Abstract
Dictyostelium actin was shown to become phosphorylated on Tyr-53 late in the developmental cycle and when cells in the amoeboid stage are subjected to stress but the phosphorylated actin had not been purified and characterized. We have separated phosphorylated and unphosphorylated actin and shown that Tyr-53 phosphorylation substantially reduces actin's ability to inactivate DNase I, increases actin's critical concentration, and greatly reduces its rate of polymerization. Tyr-53 phosphorylation substantially, if not completely, inhibits nucleation and elongation from the pointed end of actin filaments and reduces the rate of elongation from the barbed end. Negatively stained electron microscopic images of polymerized Tyr-53–phosphorylated actin show a variable mixture of small oligomers and filaments, which are converted to more typical, long filaments upon addition of myosin subfragment 1. Tyr-53–phosphorylated and unphosphorylated actin copolymerize in vitro, and phosphorylated and unphosphorylated actin colocalize in amoebae. Tyr-53 phosphorylation does not affect the ability of filamentous actin to activate myosin ATPase.
Keywords: actin polymerization, Dictyostelium, phosphorylated actin
Transfer of Dictyostelium amoebae from nutrient to nonnutrient medium initiates a 24-hour developmental cycle (1) in which the amoebae aggregate and differentiate to form multicellular organisms that mature to fruiting bodies containing stable spores from which, when they are placed in nutrient medium, amoebae germinate. Several laboratories (2–4) reported tyrosine phosphorylation of actin [phosphotyrosine actin (pY-actin)] correlated with rearrangements of the actin cytoskeleton during the developmental cycle. pY-actin appears late in maturing spores, i.e., ≈24 h into the developmental cycle, reaches a maximum level at ≈36 h, at which time ≈50% of the actin is phosphorylated (3), remains constant for ≈20 days, at 22°C, and then decreases, disappearing entirely by 30 days, at which time the spores are no longer viable (3). When viable spores are placed in nutrient medium, pY-actin is dephosphorylated, with a half-life of ≈5 min (3), before spore swelling and germination (2, 4).
Although vegetative amoebae in nutrient medium contain little or no pY-actin (5, 6), phosphorylation transiently increases (for ≈20–25 min) when amoebae are transferred from nonnutrient to nutrient medium (7), with concurrent changes in cell shape, for example, loss of pseudopods, rounding up of the previously elongated cells, and weakened adherence to the substratum (7). Tyrosine phosphorylation also occurs when vegetative amoebae in nutrient medium are exposed to phenylarsine oxide (PAO) (5), an inhibitor of phosphotyrosine phosphatase, or are subjected to stress, for example, inhibition of oxidative phosphorylation (8, 9) or elevated temperature (9), with parallel changes in cell shape similar to the changes that occur when cells are transferred from nonnutrient to nutrient medium.
Importantly, phosphorylation occurs uniquely at Tyr-53 (9). Thus, phosphorylation of actin at Tyr-53 is associated with, but not necessarily responsible for, cytoskeletal reorganization and shape changes in vegetative cells induced by changes in nutritional status or stress and with spore dormancy and viability. To begin to understand the molecular basis of these biological events, we have characterized the polymerization properties of highly purified pY53-actin and its ability to inhibit DNase I and activate myosin ATPase.
Results
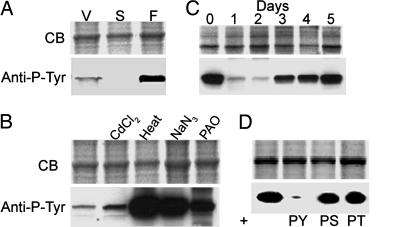
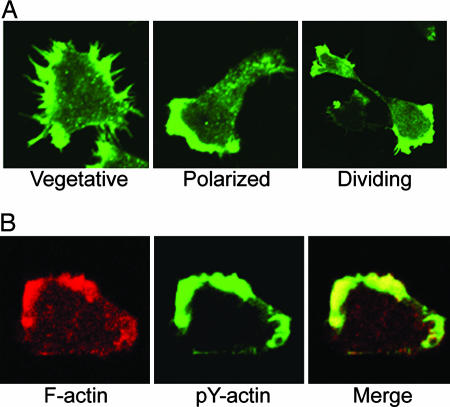
First, we confirmed the earlier reports that pY-actin is present at low levels in growing vegetative cells, essentially disappears when the amoebae are starved, and reappears at relatively high levels in mature fruiting bodies (Fig. 1A). We also confirmed that CdCl2 slightly increases, and heat, NaN3, and PAO greatly increase the level of actin tyrosine phosphorylation (Fig. 1B). In addition, we found that the amount of pY-actin substantially increases in vegetative cells after 3–5 days in culture medium (Fig. 1C), i.e., when cells reach maximum density. That the phosphorylation is tyrosine-specific was confirmed by the fact that immunoblotting with phosphotyrosine-specific antibody was blocked by excess pTyr but not by excess pSer or pThr (Fig. 1D). Like nonphosphorylated actin, pY-actin localizes to pseudopods and filopods in vegetative cells, where it colocalizes with unphosphorylated actin (Fig. 2).
Fig. 1.
Conditions that increase pY-actin in Dictyostelium. (A) Relative amounts of pY-actin in vegetative (V) and starved (S) amoebae and fruiting bodies (F) by immunoblotting with anti-pY antibody. (B) Increase of pY-actin in cells exposed to CdCl2, heat, NaN3, or PAO. (C) Increase in pY-actin with days in culture. (D) Specificity of anti-phosphotyrosine antibody. Immunoblotting of phosphorylated actin is blocked by 1 mM pY but not by 1 mM pS or 1 mM pT. CB, Coomassie blue; pY, phosphotyrosine; pS, phosphoserine; pT, phosphothreonine.
Fig. 2.
Colocalization of unphosphorylated and pY-actin. (A) Localization of pY-actin in vegetative, polarized, and dividing cells. (B) Localization of total F-actin (rhodamine phalloidin, red) and pY-actin (anti-pY, green).
Purification and Separation of Phosphorylated and Unphosphorylated Actin.
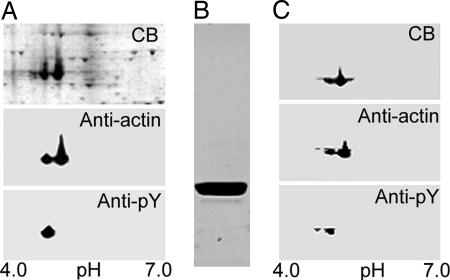
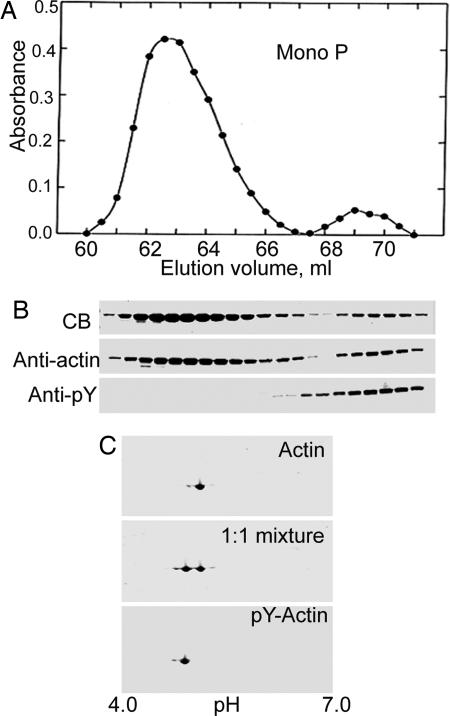
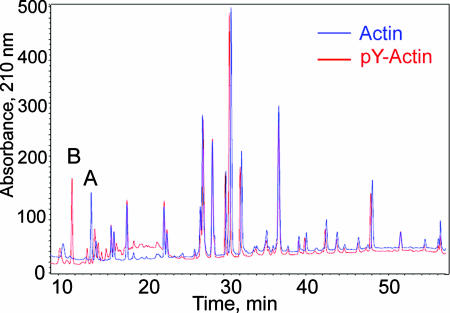
Actin purified by conventional methods is a mixture of phosphorylated and unphosphorylated actin (Fig. 3), which can be separated by chromatography on Mono P (5) (Fig. 4). HPLC-mass spectrometry (Table 1) confirmed the identifications of the two fractions in Fig. 4A as N-acetylated unphosphorylated and monophosphorylated actin, which was further supported by the loss of 80 mass units from phosphorylated actin and no change in the mass of unphosphorylated actin, when incubated with phosphatase (Table 1). Phosphorylation of Tyr-53 was confirmed by HPLC-mass spectrometry of LysC-digests of unphosphorylated and phosphorylated actin (Fig. 5). The only differences between the two spectra are the absence of peak A and the appearance of peak B in the digest of phosphorylated actin. Peptides A and B had the predicted masses for the unphosphorylated and phosphorylated peptides, respectively, spanning residues 51–61 (DSYVGDEAQSK) (Table 1).
Fig. 3.
Large-scale purification of actin. (A) Two-dimensional gel of total cell lysate stained with Coomassie blue (CB), and anti-actin and anti-pY antibodies. (B) CB-stained gel of SDS/PAGE of purified actin before separation of phosphorylated and unphosphorylated actin. (C) Two-dimensional gel of sample in B stained with CB and anti-actin and anti-pY-antibodies.
Fig. 4.
Separation of pY-actin and unphosphorylated actin. (A) Elution profile of Mono P column of the actin sample shown in Fig. 3. (B) SDS/PAGE of fractions eluted between 60.5 and 70.5 ml stained with Coomassie blue (CB) and anti-actin and anti-pY antibodies. (C) Two-dimensional gel of pooled fractions of unphosphorylated actin (Actin) and P-actin and a 1:1 mixture of the purified proteins.
Table 1.
Mass spectroscopic identification of pY53-actin
| Mass, Da |
||
|---|---|---|
| Observed | Calculated | |
| N-acetyl-actin | 41,658.37 | 41,657.68 |
| N-acetyl-actin plus phosphatase | 41,657.47 | 41,657.68 |
| N-acetyl-pY53-actin | 41,737.84 | 41,737.66 |
| N-acetyl-pY53-actin plus phosphatase | 41,657.35 | 41,657.68 |
| Peptide A | 1,197.52 | 1,197.52* |
| Peptide B | 1,277.49 | 1,277.48 |
*Assumes peptide A is residues 51–61, DSYVGDEAQSK, and peptide B is peptide A phosphorylated on Y53. The peptide masses are monoisotopic.
Fig. 5.
HPLC of LysC-digests of unphosphorylated actin (Actin) and pY-actin (P-Actin). A and B identify the only peptides that differ in the two samples.
Properties of pY53-Actin.
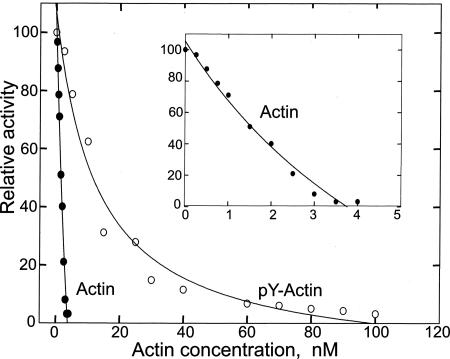
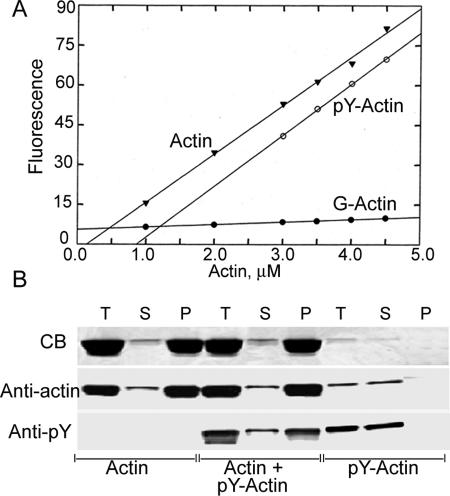
Tyr-53 phosphorylation greatly reduces actin's ability to inhibit pancreatic DNase I activity (Fig. 6), with an IC50 of 13 nM vs. an IC50 of 1.5 nM for unphosphorylated actin. Also, Tyr-53 phosphorylation increases the apparent critical concentration for polymerization (Fig. 7A) from 0.5 to 1.25 μM. Phosphorylated and unphosphorylated actin copolymerize, as shown by the incorporation of pY53-actin into filaments of unphosphorylated actin at a concentration of pY53-actin (0.2 μM) well below its critical concentration (Fig. 7B).
Fig. 6.
Concentration dependence of inhibition of pancreatic DNase I by unphosphorylated actin and pY53-actin. (Inset) Amplification of the region between 0 and 4 nM unphosphorylated actin.
Fig. 7.
Critical concentrations and copolymerization of unphosphorylated actin and pY53-actin. (A) Critical concentration curves. Actin and pY53-actin at the indicated concentrations were incubated for 24 h at room temperature. (B) Unphosphorylated actin (8 μM), p53-actin (0.2 μM), or a mixture of unphosphorylated actin (7.8 μM) and pY53-actin (0.2 μM) were incubated at room temperature for 24 h and centrifuged at 240,000 × g for 1 h in a Beckman (Fullerton, CA) TL100 centrifuge. SDS/PAGE aliquots of the supernatants (S), pellets (P), and total sample before centrifugation (T) were stained with Coomassie blue and anti-actin and anti-pY-antibodies.
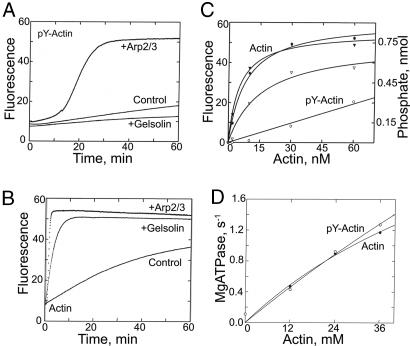
In the absence of nucleating seeds, pY53-actin polymerizes much more slowly than unphosphorylated actin (control curves in Fig. 8 A and B). Addition of gelsolin-unphosphorylated actin seeds, which nucleate polymerization by binding to barbed ends of nascent filaments, accelerates elongation of unphosphorylated actin (Fig. 8B) but inhibits elongation of pY53-actin (Fig. 8A), indicating that pY53-actin can add only very slowly, if at all, to the pointed ends of actin filaments. On the other hand, Arp2/3-VCA (verpolin, cofilin, and acidic domains of human wASP), which generates new barbed ends by binding to existing filaments, substantially accelerates polymerization of pY53-actin (Fig. 8A), indicating that pY53-actin can add to the barbed ends of filaments, albeit apparently more slowly than unphosphorylated actin (Fig. 8B).
Fig. 8.
Polymerization kinetics and myosin ATPase activation of unphosphorylated actin and pY53-actin. (A) Four-micromolar pY53-actin was polymerized alone (control) or in the presence of 20 nM gelsolin or 30 nM Arp2/3-VCA. (B) Unphosphorylated actin (4 μM) polymerized alone or in the presence of 20 nM gelsolin or 30 nM Arp2/3-VCA. (C) Correlation between ATP hydrolysis (circles) and spectrin–actin- (1.3 nM) nucleated polymerization (triangles) of 7.6 μM unphosphorylated actin (filled symbols) and 7.6 μM pY53-actin (open symbols). (D) ATPase activity of 150 nM Dictyostelium myosin II S1 as a function of the concentration of unphosphorylated and pY53-phosphorylated actin.
Hydrolysis of actin-bound ATP occurs after the addition of actin monomer to the growing filament (10), but, under normal polymerization conditions in vitro, the two events are almost concurrent. However, ATP hydrolysis accompanying spectrin-unphosphorylated actin-nucleated polymerization of pY53-actin is much slower than for unphosphorylated actin (Fig. 8C). Even as polymerization neared completion, approximately half of the subunits of pY53-actin filaments still had bound ATP. Note that polymerization of pY53-actin is accelerated by spectrin–actin seeds (compare Fig. 8C with control curve in Fig. 8A), which nucleate polymerization by binding to the pointed end, thus blocking pointed-end elongation while making more barbed ends available for growth. In contrast to the differences in their polymerization properties, we observed no difference in the activation of myosin II subfragment 1 (S1) ATPase by polymerized phosphorylated and unphosphorylated actin (Fig. 8D).
Electron Microscopy of Polymerized Actin.
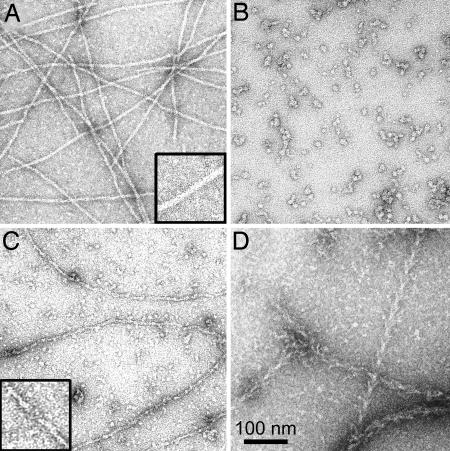
In contrast to the typical long (>2 μm) filaments of unphosphorylated actin (Fig. 9A), negatively stained images of polymerized pY53-actin revealed either short, 15- to 16-nm bars (Fig. 9B) or a mixture of short bars and filaments with obvious breaks (Fig. 9C). Because the reduced viscosities of unphosphorylated and pY53-actin were the same, 6.8 and 6.6 dl/g, respectively, it seems most likely that pY53-actin forms unstable filaments that partially or completely disassemble to small rods during negative staining. Polymerized pY53-actin is stabilized by myosin S1, appearing as typical long, decorated filaments (Fig. 9D). Filaments of pY53-actin are also stabilized by phalloidin (data not shown).
Fig. 9.
Electron microscopic images of polymerized unphosphorylated and pY53-actin. (A) Filaments of unphosphorylated actin. (B) Short bars of polymerized pY53-actin. (C) Mixture of short bars and filaments of pY53-actin. (D) Same sample of pY53-actin as in C, but with the addition of myosin S1. Insets in A and B are enlarged 2-fold.
Discussion
Tyr-53 is immediately adjacent to the hydrophobic DNase I-binding loop (11), residues 40–50 (12) of subdomain 2, which is also implicated in actin–actin (12–14) interactions. Several studies have shown that proteolytic cleavage (15, 16) of the DNase I-binding loop or chemical modification of neighboring residues Lys-61 (17–19) and Tyr-69 (20) reduce actin's affinity for DNase I and inhibit polymerization, which is reversed by phalloidin. More directly relevant to this article, chemical modification of Tyr-53 by reaction with diazonium tetrazole also blocks polymerization (21), which is not reversed by phalloidin (20). The effects of phosphorylation of Tyr-53 reported in this article are similar to the effects of chemical modification of Tyr-53 and neighboring residues, at least to the much more limited extent to which the chemically modified actins have been studied. The important difference, of course, is that Tyr-53 phosphorylation of Dictyostelium actin is a regulated, reversible biological response to developmental and environmental signals.
From the data in Fig. 8, Tyr-53 phosphorylation seems principally to inhibit nucleation and elongation at the pointed end, which could be due to an increase in the dissociation rate or a decrease in the association rate constant at the pointed end or both. When polymerization was induced by Arp2/3-VCA, which initiates elongation of branching filaments after binding to existing filaments (22), both nucleation and elongation of pY53-actin filaments were appreciably slower than for unphosphorylated actin. This finding might suggest that Arp2/3-VCA has reduced affinity for the pointed end of pY53-actin or Arp2/3-VCA binds more poorly to filaments of pY53-actin or both. However, the much slower nucleation (control curves in Fig. 8 A and B) and, therefore, the reduced number of pY53-actin filaments, which are a prerequisite for Arp2/3-VCA-induced branching, might also contribute to the slower elongation rate. This explanation would be consistent with the autocatalytic nature of Arp2/3-VCA-induced elongation in Fig. 8A. Because the extent of Arp2/3-VCA-nucleated polymerization was very similar for phosphorylated and unphosphorylated actin, Tyr-53 phosphorylation apparently has little or no effect on the critical concentration at the barbed end. The initial rates of spectrin–actin-nucleated elongation of phosphorylated and unphosphorylated actin (Fig. 8C) indicate that the association rate constant at the barbed end for pY53-actin is ≈20–25% of the association rate for unphosphorylated actin and, if the critical concentrations are the same, there would be a similar difference in the dissociation rate constants.
How might phosphorylation of Tyr-53 affect actin polymerization? Crystallographic studies of monomeric actin indicate that release of the γ-phosphate of ATP initiates a series of conformational changes that results in conversion of the disordered DNase I-binding loop of G-ATP-actin to an α-helix in G-ADP-actin (23, 24). Molecular dynamic simulations of actin oligomers and filaments (25) are consistent with a model in which the helical loop of the ADP-actin subunit weakens actin–actin interactions in trimeric nuclei and in F-actin, leading to destabilization of both. Tyr-53 phosphorylation might lock the DNase I-binding loop in a conformation that both inhibits ATP hydrolysis accompanying polymerization and weakens actin–actin interactions, particularly at the pointed end. The latter could inhibit nucleation, increase the critical concentration at the pointed end, inhibit interaction between actin and Arp2/3-VCA, and destabilize filaments.
Inhibition of actin polymerization, destabilization of actin filaments, and inhibition of Arp2/3-VCA-induced branching are consistent with the morphological changes that accompany phosphorylation of actin in live amoebae, but tyrosine phosphorylation might also induce cytoskeletal changes by affecting the interaction of actin with proteins other than Arp2/3. In addition, the slower rate of ATP hydrolysis by polymerized pY53-actin might significantly reduce treadmilling, which is important for actin-based motile processes.
On the other hand, the properties of purified pY53-actin would seem to be inconsistent with the high content of pY53-actin (50%) in polymerized actin in viable spores. However, the actin in spores is in 2-μm-long, 100- to 200-nm-wide rods, each containing six hexagonally cross-linked tubules, each of which contains three actin filaments (4, 26, 27). The unknown actin-binding proteins in these structures might interact preferentially with pY53-actin.
The increase in pY53-actin when vegetative amoebae are exposed to PAO is detectable within 10 min or less, and reaches a maximum of ≈40% of total actin in ≈60 min (unpublished observations). The simplest explanation would be that PAO inhibits an actin tyrosine phosphatase, thus disrupting the steady-state balance between actin phosphorylation and dephosphorylation. Alternatively, PAO may affect the level of tyrosine phosphorylation of a regulator protein, which, depending on its phosphorylation state, either activates or represses the actin tyrosine kinase. The Dictyostelium genome contains perhaps three protein tyrosine kinases (28) and 65–70 tyrosine kinase-like kinases (28, 29). Three tyrosine kinases, Zak1, Zak2, and PkyA, can be eliminated as possible components of the actin tyrosine phosphorylation pathway, because addition of PAO to their respective null cells increases pY53-actin just as in wild-type cells (unpublished results).
Does pY53-actin occur in other organisms? Thus far, we have found no evidence for pY-actin in mammalian cells (HeLa, Cos7, NIH 3T3, HEK293), even when exposed to stress or addition of PAO. pY-actin is present in the amoeboid stage of Acanthamoeba castellanii and is greatly increased in Acanthamoeba cysts (unpublished results), which are biologically analogous to Dictyostelium spores, and tyrosine phosphorylated actin has been reported in the plant Mimosa pudica L., where it seems to inhibit the bending of petioles that occurs upon contact (30, 31).
Materials and Methods
Cell Culture.
AX3 cells were grown in suspension culture in HL5 medium at 21°C. Cells were starved by washing and resuspending in 17 mM phosphate buffer (pH 6.2) for 4 h and developed to fruiting bodies as described (32). Actin tyrosine phosphorylation was increased by the addition of 100 μM CdCl2, 1 mM NaN3, or 45 μM PAO (Sigma, St. Louis, MO) (final concentrations) or heating at 37°C for 60 min.
Purification of Phosphorylated and Unphosphorylated Actin.
Dictyostelium (AX3) amoebae were grown in 20 liters of HL5 medium in 20 4-liter flasks at room temperature to a cell density of 1 × 107 cells per ml, 45 μM PAO and 1 mM NaN3 (final concentrations) were added, and the cells were harvested by centrifugation after 40 min. The cells (typically 300 g) were washed with 10 mM Tris (pH 7.5), resuspended in 2 ml/g of G-buffer (3 mM imidazole, pH 7.5, 0.1 mM CaCl2, 0.5 mM ATP, 1 mM DTT, and 1 mM PMSF) containing Roche (Indianapolis, IN) applied protease inhibitor (1 tablet per 50 ml), and stored at −70°C. As needed, 150 g of frozen cells was thawed in an additional 300 ml of G-buffer containing protease inhibitor, ruptured in a Parr bomb, and centrifuged at 160,000 × g for 1 h, and the supernatant was chromatographed on a DEAE 52 column as described (33). Fractions shown to contain actin by SDS/PAGE were pooled, and actin was precipitated by adding solid NH4SO4 to 50% saturation. NH4SO4 precipitation was used, rather than polymerization (33), to avoid loss of phosphorylated actin, which polymerizes less well than unphosphorylated actin. Protein was collected by centrifugation, dissolved in 20 ml of G-buffer, and centrifuged at 90,000 × g for 10 min, and the supernatant (in 8-ml aliquots) was chromatographed on a 26 × 60 Sephacryl HR column. Fractions containing actin were further purified and concentrated by FPLC on a Mono Q HR 10/10 column, and fractions containing actin were pooled and the actin precipitated with 50% saturated NH4SO4. The actin was dissolved in 16 ml of G-buffer and clarified by centrifugation, and the supernatant was chromatographed again on a 26 × 60 Sephacryl HR column.
Separation of Phosphorylated and Unphosphorylated Actin.
Phosphorylated and unphosphorylated actin were separated by FPLC on a Mono P 5/200 GL column as described (9), with some essential modifications. For complete separation, no more than 2–2.5 mg of protein was applied to a column equilibrated with 10 mM Tris (pH 8.0) containing 0.1 mM CaCl2, 0.5 mM ATP, and 1 mM DTT. Actin was eluted with a gradient of 0–500 mM KCl in 120 ml of the same buffer, collecting fractions of 0.5 ml, and fractions containing unphosphorylated and phosphorylated actins were identified by SDS/PAGE and immunoblotting. The phosphorylated actin from multiple columns was pooled, dialyzed against G-buffer, and concentrated by binding to and eluting from a Mono P column. Phosphorylated and unphosphorylated actin (5 and 25 mg, respectively, from 300 g of cells) were dialyzed against G-buffer overnight in a cold room.
Mass Spectrometric Analysis.
Alkylation and Lys-C digestion of actin and pY-actin were performed as described (34). The parent molecules and digests were analyzed by reverse-phase chromatography on a narrow-bore C18 column (Vydac, Hesperia, CA) and by mass spectrometric analysis as described (35), except that the instrument was an Agilent Technologies (Palo Alto, CA) model 1100 HPLC coupled to an Agilent Technologies model G1969A mass spectrometer with a time-of-flight detector. The flow rates of the column eluate and of the added neat acetic acid were both 20 μl/min.
Actin Polymerization.
pY53-actin and unphosphorylated actin, at the indicated concentrations, containing 4% pyrene-labeled actin, with or without addition of spectrin–actin (36), gelsolin (Sigma)-actin seeds (37) or Arp2/3-VCA (Cytoskeleton, Denver, CO) (38), was polymerized at room temperature in G-buffer by the addition of 100 mM KCl and 2 mM MgCl2 (final concentrations). The increase in fluorescence was measured with excitation at 365 nm and emission at 407 nm.
ATPase Activity.
Actin-activated ATPase activity of Dictyostelium myosin II S1 (32), 150 nM, was assayed in 20 mM imidazole (pH 7.5), 4 mM MgCl2, 25 mM KCl, and 2 mM [32P]ATP by measuring the production of 32Pi (39) after incubation at 30°C for 10 min, during which period, the reaction rates were constant.
Electrophoresis.
For SDS/PAGE, cells were harvested by centrifugation, suspended in an equal volume of buffer (50 mM KCl, 20 mM Hepes, pH 7.5, 1 mM DTT, and 0.1 mM PMSF containing Roche protease inhibitor; 1 tablet per 50 ml) and 2× SDS sample buffer, and equivalent samples were electrophoresed on 10% gels (40). For 2D electrophoresis, which was performed according to the manufacturer's procedure (80-6429-60, edition AB; Amersham Biosciences, Piscataway, NJ) and (3), samples (250 μl) containing 20 μg of total cell protein or 5 μg of purified actin were separated on a 13-cm, pH 4–7 DryStrip (Cat No. 17-6001013; Amersham Biosciences) and, in the second direction, on a 10% Tris-glycine 2D-well minigel (EC 6076 Box; Invitrogen, Carlsbad, CA). Gels were either stained with Coomassie blue or transferred electrophoretically to nitrocellulose membranes. Blots were reacted with 1,000-fold-diluted phosphotyrosine monoclonal antibody (clone 4G-10; Upstate Biotechnology, Lake Placid, NY) or anti-actin antibody (A-2066; Sigma). Two different antigens were detected simultaneously on the same blot by using secondary antibodies labeled with goat IRDye 800 anti-rabbit IgG (Rockland Immunochemicals, Gilbertsville, PA) and Alexa Fluor 680 goat anti-mouse IgG (Molecular Probes, Carlsbad, CA), both at 10,000-fold dilution, visualized at 800 and 700 nm, respectively, with the Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE).
Fluorescence Microscopy.
Indirect immunofluorescence microscopy was performed as described (41). For localization of pY-actin, cells were fixed for 5 min in −20°C methanol containing 1% formalin, incubated with 100-fold-diluted anti-phosphotyrosine monoclonal antibody (4G-10; Upstate Biotechnology), followed by 750-fold-diluted FITC-conjugated goat anti-mouse IgG (Molecular Probes). For localization of F-actin and pY-actin after staining with antibodies, cells were incubated with 500-fold-diluted rhodamine phalloidin (R-415; Molecular Probes). Confocal microscopy was performed on a Zeiss (Thornwood, NY) LSM-510 laser scanning fluorescence microscope equipped with a Plan apoX63 oil objective.
Electron Microscopy.
Actin was polymerized in the presence of 100 mM KCl, 2 mM MgCl2, and 5 mM imidazole (pH 7.5). One drop of the filament suspension (0.1–0.5 mg/ml) was placed on a formvar/carbon-coated, 400-mesh copper grid previously glow discharged for 30 s in an EMScope TB500 (Emscope Laboratories, Ashford, U.K.). After 1 min, the excess liquid was absorbed with filter paper. The grid was stained with a drop of 0.5–0.1% aqueous uranyl acetate for 30 s, the excess stain was removed with filter paper, and the sample was observed in a JEM-1200EX II transmission electron microscope (JEOL, Tokyo, Japan).
Other Procedures and Materials.
Dictyostelium S1 was purified by FLAG-affinity chromatography as described (32) and stored in liquid N2 until use. S1 concentrations were determined by absorption at 280 nm by using an extinction coefficient of 0.7 cm2/mg and by the Bradford assay, with the same results. Actin concentrations were determined by absorption at 290 nm by using an extinction coefficient of 0.62 cm2/ml. Pyrene (Molecular Probes) labeling of actin was carried out as described (42). DNase I activity was assayed by monitoring OD260 after adding Sigma DNA (50 μg/ml) to a solution containing 0.7 nM Sigma DNase I (final concentrations) and various concentrations of G-actin in 10 mM Tris (pH 8), 1 mM MgCl2, 0.1 mM CaCl2, and 1 mM NaN3. Viscosities were measured at 24°C in a falling-ball viscometer containing 2 ml of actin polymerized overnight at 24°C in 100 mM KCl and 2 mM MgCl2. Buffer flow time was 65 s.
Acknowledgments
We thank Sally Zigmond (University of Pennsylvania, Philadelphia, PA) for spectrin; Lin Lin (National Heart, Lung, and Blood Institute) for the mammalian cell lines; Alan R. Kimmel (National Institute of Diabetes and Digestive and Kidney Diseases) for the Zak1-, Zak2-, and PkyA-null cells; Nancy Wehr for assistance in mass spectroscopy; and Marie-France Carlier for helpful comments on the manuscript.
Abbreviations
- PAO
phenylarsine oxide
- pY-actin
phosphotyrosine actin
- S1
myosin II subfragment 1
- VCA
verpolin, cofilin, and acidic domains of human wASP
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Loomis WF. Dictyostelium discoideum: A Developmental System. New York: Academic; 1975. [Google Scholar]
- 2.Gauthier ML, Lydan MA, O’Day DH, Cotter DA. Cell Signal. 1997;9:79–83. doi: 10.1016/s0898-6568(96)00113-1. [DOI] [PubMed] [Google Scholar]
- 3.Kishi Y, Clements C, Mahadeo DC, Cotter DA, Sameshima M. J Cell Sci. 1998;111:2923–2932. doi: 10.1242/jcs.111.19.2923. [DOI] [PubMed] [Google Scholar]
- 4.Sameshima M, Kishi Y, Osumi M, Minamikawa-Tachino R, Mahadeo D, Cotter DA. J Struct Biol. 2001;136:7–19. doi: 10.1006/jsbi.2001.4424. [DOI] [PubMed] [Google Scholar]
- 5.Schweiger A, Mihalache O, Ecke M, Gerisch G. J Cell Sci. 1992;102:601–609. doi: 10.1242/jcs.102.3.601. [DOI] [PubMed] [Google Scholar]
- 6.Howard PK, Sefton BM, Firtel RA. Cell. 1992;71:637–647. doi: 10.1016/0092-8674(92)90597-6. [DOI] [PubMed] [Google Scholar]
- 7.Howard PK, Sefton BM, Firtel RA. Science. 1993;259:241–244. doi: 10.1126/science.7678470. [DOI] [PubMed] [Google Scholar]
- 8.Jungbluth A, von Arnim V, Biegelmann E, Humbel B, Schweiger A, Gerisch G. J Cell Sci. 1994;107:117–125. doi: 10.1242/jcs.107.1.117. [DOI] [PubMed] [Google Scholar]
- 9.Jungbluth A, Eckerskorn C, Gerisch G, Lottspeich F, Stocker S, Schweiger A. FEBS Lett. 1995;375:87–90. doi: 10.1016/0014-5793(95)01165-b. [DOI] [PubMed] [Google Scholar]
- 10.Carlier M-F, Pantaloni D, Korn ED. J Biol Chem. 1984;259:9983–9986. [PubMed] [Google Scholar]
- 11.Holmes KC, Popp D, Gebhard W, Kabsch W. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 12.Milligan RA. Proc Natl Acad Sci USA. 1996;93:21–26. doi: 10.1073/pnas.93.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenz M, Poole KJV, Popp D, Rosenbaum G, Holmes KC. J Mol Biol. 1993;234:826–836. doi: 10.1006/jmbi.1993.1628. [DOI] [PubMed] [Google Scholar]
- 14.Sutoh K. Biochemistry. 1984;23:1942–1946. doi: 10.1021/bi00304a009. [DOI] [PubMed] [Google Scholar]
- 15.Khaitlina SY, Moraczewska J, Strzelecka-Golaszewska H. Eur J Biochem. 1993;218:911–920. doi: 10.1111/j.1432-1033.1993.tb18447.x. [DOI] [PubMed] [Google Scholar]
- 16.Khaitlina SY, Strzelecka-Golaszewska H. Biophys J. 2002;82:321–334. doi: 10.1016/S0006-3495(02)75397-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burtnick LD. Biochim Biophys Acta. 1984;791:57–62. doi: 10.1016/0167-4838(84)90281-4. [DOI] [PubMed] [Google Scholar]
- 18.Miki M. Eur J Biochem. 1987;164:229–235. doi: 10.1111/j.1432-1033.1987.tb11015.x. [DOI] [PubMed] [Google Scholar]
- 19.Combeau C, Carlier M-F. Biochemistry. 1992;31:300–309. doi: 10.1021/bi00116a041. [DOI] [PubMed] [Google Scholar]
- 20.Miki M, Barden JA, dos Remedios CG, Phillips L, Hambly BD. Eur J Biochem. 1987;165:125–130. doi: 10.1111/j.1432-1033.1987.tb11202.x. [DOI] [PubMed] [Google Scholar]
- 21.Bender N, Fasold H, Kenmoku A, Middelhoff G, Volk K-E. Eur J Biochem. 1976;64:215–218. doi: 10.1111/j.1432-1033.1976.tb10290.x. [DOI] [PubMed] [Google Scholar]
- 22.Egile C, Rouiller I, Xu X-P, Volkmann N, Li R, Hanein D. PloS Biol. 2005;3:1902–1909. doi: 10.1371/journal.pbio.0030383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otterbein LR, Graceffa P, Dominguez R. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- 24.Graceffa P, Dominguez R. J Biol Chem. 2003;278:34172–34180. doi: 10.1074/jbc.M303689200. [DOI] [PubMed] [Google Scholar]
- 25.Chu J-W, Voth GA. Proc Natl Acad Sci USA. 2005;102:13111–13116. doi: 10.1073/pnas.0503732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sameshima M, Chijiiwa Y, Kishi Y, Hashimoto Y. Cell Struct Funct. 1994;19:189–194. doi: 10.1247/csf.19.189. [DOI] [PubMed] [Google Scholar]
- 27.Sameshima M, Kishi Y, Osumi M, Mahadeo D, Cotter DA. Cell Struct Funct. 2000;25:291–295. doi: 10.1247/csf.25.291. [DOI] [PubMed] [Google Scholar]
- 28.Kimmel AR. In: Dictyostelium Genomics, Loomis WF, Kuspa A, editors. Norfolk, UK: Horizon Scientific; 2005. pp. 211–234. [Google Scholar]
- 29.Goldberg JM, Manning G, Liu A, Fey P, Pilcher KE, Xu Y, Smith JL. PloS Genet. 2006;2:291–303. doi: 10.1371/journal.pgen.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kameyama K, Kishi Y, Yoshimura M, Kanzawa N, Sameshima M, Tsuchiya T. Nature. 2000;407:37. doi: 10.1038/35024149. [DOI] [PubMed] [Google Scholar]
- 31.Kanzawa N, Hoshino Y, Chiba M, Hoshino D, Kobayashi H, Kamasawa N, Kiski Y, Osumi M, Sameshima M, Tsuchiya T. Plant Cell Physiol. 2006;47:531–539. doi: 10.1093/pcp/pcj022. [DOI] [PubMed] [Google Scholar]
- 32.Liu X, Shu S, Kovacs M, Korn ED. J Biol Chem. 2005;280:26974–26983. doi: 10.1074/jbc.M504453200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gordon DJ, Eisenberg E, Korn ED. J Biol Chem. 1976;251:4778–4786. [PubMed] [Google Scholar]
- 34.Kang DK, Jeong J, Drake SK, Wehr NB, Rouault TA, Levine RL. J Biol Chem. 2003;278:14857–14864. doi: 10.1074/jbc.M300616200. [DOI] [PubMed] [Google Scholar]
- 35.Taggart C, Cervantes-Laurean D, Kim G, McElvaney NG, Wehr N, Moss J, Levine RL. J Biol Chem. 2000;275:27258–27265. doi: 10.1074/jbc.M004850200. [DOI] [PubMed] [Google Scholar]
- 36.Brenner SL, Korn ED. J Biol Chem. 1980;255:1670–1676. [PubMed] [Google Scholar]
- 37.Coué M, Korn ED. J Biol Chem. 1985;260:15033–15041. [PubMed] [Google Scholar]
- 38.Boujemaa-Paterski R, Gouin E, Hansen G, Samarin S, Le Clainche C, Didry D, Dehoux P, Cossart P, Kocks C, Carlier M-F, Panataloni D. Biochemistry. 2001;40:11390–11404. doi: 10.1021/bi010486b. [DOI] [PubMed] [Google Scholar]
- 39.Pollard TD, Korn ED. J Biol Chem. 1973;248:4682–4690. [PubMed] [Google Scholar]
- 40.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 41.Shu S, Liu X, Korn ED. Proc Natl Acad Sci USA. 2003;100:6499–6544. doi: 10.1073/pnas.0732155100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooper JA, Walker SB, Pollard TD. J Muscle Res Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]