Fig. 1A). The decline in water content was gradual over the period of seed development (Fig. 1B), with greater than 50% water loss between 23 and 48 DAP. Under the high humidity conditions, water content of seeds from plants exposed to the freezing treatment was the same as controls, yet the freeze-exposed seeds retained higher Chl levels. This result demonstrates that these two factors, freeze and water loss, affect Chl degradation independently and can be separated experimentally.
Pheide a Accumulated in Maturing Canola Seeds after Freeze Exposure
To determine candidate steps in the Chl degradation of canola seed that may be sensitive to freeze exposure, we investigated the effects of freeze exposure on pools of Chl degradation catabolites during canola seed development (Fig. 3). The catabolites were separated by HPLC based on their polarity in organic solvent and were quantified by fluorescence spectroscopy, which has the sensitivity required to detect Chl catabolites in the trace amounts normally present. The concentration of each catabolite was determined from the fluorescence intensity data using equations developed for the quantification of tetrapyrrole moieties (Rebeiz, 2002).
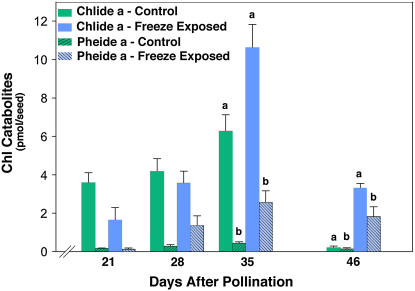
Figure 3.
The effect of freezing on pools of Chl degradation catabolites in canola seeds during development. Seeds from canola plants exposed to a 6-h freeze on 20 DAP as described in Figure 1, accumulated Chlide a and Pheide a during seed development resulting in statistically significant differences (P < 0.05 [a, b]) in control seeds in both compounds by 35 DAP and thereafter.
Freeze exposure on 20 DAP had little effect on catabolite pool sizes until 8 d later. By 28 DAP, there was a 3- to 4-fold freeze-induced increase in Pheide a levels. The accumulation of Pheide a in the freeze-treated seeds became more exaggerated as seed development progressed, showing a nearly 10-fold increase compared to control by 46 DAP (Fig. 3). During the later stages of seed development, 35 DAP and after, freeze treatment induced chlorophyllide (Chlide) a accumulation and, at seed maturity (i.e. 46 DAP), the percent increase of Chlide a exceeded that of Pheide a. The increased levels of Pheide a, and eventually Chlide a, in freeze-exposed seeds suggested that freezing interferes in some fashion with PaO function. That Pheide a accumulation preceded Chlide a accumulation suggested that a progressive feedback within the degradation pathway had developed.
In principle, a decrease in the products of PaO would also be anticipated if the increase in Pheide a in freeze-treated seeds is due to a decrease in PaO activity. However, in senescing leaves, pFCC, the product of the PaO/RCCR reaction, is present in exceedingly low and difficult-to-quantify amounts, and FCCs and RCCs do not accumulate to detectable levels. Noncolored catabolites are the only products downstream of PaO that accumulate in senescing leaves in canola (Pružinská et al., 2005). Neither FCCs nor RCCs accumulated to detectable levels in either control or freeze-treated seeds (data not shown). Thus, while we were unable to confirm that a decrease in product of the PaO reaction accompanied the increase in substrate, the lack of accumulation of downstream product is strong evidence that freeze treatment had no significant effect on Chl degradation reactions downstream of PaO.
Canola Found to Have Two PaO Genes with High Homology to AtPaO
The gene sequence of PaO from Arabidopsis was used to clone and identify the orthologous genes in canola. Two different cDNA clones of PaO were isolated from senescing canola leaves (Supplemental Fig. 1), which will be referred to as BnPaO1 and BnPaO2. The codon-derived amino acid sequences had 92% identity with AtPaO. In comparison to each other, the derived amino acid sequences of the two canola clones were 98% identical. Like PaO from maize and Arabidopsis (Gray et al., 2004), the two canola genes contain a conserved Rieske iron-sulfur domain and a mononuclear iron-binding domain as well as two predicted transmembrane domains.
There are two notable differences in protein sequence at the N-terminal region of BnPaO1, BnPaO2, and AtPaO, possibly due to an insertion/deletion event postdating the divergence of Arabidopsis and canola (Supplemental Fig. 2). Alternatively, as canola is an allotetraploid, BnPaO1 and BnPaO2 could be derived from the two ancestral genomes of canola. BnPaO2 has an additional Ser residue (Ser-29) when compared to BnPaO1 and AtPaO. In the same region, AtPaO has two Thr residues (Thr-27 and Thr-28) positioned where both BnPaO clones have an Asn and a Ser residue (Asn-27 and Ser-28). Both BnPaO clones have an extra Ala residue (Ala-30) not found in AtPaO. Furthermore, in the 75 to 78 region, BnPaO2 is completely missing a sequence of G-D-K-E found in both AtPaO and BnPaO1. Because BnPaO2 has an additional Ser residue (Ser-29) and is missing Gly-75, Asp-76, Lys-77, and Glu-78, collectively this protein has three fewer amino acids than BnPaO1. AtPaO has one less amino acid than BnPaO1 due to the missing Ala residue (Ala-30).
While the expression of BnPaO2 was measurable in seeds throughout seed development, BnPaO1 transcripts were detectable only during early seed development. At 8 to 10 DAP, BnPaO2 transcripts showed nearly 5.5-fold higher levels of expression when compared to BnPaO1 transcripts, and BnPaO2 transcripts at 8 to 10 DAP were expressed at similar levels to 21 DAP canola seeds (data not shown). BnPaO2 transcripts accumulated as seed development progressed with >10-fold increase from 21 to 41 DAP, and the accumulation was not affected by the 6-h freezing exposure on 20 DAP (Fig. 4A). The expression of both BnPaO transcripts was readily detectable in senescing canola leaves (data not shown).
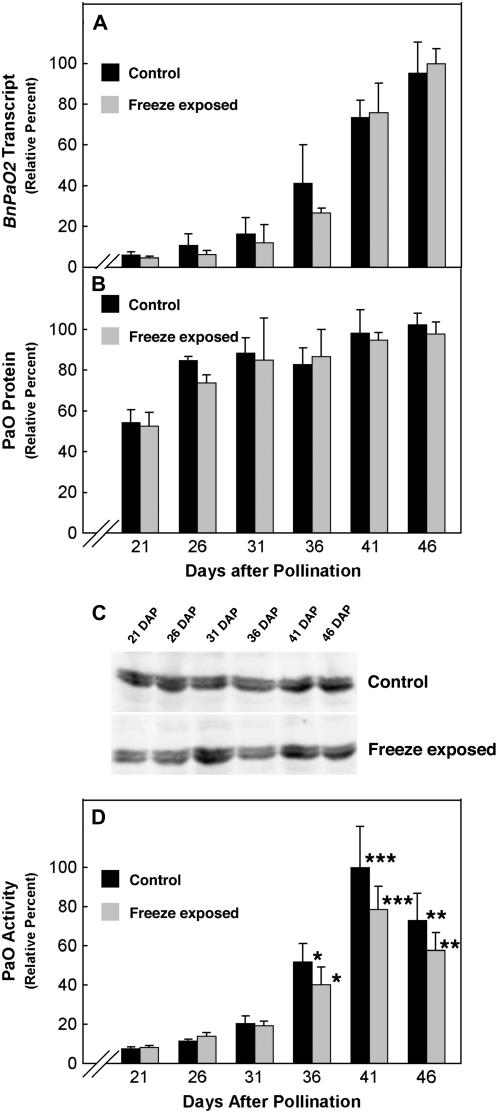
Figure 4.
Comparison of the effects of freeze exposure on PaO transcript, protein, and activity levels during canola seed development. A, BnPaO transcript levels increased nearly 15-fold during canola seed development between 21 and 46 DAP. The increase in PaO transcripts was similar in freeze-exposed canola seeds. B, PaO protein expression was determined from immunoblots of canola seed membrane fractions. Protein levels increased only 2-fold over the period of seed development and there were no significant effects of freeze exposure on PaO protein content. The units for protein expression is intensity/pixel. C, PaO was detected with polyclonal antibodies from the maize LLS1 (PaO) protein. Equal amounts of protein from membrane fractions of canola seeds were loaded in each lane. A PaO doublet was routinely observed using 7% polyacrylamide SDS-PAGE. D, PaO activity profile was induced about 10-fold during seed maturation. The increase in PaO activity between 21 and 31 DAP was similar in control and freeze-exposed seeds. In the later stages, induction of PaO activity was impaired in freeze-exposed seeds compared to control seeds (P < 0.01 [*]; P < 0.001 [**]; P < 0.0001 [***]).
Canola Seed PaO Shown to Be Regulated by a Freezing-Sensitive, Posttranslational Mechanism
The amount of PaO protein from the membrane fraction of developing canola seeds was measured by immunoblot analysis and quantified by infrared imaging. PaO protein levels increased only about 2-fold (Fig. 4B) over the period of seed development in which BnPaO2 transcripts increased >10-fold (Fig. 4A). Freezing exposure given on 20 DAP did not affect PaO protein content at any subsequent point during seed development. The immunoreactive complexes of PaO protein from canola seed resolved into a doublet on miniblots using 10% or lower acrylamide, differing in apparent molecular mass by approximately 0.5 kD (Fig. 4C). Only the lighter, bottom band showed increasing intensity as the seeds matured. No effect of freezing was evident for either PaO band.
Whereas PaO protein levels only doubled between 21 and 41 DAP and were unaffected by freezing exposure, PaO activity increased more than 10-fold over this period, and this induction was suppressed >20% by freezing (Fig. 4D). That the increase in PaO activity was at least 5 times greater than the increase in PaO protein implies strong posttranslational regulation of PaO during seed maturation.
Evidence for Dynamic Phosphorylation of Canola Seed PaO
Analysis of the BnPaO2 codon-derived amino acid sequence revealed two potential Ser/Thr calcium-dependent protein kinase (CDPK) phosphorylation recognition sites at Ser-18 and Thr-402 (Supplemental Fig. 1). These candidate phosphorylation sites are fully conserved in BnPaO1 and AtPaO. To investigate if there were changes in PaO phosphorylation corresponding to the changes we had observed in PaO activity, we used immobilized metal affinity chromatography (IMAC), which separates phosphorylated and nonphosphorylated proteins by binding the phosphorylated form, followed by immunoblot analysis.
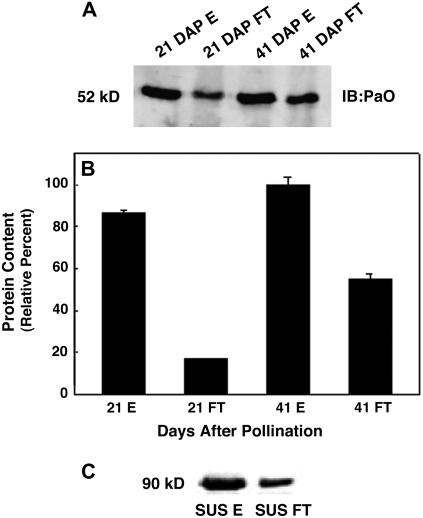
PaO phosphorylation was measured at 21 DAP when significant PaO protein was present (Fig. 4B) but activity was low (Fig. 4D) and compared with 41 DAP when PaO protein and activity levels were greatest. At 21 DAP, PaO was detected in both phosphorylated and nonphosphorylated fractions; however, the phosphorylated fraction contained nearly 5-fold higher amounts of PaO protein (Fig. 5A). At 41 DAP, phosphorylated PaO proteins did not show a significant increase from 21 DAP; however, the nonphosphorylated fraction increased by 3-fold, showing that the increase in PaO content between 21 and 41 DAP could be nearly accounted for by the nonphosphorylated form. Neither antiphos-Thr nor antiphos-Ser antibodies (Zymed) reacted with PaO from either fraction (data not shown).
Figure 5.
IMAC revealed dynamic PaO phosphorylation during canola seed development. PaO phosphorylation was determined at 21 DAP when PaO protein was one-half of maximum but activity very low and at 41 DAP when both PaO protein and activity were highest. A PhosphoProtein Purification kit (Qiagen) was used to separate phosphorylated (elute [E]) and nonphosphorylated (flow through [FT]) canola seed membrane proteins. The PaO fractions were identified and quantified by immunoblot and infrared imaging. A, Solubilized total membrane fractions containing PaO were isolated in combination with protease and phosphatase inhibitors. Protein expression was measured based on isolated membrane fractions of canola seeds. B, PaO protein levels increased over the period of seed development. Quantification of the blot from A demonstrated that at 21 DAP, the eluted phosphorylated fraction (21 E) contained almost 5-fold higher amounts of PaO protein than the flow through nonphosphorylated fraction (21 FT). At 41 DAP, while phosphorylated PaO proteins (41 E) showed only a modest increase from 21 DAP, the nonphosphorylated fraction (41 FT) increased by >3-fold. PaO was detected with polyclonal antibodies from the maize LLS1 (PaO) protein. Equal amounts of protein from membrane fractions of canola seeds were loaded in each lane. The units for protein expression are intensity/pixel. C, Separation of phosphorylated (SUS E) and nonphosphorylated (SUS FT) maize leaf SUS.
As validation of this approach to investigate PaO phosphorylation, we used IMAC columns with sucrose synthase (SUS), which has two well-known phosphorylation sites at Ser-15 and Ser-170 (Huber and Huber, 1996; Winter et al., 1997; Hardin et al., 2004). Phosphorylated SUS was isolated from basal elongating maize leaf tissue and purified by anion-exchange chromatography. After separation on the phosphoprotein affinity column, a SUS phosphospecific antibody detected phosphorylated SUS in both the fraction initially immobilized and subsequently eluted from the column with phosphate as well as in the flow-through fraction that did not bind to the column (Fig. 5C). The proportion of phosphorylated SUS in the bound versus unbound fraction was 5.6 to 1 when 0.5 mg of SUS was loaded on the column but only 2.45 to 1 when loading was increased to 1 mg. This result is indicative of exceeding the phosphoprotein binding capacity of the column, which is reported to have a maximum phosphoprotein binding capacity of 0.5 mg. Nevertheless, because the PaO-containing lysate loaded on the IMAC column contained only 10% to 20% of the total phosphoprotein of the SUS samples, overloading, and thus contamination, of the unbound fraction with phospho-PaO was unlikely.
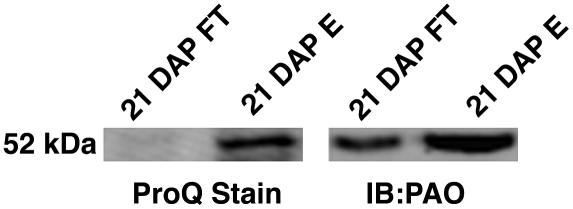
To further verify the phosphorylation of PaO, Pro-Q Diamond-blot staining (Invitrogen) was used to scrutinize the phosphorylated and nonphosphorylated IMAC fractions collected at 21 DAP (Fig. 6). As expected, Pro-Q analysis showed no bands representing phosphorylated proteins, including PaO, in the IMAC flow-through fraction, which should contain only nonphosphorylated proteins. However, a 52-kD band corresponding to PaO (western blot) was shown to align with a Pro-Q staining protein of a similar running molecular weight in the phosphorylated fraction eluted from the IMAC column.
Figure 6.
Detection of dynamic PaO phosphorylation by Diamond Pro-Q staining. Duplicates of the 21 DAP phosphorylated and nonphosphorylated fractions from IMAC were separated on SDS-PAGE and transferred to polyvinylidene difluoride membrane and the membrane was cut in half. One half was used for an immunoblot with polyclonal antibodies for PaO (IB:PaO), and the other half was used for Pro-Q Diamond staining (ProQ Stain). The two halves were realigned based on a ladder that was cut in middle between the two half blots. Pro-Q analysis showed no bands representing phosphorylated proteins, including PaO, in the IMAC flow through fraction containing nonphosphorylated proteins. However, a 52-kD band corresponding to PaO was shown to align with a Pro-Q staining protein of a similar running molecular weight in the phosphorylated fraction eluted from the IMAC column.
DISCUSSION
There have been numerous demonstrations that the inhibition of PaO activity during leaf senescence leads to the accumulation of Pheide a and the inhibition of Chl degradation. In pao1, the insertional knockout mutant of Arabidopsis, approximately 80% of the Chl is retained during dark-induced leaf senescence; Chl that is degraded is largely accounted for by Pheide a accumulation in the leaf because no further downstream products can be detected (Pružinská et al., 2005). Similarly, senescing leaves of stay-green mutants of Festuca pratensis and Lolium temulentum accumulate Chlide and Pheide a, and reduced PaO activity has been shown to be the biochemical defect in these mutants (Vicentini et al., 1995; Roca et al., 2004). In freeze-exposed canola seeds, the induction of PaO activity later in seed development is impaired but not obliterated (Fig. 4D), resulting in a leaky phenotype where Pheide a accumulates (Fig. 3) and Chl degradation is slowed (Fig. 2) but not stopped. It might be expected that the 20% lower maximum induction of PaO activity in freeze-exposed maturing canola seeds (Fig. 4D) would prolong but not prevent full clearing of Chl from the seeds. However, another factor that can ultimately limit Chl clearing from the maturing seed is seed moisture content, and we believe that the intersection of these two control mechanisms is the cause of the green seed problem. When the moisture content of canola seeds dips below approximately 40%, many aspects of seed metabolism, including Chl degradation (Green et al., 1998), come to a halt. Under field conditions, freeze exposure may enhance the rate of seed desiccation (Green et al., 1998), further exacerbating the effects of the impaired induction of PaO activity on Chl degradation that are evident even when accelerated desiccation is prevented (Figs. 1B and 4D).
In this work, as with all cases in which PaO activity has been shown to be impaired (Hilditch et al., 1989; Bachmann et al., 1994; Pružinská et al., 2003, 2005; Tanaka et al., 2003; Roca et al., 2004), it is clear that a regulatory mechanism limiting Chl metabolism is engaged that feeds back, ultimately preventing the removal of Chl from the thylakoid membrane. That a strong feedback control on Chl degradation is necessary (Takamiya et al., 2000; Hörtensteiner, 2006) is evident in the lesion-mimic phenotype of PaO mutants such as acd1 in Arabidopsis (Greenberg and Ausubel, 1993) and lls1 in maize (Gray et al., 1997), where accumulation of even small amounts of visible light-absorbing Chl metabolites is extremely phototoxic. While no necrotic patches were observed on freeze-treated canola seeds, this is likely because the seeds were exposed to only low light intensities. Thus, as previously observed in the leaves of PaO mutants, it is apparent that the freeze treatment of canola seeds also indirectly affects Chl degradation processes upstream of PaO.
The profiles of PaO transcript level, protein content, and activity all qualitatively correlated with the progression of Chl degradation during seed development. However, PaO protein increased only 2-fold over the measured period of seed development, while transcript levels indicated >10-fold increase in expression. Some of the apparent discrepancy could be explained if PaO protein were highly stable compared to PaO transcript. If so, the rate of newly synthesized PaO protein could track the transcript level, yet the amount of new protein would be small in comparison to the accumulated stable pool. Indeed, PaO protein levels were one-half their maximum level at 21 DAP (Fig. 4B), prior to any measurable losses of Chl (Fig. 2) and when BnPaO transcript levels (Fig. 4A) were low. Beyond the dependence of PaO expression on transcript abundance, it is evident from our results that posttranslational control of the induction of PaO activity accompanies the degreening of canola seed. Our data showed that PaO protein content and activity have starkly different profiles during canola seed development. Whereas freeze exposure caused a statistically significant >20% reduction in the induction of PaO activity during 36 to 46 DAP, freezing was without any significant effect on PaO transcript or protein amounts.
Although the posttranslational regulatory mechanism is not yet known, it appears likely that reversible protein phosphorylation is involved. Using both IMAC and Pro-Q Diamond-blot staining, we demonstrated a correlation between PaO dephosphorylation and increasing PaO activity during seed maturation. The stoichiometry of phosphorylated/dephosphorylated PaO decreased from 5 to 1 on 21 DAP to 2 to 1 on 41 DAP, with the dephosphorylated form of PaO increasing more than 3-fold over this interval. The 10-fold increase in PaO activity (Fig. 4D) illustrates that the observed change in PaO phosphorylation is large enough to have played a significant, although perhaps not exclusive, role in the posttranslational activation of this enzyme.
There are two CDPK recognition sites in BnPaO1, BnPaO2, and AtPaO protein sequences (Supplemental Fig. 1). The first CDPK site is located within the putative chloroplast target sequence, most likely cleaved once the protein is translocated; thus, this CDPK site is not likely to be involved in regulation of the enzyme in the chloroplast. The common CDPK consensus phosphorylation site is φ-x-Basic-x-x-S/T, where the underlined Ser or Thr is phosphorylated, x is any residue, and φ is a hydrophobic residue (Huang and Huber, 2001). Although CDPKs are reported to associate with various membranes within the cell (Harper et al., 2004), it is not yet known whether CDPKs are found in the chloroplast, where it would presumably need to be located to phosphorylate PaO, which is located on the inner chloroplast envelope. However, ChloroP (http://www.cbs.dtu.dk/services/ChloroP/) searches indicate that 10 of the 34 putative CDPKs found in Arabidopsis contain hypothetical chloroplast targeting sequences. Computer analysis of the codon-derived PaO protein sequences revealed other possible phosphorylation sites at various Ser and Thr residues (http://www.cbs.dtu.dk/services/NetPhos). It is interesting to note that the two major differences near the N-terminal region of the two forms of BnPaO (Supplemental Fig. 2) remove a Ser residue (Ser-29) in BnPaO1, a potential Ser phosphorylation sites as well as four amino acids (G-D-K-E).
That the freezing episode and any decrease in the observed rate of Chl degradation or PaO activity can be separated by more than a week indicates that freezing does not interfere directly with the PaO protein but with the program controlling Chl clearing from the seed. Since freezing appears to interfere with the activation of PaO by dephosphorylation, the delayed effect may be mediated at the level of PaO phosphorylation/dephosphorylation. Cold stress may indirectly lead to changes in protein phosphorylation and significant changes in CDPK activity by affecting the fluctuations in cytosolic Ca2+ levels (Martin and Busconi, 2001; Cheng et al., 2002). It has been shown in various species, including alfalfa (Medicago sativa) and rice (Oryza sativa), that CDPK activity can be induced by low temperature (Monroy and Dhindsa, 1995), which in the case of PaO would be expected to increase the ratio of phosphorylated/dephosphorylated PaO and thereby decrease activity.
The doublet seen in the immunoblots of PaO protein from canola seeds (Fig. 4C) is most likely due to the presence of both BnPaO1 and BnPaO2 proteins in the sample. The two distinct clones of PaO were isolated and identified from leaves of canola, differing by 526 D, which would account for the difference between the two bands. Although BnPaO1 transcript was expressed only early in seed development, it seems likely that its protein product persisted after BnPaO1 transcript disappeared. Only BnPaO2 transcript was detected at later stages in seeds. Interestingly, the doublet is not seen when the protein has been isolated on IMAC columns (Figs. 5 and 6). The protein retained on the IMAC column (i.e. phosphorylated form) corresponds to the lighter band of the doublet and is the band that increases during seed development (Fig. 4C). That the doublet is not seen when the sample is run over the IMAC column (Figs. 5 and 6) could support the notion that phosphorylation of BnPaO2 plays a role in the regulation of PaO activity in canola seed. Another possibility is that the doublet is due to differential posttranslational modification, including phosphorylation. However, because a single phosphorylation will add only 80 D, it seems unlikely that even multiple sites of reversible phosphorylation could alone account for the approximately 500 D difference estimated from electrophoretic mobility on SDS-PAGE.
The extent to which posttranslational control of PaO operates in leaf senescence is uncertain. Pružinská et al. (2005) showed a quantitative correlation among PaO activity, transcript level, and protein level, thereby demonstrating that, unlike the situation we found in canola seeds, posttranslational control was not necessary to explain PaO regulation during dark-induced senescence of Arabidopsis leaves. Indeed, the original suggestion of the possible involvement of phosphorylation in PaO regulation (Pružinská et al., 2003), the inhibition of PaO activity by phosphatase treatment, is the opposite response that we would expect and was subsequently not reproduced (Pružinská et al., 2005). While these observations do not eliminate a possible role for phosphorylation in PaO regulation during Arabidopsis leaf senescence, they do indicate some important differences in the overall regulation of Chl degradation in senescing leaves and maturing seeds. Indeed, freezing does not interfere with the timing or extent of Chl degradation in dark-induced leaf senescence system of either Arabidopsis or canola (data not shown).
CONCLUSION
Freezing exposure of developing canola seeds hinders the programmed degreening of the seed by interfering with the posttranslationally controlled induction of PaO activity. Although the rate of Chl degradation is slowed by only approximately 20%, the inhibition is sufficient to prevent Chl from fully clearing from the seed before seed moisture content dips below the threshold at which seed metabolism is suspended. The mechanism of the posttranslational control is unknown, but the increase in PaO activity during seed maturation corresponds to a decrease in the phosphorylation of the PaO enzyme. Canola has two highly homologous PaO genes that contain two candidate CDPK phosphorylation sites.
MATERIAL AND METHODS
Plant Material
Canola (Brassica napus L. cv Westar) seeds were germinated in moist vermiculite. Two-week-old seedlings were transplanted to 12-inch pots containing Sunshine Mix LC1 soil (SunGro Horticulture) and grown in growth chambers at 12-h photoperiod of 450 μmol photons m−2 s−1 and 22°C-day/16°C-night thermoperiod with a relative humidity of 70%. Canola plants were fertilized weekly with 20-20-20 Peters Professional fertilizer (United Industries). A set of 10 to 12 canola plants was grown for each of the control and freeze condition experiments. After bolting and prior to flowering, inflorescences with similar maturity were chosen from each canola plant for hand pollination. The tip of each flowering bud was cut open and hand pollinated. Each pollination was marked on the stems of the flower bud. When collecting samples, siliques were randomly chosen from different inflorescences of each plant and pooled.
Freeze treatment was on whole plants in pots at 20 DAP in a darkened, controlled-environment chamber initially set at 22°C with high humidity. The temperature was decreased 5°C/h until reaching −4°C, where the temperature was held for 6 h and then increased 5°C/h until reaching the initial temperature of 22°C. Chamber conditions were then reset to normal growth conditions. Seeds were collected at preset intervals throughout the completion of seed development.
RNA Isolation
Seeds harvested at various developmental stages were ground in liquid nitrogen, and total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer's protocol. RNA quality was checked on 1% Tris-acetate EDTA agarose gel and the A260 was determined. RNA (20 μg) was mixed with RQ1 DNase (Promega), buffer, RNasin, and water to a total volume of 50 μL and DNase treated following the manufacturer's protocol. One to two micrograms of DNase-treated total RNA was used as a template for cDNA synthesis using manufacturer's protocol (Invitrogen).
PCR
First-strand cDNA synthesis was performed using 1 to 2 μg DNase-treated RNA and Oligo(dT)18 primer according to manufacturer's instructions (Invitrogen). Quantitative real-time reverse transcription-PCR used QuantiTect SYBR Green PCR kit (Qiagen) with Cepheid SmartCycler according to the manufacturer's suggestions. Actin-3 was used as internal control. The primers used for amplifications were: BnPaO (i.e. BnPaO1 and BnPaO2), forward, 5′-GAAGCTCGCGCTGTTAAATC-3′, reverse, 5′-CCCTTTGAATTGTCACCGTT-3′; BnPaO1, forward, 5′-ACGGCGGAGATAAGGAAGAA-3′, reverse, 5′-CTCGACCCAGGAGCTGAA-3′; BnPaO2, forward, 5′-GACGGAAACTTCTCGACAGC-3′, reverse, 5′-TTGAACTCAGACCCTTCTTCG-3′; actin-3, forward, 5′-ATGGTTAAGGCTGGTTTTGCT-3′, reverse, 5′-ATCCTTCTGTCCCATTCCAAC-3′. All primers used were within the optimal amplicon range between 100 and 200 bp. For each gene, a range of six dilutions of genomic DNA of known concentration was amplified under the same conditions as the cDNA samples and then used as the standard curve to determine the number of cDNA molecules present in the experimental samples. At least four values were produced for each sample and repeated independently at least twice.
Cloning of PaO from Canola
The primers for cloning PaO from canola leaves 5 d after darkening were designed based on the open reading frame sequence of At3g44880: forward, 5′-ATGTCAGTAGTTTTACTCTCTTCT-3′, reverse, 5′-TCGATTTCAGAATGTACATAATCT-3′. PaO cDNA corresponding to the size of the open reading frame, approximately 1,600 bp, was cloned using a commercial cloning kit, pDrive Cloning Vector (Qiagen). Multiple colonies were sequenced from both directions with internal primers, M13 reverse and M13 forward (−20). The canola PaO open reading frame was completely sequenced in both directions using an automated DNA sequencing system, ABI 373A DNA sequencer (Applied Biosystems). Sequencher 4.5 (Genecodes) was used to align sequences, view chromatograms, and edit sequences at the Biotechnology Center of University of Illinois at Urbana-Champaign.
Isolation of Plastid (Gerontoplast) Membranes
Canola seeds were homogenized in 5 mL/g fresh weight of a medium containing 400 mm sorbitol, 25 mm tricine-KOH, pH 8.0, 2 mm EDTA, 1 mm MgCl2, 0.1% bovine serum albumin (w/v), 5 mm polyethylene glycol 4,000, and 10 mm cysteamine-HCl using a chilled mortar and pestle. After filtration through a layer of nylon membrane, the homogenate was centrifuged at 10,000g for 4 min. The membrane pellet was resuspended with the above medium without EDTA, MgCl2, and bovine serum albumin, corresponding to 2 mL/g fresh weight leaf tissue and centrifuged at 10,000g for 4 min. The supernatant was removed and pellet frozen in liquid nitrogen and stored at −80°C.
Isolation of Phosphorylated PaO Membrane Fractions
Membrane fractions from chloroplasts were isolated as described above with the addition of the following: 1 μm E64 Cys protease inhibitor, 0.1 μm Microcystin-LR, 1 mm 4-(2-aminoethyl) benzenesulphonyl fluoride, 1 mm p-aminobenzophenone, 5 mm caproic acid, 10 μm leupeptin, 1 mm dithiothreitol, 1 mm NaF, 1 mm NaVO4, 1 mm EDTA, and 1 mm EGTA. The membrane pellet was resuspended in 2 μm E64 Cys protease inhibitor, 0.5 μm Microcystin-LR, 10 μm MG132 Mycoplasma genitalium proteasome inhibitor, 1 mm 4-(2-aminoethyl) benzenesulphonyl fluoride, 1 mm p-aminobenzophenone, 5 mm caproic acid, 5 μm leupeptin, 10 mm dithiothreitol, 20 mm NaF, 1 mm NaVO4, 5 mm EDTA, 1 mm EGTA, 10 mm NaMO4, and 5 μg/μL SBT1 subtilisin-like Ser protease.
Extraction of Chl and Chl Catabolites
Chl was extracted from ground canola seeds with 500 μL of N, N′-dimethylformamide in a microfuge tube using a mini plastic pestle. After three subsequent washings with 300 μL N, N′-dimethylformamide, the homogenate was centrifuged at 12,000g for 2 min at room temperature. The pellet was then extracted further with 300 μL N, N′-dimethylformamide and the pooled supernatants adjusted to a final volume of 2 mL. The Chl content of the seeds was determined spectrophotometrically using the specific absorption coefficients for Chl a and b of Porra and Grimme (1974).
Chl catabolites were separated by HPLC based on their polarity in organic solvent and were quantified by fluorescence spectroscopy according to published procedures (Rebeiz, 2002; Pružinská et al., 2003, 2005). The concentration of each catabolite was determined from the fluorescence intensity data using equations developed for the quantification of tetrapyrrole moieties (Rebeiz, 2002).
Isolation of PaO and RCCR
The chloroplast membrane pellet (equivalent to 25 g fresh weight), isolated as described above, was resuspended in 1.25 mL of 25 mm Tris-MES, pH 8.0, and centrifuged twice at 12,000g for 5 min at 4°C to remove debris. The supernatant containing RCCR was transferred to a new tube and stored at −80°C until used for PaO assay.
Following the removal of soluble proteins, the membrane pellet was washed three times in 5 mL 25 mm Tris-MES, pH 8.0, and centrifuged at 12,000g for 5 min followed by the removal of the supernatant. The washed membrane pellets were then resuspended in 750 μL Tris-MES, pH 8.0, and mixed with Triton X-100 to a final concentration of 1%. The membrane proteins were solubilized by shaking for 30 min at 4°C and centrifuged at 10,000g for 5 min. The supernatant containing the solubilized membrane proteins was used for PaO assay.
PaO Assays
PaO activity was assessed by using a coupled PaO/RCCR assay according to established protocols (Hörtensteiner et al., 1995; Rodoni et al., 1998; Pružinská et al., 2003). The assay contained 25 μL of enzyme preparation (PaO) and 10 μL RCCR, supplemented with 2 mm Pheide a, 10 μg ferredoxin, 1 mm NADPH, 2 mm Glc-6-P, and 50 mU Glc-6-P dehydrogenase in a total volume of 50 μL (Rodoni et al., 1998). As a source of RCCR, either stromal protein isolates as described above or Arabidopsis (Arabidopsis thaliana) RCCR expressed in Escherichia coli (Pružinská et al., 2005) was used. PaO assays were stopped after 1 h by the addition of 80 μL of methanol followed by centrifugation at 12,000g for 2 min to remove debris. The resulting supernatant was applied to a Waters HPLC system (600E System Controller, 700 Satellite Wisp; Waters) using an isocratic gradient with 50 mm potassium phosphate, pH 7.0/methanol (1:2 v/v) as the solvent. Products were identified by retention time on a ODS Hypersil reverse phase column (250 × 4.6 mm, 5 μm particle size; Agilent Technologies) and detected by fluorescence (excitation 320 nm, emission 450 nm) using a Hitachi Fluorescence Spectrophotometer (F-1260; Hitachi High Technologies America) or UV absorption (320 nm) using a Waters 486 Tunable Absorbance Detector.
Protein Isolation and Immunoblot Analysis
Membrane proteins were extracted from seeds (Pružinská et al., 2003) and quantified by Bradford analysis. Proteins (10 μg) were separated on a 10% SDS-polyacrylamide gel and blotted onto nitrocellulose membrane. The membranes were blocked for 1 h at room temperature with blocking buffer (LI-COR). The membranes were incubated in primary antibody against monoclonal or polyclonal antibodies from the maize (Zea mays) LLS1 (PaO) protein. The antibodies recognize the PaO protein in different monocot and dicot species, including Arabidopsis and canola. After washing in phosphate-buffered saline Tween 20, blots were incubated for 1 h at room temperature with goat anti-mouse IgG or goat anti-rabbit IgG. The immunoreactive complexes were visualized by fluorescence emission and quantified with a LI-COR Odyssey (LI-COR) infrared imaging system.
Phosphoprotein Detection Using Pro-Q Diamond-Blot Staining and IMAC
Phosphoproteins were detected on polyvinylidene difluoride membranes using Pro-Q Diamond-blot staining protocol (Invitrogen). A Peppermint Stick phosphoprotein standard was used where 1 μL corresponded to 0.5 μg. Images were acquired on a Typhoon 8600 Variable Mode Imager (Amersham Pharmacia Biotech) following Pro-Q, with 532-nm laser, 580-nm bandpass filter at normal sensitivity, and a photomultiplier tube voltage of 300. IMAC separation of phosphorylated and nonphosphorylated PaO was accomplished using Qiagen PhosphoProtein Purification kit (Qiagen) according to manufacturer's instructions.
Statistical Analysis
All data were analyzed by a mixed model ANOVA (PROC MIXED; SAS Institute, 1996) with treatment as a fixed factor, time as a repeated factor, and a compound symmetry covariance structure. Preplanned comparisons of means for each time point were analyzed with linear contrasts.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number DQ388373 for BnPaO1 and DQ388372 for BnPaO2.
Supplementary Material
Acknowledgments
We are grateful to Dr. John Gray for providing antibodies for PaO and Dr. Steven Huber for supplying SUS proteins and antibodies for SUS. We thank Dr. Adriana Ortiz-Lopez for her contributions to the initial stages of this research. We acknowledge Kateri Duncan, Dr. Shane Hardin, Dr. Aleel Grennan, and Qingiu Gong for their contributions to this research and Dr. Aleel Grennan for her expert help with the manuscript.
This work was supported in part by the Integrative Photosynthesis Research Training Grant from the Department of Energy (grant no. DEFGO2–92ER20095), funded under the Program for Collaborative Research in Plant Biology, and by the Swiss National Science Foundation (grant no. 3100A0–105389).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Donald R. Ort (d-ort@uiuc.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bachmann A, Fernandez-Lopez J, Ginsburg S, Thomas H, Bouwkamp JC, Solomos T, Matile P (1994) Stay-green genotypes of Phaseolus vulgaris L.: chloroplast proteins and chlorophyll catabolites during foliar senescence. New Phytol 126: 593–600 [Google Scholar]
- Benedetti CE, Arruda P (2002) Altering the expression of the chlorophyllase gene ATHCOR1 in transgenic Arabidopsis caused changes in the chlorophyll-to-chlorophyllide ratio. Plant Physiol 128: 1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SH, Willmann MR, Chen HC, Sheen J (2002) Calcium signaling through protein kinases: the Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129: 469–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Kolacna L, Rawsthorne S (1996) Photosynthesis by developing embryos of oilseed rape (Brassica napus L.). J Exp Bot 47: 1763–1769 [Google Scholar]
- Gray J, Close PS, Briggs SP, Johal GS (1997) A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 89: 25–31 [DOI] [PubMed] [Google Scholar]
- Gray J, Wardzala E, Yang M, Reinbothe S, Haller S, Pauli F (2004) A small family of LLS1-related non-heme oxygenases in plants with an origin amongst oxygenic photosynthesizers. Plant Mol Biol 54: 39–54 [DOI] [PubMed] [Google Scholar]
- Green BR, Singh S, Babic I, Bladen C, Johnson-Flanagan AM (1998) Relationship of chlorophyll, seed moisture and ABA levels in the maturing Brassica napus seed and effect of a mild freezing stress. Physiol Plant 104: 125–133 [Google Scholar]
- Greenberg JT, Ausubel FM (1993) Arabidopsis mutants compromised for the control of cellular damage during pathogenesis and aging. Plant J 4: 327–341 [DOI] [PubMed] [Google Scholar]
- Hardin SC, Winter H, Huber SC (2004) Phosphorylation of the amino terminus of maize sucrose synthase in relation to membrane association and enzyme activity. Plant Physiol 134: 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JE, Breton G, Harmon A (2004) Decoding Ca2+ signals through plant protein kinases. Annu Rev Plant Biol 55: 263–288 [DOI] [PubMed] [Google Scholar]
- Hilditch PI, Thomas H, Thomas BJ, Rogers LJ (1989) Leaf senescence in a non-yellowing mutant of Festuca pratensis: proteins of photosystem II. Planta 177: 265–272 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S (2006) Chlorophyll degradation during senescence. Annu Rev Plant Biol 57: 55–77 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S, Vicentini F, Matile P (1995) Chlorophyll breakdown in senescent cotyledons of rape, Brassica napus L.: enzymatic cleavage of pheophorbide a in vitro. New Phytol 129: 237–246 [DOI] [PubMed] [Google Scholar]
- Huang JZ, Huber SC (2001) Phosphorylation of synthetic peptides by a CDPK and plant SNF1-related protein kinase: influence of proline and basic amino acid residues at selected positions. Plant Cell Physiol 42: 1079–1087 [DOI] [PubMed] [Google Scholar]
- Huber SC, Huber JL (1996) Role and regulation of sucrose-phosphate synthase in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47: 431–444 [DOI] [PubMed] [Google Scholar]
- Jacob-Wilk D, Holland D, Goldschmidt EE, Riov J, Eyal Y (1999) Chlorophyll breakdown by chlorophyllase: isolation and functional expression of the Chlase1 gene from ethylene-treated Citrus fruit and its regulation during development. Plant J 20: 653–661 [DOI] [PubMed] [Google Scholar]
- Johnson-Flanagan AM, Thiagarajah MR (1990) Degreening in canola (Brassica napus cv. Westar) embryos under optimum conditions. J Plant Physiol 136: 180–186 [Google Scholar]
- Kariola T, Brader G, Li J, Palva ET (2005) Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell 17: 282–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levadoux WL, Kalmokoff M, Pickard M, GrootWassink J (1987) Pigment removal from canola oil using chlorophyllase. J Am Oil Chem Soc 64: 139–144 [Google Scholar]
- Mach JM, Castillo AR, Hoogstraten R, Greenberg JT (2001) The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci USA 98: 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ML, Busconi L (2001) A rice membrane-bound calcium-dependent protein kinase is activated in response to low temperature. Plant Physiol 125: 1442–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile P, Hörtensteiner S, Thomas H (1999) Chlorophyll degradation. Annu Rev Plant Physiol Plant Mol Biol 50: 67–95 [DOI] [PubMed] [Google Scholar]
- Matile P, Schellenberg M (1996) The cleavage of pheophorbide a is located in the envelope of barley gerontoplasts. Plant Physiol Biochem 34: 55–59 [Google Scholar]
- Monroy AF, Dhindsa RS (1995) Low-temperature signal transduction: induction of cold acclimation-specific genes of alfalfa by calcium at 25°C. Plant Cell 7: 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhuber M, Berghold J, Breuker K, Hörtensteiner S, Kräutler B (2003) Breakdown of chlorophyll: a nonenzymatic reaction accounts for the formation of the colorless “nonfluorescent” chlorophyll catabolites. Proc Natl Acad Sci USA 100: 6910–6915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Grimme LH (1974) A new procedure for the determination of chlorophylls a and b and its application to normal and regreening Chlorella. Anal Biochem 57: 255–267 [DOI] [PubMed] [Google Scholar]
- Pružinská A, Tanner G, Anders I, Roca M, Hörtensteiner S (2003) Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc Natl Acad Sci USA 100: 15259–15264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pružinská A, Tanner G, Aubry S, Anders I, Moser S, Müller T, Ongania KH, Kräutler B, Youn JY, Liljegren SJ, et al (2005) Chlorophyll breakdown in senescent Arabidopsis leaves: characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol 139: 52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz CA (2002) Analysis of intermediates and end products of the chlorophyll biosynthetic pathway. In AG Smith, M Witty, eds, Heme, Chlorophyll, and Bilins: Methods and Protocols. Humana Press, Totowa, NJ, pp 111–155
- Roca M, James C, Pružinská A, Hörtensteiner S, Thomas H, Ougham H (2004) Analysis of the chlorophyll catabolism pathway in leaves of an introgression senescence mutant of Lolium temulentum. Phytochemistry 65: 1231–1238 [DOI] [PubMed] [Google Scholar]
- Roca M, Minguez-Mosquera MI (2003) Involvement of chlorophyllase in chlorophyll metabolism in olive varieties with high and low chlorophyll content. Physiol Plant 117: 459–466 [DOI] [PubMed] [Google Scholar]
- Rodoni S, Mühlecker W, Anderl M, Kräutler B, Moser D, Thomas H, Matile P, Hörtensteiner S (1997) Chlorophyll breakdown in senescent chloroplasts: cleavage of pheophorbide a in two enzymatic steps. Plant Physiol 115: 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodoni S, Schellenberg M, Matile P (1998) Chlorophyll breakdown in senescing barley leaves as correlated with pheophorbide a oxygenase activity. J Plant Physiol 152: 139–144 [Google Scholar]
- Ruuska SA, Schwender J, Ohlrogge JB (2004) The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiol 136: 2700–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender J, Goffman F, Ohlrogge JB, Shachar-Hill Y (2004. a) Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432: 779–782 [DOI] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge JB, Shachar-Hill Y (2004. b) Understanding flux in plant metabolic networks. Curr Opin Plant Biol 7: 309–317 [DOI] [PubMed] [Google Scholar]
- Shioi Y, Tomita N, Tsuchiya T, Takamiya K (1996) Conversion of chlorophyllide to pheophorbide by Mg-dechelating substance in extracts of Chenopodium album. Plant Cell Physiol 34: 41–47 [Google Scholar]
- Takamiya K, Tsuchiya T, Ohta H (2000) Degradation pathway(s) of chlorophyll: What has gene cloning revealed? Trends Plant Sci 5: 426–431 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Hirashima M, Satoh S, Tanaka A (2003) The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: Inhibition of the pheophorbide a oxygenase activity does not lead to the “stay-green” phenotype in Arabidopsis. Plant Cell Physiol 44: 1266–1274 [DOI] [PubMed] [Google Scholar]
- Tsuchiya T, Ohta H, Okawa K, Iwamatsu A, Shimada H, Masuda T, Takamiya K (1999) Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: finding of a lipase motif and the induction by methyl jasmonate. Proc Natl Acad Sci USA 96: 15362–15367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicentini F, Hörtensteiner S, Schellenberg M, Thomas H, Matile P (1995) Chlorophyll breakdown in senescent leaves: identification of the biochemical lesion in a stay-green genotype of Festuca pratensis Huds. New Phytol 129: 247–252 [DOI] [PubMed] [Google Scholar]
- Willms JR, Salon C, Layzell D (1999) Evidence for light-stimulated fatty acid synthesis in soybean fruit. Plant Physiol 120: 1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter H, Huber JL, Huber SC (1997) Membrane association of sucrose synthase: changes during the graviresponse and possible control by protein phosphorylation. FEBS Lett 420: 151–155 [DOI] [PubMed] [Google Scholar]
- Wüthrich KL, Bovet L, Hunziker PE, Donnison IS, Hörtensteiner S (2000) Molecular cloning, functional expression and characterization of RCC reductase involved in chlorophyll catabolism. Plant J 21: 189–198 [DOI] [PubMed] [Google Scholar]
- Yao N, Greenberg JT (2006) Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell 18: 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HY, Vasanthan T, Wettasinghe M (2004) Dry matter, lipids, and proteins of canola seeds as affected by germination and seedling growth under illuminated and dark environments. J Agric Food Chem 52: 8001–8005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.